Abstract
Background
Malaria remains a global health problem and the majority of deaths are caused by Plasmodium falciparum parasites. Due to the rapid emergence of drug-resistant strains, novel avenues of research on the biology of the parasite are needed. The massive proliferation of asexual, intra-erythrocytic parasites every 48 h could kill the human host prior to transmission of slow-developing gametocytes to the mosquito vector. A self-induced P. falciparum programmed cell death mechanism has been hypothesized to maintain this balance between the parasite and its two hosts, but molecular participants of the cell death pathway in P. falciparum have not been characterized. Proteins with SWIB/MDM2 domains play a key role in metazoan programmed cell death and this study provides the first evaluation of two parasite SWIB/MDM2 homologues, PF3D7_0518200 (PfMDM2) and PF3D7_0611400 (PfSWIB).
Methods
The function of these proteins was assessed by predicting their structural topology with the aid of bioinformatics and determining their location within live transgenic parasites, expressing green fluorescent protein-tagged PfMDM2 and PfSWIB under normal and elevated temperatures, which mimic fever and which are known to induce a programmed cell death response. Additionally, P. falciparum phage display library technology was used to identify binding partners for the two parasite SWIB/MDM2 domains.
Results
Structural features of the SWIB/MDM2 domains of PfMDM2 and PfSWIB, suggested that they are chromatin remodelling factors. The N-terminal signal sequence of PfMDM2 directed the protein to the mitochondrion under both normal and heat stress conditions. Plasmodium falciparum phage display library technology revealed that the C-terminal SWIB/MDM2 domain of PfMDM2 interacted with a conserved protein containing a LisH domain. PfSWIB localized to the cytoplasm under normal growth conditions, while approximately 10 % of the heat-stressed trophozoite-stage parasites presented a rapid but short-lived nuclear localization pattern. Two PfSWIB binding partners, a putative Aurora-related kinase and a member of the inner membrane complex, were identified.
Conclusion
These novel data provide insight into the function of two parasite SWIB/MDM2 homologues and suggest that PfMDM2 plays a role within the mitochondrion and that PfSWIB is involved in a stage-specific, heat-stress, response pathway.
Electronic supplementary material
The online version of this article (doi:10.1186/s12936-015-1065-9) contains supplementary material, which is available to authorized users.
Keywords: SWIB/MDM2, Plasmodium falciparum, Heat stress response, Malaria, Programmed cell death, Apoptosis
Background
The asexual intra-erythrocytic life cycle of P. falciparum lasts approximately 48 h, from invasion to egress, whereby a single merozoite can give rise to as many as 32 new merozoites, while the sexual gametocytes, which are transmitted to the Anopheles mosquito host, require up to 12 days to reach maturity [1]. In light of this, if all the newly formed merozoites were to invade erythrocytes every 48 h, the human host may die before gametocyte maturation has occurred. As the human immune system removes the parasite poorly there must be another means of regulation to protect the host from premature death. One such mechanism has been hypothesized as parasite self-induced, programmed cell death (PCD) [2].
Apoptotic features in the malaria parasite were first described in 1997 [3, 4], with the ‘crisis form’ of the parasite hypothesized as a PCD marker [4]. Subsequently, numerous studies have documented apoptosis and autophagy markers, including DNA laddering, loss of mitochondrial membrane potential, apoptotic body formation, and cytoplasmic vacuolization during various life stages of Plasmodium in response to a variety of stress stimuli [5]. Markers, such as DNA fragmentation and mitochondrial dysregulation, have been noted in cultured parasites under normal, non-limiting conditions, which suggests an intrinsic property [6]. To date, no experimentally proven PCD machinery has been described in the parasite, although several candidate genes have been identified by bioinformatics, including metacaspases [7] and SWIB/MDM2 domains [8].
The mammalian MDM2 protein, originally identified in transformed mice fibroblasts, contains several functional domains, including a SWIB/MDM2 domain [9, 10]. This anti-apoptotic protein is located primarily within the nucleus of unstressed cells where it binds to p53 via its N-terminal region, containing the SWIB/MDM2 domain. This interaction prevents p53 binding to DNA and induces the nuclear export, ubiquitination and proteasome-dependent degradation of p53 [11]. Under genotoxic conditions, numerous processes occur to stabilize p53, including MDM2 phosphorylation to prevent its association with p53, which brings about cell cycle arrest and, if required, cell death [11].
SWIB/MDM2 domains have also been identified in several other proteins and protein complexes, one such being the 2 MDa multi-subunit nuclear assembly, the SWI/SNF complex [12]. This ATP-dependent chromatin remodelling complex and transcriptional regulator, originally discovered in yeast, binds to DNA and hydrolyses ATP, which alters chromatin structure through nucleosome sliding and histone octomer insertion and/or ejection. The complex is composed of constant units, believed to be core functional units and includes the Swp73p/SNF12 protein containing a SWIB/MDM2 domain, as well as other apparently variable units, proposed to facilitate a degree of specificity and/or functionality [12]. The complex is involved in various stress response pathways, including exposure to elevated temperatures, heavy metals and metabolic inhibitors [13, 14]. In humans, a homologue of the SWI/SNF complex has also been shown to associate with p53 and regulate its activities, facilitating cell cycle halting and fine tuning the balance between repair and apoptosis induction [15–17]. The BAF60a protein of the complex is responsible for p53 binding, not through its C-terminal SWIB/MDM2 domain but rather directly via an N-terminal region [17].
SWIB/MDM2 domains participate in activities such as protein–protein [18] and chromatin-related interactions [19], but their precise functional role(s) in the cell are poorly characterized.
The P. falciparum genome encodes two putative SWIB/MDM2 domain-containing proteins: PF3D7_0518200, SWIB/MDM2 domain-containing protein, putative [PlasmoDB: PF3D7_0518200], which will be designated as PfMDM2 in this study; and PF3D7_0611400, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin [PlasmoDB: PF3D7_0611400], which will be designated as PfSWIB.
This study aimed to assess the two P. falciparum SWIB/MDM2 homologues. Under normal and heat stress conditions PfMDM2 localized to the mitochondrion, while the cytoplasmic PfSWIB protein underwent a short-lived nuclear redistribution after heat stress. Biopanning revealed one or more binding partners for the SWIB/MDM2 domains of these two P. falciparum homologues, suggestive of transcriptional or stress pathway involvement.
Methods
Bioinformatics
Amino acid sequences of proteins containing SWIB/MDM2 domains were collected from a variety of prokaryotic and eukaryotic species—Homo sapiens [NCBI: ACX31156]; Canis lupus familiaris [NCBI: BAB11975]; Mus musculus [NCBI: AAB09030]; Sus scrofa [NCBI: ABV09038]; Gallus gallus [NCBI: AAF04192]; Danio rerio [NCBI: NP_571439.2]; Pongo abelii [NCBI: NP_001124685.1]; Callorhinchus milii [NCBI: AEW46991]; Xenopus laevis [NCBI: NP_001086070]; Arabidopsis thaliana [NCBI: AAP68331.1]; Toxoplasma gondii [NCBI: EEB00952.1]; Taranis japonicus [NCBI: ACS36127.1]; Caenorhabditis elegans [NCBI: CAA87424.1]; Grosmannia clavigera [NCBI: EFW99809.1]; Drosophila melanogaster [NCBI: AAF48235.1]; Solanum chacoense [NCBI: ABE11612.1]; Homo sapiens [NCBI: ACX31156.1]; Zea mays [NCBI: NP_001148297.1]. These were used for multiple EMBL-EBI clustal omega sequence alignments [20] against the two malaria proteins of interest. Multiple sequence alignments were expressed graphically using BioEdit Sequence Alignment Editor [21]. Percentage identity and similarity was calculated using EMBOSS Needle [22].
Tertiary structure analysis
Structural modelling of the malaria proteins was conducted with the online servers of the Swiss Model Workspace Automatic Modelling Mode [23], PHYRE2 [24] and ESyPred3D Web Server 1.0 [25]. The following crystal and solution structures were used: the Xenopus laevis MDM2 SWIB/MDM2 domain [PDB id: 1YCQ chain A] [9], the Mus musculus SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily D member 1 SWIB/MDM2 domain [PDB id: 1UHR] [26], and the Arabidopsis thaliana SWIB/MDM2 domains [PDB id: 1V31 and 1V32] [27, 28]. Graphical display, orientation and colouring of various PDB files were conducted using the PyMOL Molecular Graphics System [29]. The quality of all the three-dimensional models was assessed by the QMEAN Server [30] and the best candidate models presented.
Localization prediction
Various online prediction algorithms were employed to determine the likely cellular compartment to which the proteins would localize. These included cNLS Mapper [31]; MitoProt II—v1.101 [32]; NucPred [33]; PredSL; PSORT Prediction; PREDOTAR V1.03; PlasmoDB—PlasmoAP Results [34]; PATS Version 1.2.1 [35, 36]; PlasMit [37] and iPSORT Prediction.
