Abstract
Strong disruptive ecological selection can initiate speciation, even in the absence of physical isolation of diverging populations. Species evolving under disruptive ecological selection are expected to be ecologically distinct but, at least initially, genetically weakly differentiated. Strong selection and the associated fitness advantages of narrowly adapted individuals, coupled with assortative mating, are predicted to overcome the homogenizing effects of gene flow. Theoretical plausibility is, however, contrasted by limited evidence for the existence of rugged adaptive landscapes in nature. We found evidence for multiple, disruptive ecological selection regimes that have promoted divergence in the sympatric, incipient radiation of ‘sharpfin’ sailfin silverside fishes in ancient Lake Matano (Sulawesi, Indonesia). Various modes of ecological specialization have led to adaptive morphological differences between the species, and differently adapted morphs display significant but incomplete reproductive isolation. Individual fitness and variation in morphological key characters show that disruptive selection shapes a rugged adaptive landscape in this small but complex incipient lake fish radiation.
Keywords: adaptive radiation, sympatric speciation, ecological speciation, adaptive landscapes, disruptive selection, Telmatherina
1. Introduction
Disruptive ecological selection favours adaptation to distinct environments [1,2], and may promote speciation, even in the absence of physical isolation [2–6]. Fitness advantages of well-adapted phenotypes arise by local adaptation, a phenomenon that is common in both plants and animal taxa (reviewed in [7]), including case studies of ecological speciation, such as sticklebacks, Darwin's finches and Heliconius butterflies [8–10].
The fitness consequences of phenotypic variation in a given environment can be conceptualized with adaptive landscapes [1,11,12]. Phenotypes that are well adapted with respect to key evolutionary traits tend to occupy peaks in the adaptive landscape, whereas poorly adapted individuals occur in lower regions of the landscape [1,2,11–13]. Adaptive landscape topologies may range from uniform (lack of selection) or unimodal (stabilizing selection) to bi- or multimodal (disruptive selection) distributions [2,12,13]. In the simplest case, such a landscape is based on fitness estimation over variation in one single phenotypic trait, such as the beak in birds [14,15]. However, it is likely that selection frequently affects more than a single structure, and adaptive landscapes can also be used to explore more complex, multivariate axes of phenotypic trait variation [1,11,13,16].
Adaptive radiations provide examples for the rapid evolution of conspicuous adaptations to alternative food resources and habitats [1]. Disruptive ecological selection may affect variation in multiple key traits, and gain additional momentum with the evolution of increasing species diversity [11]. The adaptive landscape of an adaptive radiation can therefore be expected to be complex and rugged, sensu Nosil [2, p. 58] (i.e. with multiple distinct peaks and deep valleys) [1,11–13].
Empirical evidence for the theoretical prediction that rugged adaptive landscapes facilitate the evolution of species flocks remains still limited, certainly for adaptive radiations in which taxa evolved in sympatry. A notable exception is a study [17] of the small Cyprinodon pupfishes radiation on San Salvador islands, for which a direct link between phenotype and fitness is established, and which supports the role of rugged adaptive landscapes in adaptive radiation. However, this example is based on hybrids that were bred in cages or the laboratories, and from species that rarely interbreed in nature [18]. Therefore, evidence that rugged adaptive landscapes promote adaptive radiation in nature remains largely lacking.
Here we study a radiation of sailfin silversides that constitutes a promising system to examine the relation between the shape of adaptive landscapes and adaptive radiation. More precisely, we test whether ecologically based disruptive selection can drive the evolution of rugged adaptive landscapes under sympatric conditions.
The small, but morphologically complex fish radiation of ‘sharpfins' (Telmatherinidae: Telmatherina) from ancient graben-Lake Matano (Central Sulawesi, Indonesia) is the most diverse lineage of sailfin silversides in the Malili Lakes System [19]. The species flock is endemic to Lake Matano [20] and consists of several ecological specialists with conspicuous morphological and behavioural adaptations to various ecological niches [21–23]. However, specific morphotypes do not form well-sorted clades in phylogenetic analyses based on mitochondrial markers [24]. High levels of interspecific gene flow [25] and direct evidence for hybridization [24,26] have been observed, but morphotypes nevertheless mate largely assortatively [25,27]. As such, reproductive isolation among Lake Matano's Telmatherina can be considered to be substantial but incomplete. The onset of sharpfin diversifications commenced approximately 0.9 Ma [28].
Using a random sample of sharpfins, we perform integrated analyses of an estimator for individual fitness, an array of morphological traits that influence ecological specialization, the habitat context and stomach contents. We estimate the adaptive landscape underlying the sharpfin radiation and present signatures of ecologically based disruptive selection in the light of population structure, habitat use and trophic ecology.
2. Material and methods
(a). Sampling
We pre-defined a total of 160 sampling locations, distributed equally (approx. every 500 m) all around Lake Matano's shoreline (electronic supplementary material, figure S1a). Sampling locations were fished in a random order between May and August 2010, by placing multimesh gillnets at depths of 0.75, 1.5, 3, 4.5 and 6 m. Gillnets were placed consistently for 20 min; fishes were euthanized in MS222, and the gutted body and liver were weighed with a precision of 0.001 g. Subsequently, a muscle tissue sample was stored in RNAlater (Qiagen), whereas specimens were transferred to 80% ethanol. Only mature individuals (determined by fin shape and colour in males, and gonad state in females) were included in the analyses (n = 1060).
(b). Ecomorphological traits
We analysed variation in overall body shape and head shape with geometric morphometrics from digital X-ray images [29]. Homologous landmarks (15 and 13 for the body and head, respectively; electronic supplementary material, figure S1b) were digitized in tpsDig2 [30]. GLS procrustes superimposition coordinates [29] were calculated for each dataset in PAST [31] to reduce effects of size and position. The first right gill arches were dissected and measured with ImageJ [32] (electronic supplementary material, figure S1b). Principal component analyses (PCA) were carried out independently for each geometric morphometric dataset and the relative lengths of the measured gill traits (residuals of a linear regression of measured distances by the length of upper arch).
(c). Phenotypic structure
Principal components (PCs) of the shape analyses and the gill trait measurements were scored to their true rank, which is defined here as the number of PCs, explaining together 99% of the total variance in each dataset. The datasets were then scored by multiplying each PC's residual by the square root of the rank divided by the variance of the dataset [33]. We combined the PCs of the shape analyses and the gill trait measurements in a k-mean clustering approach, and in a discriminant principal component analysis (DAPC; R package adegenet [34,35]), to examine how the various morphotypes in the radiation are structured in morphospace. Overfitting during the DAPC was avoided by retaining less than one-third of the original number of PCs (18 of 56) [34].
Identifying the actual number of clusters with a statistical procedure (e.g. BIC support values for various scenarios) is difficult [34]. Here, the BIC values indicate that 5–11 clusters can appropriately summarize the data. To estimate the actual number of clusters within the obtained range we applied the NbClust function [36,37] to the dataset. This function uses 22 criteria to determining the number of clusters and indicated that five clusters (morphotypes) summarize the data most adequately (electronic supplementary material, table S1).
(d). Stomach content analysis
Individual stomach contents were analysed following Pfaender et al. [23,38]. Food items of the gastrointestinal tract between oesophagus and pylorus were determined to the most accurate taxonomic level feasible and their relative volumetric proportions (in %) were estimated. The 933 individuals containing food items in their stomach were included in the stomach content analysis. Strong negative correlations between some types of content were observed, especially between molluscs, terrestrial insects, copepods and fish eggs. Therefore, a PCA was applied to the correlation matrix of the proportional stomach contents to reduce multicollinearity. We applied statistical tests (ANOVAs and Tukey's post hoc tests when variances were equal; Welch tests with Dunnet T3 post hoc tests when they were unequal) on individual PCs to test for pairwise differentiation between morphotypes in feeding ecology.
(e). Habitat differentiation
We examined the habitats where fish were caught to identify habitat parameters that may explain differences in the abundance of sharpfin morphotypes. Four habitat parameters were included in the canonical correspondence analysis (CCA; R package vegan [39]): dominant substrate at the general sampling location (rock, gravel, sand); substrate directly at the gillnet (soft to hard in %); bathymetric slope where the gillnet was positioned (5° steps); and sedimentation, which is related to surrounding canopy coverage (low, medium, high). Significance of the habitat–morphotype associations and individual canonical axes were calculated using the anova.cca function [39] with 11 000 permutations per step. Each placed gillnet was treated as a single catch event.
(f). Amplified fragment length polymorphism procedure
DNA was extracted from muscle tissue (NucleoSpin 96; Machery and Nagel) of the 1060 individuals, following the manufacturer's protocol. Quality of DNA was checked on 1% agarose gels; DNA concentrations were measured with a NanoDrop 200 (Thermo Scientific) and adjusted to 25 ng µl−1. We applied a slightly modified AFLP method described by Vos [40] (see the electronic supplementary material, notes and [25,24] for details). In total, 1218 polymorphic AFLP markers were retained.
(g). Genetic differentiation and population structure
To test how the identified phenotypic clusters relate to genetic population structure, a DAPC and reassignment test (see detailed description above) were applied to the retained AFLP markers. Analysis of molecular variance (AMOVA [41]), implemented in Arlequin v. 3.5.1.2 [42], was used to investigate the structure of the genetic variation among phenotypic clusters.
(h). Loci under selection
Allele distribution models are a powerful method to identify alleles under selection from AFLP datasets [43,44]. They use multiple univariate logistic regressions to test the relation between the frequency of AFLP loci and phenotypic variables. We applied ADMs in MATSAM [43,44] on all retained loci and the PC residuals from the ecomorphological trait analyses, centroid size, sex and sample location to test for correlation. p-values resulting from multiple tests have been adjusted with the sequential goodness-of-fit correction (SGoF [45]). The correction controls for the family-wise error rate, which is suitable given that a large number of tests are performed [45].