Parasite culture
The 3D7 strain of P. falciparum was cultured according to a slightly modified form of the standard protocol [38]. Briefly, parasites were maintained at 5 % haematocrit in culture medium (RPMI 1640 (Gibco BRL, Life Technologies Corp, CA, USA), supplemented with 0.5 % Albumax II (Life Technologies Corp), 0.21 % sodium bicarbonate, 50 mg/L gentamycin and 50 mg/L hypoxanthine). The culture medium was changed daily and optimal pH was maintained by daily gassing of the culture with 2 % O2, 5 % CO2 and 93 % N2. Sorbitol treatment was employed for asexual parasite synchronization [39].
Ethics clearance was obtained for this project. Ethics number M13-05-69; The University of the Witwatersrand; Committee for Research on Human Subjects (medical).
PCR amplification and cloning
Genomic DNA was extracted from infected erythrocytes [40] and used for the amplification of various genes for both recombinant protein expression and transgenic parasite production. The primers, containing appropriate restriction endonuclease sites, are presented in the Additional files 1 and 2. PCR products and vectors were digested with the appropriate FastDigest® restriction endonucleases (Thermo Fisher Scientific, Inc., Ueberlingen, Germany). The genes or gene domains were subsequently cloned into pARL2-GFP (donated by Dr Jude Przyborski, Marburg, Germany), pGEX-4T-2 or pET-15b vectors depending on the downstream application. After ligation, the pARL2-GFP constructs were used for the transformation of Escherichia coli XL10-Gold® ultracompetent cells (Stratagene, CA, USA), while the Rosetta™ 2 (DE3) Escherichia coli line (Novagen Inc., WI, USA) was used for the pGEX-4T-2 and pET-15b constructs.
Recombinant His- and GST-tagged protein expression and purification
Transformed colonies were grown in Overnight Express™ Instant TB Medium (Novagen Inc.), containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol, for 22 h at 250 rpm, at room temperature (~20 °C). The Escherichia coli cells were lysed by freeze-thawing and sonication and protein purification was conducted using the MagneGST™ and MagneHis™ kits (Promega, WI, USA), according to the manufacturer’s specifications. Depending on the downstream application, the purified fusion proteins could either be eluted from or retained on the magnetic beads. GST-tagged recombinant proteins were eluted in 150 μl of GST-Elution buffer (500 mM L-Glutathione, 500 mM NaCl, 50 mM Tris–HCl, pH 8.1) while the His-tagged recombinant proteins were eluted in 100 μl of His-Elution buffer (50 mM Na2HPO4/NaH2PO4 buffer, 150 mM NaCl, 0.5 M imidazole, pH 7.5 for PfLisH and pH 7.8 for PfARK3). Protein concentration was determined using a bovine serum albumin (BSA) standard curve and molecular weight was assessed relative to a red cell membrane marker.
The eluted GST- and His-fusion proteins were dialysed at 4 °C against three changes of Tris buffered saline (TBS) (50 mM Tris–HCl, 150 mM NaCl, pH 7.5) or sodium phosphate buffer (50 mM Na2HPO4/NaH2PO4 buffer, 150 mM NaCl, pH 8), using a 10 kDa Slide-A-Lyzer MINI dialysis unit (Pierce Biotechnology Inc, IL, USA) for 30 min. The protein samples were then assessed by 12 % SDS–polyacrylamide gel electrophoresis (SDS-PAGE) [41] and immunoblotting, using a 1:25,000 Anti-GST horseradish peroxidase (HRP) conjugated primary antibody (Amersham Biosciences Ltd, Little Chalfont, UK) or 1:1200 Anti-His HRP conjugate primary antibody (Qiagen GmbH, Hilden, Germany).
Biopanning against Plasmodium falciparum phage display libraries
Plasmodium falciparum phage display, mixed stage libraries were used according to a previous protocol, with slight modifications [42]. Briefly, a pre-screening step to eliminate background was carried out whereby the starting library (~1 × 107 pfu/ml) was mixed with MagneGST beads containing at least 8 μg of recombinant GST, at room temperature for 1 h. The unbound phage were mixed with MagneGST beads bound to at least 8 μg of recombinant GST-PfMDM2 or GST-PfSWIB protein, for 1 h at room temperature. The beads were removed, washed five times with 2 ml of 0.05 % Tween-TBS for 10 min, and then added to 50 ml of log phase BLT5403 cells and incubated at 37 °C until lysis was noted. The lysate was then used as the starting library for the next round of biopanning. In total, four sequential rounds of biopanning were conducted to enrich for phage binding specifically to the recombinant PfMDM2 and PfSWIB proteins. After the final round, these phage were used for PCR analysis. The empty cassette PCR product was 216 bp and thus plaque PCR products ≥300 bp were sent for sequencing at Inqaba Biotec™, Johannesburg, South Africa.
In vitro binding assays
In vitro binding assays were conducted to confirm the interactions identified using biopanning. One μg of the dialysed recombinant His-fusion binding partner protein, re-attached to 5 μl MagneHis beads, was exposed to increasing concentrations of the GST-tagged SWIB/MDM2 partner for 1 h at room temperature in a total volume of 150 μl TBS. The beads were washed three times in TBS and then the protein complexes were subjected to SDS-PAGE [40]. Control reactions were conducted as described above using equivalent amounts of recombinant GST protein as well as heat denatured (70 °C for 15 min) SWIB/MDM2 proteins to compensate for non-specific binding. The experiments were conducted in a reciprocal fashion, whereby dialysed recombinant GST-PfMDM2 proteins were re-attached to MagneGST beads and exposed to increasing concentrations of the His-tagged binding partners. The resultant interactions were then assessed by 12 % SDS-PAGE and visualized by immunoblotting, as described above, or with Coomassie blue (0.05 % Coomassie Brilliant Blue R-250 (w/v), 25 % Isopropanol (v/v), 10 % acetic acid (v/v)). Protein concentrations were determined by densitometry, relative to a BSA standard, in the case of the latter technique.
Transgenic parasite generation
Transfections were performed on synchronized ring-stage 3D7 P. falciparum parasites in a 2 mm BioRad Gene Pulser® Cuvette (Bio-Rad Laboratories, CA, USA) with a minimum of 100 μg of plasmid DNA by electroporation using the Bio-Rad GenePulse Xcell™ electroporator (Bio-Rad Laboratories), at 310 V, with a resistance of 950 μF and a time of less than 15 ms. Positive WR99210 drug selection, at a final concentration of 2 nM, was applied and maintained after the first cycle of growth, as parasites which had taken up the pARL2-GFP construct would carry the human dihydrofolate reductase gene, providing resistance to the drug. The transgenic parasites were noted in cultures from 25 to 43 days after transfection.
Heat shock analysis
Synchronized trophozoite stage transgenic parasites were exposed to 41 °C, a temperature equivalent to malaria-induced febrile illness, for 2 h and then returned to 37 °C. Previous work has indicated that such exposure induces a time-dependent apoptosis-like death mechanism [43]. The stressed parasites were viewed 30 min after the end of heat stress and then at set time points by fluorescence microscopy.
Microscopy
For nuclear staining, parasites were treated with DAPI (Sigma-Aldrich Corp, MI, USA), at a final concentration of 0.2 μg/ml, or Hoechst 33258 pentahydrate (Invitrogen, CA, USA), at a final concentration of 6 μg/ml and incubated for 5 min at room temperature or 2 h at 37 °C, respectively, and then washed twice in culture medium.
To stain mitochondria, MitoSOX™ Red (Molecular Probes, Invitrogen, CA, USA) was used, which is selectively targeted to the mitochondria of living cells and then oxidized by mitochondrial superoxides [44] into a red fluorescent form. To verify that MitoSOX stains the mitochondria of P. falciparum, co-localization with MitoTracker Green FM (Molecular Probes, Invitrogen, CA, USA) was confirmed (see Additional file 3). This green fluorescent stain accumulates in the active mitochondria of P. falciparum [45], but it has almost identical emission and excitation properties as GFP, which prevented its use in this study. MitoSOX™ was added to 300 μl of cultured wild type 3D7 P. falciparum parasites at a final concentration of 0.5 μM and incubated for 15 min at 37 °C. Cells were washed twice with culture medium before visualization at 1000× magnification, using the BX41 Olympus Microscope system. The system included a U-MWU2 filter (excitation between 330 and 385 nm and emission above 410 nm, for DAPI visualization), a U-MWB2 filter (excitation between 460 and 490 nm and an emission above 510 nm, for GFP and MitoTracker Green FM visualization), U-MWG2 filter (excitation between 510 and 550 nm and emission maximum at 590 nm, for MitoSOX™ visualization), a U-25ND25 Olympus neutral density filter, an Olympus DP72 camera; and CellSense Dimensions 1.7 Software (Olympus Optical Corp Ltd, Tokyo, Japan). Images were combined and processed uniformly with Adobe Photoshop 7.0. The overlap coefficient (R) was determined for five parasites for each experimental procedure and averaged with the aid of Image J [46].