(i). Geographical differentiation
Pairwise least-cost paths between the sampling locations were calculated from a resistance map with Circuitscape [46]. As sharpfins inhabit the littoral zones of Lake Matano [19,24], we clipped out areas deeper than 50 m to avoid misleading interpretations of the shortest possible way across the lake for these fishes. We examined correlations between geographical distance (pairwise least-cost paths) and genetic (Euclidean) distance with Mantel tests (R package ade4 [47]) within morphotypes.
(j). Fitness proxies
Estimating fitness is challenging, especially in large natural populations. For fishes, various proxies have been applied to quantify lifetime reproduction and survivorship [48] (see also [8,48,49]). Here, we use the relative liver weight as proxy for individual fitness, because the liver is an important fat-storage organ in fishes [50] and is frequently used as an estimator for individual condition in fishes (e.g. [51,52]). Moreover, individual condition is tightly correlated with reproductive capacity in other fish species (e.g. [53–56]). Hence, it is reasonable to assume that relative liver mass is indicative for the individual energy status in sailfin silversides, and that an enhanced energy status enables an individual to produce larger numbers of viable offspring, thus leading to greater individual fitness. Relative liver weight was calculated as the residuals of a generalized linear model constructed with data on sex, body size and the ratio between log-transformed liver weight and gutted body weight. We tested the reliability of our fitness proxy by investigating the correlation between relative liver mass and reproductive capacity (the ratio between log-transformed gonad weight and gutted body weight) with a subset of 127 female sharpfins. Because female sharpfins spawn promiscuous numerous times per day (J.P. & F.H. 2012–2014, personal observation), without distinct spawning season female gonad mass appears a reliable estimator of reproductive capacity [27].
The use of relative liver mass as a general proxy for individual fitness does not imply that all potential fitness components are adequately covered by it. Absolute size and phenotypic proportions such as body depth vary strongly among the morphotypes (see results below and [23]). To test for potential differences in how our proxy represents fitness for various morphotypes, we applied an ANOVA to the residuals of log liver weight versus log body weight; the test result did not show significant differences (F4,1032 = 1.902, p = 0.108).
(k). Adaptive landscapes
To test for a signal of disruptive selection along major axes of morphological variation, we applied non-parametric regressions without an a priori assumption about the shape of the fitness surface. Therefore, we used the generalized additive models (GAM) routine of R package mgcv [34,57], with the fitness proxy as dependent variable and the linear discriminant axes (LDs) obtained from the linear discriminant analysis (implemented in the DAPC) of the ecomorphological PCs as predictor variables. The GAM models were calculated without splines, with cubic splines, and with thin plate splines. The final model was chosen based on the lowest Akaike information criterion values (electronic supplementary material, table S3f). Splines were smoothed via generalized cross-validation and an approximation of the significance of smoothing terms was calculated following Wood [58]. Trait axes with significant smoothing according to the GAM analyses were visualized as pairwise combinations of two axes using the Tps function and its incorporated plot function (R package fields [59]). The smoothing parameter for the plot function minimizes the generalized cross-validation score, and incorporates the estimated degrees of freedom from the GAM analyses. To test whether sharpfin population means plot in regions of higher fitness than expected by chance, we compared the observed fitness with random simulations of mean fitness. Using the runif function in R [34], 1000 population means and 10 000 individuals were drawn from the adaptive landscape. Individual fitness values were obtained from the landscape, using the raster and extract functions (R package raster) [60]. A pairwise comparison of each population and randomly generated means was carried out, with an ANOVA and Tukey's post hoc test with individuals falling in a range of a standard error of 0.75 around random means. To adjust p-values given the large number of post hoc tests SGoF [45] was applied. To test whether a morphotype's mean fitness differed significantly from the population of random fitness means, a bimodal test was carried out in R [34]. Tests for which observed fitness mean was significantly greater than the pool of random fitness means were treated as positives and others as negatives. To identify whether morphotype means occupy an average peaks in the adaptive landscape, we compared (using methods described above) fitness values from the regions in the landscape that are occupied by two morphotypes with the values obtained in intermediate regions.
3. Results
(a). Phenotypic diversity and ecology
Cluster analyses of the observed morphological variation in ecomorphological candidate traits (electronic supplementary material, figure S1b) [23,61–63] support five morphotype groups (figure 1; electronic supplementary material, table S1; total assignment probability: 83.28%), which largely match the nominal sharpfin species [19,64]. The morphotype Telmatherina sp. ‘thicklip’ (figures 1 and 2a) is very distinct, whereas the other four morphotypes (i.e. the nominal taxa T. opudi, T. sarasinorum, T. wahjui and T. sp. ‘elongated’) partially overlap in morphospace (figure 2a). Our analysis does not confirm the existing hypothesis that two, fish-feeding sharpfins exist (T. abendanoni and T. sp. ‘elongated’) [19]. However, this result may be an artefact, because the fish-feeding morphs are rare in nature (and hence rare in our sampling), and because both morphs are very similar in comparison to the greater differences between various feeding types [19]. Therefore, further investigation is required in this respect. Because our samples contained mainly the ‘elongated’ fish-feeding morph we refer to this fifth morphotype as T. sp. ‘elongated’.
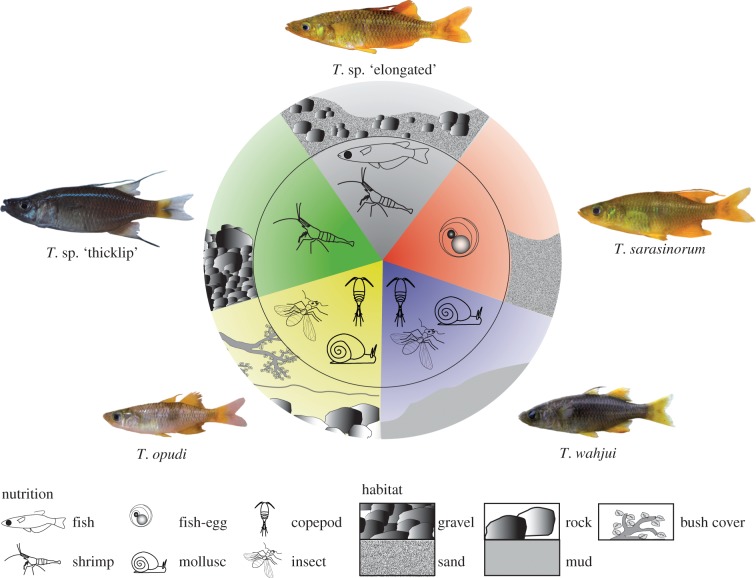
Figure 1.
Lake Matano's sharpfin Telmatherina, and key aspects of their ecological differentiation. Details on food sources and habitat use are described in the text and electronic supplementary material, figures S2 and S3.
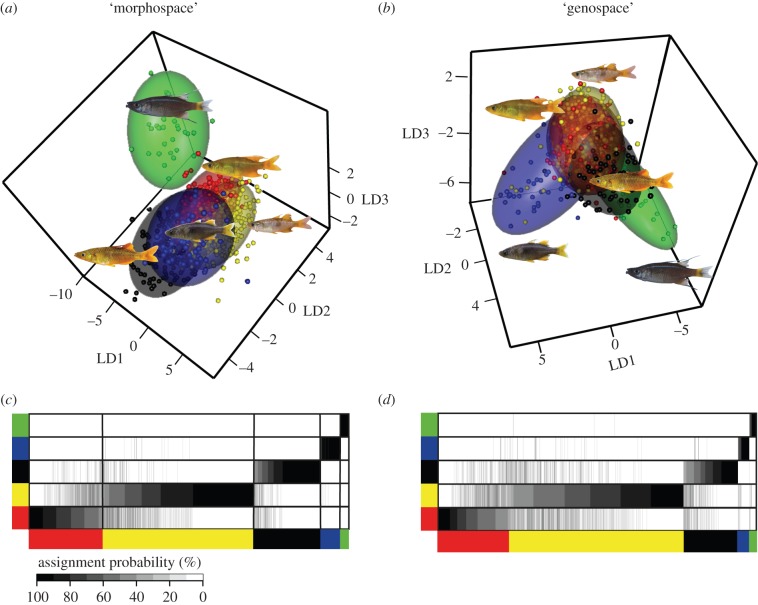
Figure 2.
(a) Morphological and (b) genetic differentiation between sharpfins with (c,d) assignment probabilities of individuals to their respective clusters. Scatterplots show the first three linear discriminant axes of the discriminant principal component analyses. Convex hulls in (a) and (b) represent 95% confidence ellipses. Heat maps display the individual assignment probability to the respective (c) phenotypic and (d) genotypic cluster on the y-axis as vertical thin lines for each individual. Assignment probabilities to a given group are displayed in greyscale from white (0%) to black (100%). Clusters are colour-coded in all panels according to the scheme displayed in figure 1.
As expected for an emerging adaptive radiation [1,2,65], the five morphotypes differ significantly in trophic ecology and habitat use (figure 1; electronic supplementary material, figures S2 and S3; CCA: F7,272 = 2.535, p = 0.021, based on 9999 permutations; CCA1 p < 0.001, 9999 permutations; CCA2 p < 0.05, 19 999 permutations). Telmatherina sarasinorum feeds on fish eggs of the roundfin Telmatherina [19], and hence occupies predominantly sandy habitats that serve as spawning grounds for these roundfins. The slightly smaller T. opudi feeds on insects and zooplankton; it prefers a hard substrate of inshore habitats that provide shelter, preferably covered by terrestrial plants. Predominantly fish-feeding sharpfins also prey on shrimps at sand and gravel habitats. Telmatherina sp. ‘thicklip’ forage nearly exclusively on shrimps on gravel fields, whereas T. wahjui inhabit the shallows of muddy areas (figure 1; electronic supplementary material, figures S2 and S3), where they feed on insects, molluscs and zooplankton.