Results
Plasmodium falciparum SWIB/MDM2 domains conform to a helical cleft topology
Two broad groups of SWIB/MDM2 domains have been identified. The first group is involved in p53 binding and identified in MDM2 proteins, such as that found in the Xenopus laevis MDM2 protein [9]. The second group is identified in a variety of eukaryotic proteins and participates in chromatin remodelling, transcriptional regulation and unknown functions, such as that found in the Saccharomyces cerevisiae SNF12 protein [NCBI:CAA96302.1] and the Arabidopsis thaliana At1g31760 protein [NCBI:At1g31760] for example [19]. The Plasmodium Database [47] was screened using the BlastP search function for SWIB/MDM2 domain homologues. No matches were found relative to the domain identified in MDM2 proteins; however, low scoring matches were found to the SWIB/MDM2 domains document in non-MDM2 proteins. Two of these proteins, expressed by the parasite, are annotated as having SWIB/MDM2 domains—PfSWIB and PfMDM2—and these were selected for subsequent analysis.
Predicted cellular localization signal sequences, as well as predicted functional domains within the two P. falciparum SWIB/MDM2 homologues are presented in Fig. 1. PfMDM2 is annotated to contain a C-terminal SWIB/MDM2 domain, constituting most of the protein [48]. The same putative domain was annotated in the N-terminal region of PfSWIB and was the only functional domain identified within this large protein of 830 amino acids [48]. Both domains were surrounded by a variety of monopartite and bipartite nuclear signal sequences predicted by different algorithms. A mitochondrial localization sequence was identified at the N-terminus of PfMDM2.
Fig. 1.

Signal sequence and domain predictions for PfMDM2 and PfSWIB. Signal sequences and functional domains within a full-length PfMDM2 and b the N-terminus of PfSWIB (first 420 amino acids). The red line indicates a putative mitochondrial localization signal sequence identified by the iPSORT Prediction algorithm with LLRTNIFSA being of critical importance; the blue lines indicate putative nuclear localization sequences identified by cNLS Mapper; the black lines indicate putative nuclear localization signal sequences identified by NucPred; the purple lines indicate the putative SWIB/MDM2 domains as defined on PlasmoDB. The green line represents the truncated form of the recombinant PfMDM2 protein; while the orange line represents the recombinant region expressed for PfSWIB
To further assess these candidates, primary sequence alignments were conducted using Clustal Omega while percentage identity and similarity was calculated using EMBOSS Needle. The SWIB/MDM2 domain has been retained during evolution [18]; therefore, to strengthen the analysis, proteins from several species ranging from Homo sapiens and other metazoans to unicellular organisms, such as Toxoplasma gondii, were used for this evaluation. The MDM2 domain of the X. laevis MDM2 protein shared 17.5 % identity and 35 % similarity and 19.2 % identity and 38.5 % similarity to the annotated domains of PfMDM2 and PfSWIB, respectively. The binding of MDM2 to p53 is mainly the result of van der Waals forces, facilitated by a high proportion of aromatic and hydrophobic residues within the SWIB/MDM2 domain [9, 49]—42.1 % in Xenopus laevis for example. The P. falciparum SWIB/MDM2 domains have a slightly lower hydrophobic and aromatic amino acid residue composition (39.4 % for PfMDM2 and 40.6 % for PfSWIB). Of the 16 residues marked as critical for p53 binding in the MDM2 domain (Fig. 2), only one is identical in each P. falciparum protein—Ile83 in PfMDM2 and Asp297 in PfSWIB—but there is a degree of semi-conservation. Furthermore there is a high degree of conservation of hydrophobic and/or aromatic residues (Fig. 2) that may facilitate a suitable environment for protein–protein interactions. The P. falciparum homologues could deviate in essential amino acids due to potential sequence and structural differences in their binding partner(s), relative to the metazoan p53 protein.
Fig. 2.
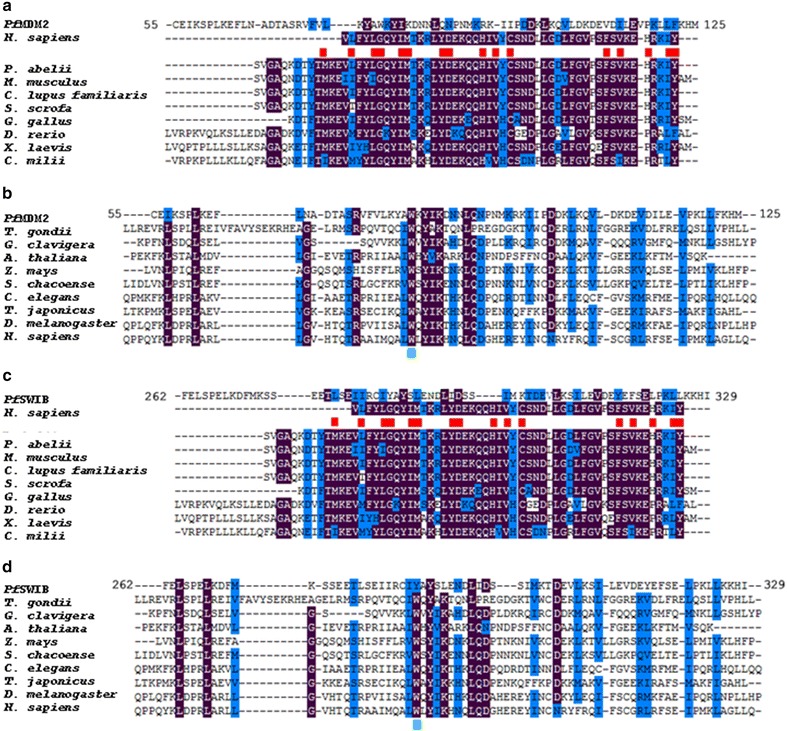
Clustal Omega primary amino acid sequence alignments for PfMDM2 and PfSWIB to various SWIB/MDM2 domains. Alignment of the PfMDM2 SWIB/MDM2 to SWIB/MDM2 domains identified in a MDM2 proteins and b other proteins. Alignment of the PfSWIB SWIB/MDM2 to SWIB/MDM2 domains identified in c MDM2 proteins and d other proteins. The red blocks indicate critical amino acid residues for p53 binding [9, 49], which show limited conservation in the two parasite domains. The green blocks highlight a previously described Trp residue conserved in SWIB/MDM2 domains, not found in MDM2 proteins [18]. Purple blocks indicate identical amino acids; blue blocks indicate similar amino acids; threshold set at 80 %
Similar to the Xenopus laevis protein, the MDM2 domain identified in the Arabidopsis thaliana SWI/SNF complex component SNF12 homolog [NCBI: Q9FMT4] shared 18.1 % identity and 36.1 % similarity and 24 % identity and 40.0 % similarity to the annotated domains of PfMDM2 and PfSWIB respectively. The residues of SWIB/MDM2 domains involved in chromatin remodelling and transcriptional processes are unknown. However, SWIB/MDM2 domains in MDM2 proteins have a conserved Gly residue whereas the domains in non-MDM2 proteins have retained a Trp residue in this position [18]. The Trp residue was conserved in PfMDM2 (Trp80 marked in green in Fig. 2) but not PfSWIB.
Overall sequence similarity was greater than identity in all these alignments, which correlates to a greater likelihood of homology as certain residue exchanges may have little or no effect on tertiary structure and/or protein function [50]. Protein structure, as opposed to sequence, often shows greater conservation during evolution [50, 51] and was, therefore, characterized for both P. falciparum SWIB/MDM2 homologues.
Neither P. falciparum SWIB/MDM2 domains has been crystallized. Predicted tertiary structures were generated using several online tools—the Swiss Model Workspace Automatic Modelling Mode [23], PHYRE2 [24] and ESyPred3D Web Server 1.0 [25]. Predictions were made relative to the server’s inherent template databases or to user-defined templates. All algorithms predicted a helical cleft topology, comparable to that of crystallized and soluble structures of SWIB/MDM2 domains, for both malaria proteins. The most reliable model for each, based on Q-mean analysis whereby a score closer to 1 represents a more reliable model, showed that PfMDM2 (Fig. 3e) and PfSWIB (Fig. 3f) had good Q-mean values and were similar to crystallized or soluble structures of SWIB/MDM2 domains (Figs. 3a–c). They aligned well when overlaid (Fig. 3g–h). However, both of these predicted structures lacked the presence of beta-sheets, involved in capping the classical SWIB/MDM2 ‘twisted cleft’ topology [9]; although, the SWIB/MDM2 domain of the yeast Swp73p/SNF12 protein [NCBI: P53628] also lacked beta sheets when modelled by the PHYRE2 algorithm (Fig. 3d). The modelled domain of PfMDM2 was composed of residues 56–124, which was two amino acids smaller than the annotated SWIB/MDM2 domain (residue 55–125).
Fig. 3.
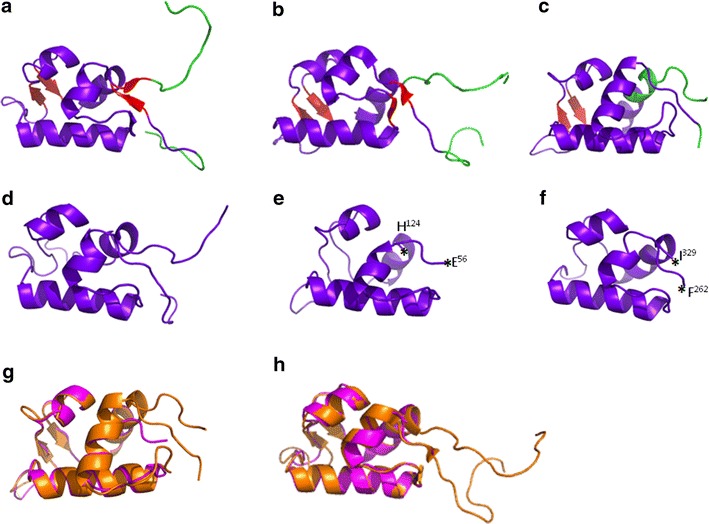
Molecular modelling of PfMDM2 and PfSWIB. Purple and red regions denote amino acids comprising the SWIB/MDM2 domain while green regions denote flanking regions. Purple regions represent alpha helices and random coils while red regions specifically highlight the beta sheets of the structure. a Solution structure of the SWIB/MDM2 domain of Mus musculus SWI/SNF related, matrix-associated, actin-dependent regulator of chromatin subfamily D member 1 protein (1UHR); b Solution structure of the Arabidopsis thaliana SWIB/MDM2 domain of the hypothetical protein At5g14170 (1V31); c Crystal structure of the Xenopus laevis SWIB/MDM2 domain of the E3 ubiquitin-protein ligase MDM2 protein (1YCQ, chain a); d Putative SWIB/MDM2 domain of yeast SNF12 modelled against 1UHR using the PHYRE2 modelling algorithm; e Putative SWIB/MDM2 domain of PfMDM2 modelled against 1YCQ using the EsyPred modelling algorithm, Qmean score 0.835, 13.7 % identity, annotated residues denote the start and end of the modelled domain, which are indicated by stars; f Putative SWIB/MDM2 domain of PfSWIB modelled simultaneously against 1UHR, 1V31 and 1V32 (solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana) using the PHYRE2 modelling algorithm, Qmean score 0.745, 28, 24 and 28 % identity to each template respectively, annotated residues denote the start and end of the modelled domain, which are indicated by stars. g Overlay of the modelled PfMDM2 domain (magenta) against 1YCQ (orange); h Overlay of the modelled PfSWIB domain (magenta) against 1V31 (orange)
PfMDM2 localized to the mitochondria under normal and heat stress conditions
The episomal pARL2-GFP expression system utilizes the crt promoter, allowing for continuous high level PfMDM2-GFP expression during all stages of the asexual intra-erythrocytic cycle, to aid in localization studies. However, focus was placed on the late asexual stages, since the transcriptional profile of PfMDM2 showed the highest mRNA expression in late trophozoites and schizonts, and current proteomic data has documented the presence of the protein in schizonts and salivary gland sporozoites [52, 53].
As presented in Fig. 4, a discrete, non-nuclear, localization pattern was observed in all the intra-erythrocytic asexual life stages. The number of GFP focal points increased with an increase in the nuclei of late stage schizonts (Fig. 4a) and MitoSOX confirmed mitochondrial localization. The correlation coefficient always greater than 0.9 (Fig. 4b; correlation coefficient (R) 0.94). The truncated PfMDM2-GFP protein lacking the N-terminal section and the predicted mitochondrial localization signal sequence (Fig. 1) remained in the cytoplasm (Fig. 4a). Although the protein was predicted to contain several strong nuclear localization sequences, the experimental data revealed that these sequences were inactive under the described conditions.
Fig. 4.
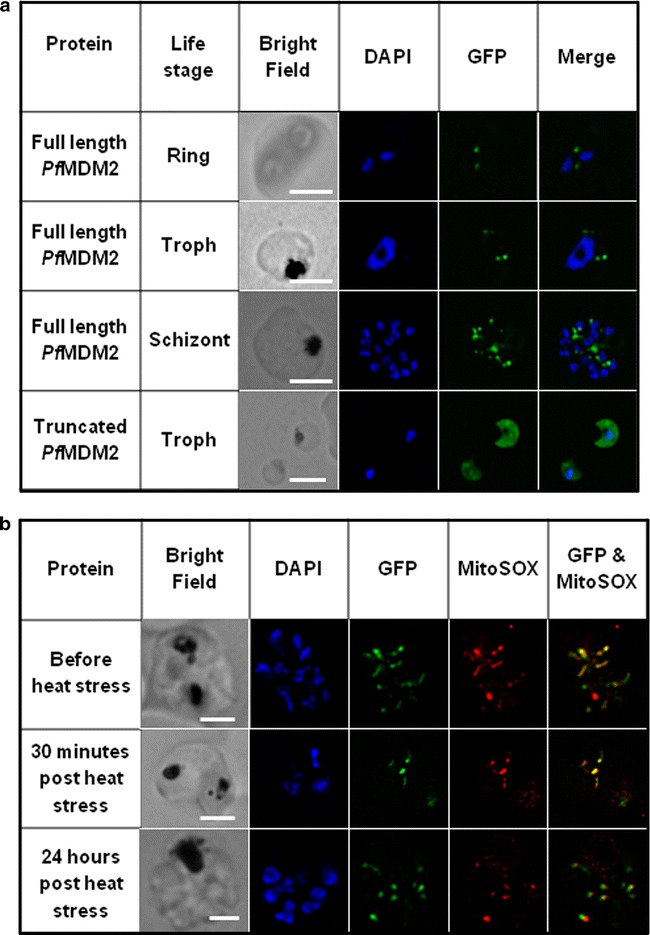
Mitochondrial localization of GFP-tagged PfMDM2 within live Plasmodium falciparum parasites, directed by the protein’s N-terminus. a Live imaging of the PfMDM2-GFP transgenic P. falciparum parasites under normal conditions. A distinct dot-like GFP signal was observed for PfMDM2, adjacent to the blue DAPI-stained nucleus, increasing in number with the progression of the life cycle. The truncated PfMDM2 protein lacking the N-terminus showed cytoplasmic distribution of the GFP signal. b Live co-localization of GFP-tagged PfMDM2 protein to the mitochondria of late schizont stage P. falciparum parasites under normal conditions and 30 min and 24 h after the termination of heat stress (41 °C for 2 h). R ring stage, T trophozoite stage, S schizont stage. Scale bar in bright field represents 2.5 μm
Heat-stress has previously been shown to induce PCD markers in P. falciparum [43]. The full-length PfMDM2-GFP protein was retained (correlation coefficient always greater than 0.9) within the mitochondria of late stage parasites, 30 min after the termination of heat stress at 41 °C for 2 h (Fig. 4b; correlation coefficient (R) 0.93). After 24 h (Fig. 4b) the red MitoSOX™ staining pattern was similar to that of PfMDM2-GFP but when overlaid the signals appeared to have shifted slightly; however, analysis with Image J still showed that co-localization was retained (correlation coefficient (R) 0.92). This shift may have been the result of the rapid cellular movement of the parasite or could indicate that the protein had moved out of the mitochondrion. The surviving parasites were, as expected, delayed in their development and were still in the late trophozoite stage.
PfSWIB shuttled between the cytoplasm and the nucleus under heat stress
Transcriptome analysis revealed that PfSWIB mRNA was expressed throughout the intra-erythrocytic life stages. Current proteomic data indicate that the protein was only detected in the late erythrocytic life stages, gametocytes and salivary gland sporozoites; however, it could still be expressed throughout the life cycle as the data are not complete [52, 53]. The use of the episomal pARL2-GFP expression system utilizing the crt promoter, would allow for continuous high level PfMDM2-GFP expression during all stages of the asexual intra-erythrocytic cycle, to aid in this localization study. Focus was, therefore, again placed on late asexual intra-erythrocytic parasites, which showed cytoplasmic localization of PfSWIB-GFP under normal growth conditions (Fig. 5). When PCD was induced by exposure to 41 °C for 2 h, there was a redistribution of PfSWIB-GFP to the nucleus, 30 min after termination of heat stress (correlation coefficient (R) always greater than 0.9). This nuclear GFP signal was only noted in roughly 10 % of the stressed trophozoites and is likely due to the activation of one of the protein’s predicted nuclear localization signals. From 2 h (Fig. 5) onwards after the termination of heat stress no nuclear signal was evident within the population. Nucleolar and/or chromatin localization has been documented for SWIB/MDM2 homologues in response to stress [11, 13, 54], although, in this study such discrete localization could not be determined.