Morphological differentiation among sharpfin morphotypes (figures 2a and 3) is consistent with adaptations predicted by their respective ecological niches [23,38,62,63,66]. These include short and widely spaced gill rakers in fish and shrimp predators, and long and closely spaced gill rakers in sharpfins feeding on fish eggs, zooplankton or insects (figures 2a and 3). Deep bodies, known to increase manoeuverability in fishes [63], are characteristic for the shrimp- and fish-egg-feeders, whereas slender shapes occur predominantly in fish predators and zooplankton-feeders (figures 1, 2a and 3). Pointed snouts are typically found in ‘pick-feeding’ fish species, as fish-egg-, zooplankton- and mollusc-feeders. The jaws of shrimp-feeders are, by contrast, compact and the lips are pronounced. The rather gracile and elongated jaws of fish-feeders indicate a modification for a fast movement, suggesting a ram-feeding mode of foraging (figures 1, 2a and 3) [38,62].
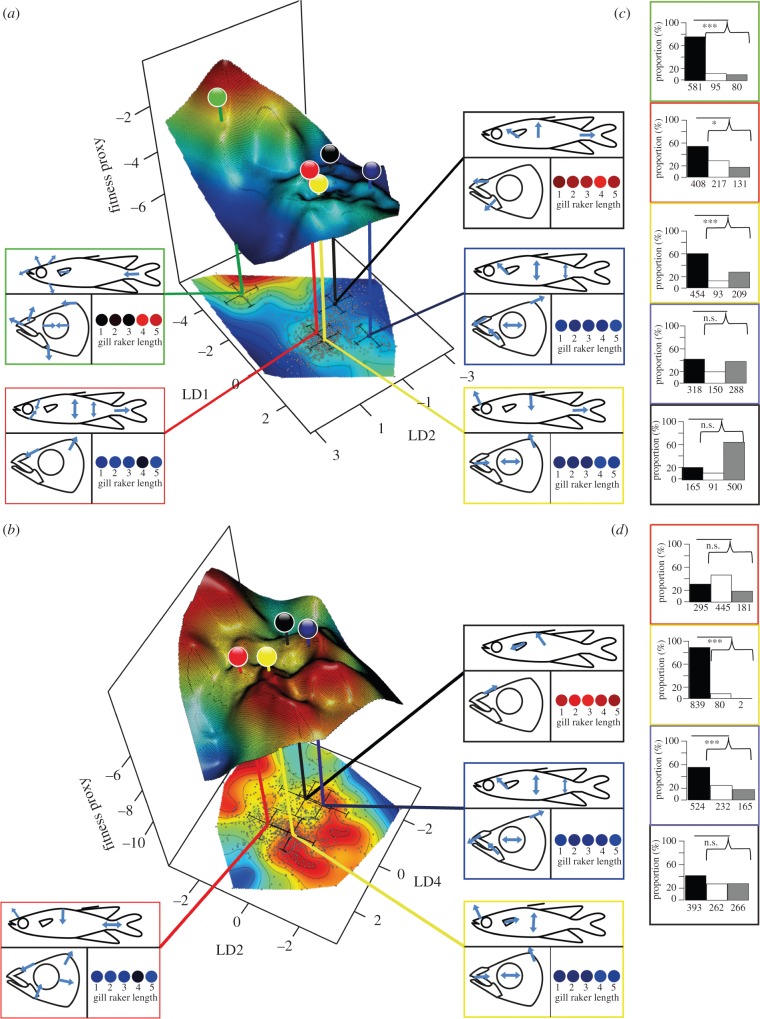
Figure 3.
Adaptive landscapes of sharpfins (a,b) and significance of the occupied peaks versus random sampling (c,d). Two axes (LD1 and LD2 in a; LD2 and LD4 in b) represent ecomorphological differentiation, whereas the third represents the fitness proxy, (a) for the entire radiation (five morphotypes) and (b) excluding T. sp. ‘thicklip’. Plots (a,b) show thin plate splines, with smoothing parameters obtained from minimizing the general cross-validation scores. Positions of individuals are plotted as grey dots on the contour map, whereas species means are colour-coded as in figure 1. Species means and standard deviations were calculated from individuals with an assignment probability of greater than 95% to the respective morphotype. Statistical details are provided in the electronic supplementary material, table S3. Comparative representations of studied ecomorphological traits (shape of body, head and gills) for the mean of each species are provided in insets. Direction and length of arrows in these insets indicate how each species differs from the center of the morphospace occupation. Variation in gill raker length is displayed as a colour gradient from red (short) to blue (long and gracile). Barplots (c,d) show the proportion of tests (number on the x-axis) with a significantly higher (black bars), not significantly different (white bars) or significantly lower (grey bar) fitness of sharpfins than random means. Significantly enhanced fitness of sharpfin species is indicated by lines above barplots.
(b). Genotypic differentiation in sympatry
Sympatric speciation predicts weak genetic differentiation among diverging populations, restricted only to small proportions of the genome [67,68]. The morphotypes described above are generally well supported by our predominantly nuclear AFLP data as the probability for assignment to the correct morphotype is approximately 71%. However, this percentage also indicates that mismatches occur frequently, which relates to substantial overlap between the species in morphology and genetics (figure 2c,d). Genetic clusters are differentiated significantly in nearly all pairwise Fst comparisons (electronic supplementary material, table S2a,b). Variation in ecomorphological traits correlates well with variation in 13% of the analysed nuclear markers (electronic supplementary material, table S2c). Removal of these putatively selective loci decreases the success of assignment to the correct morphotype markedly (52.29%). This finding suggests that sharpfin genetic population structure is maintained by comparatively small proportions of nuclear loci. We did not detect any spatial effects in population structure (electronic supplementary material, table S2d) and all morphotypes except for T. sp. ‘thicklip’ were found evenly distributed around Lake Matano's shoreline (electronic supplementary material, figure S1a).
(c). Adaptive landscapes
Disruptive selection is predicted to shape rugged adaptive landscapes during speciation in the face of gene flow [1,2,69]. As expected, relative liver weight is correlated with female gonad mass, and thus reproductive capacity in sharpfins (n = 127♀; t 9.42; R2 41.5%; p < 0.001). The finding indicates that our earlier assumption that relative liver weight is a good proxy for individual fitness in sharpfins much as in other fishes [53–56] is justified. Upon comparing relative liver weight with variation in the size-independent morphological dataset in the GAM (R2 5.85%; effective degrees of freedom: 11.57; generalized cross-validation: 0.191; electronic supplementary material, table S3), three LDs (LD1, LD2 and LD4) revealed a significant correlation. Resulting landscapes (figure 3; electronic supplementary material, figure S4) show multiple local maxima. Individual species generally plot near local fitness peaks in the landscape constructed with GAMs from the two main axes of morphological variation (LD1 versus LD2) and the fitness proxy (figure 3a). Some individuals do not appear to be particularly well adapted, but the mean fitness of three of the five species significantly exceeds random expectations (figure 3c). As mentioned, the shrimp-feeding T. sp. ‘thicklip’ is morphologically very different from the other species, and its fitness peak is separated from the region of enhanced fitness occupied by the four other species by a deep valley (figure 3a; electronic supplementary material, S5a). In the landscape LD2 versus LD4 constructed from the complete dataset, the macroinvertebrate-feeder T. opudi occupies a significant fitness peak (electronic supplementary material, figure S4a,c), which is separated by a significant valley from fish-feeders (T. sp. ‘elongated’; electronic supplementary material, figure S5a).
Zooming in to the region of the adaptive landscape occupied by T. opudi, T. wahjui, T. sarasinorum and T. sp. ‘elongated’ (figure 3b; only LD2 and LD4 correlate significantly with fitness in this reduced dataset; electronic supplementary material, table S4d) highlights that two of the four trophic specialists (T. opudi and T. wahjui) occupy local fitness peaks with significantly enhanced fitness (figure 3d), although higher fitness peaks occur nearby in the landscape. In contrast with the complete dataset, the conspicuously wide-headed T. wahjui is situated in a region of significantly enhanced fitness (figure 3b,d). However, T. opudi inhabits the most distinct fitness peak in that landscape, separated by significant valleys from T. wahjui and T. sp. ‘elongated’ (figure 3b; electronic supplementary material, figure S5b).
Additional valleys appear plausible from visual impression (figure 3; electronic supplementary material, figure S4), and in several cases (LD1 versus LD2: T. wahjui; figure 3c; LD2 versus LD4: T. wahjui, T. sp. ‘elongated’, T. sp. ‘thicklip’; electronic supplementary material, figure S4c; reduced data: T. sarasinorum, T. sp. ‘elongated’; figure 3b; LD1 versus LD4: T. opudi, T. sp. ‘thicklip’), the number of individuals with fitness proxies exceeding neutral expectations is higher than that of sharpfins with below-average fitness; these cases are, however, not supported in rigorous binomial tests.
Differences in gill raker length dominate the morphological variance, explained by LD1, whereas changes in body and head shape are summarized in the following axes (LD2 and LD4; figure 3a,b; electronic supplementary material, figure S4a,b). Therefore, disruptive selection is significant in gill traits, where species means tend to peak in the adaptive landscape with either short and wide-spaced or long and narrow-spaced gill rakers (figure 3a,b). Signal for disruptive selection in head shape is especially prominent along axes of variation in snout proportions (figure 3a,b). Finally, different kinds of body shapes are associated to high fitness proxy values: deep-bodied fishes with a short caudal peduncle, individuals with conspicuously oval body shapes and conspicuously slender sailfins with either an elongated or a short caudal peduncle (figure 3a,b).
4. Discussion
Although ecological speciation is considered a major driver of diversification [2,65], our knowledge on how ecological factors have contributed to lineage divergence and speciation in complex radiations remains limited [2,70]. Here, we document that ecological opportunities (such as different food sources in sharpfin silversides) promote radiation by creating complex, rugged adaptive landscapes to which populations adapt. Adaptation to ecological opportunities is driven by selection, which can be disruptive in rugged adaptive landscapes.