Fig. 5.
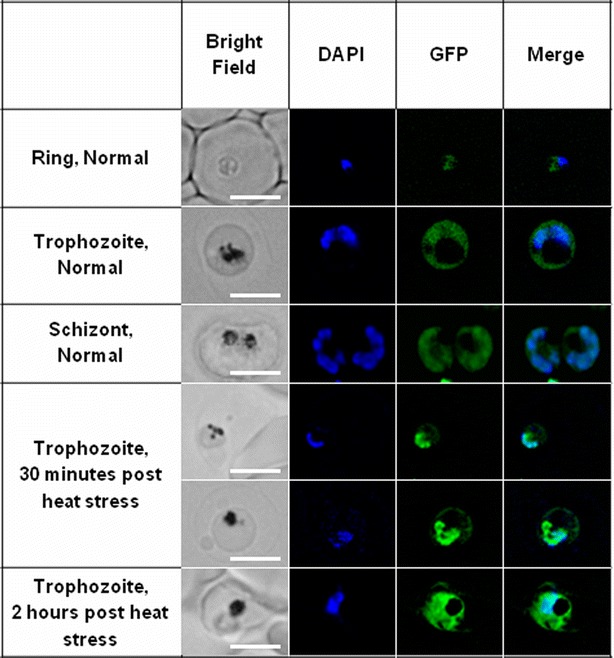
Live cellular localization of GFP-tagged PfSWIB within Plasmodium falciparum. A distinct cytoplasmic GFP signal was observed in all life stages under normal conditions,whereas exposure to 41 °C for 2 h resulted in nuclear localization, 30 min after termination of heat stress. This redistribution was short lived since a nuclear signal was absent 2–24 h afterwards. R ring stage, T trophozoite stage, S schizont stage. Scale bar in bright field represents 2.5 μm
Interactions of the SWIB/MDM2 domains of PfMDM2 and PfSWIB
The SWIB/MDM2 domains of PfMDM2 and PfSWIB were expressed and purified as recombinant GST-fusion proteins. SDS-PAGE and immunoblotting revealed that they migrated at ~33 and ~42 kDa, roughly their correct molecular masses of 36 and 44 kDa (Fig. 6a), respectively. PfMDM2 expressed as a full-length protein, but a small amount of a truncated form of ~24.5 kDa, not much bigger than the 23 kDa GST tag alone, was also produced.
Fig. 6.
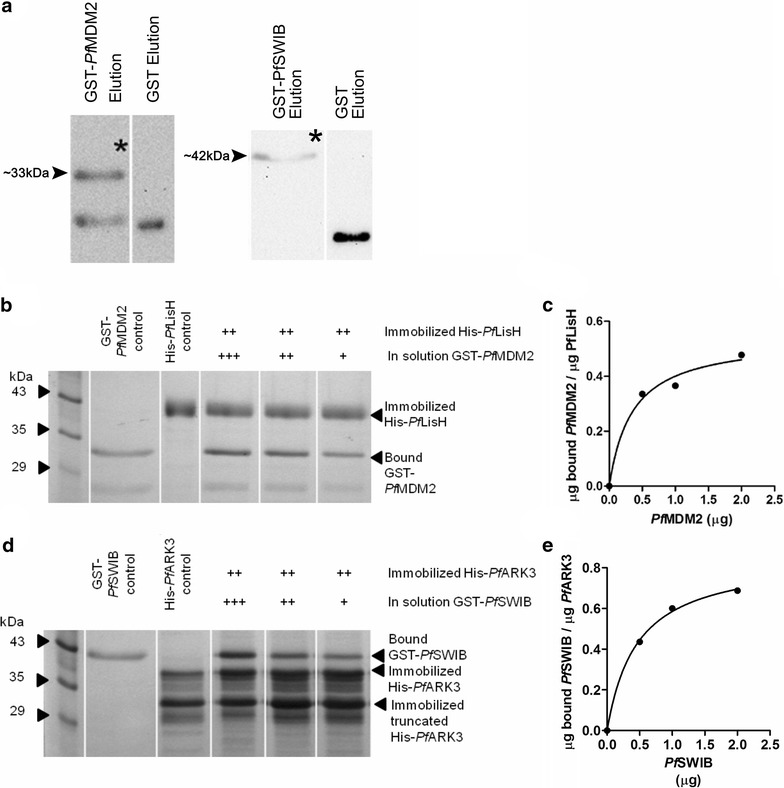
In vitro binding assays showed a concentration dependent interaction between PfLisH and PfMDM2. Representative images of a isolated and purified recombinant GST-PfMDM2 and GST-PfSWIB, migrating at ~33 and ~42 kDa, respectively, and identified by Western blot analysis using an anti-GST antibody (marked with star); b stained SDS polyacrylamide gels and c binding curves showing the concentration-dependent association between a constant amount of immobilized PfLisH and increasing amounts of PfMDM2; and d stained SDS polyacrylamide gels and e binding curves showing the concentration-dependent association between a constant amount of immobilized PfARK3 and increasing amounts of PfSWIB
The two SWIB/MDM2 recombinant domains were used for binding partner identification by phage display library technology (Table 1)—of the binding partners identified, three were assessed further. A central region of the PF3D7_1303400 protein [PlasmoDB: PF3D7_1303400] bound to PfMDM2. This conserved 1022 amino acid protein of unknown function contains an N-terminal Lissencephaly type-1-like homology (LisH) domain [48], situated 466 amino acids upstream from the PfMDM2 binding site and this protein will be designated as PfLisH. Bioinformatic predictions revealed a strong possibility of nuclear localization for this protein.
Table 1.
Binding partners for PfMDM2 and PfSWIB identified by biopanning of Plasmodium falciparum phage display libraries
| Protein | Identity of the binding partner (total number of amino acids) | Amino acid sequence of binding domain (location in protein) | Annotated domains within binding partner (location in protein) | Region of the binding partner cloned into a His-tag vector |
|---|---|---|---|---|
| PfMDM2 | PF3D7_1303400—conserved Plasmodium protein, unknown function (1022) |
KKKKKKEQTNEG KKSVKGINKKDK KRNSKVESKK (505–538) |
LisH domain (8–34) |
485–664 |
| PfSWIB | PF3D7_1356800—serine/threonine protein kinase, putative (ARK3) (4044) |
IYEKVNIDNDKV KKKNLHSINDKK IKINKTFMNEKD MKGNNRKKYNTE KRDNIKRNENDN EKK (788–847) |
Ser/Thr protein kinase domain, putative (1282–1528) |
601–922 |
| PfSWIB | PF3D7_1003600—membrane skeletal protein IMC1-related (ALV5) (281) |
PKTIIQEKIIHV PKNVTHIVEK IVEVPEVKYIEK IVEVPHIHYKNK YVPK (58–107) |
Inner membrane complex protein domain (58–153) |
2–241 |
Two binding partners were identified for PfSWIB. Firstly, an N-terminal region of PF3D7_1356800 [PlasmoDB: PF3D7_1356800], a putative serine/threonine protein kinase (Aurora-related kinase 3 (PfARK3)) [48] was identified and the binding domain was 434 amino acids upstream of the putative kinase domain. Bioinformatic analysis as part of this study suggested nuclear localization for this protein. Secondly, an N-terminal region, correlating to part of the inner membrane complex domain of PF3D7_1003600 [PlasmoDB: PF3D7_1003600], a membrane skeletal protein (Alveolin 5 (PfALV5)) [48] was identified, which localizes to the inner membrane complex of the parasite [55].
The biopanning-identified binding domains of these three proteins were directionally cloned into the pET-15 vector (Table 1). Only His-PfLisH and PfARK3 expressed as soluble recombinant proteins in sufficient quantity to verify the biopanning interactions. The His-PfLisH protein was identified by immunoblotting using an antibody directed against the His-tag and it migrated at ~41 kDa, roughly double its expected molecular mass of 23.5 kDa (Fig. 6b, lane 3). The construct contained the correct sequence, so the aberration was probably due to the two low complexity regions in the recombinant protein, which affected the binding of SDS [56]. The His-PfARK3 protein (Fig. 6d, lane 3), migrated at its appropriate molecular weight of 40 kDa and a prominent truncated form of ~31 kDa, still containing the N-terminal His tag, was also expressed. Based on the difference in molecular weight, the truncated protein would likely have lost about 75 C-terminal amino acids, including only about three amino acids from the binding domain identified by biopanning. In vitro binding assays were conducted with the GST-tagged protein in solution and the immobilized His-tagged protein, and also in a reciprocal manner. Two independent binding assays for each protein set confirmed a concentration-dependent association between PfMDM2 and PfLisH, as well as PfSWIB and PfARK3 (Fig. 6). Heat denatured ligand and GST protein alone were included as negative controls to ensure the specificity of binding. The controls revealed <25 % non-specific binding.