Our sampling of Lake Matano's sharpfins reveals the presence of five, mainly sympatric morphotypes (figure 2; electronic supplementary material, figure S1). Variation in morphological key traits, including the shape of the head, body and gill rakers, is directly related to different feeding strategies, behaviour and habitat use (figures 1 and 2; electronic supplementary material, figures S2 and S3). All morphotypes relate to ecologically and genetically largely differentiated species, from deep-bodied shrimp-feeders, inhabiting gravel fields, to smaller near-shore insectivores and slender fish-feeders dwelling with shoaling juveniles (figure 1). Genome-wide surveys of polymorphism demonstrate that the variation in ecomorphological traits is correlated with variation in a restricted number of genetic markers (electronic supplementary material, table S2c). These findings clearly meet predictions for an ongoing radiation, evolving sympatrically under ecological selection pressure [68,70]. The incipient nature of this adaptive radiation, however, also results in overlaps in morphology and genetic variability between four of the five morphotypes, and intermediate, usually less fit individuals remain relatively common (figures 2 and 3; electronic supplementary material, figure S5).
A central aspect of this study is the significant signal of disruptive selection along major axes of morphological variation (electronic supplementary material, table S3; figure 3), resulting in complex adaptive landscapes with multiple peaks (figure 3; electronic supplementary material, figure S4). As predicted [2,12], the five morphotype means occupy several regions of high fitness (figure 3). Interestingly, valleys between these fitness peaks are, with exception of T. sp. ‘thicklip’, not very deep, but frequently reflect regions of significant lower fitness (figure 3; electronic supplementary material, figure S5). This suggests a general lower fitness of individuals intermediate to the population means (i.e. hybrids) compared with the population means. Incomplete speciation, possibly in combination with adaptation that arises from combined divergence in several traits, or traits that are not quantified here, provides a possible explanation for a lack of significant valleys between some of the phenotypic less distinct sharpfins. Nevertheless, sharpfins clearly fulfil predictions for ‘rugged’ adaptive landscapes in an emerging adaptive radiation, driven by disruptive ecological selection.
Selective regimes may vary over time, as reported, for example, from postglacial stickleback systems [71]. We also cannot exclude that the adaptive landscapes reconstructed here, and the selective regimes that shape divergence of the sharpfin flock, may change through time. However, there are no apparent hints for this assumption. The environment of Lake Matano is considered long-term stable [20]. Furthermore, correlations among traits and sources of nutrition remained consistent across studies that are based on material from different years (cf. [23]). Finally, the present samples originate from a total of 160 locations, sampled over a period of four months, a design that largely prevents dominance of local or single effects. All this suggests that the snapshot view of the present study is not an exception.
Disruptive ecological selection has been identified as a causal root of population divergence also in other lake fishes, including, for example, species pairs of sticklebacks or coregonids, diverging in ecomorphological traits along the benthic–limnetic axis [8,49,72,73]. In contrast with the comparatively frequent examples for bimodal population divergence, the importance of disruptive ecological selection in complex species flock formation has only rarely been demonstrated [17,68]. Among the available examples are two bird radiations, Geospiza finches and Loxia crossbills, where disruptive selection acts on beak size [14,15]. In fishes, the adaptive landscapes of the Cyprinodon pupfish radiation [17] show patterns similar to those detected in the sharpfin radiation, though landscape topographies are more complex in sharpfins. This complexity largely fits the presence of five phenotypic groups in sharpfins, compared with only two ecological specialists and one generalist in the pupfish example [17,18].
The natural occurrence of interbreeding between sailfin morphospecies, largely separated by divergent ecological selection, provides a plausible explanation for both the complex adaptive landscapes and the porous population boundaries observed. Substantial but incomplete reproductive isolation in the sharpfin flock is apparent from the weak, but significant population genetic structure (electronic supplementary material, table S2). Assortative mating has been reported from most of Lake Matano's Telmatherina species, including three of the five sharpfins (T. sarasinorum, T. wahjui, T. sp. ‘thicklip’; see [21,22,27,24]). Together with significant differences in habitat use (figure 1; electronic supplementary material, figure S3), genetic population structure and assortative mating provide strong arguments to reject the hypothesis of only one hypervariable species, and are in agreement with very similar patterns reported from its sister clade, Lake Matano's ‘roundfins' [25]. In both Telmatherina clades, roundfins and sharpfins [24], divergence apparently follows ecological selection pressure under sympatric conditions, leading to the consensus view that the ‘Matano flock’ probably represents an early stage of adaptive radiation [1,2,5].
However, evidence for early stages of species flock formation appears surprising in the light of recent estimations for the age of divergence in sailfin silversides. Stelbrink et al. [28] applied different molecular clock approaches, and argued that the initial divergence of Telmatherina within Lake Matano, leading to the formation of sharpfins and roundfins, dates back approx. 1.9 Myr. This is in general agreement with the estimated geological age of the lake [20], and implies ages of approx. 1 Myr for the onset of divergence within both sharpfins and roundfins [28].