Discussion
SWIB/MDM2 proteins have been localized to the mitochondria, nucleus, chloroplast, and cytoplasm within a variety of organisms and their functional pro-survival and stress response roles in the nucleus are well documented [11, 13, 14, 17, 18, 19, 54]. However, nothing is known about the functions of the P. falciparum protein homologues. To investigate this, several molecular and bioinformatic analyses were performed, including the localization of GFP-tagged PfMDM2 and PfSWIB in transgenic parasites under both normal and heat stress conditions, and biopanning to identify binding partners.
The predicted tertiary structures of PfMDM2 and PfSWIB show similarities to SWIB/MDM2 domains
Bioinformatic analysis of SWIB/MDM2 domains identified in a variety of organisms has led to the idea that all extant forms share a common evolutionary ancestor with similar protein–protein interactions [18]. Crystallized SWIB/MDM2 domains exhibit ‘twisted cleft’ topologies constituted by four helices, creating a ‘barrel’, capped by one or more beta sheets [9, 26, 27, 28]. The SWIB/MDM2 domain is lined with numerous hydrophobic and aromatic residues (46.8 % of the amino acids for human MDM2), to create a suitable environment for the interaction with the transactivation domain of p53, mainly through van der Waals forces [9]. Two of the helices and one of the beta sheets of the ‘twisted cleft’ are involved directly in p53 binding [9].
The P. falciparum SWIB/MDM2 domains have similar, although slightly lower, hydrophobic and aromatic amino acid composition (39.4 % for PfMDM2 and 40.6 % for PfSWIB) and were predicted to form the ‘barrel’ of the classical SWIB/MDM2 structure but lacked beta-sheets. Modelling of the yeast SWIB/MDM2 domain, involved in chromatin remodelling [57], gave a similar result, suggesting that the ‘capping’ portion of the structure is not common to all SWIB/MDM2 domains. These data suggest that the parasite proteins are SWIB/MDM2 homologues.
The role of Pf MDM2 in the mitochondrion
This study revealed mitochondrial localization of PfMDM2 under normal and heat stress conditions (Fig. 3), indicating that PfMDM2 is not implicated in a nuclear heat-induced PCD response. However, PfMDM2 localization could not be assessed in those parasites which had died and a role of the protein in PCD of that sub-population could not be evaluated. In Arabidopsis, two out of six group I SWIB proteins, At1g31760 [NCBI: ABD59092.1] and At2g35605 [NCBI: AEC09127.1], containing only a SWIB/MDM2 domain, have been localized to the mitochondria, but their functions are unknown [19]. The P. falciparum genome shares similarities with Arabidopsis thaliana [58] and EMBOSS Needle alignment of the SWIB/MDM2 domain of PfMDM2 revealed strong conservation of residues in At1g31760 (39 % identity and 51.9 % similarity) and At2g35605 (43.3 % identity and 55.8 % similarity). Thus, it can be speculated that PfMDM2 plays a similar role to At1g31760 or At2g35605 within the mitochondrion. PfMDM2 could be involved in transcriptional regulation, as part of a larger complex, since the mitochondrial genome codes for three genes [44].
A single phage display interaction (PfLisH) was documented for PfMDM2, which could indicate that this domain is limited in its interactions or it could reflect the fact that the phage display libraries used are not representative of the entire transcriptional profile of the parasite throughout its intra-erythrocytic life cycle [59]. In addition, immobilization of the P.falciparum SWIB/MDM2 domain on magnetic beads may limit its interaction with the phage. Finally, the SWIB/MDM2 domain on its own may not be sufficient for interactions that require the entire protein or specific post-translational modifications [59].
The PfLisH mRNA and protein expression profile coincides with that of PfMDM2 [52, 53], but the cellular location of PfLisH is unknown, although bioinformatics strongly suggests nuclear localization, which makes an in vivo association under heat stress unlikely. PfMDM2 has several predicted nuclear localization signals, possibly allowing it to traffic to the nucleus under other stressful conditions, providing an opportunity to interact with PfLisH.
LisH motifs have been identified in 114 eukaryotic proteins from yeast to humans [60], and this commonly N-terminal domain participates in a variety of cellular processes, including transcriptional regulation, although it does not possess any known DNA binding motifs [61]. Of interest are the human TBL1 and the yeast Sif2p proteins. TBL1 interacts with the N-terminal SANT (SWI3/ADA2/N-CoR/TFHIIB) domains of the nuclear receptor co-repressor protein which is present in a variety of chromatin-associated complexes, including the SWI/SNF complex [62, 63].
If PfMDM2 does move to the nucleus under specific conditions it could be hypothesized that PfLisH and PfMDM2 interact as part of a larger gene transcription complex. In the case of yeast Sif2p, its deletion increases mortality in response to starvation, especially under heat stress, which supports a role in gene-regulated stress resistance [64]. Therefore, a potential interaction of PfMDM2 with PfLisH in the nucleus may reflect a response to a physiological stress other than heat.
To live or let die: that is the question for PfSWIB
This study highlighted the unexpected localization of PfSWIB to the cytoplasm under normal growth conditions; the rapid but short-lived nuclear localization in response to elevated temperatures (Fig. 5), and a novel binding partner (Fig. 6; Table 1). These data suggest three possible scenarios pertaining to the function of PfSWIB.
Could PfSWIB be involved in parasite survival?
Previous work has documented that only a small number of late-stage parasites, undergoing heat stress, survive in vitro 24 h later [43], and this may correlate to the small number of trophozoite parasites showing a nuclear PfSWIB-GFP signal. Elevated temperatures may activate one or more of the predicted nuclear localization signals, possibly by phosphorylation which is known to regulate nuclear import in other proteins [65]. The binding partner, PfARK3, is a putative atypical aurora-related serine/threonine protein kinase [66] and has a mRNA and protein expression pattern correlating to the late intra-erythrocytic asexual life stages—coinciding with the expression of PfSWIB [52, 53]. PfARK3 lacks any annotated signal sequences but bioinformatic analysis in this study suggests nuclear localization, although such predictions should be treated with caution.
The human Aurora kinase A aids in inactivating and promoting the degradation of p53 through phosphorylation [67, 68], and a putative p53 homologue has been identified in the parasite genome [8]. It could thus be speculated that PfSWIB and PfARK3 participate in preventing p53-mediated apoptosis in the nucleus in response to heat stress. Alternatively, PfSWIB may influence parasite survival through transcriptional regulation by directing the phosphorylation activities of PfARK3, as human Aurora kinase B and its yeast homologue play key roles in gene transcription through histone H3 phosphorylation [69, 70]. However, although there was a considerable drop in the parasitaemia 24 h after heat stress, more than 10 % of the population still remained, suggesting that this cellular phenomenon could be pro-survival response in a parasite sub-population.
Could PfSWIB be involved in parasite death?
The SWIB/MDM2 homologue BAF60a of the mammalian SWI/SNF complex induces p53-directed apoptosis [17] and the parasite PfSWIB protein may play a similar stage specific pro-PCD role, whereby transient nuclear localization triggers removal of those parasites. Several studies have shown that elevated temperatures >38.5 °C significantly inhibited the development and growth of intra-erythrocytic asexual parasites, which showed several features of necrosis and/or PCD [5]. This event is hypothesized to reduce the host’s parasite burden to prevent premature host death before effective parasite transmission [2, 43, 71]. Furthermore, interactions with PfARK3 may influence parasite survival through transcriptional regulation by possible histone phosphorylation [69, 70]. Fever is linked to schizont rupture and merozoite re-invasion, with the early life stages surviving in vitro heat stress better than late stages [5, 43, 71, 72]. Thus, removal of any lagging trophozoite stage parasites, each capable of giving rise to as many as 32 new parasites, could have a significant impact in decreasing the number of parasites. The schizonts, which did not show nuclear PfSWIB-GFP localization, may have passed the point of susceptibility to PfSWIB-directed PCD.
Could PfSWIB play a non-PCD role?
Although the protein conforms to a similar three-dimensional topology as other SWIB/MDM2 homologues, it does not imply a direct PCD role. A group I SWIB protein, At3g48600 [NCBI: AEE78436.1], has been documented in the cytoplasm of Arabidopsisthaliana [19]. EMBOSS Needle alignment of this protein with the SWIB/MDM2 domain of PfSWIB revealed 22.5 % identity and 42.7 % similarity, suggesting that these two proteins may share similar, as yet unknown, functions in the cytoplasm.
One of the roles of PfSWIB may be related to heat stress regulation. Firstly, the absence of Swp73p/SNF12 in yeast resulted in temperature-sensitive mutants, highlighting the involvement of this SWIB homologue in heat stress transcriptional regulation [57]. Secondly, in other organisms, similar migration patterns for heat stress participants in response to the addition or removal of heat stress, has been documented [73]. Thirdly, association between PfAVL5, a member of the inner membrane complex [55], and PfSWIB may assist in compartmentalizing the PfSWIB protein until stress induces nuclear localization, another feature of heat stress participants [74–76]. A discrete localization pattern to the inner membrane complex was not documented for PfSWIB, which may be a consequence of PfSWIB-GFP protein over-expression, using the crt-promoter driven expression system. Fourthly, the SWI/SNF complex is involved in transcriptional initiation of heat shock protein 70 genes in humans and yeast [77, 78] and PfSWIB may facilitate such transcriptional regulation [69, 70].
Conclusion
The concept of PCD in P. falciparum is supported by the presence of biochemical markers in parasites subjected to stressful conditions. However, there is currently no knowledge of the genes and pathways involved in this process. This study provides novel insight by evaluating the molecular functions of two SWIB/MDM2 homologues, PfMDM2 and PfSWIB, which are potential PCD participants. It provides the first description of their localization within the parasite and their response to elevated temperatures, which induce PCD markers in parasites and which mimic fever periods experienced by malaria patients. Novel interactions with other parasite proteins were also identified. Bioinformatics suggested that the proteins were chromatin remodelling family members, deviating slightly from the typical twisted cleft topology of this group but structurally similar to the yeast SWIB/MDM2 homologue. Unexpectedly, PfMDM2 showed N-terminal-directed mitochondrial localization under both normal and heat-induced PCD conditions. PfSWIB localized to the cytoplasm under normal conditions, but after heat stress, it revealed a short-lived nuclear localization in a sub-population of trophozoites. Interestingly, some SWIB homologues in Arabidopsis thaliana also localize to either the mitochondria or the cytoplasm, which suggests that PfMDM2 and PfSWIB may play similar roles to these proteins, as opposed to metazoan SWIB/MDM2 homologues. Based on the data from this study, it is hypothesized that PfMDM2 is involved in mitochondrial maintenance and gene expression, possibly as part of a larger transcriptional complex. Furthermore, it is postulated that PfSWIB may have a stage-specific, pro-survival function and participate in the heat stress response of the parasite.
Authors’ contributions
WAV participated in the design of the study, carried out all the experimental work and bioinformatic analysis, and drafted the manuscript. TLC conceived the study, participated in its design and coordination and aided in drafting and editing the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Dr. Sonja Lauterbach, Dr. Roberto Lanzillotti and Mr. Dale Liebenberg for kindly donating P. falciparum phage display libraries towards this study. This work is based upon research supported by the National Research Foundation (NRF), Grant 73703, and the Medical Faculty Research Endowment Fund of the University of the Witwatersrand.WAV was funded by the National Research Foundation (NRF) Grant Holder Linked Bursary (Grant 73703) and the University of the Witwatersrand Merit Award. Any opinion, findings and conclusions or recommendations expressed in this material are those of the authors and therefore the NRF do not accept any liability in regard thereto.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- PCD
programmed cell death
- MDM2
mouse double minute 2
- SWI/SNF
switch/sucrose non fermentable
- SWIB
SWI/SNF complex B
- p53
protein 53
- BAF60a
BRG1-associated factor 60a
- LisH
lissencephaly type-1-like homology (LisH)
- ARK
aurora related kinase
- AVL
alveolin
- HRP
horse radish peroxidase
- BSA
bovine serum albumin
Additional files
10.1186/s12936-015-1065-9 PCR primers for the amplification of Plasmodium falciparum genes/domains.
10.1186/s12936-015-1065-9 PCR primers for the amplification of binding partners for directional cloning into the pET-15b vector.
10.1186/s12936-015-1065-9 Live imaging of Plasmodium falciparum schizonts with Mitotracker™ FM Green and MitoSOX™ showing mitochondrial co-localization (Correlation coefficient (R) 0.91). Scale bar in bright field represents 5 μm.
Contributor Information
Warren Antonio Vieira, Phone: +27 11 717 2188, Email: warrenv@live.co.za.
Thérèsa L. Coetzer, Phone: +27 11 717 2188, Email: theresa.coetzer@nhls.ac.za
References
- 1.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 2.Deponte M, Becker K. Plasmodium falciparum—do killers commit suicide? Trends Parasitol. 2004;20:165–169. doi: 10.1016/j.pt.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava IK, Rottenberg H, Vaidya AB. Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J Biol Chem. 1997;272:3961–3966. doi: 10.1074/jbc.272.7.3961. [DOI] [PubMed] [Google Scholar]
- 4.Picot S, Burnod J, Bracchi V, Chumpitazi BF, Ambroise-Thomas P. Apoptosis related to chloroquine sensitivity of the human malaria parasite Plasmodium falciparum. Trans R Soc Trop Med Hyg. 1997;91:590–591. doi: 10.1016/S0035-9203(97)90039-0. [DOI] [PubMed] [Google Scholar]
- 5.Engelbrecht D, Durand PM, Coetzer TL. On programmed cell death in Plasmodium falciparum: status quo. J Trop Med. 2012;2012:646534. doi: 10.1155/2012/646534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mutai BK, Waitumbi JN. Apoptosis stalks Plasmodium falciparum maintained in continuous culture condition. Malar J. 2010;9(Suppl 3):S6. doi: 10.1186/1475-2875-9-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y, Wang X, Liu X, Wang Y. Data-mining approaches reveal hidden families of proteases in the genome of malaria parasite. Genome Res. 2003;13:601–616. doi: 10.1101/gr.913403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coetzer TL, Durand PM, Nedelcu AM. Genomic evidence for elements of a programmed cell death pathway in Plasmodium: exploiting programmed parasite death for malaria control? Blood. 2010;116:4226. [Google Scholar]
- 9.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 10.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-R. [DOI] [PubMed] [Google Scholar]
- 11.Shadfan M, Lopez-Pajares V, Yuan ZM. MDM2 and MDMX: alone and together in regulation of p53. Transl Cancer Res. 2012;1:88–99. [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson BG, Roberts CWM. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 13.De La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, et al. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20:2839–2851. doi: 10.1128/MCB.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shivaswamy S, Iyer VR. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol Cell Biol. 2008;28:2221–2234. doi: 10.1128/MCB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JH, Park EJ, Hur SK, Kim S, Kwon J. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair. 2009;8:29–39. doi: 10.1016/j.dnarep.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem. 2002;277:22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- 17.Oh J, Sohn DH, Ko M, Chung H, Jeon SH, Seong RH. BAF60a interacts with p53 to recruit the SWI/SNF complex. J Biol Chem. 2008;283:11924–11934. doi: 10.1074/jbc.M705401200. [DOI] [PubMed] [Google Scholar]
- 18.Bennett-Lovsey R, Hart SE, Shirai H, Mizuguchi K. The SWIB and the MDM2 domains are homologous and share a common fold. Bioinformatic. 2002;18:626–630. doi: 10.1093/bioinformatics/18.4.626. [DOI] [PubMed] [Google Scholar]
- 19.Melonek J, Matros A, Trosch M, Mock HP, Krupinska K. The core of chloroplast nucleoids contains architectural SWIB domain proteins. Plant Cell. 2012;24:3060–3073. doi: 10.1105/tpc.112.099721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clustal Omega. EMBL-EBI, Cambridge. 2015. http://www.ebi.ac.uk/Tools/msa/clustalo/Accessed 1 Oct 2014.
- 21.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 22.EMBOSS Needle. EMBL-EBI, Cambridge. 2015. http://www.ebi.ac.uk/Tools/psa/emboss_needle/. Accessed 1 Oct 2014.
- 23.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 24.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 25.Lambert C, Léonard N, De Bolle X, Depiereux E. ESyPred3D: prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- 26.Yamada K, Saito K, Nameki N, Inoue M, Koshiba S, Shirouzu M, et al. 1UHR: solution structure of the SWIB domain of mouse BRG1-associated factor 60a. RCSB Protein Data Bank. 2004. http://www.rcsb.org/pdb/explore.do?structureId=1uhr.
- 27.Yoneyama M, Tochio N, Koshiba S, Inoue M, Kigawa T, Yokoyama S. 1V32: Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana. RCSB Protein Data Bank. 2004. http://www.rcsb.org/pdb/explore/explore.do?structureId=1V32.
- 28.Yoneyama M, Tochio N, Koshiba S, Inoue M, Kigawa T, Yokoyama S. 1V31: Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g14170 from Arabidopsis thaliana. RCSB Protein Data Bank. 2004. http://www.rcsb.org/pdb/explore/explore.do?structureId=1V31.
- 29.Schrodinger LLC. The PyMOL molecular graphics system, open source project and code. http://sourceforge.net/projects/pymol/(2015). Accessed 5 Dec 2015.
- 30.Benkert P, Künzli M, Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 33.Brameier M, Krings A, Maccallum R. NucPred-predicting nuclear localization of proteins. Bioinformatics. 2007;23:1159–1160. doi: 10.1093/bioinformatics/btm066. [DOI] [PubMed] [Google Scholar]
- 34.Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, et al. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- 35.Waller RF, Keeling PJ, Donald RGK, Striepen B, Handman E, Lang-Unnasch N, et al. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc Natl Acad Sci USA. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuegge J, Ralph S, Schmuker M, Mcfadden GI, Schneider G. Deciphering apicoplast targeting signals—feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene. 2001;280:19–26. doi: 10.1016/S0378-1119(01)00776-4. [DOI] [PubMed] [Google Scholar]
- 37.Bender A, Van Dooren GG, Ralph SA, Mcfadden GI, Schneider G. Properties and prediction of mitochondrial transit peptides from Plasmodium falciparum. Mol Biochem Parasitol. 2003;132:59–66. doi: 10.1016/j.molbiopara.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 39.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. Am Soc Parasitol. 1979;65:418–420. doi: 10.2307/3280287. [DOI] [PubMed] [Google Scholar]
- 40.Cowman AF, Crabb BS, Maier AG, Tonkin CJ, Healer J, Gibson P, et al. Preparation of P. falciparum genomic DNA. In: Moll K, Ljugstöm I, Perlmann H, Scherf A, Wahlgren M, editors. Methods in Malaria Research. Fifth ed. Paris: Malaria Research and Reference Reagent Resource Center. 2008. p. 287–8.
- 41.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 42.Lauterbach S, Lanzillotti R, Coetzer T. Construction and use of Plasmodium falciparum phage display libraries to identify host parasite interactions. Malar J. 2003;2:47. doi: 10.1186/1475-2875-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engelbrecht D, Coetzer TL. Turning up the heat: heat stress induces markers of programmed cell death in Plasmodium falciparum in vitro. Cell Death Dis. 2013;4:e971. doi: 10.1038/cddis.2013.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torrentino-Madamet M, Desplans J, Travaille C, James Y, Parzy D. Microaerophilic respiratory metabolism of Plasmodium falciparum mitochondrion as a drug target. Curr Mol Med. 2010;10:29–46. doi: 10.2174/156652410791065390. [DOI] [PubMed] [Google Scholar]
- 45.Tonkin CJ, Van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, et al. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol. 2004;137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Rasband W. Image J, version 1.48v. 2015. http://imagej.nih.gov/ij/index.html. Accessed 4 Dec 2015.
- 47.PlasmoDB. The Plasmodium genomic resource. The EuPathDB Project Team. 2015. http://plasmodb.org/plasmo/. Accessed 1 Jan 2014.
- 48.Interpro. EMBL-EBI, Cambridge. 2015. http://www.ebi.ac.uk/interpro/. Accessed 4 Dec 2015.
- 49.Freedman DA, Epstein CB, Roth JC, Levine AJ. A genetic approach to mapping the p53 binding site in the MDM2 protein. Mol Med. 1997;3:248–259. [PMC free article] [PubMed] [Google Scholar]
- 50.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 51.Geourjon C, Combet C, Blanchet C, Deléage G. Identification of related proteins with weak sequence identity using secondary structure information. Protein Sci. 2001;10:788–797. doi: 10.1110/ps.30001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.López-Barragán MJ, Lemieux J, Quiñones M, Williamson KC, Molina-Cruz A, Cui K, et al. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genom. 2011;12:587. doi: 10.1186/1471-2164-12-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Florens L, Liu X, Wang Y, Yang S, Schwartz O, Peglar M, et al. Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol Biochem Parasitol. 2004;135:1–11. doi: 10.1016/j.molbiopara.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 54.Catalano A. O’day D. Nucleoplasmic/nucleolar translocation and identification of a nuclear localization signal (NLS) in Dictyostelium BAF60a/SMARCD1 homologue Snf12. Histochem Cell Biol. 2012;138:515–530. doi: 10.1007/s00418-012-0973-9. [DOI] [PubMed] [Google Scholar]
- 55.Kono M, Herrmann S, Loughran NB, Cabrera A, Engelberg K, Lehmann C, et al. Evolution and architecture of the inner membrane complex in asexual and sexual stages of the malaria parasite. Mol Biol Evol. 2012;29:2113–2132. doi: 10.1093/molbev/mss081. [DOI] [PubMed] [Google Scholar]
- 56.Tompa P. Intrinsically unstructured proteins. Trends Biol Sci. 2002;27:527–533. doi: 10.1016/S0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 57.Cairns BR, Levinson RS, Yamamoto KR, Kornberg RD. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 1996;10:2131–2144. doi: 10.1101/gad.10.17.2131. [DOI] [PubMed] [Google Scholar]
- 58.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanzillotti R, Coetzer TL. Phage display: a useful tool for malaria research? Trends Parasitol. 2008;24:18–23. doi: 10.1016/j.pt.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Emes RD, Ponting CP. A new sequence motif linking lissencephaly, Treacher Collins and oral–facial–digital type 1 syndromes, microtubule dynamics and cell migration. Hum Mol Genet. 2001;10:2813–2820. doi: 10.1093/hmg/10.24.2813. [DOI] [PubMed] [Google Scholar]
- 61.Wei Y, Jin J, Harper JW. The cyclin E/Cdk2 substrate and cajal body component p220NPAT activates histone transcription through a novel lish-like domain. Mol Cell Biol. 2003;23:3669–3680. doi: 10.1128/MCB.23.10.3669-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/S1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 63.Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 64.Cockell M, Renauld H, Watt P, Gasser SM. Sif2p interacts with the Sir4p amino-terminal domain and antagonizes telomeric silencing in yeast. Curr Biol. 1998;8:787–790. doi: 10.1016/S0960-9822(98)70304-5. [DOI] [PubMed] [Google Scholar]
- 65.Nardozzi JD, Lott K, Cingolani G. Phosphorylation meets nuclear import: a review. Cell Commun Signal. 2010;8:32. doi: 10.1186/1478-811X-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reininger L, Wilkes JM, Bourgade H, Miranda-Saavedra D, Doerig C. An essential Aurora-related kinase transiently associates with spindle pole bodies during Plasmodium falciparum erythrocytic schizogony. Mol Microbiol. 2011;79:205–221. doi: 10.1111/j.1365-2958.2010.07442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, et al. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J Biol Chem. 2004;279:52175–52182. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- 68.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 69.Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, et al. Mitotic phosphorylation of histone H3 Is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/S0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 70.Goto H, Yasui Y, Nigg EA, Inagaki M. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7:11–17. doi: 10.1046/j.1356-9597.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- 71.Porter H, Gamette MJ, Cortes-Hernandez DG, Jensen JB. Asexual blood stages of Plasmodium falciparum exhibit signs of secondary necrosis, but not classical apoptosis after exposure to febrile temperature (40 °C) J Parasitol. 2008;94:473–480. doi: 10.1645/GE-1343.1. [DOI] [PubMed] [Google Scholar]
- 72.Long HY, Lell B, Dietz K, Kremsner PG. Plasmodium falciparum: in vitro growth inhibition by febrile temperatures. Parasitol Res. 2001;87:553–555. doi: 10.1007/s004360100374. [DOI] [PubMed] [Google Scholar]
- 73.Hsu SF, Lai HC, Jinn TL. Cytosol-localized heat shock factor-binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in Arabidopsis. Plant Physiol. 2010;153:773–784. doi: 10.1104/pp.109.151225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Al-Whaibi MH. Plant heat-shock proteins: a mini review. J King Saud Univ. 2011;23:139–150. doi: 10.1016/j.jksus.2010.06.022. [DOI] [Google Scholar]
- 75.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schlesinger MJ, Aliperti G, Kelley PM. The response of cells to heat shock. Trends Biol Sci. 1982;7:222–225. doi: 10.1016/0968-0004(82)90096-2. [DOI] [Google Scholar]
- 77.Corey LL, Weirich CS, Benjamin IJ, Kingston RE. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 2003;17:1392–1401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han Q, Lu J, Duan J, Su D, Hou X, Li F, et al. Gcn5- and Elp3-induced histone H3 acetylation regulates hsp70 gene transcription in yeast. Biochem J. 2008;409:779–788. doi: 10.1042/BJ20070578. [DOI] [PubMed] [Google Scholar]