Three potential explanations for resolving this conflict appear moderately plausible. First, environmental instabilities may repeatedly shift the balance between disruptive selection and homogenizing gene flow, and may even lead to a breakdown of the barriers to gene flow and the reamalgamation of incipient species (e.g. [74,75]). However, we consider a ‘speciation reversal’ scenario rather unlikely in the case of extraordinarily deep Lake Matano, which is considered long-term stable [20,76]. There are so far no indications for significant earlier disturbances, or increased turbidity or other changes that might plausibly trigger the breakdown of reproductive barriers, despite the lake nowadays being clearly affected by human activities [77].
Second, intralake divergence may have proceeded, for unknown reasons, generally at rather low speed. This would be surprising in a species flock characterized by pronounced sexual and ecological selection pressure [78], and rates of speciation reported from other fish radiations [79]. As a third alternative, speciation processes might remain incomplete in Telmatherina of this lake. This would imply that sharpfins and roundfins remain in a stage of the speciation continuum [2,5] where disruptive ecological selection is strong enough to maintain a stable phenotypic diversity, but fails to complete the speciation process. Superficially similar cases have been reported from walking-stick insects or haplochromine cichlids (reviewed in [67]), but we are not aware of other adaptive radiations where speciation likewise remained incomplete about a million years after the onset of speciation processes.
Supplementary Material
Acknowledgements
We thank the Indonesian Institute of Sciences (LIPI) and the Kementerian Negara Riset dan Teknology (RISTEK) for the permit to conduct research in Indonesia. PT VALE Indonesia Tbk provided logistic support in Sulawesi. We thank S. Chapuis and M. Milanovic for assistance in the field, and M. Proest, R. Bach and J. Schmidt for assistance in the laboratory. For valuable discussions about previous versions of the manuscript, we thank Bert van Bocxlaer and Leon Hilgers.
Ethics
The study was carried out under research permits from the Kementerian Negara Riset dan Teknology (RISTEK), Indonesia (permit no. 992/FRP/SM/V/2010) and in cooperation with the Indonesian Institute of Sciences (LIPI). No endangered or protected species were involved. Fishes were sacrificed with MS-222. All procedures followed the Guidelines for the Use of Fishes in Research from the American Fisheries Society and the legal requirements of Indonesia and Germany.
Authors' contributions
J.P. contributed to conception and design of the study, performed fieldwork, generated the morphometric and AFLP data, and carried out most of the analyses. U.K.S. contributed to the development of the research idea, and to final stages of manuscript preparation. R.K.H. participated in fieldwork and contributed to the discussion of the results. F.H. conceived, designed and supervised this project, and contributed to data analyses. J.P. and F.H. wrote the paper.
Competing interests
We declare we have no competing interests.
Funding
This research and fieldwork was funded by a research grant of the DFG (to F.H.: HE 5707/2-1) and strongly benefitted from previous fieldwork funded by the DFG (to U.K.S.: DFG SCHL567/2).
References
- 1.Schluter D. 2000. The ecology of adaptive radiation. Oxford, UK: Oxford University Press. [Google Scholar]
- 2.Nosil P. 2012. Ecological speciation. Oxford, UK: Oxford University Press. [Google Scholar]
- 3.Dieckmann U, Doebeli M. 1999. On the origin of species by sympatric speciation. Nature 400, 354–357. ( 10.1038/22521) [DOI] [PubMed] [Google Scholar]
- 4.Nosil P. 2008. Speciation with gene flow could be common. Mol. Ecol. 17, 2103–2106. ( 10.1111/j.1365-294X.2008.03715.x) [DOI] [PubMed] [Google Scholar]
- 5.Via S. 2009. Natural selection in action during speciation. Proc. Natl Acad. Sci. USA 106, 9939–9946. ( 10.1073/pnas.0901397106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Via S. 2012. Divergence hitchhiking and the spread of genomic isolation during ecological speciation-with-gene-flow. Phil. Trans. R. Soc. B 367, 451–460. ( 10.1098/rstb.2011.0260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hereford J. 2009. A quantitative survey of local adaptation and fitness trade-offs. Am. Nat. 173, 579–588. ( 10.1086/597611) [DOI] [PubMed] [Google Scholar]
- 8.Bolnick DI, Lau OL. 2008. Predictable patterns of disruptive selection in stickleback in postglacial lakes. Am. Nat. 172, 1–11. ( 10.1086/587805) [DOI] [PubMed] [Google Scholar]
- 9.Merrill RM, Wallbank RW, Bull V, Salazar PC, Mallet J, Stevens M, Jiggins CD. 2012. Disruptive ecological selection on a mating cue. Proc. R. Soc. B 279, 4907–4913. ( 10.1098/rspb.2012.1968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendry AP, Huber SK, De Leon LF, Herrel A, Podos J. 2009. Disruptive selection in a bimodal population of Darwin's finches. Proc. R. Soc. B 276, 753–759. ( 10.1098/rspb.2008.1321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold SJ, Pfrender ME, Jones AG. 2001. The adaptive landscape as a conceptual bridge between micro-and macroevolution. Genetica 112, 9–32. ( 10.1023/A:1013373907708) [DOI] [PubMed] [Google Scholar]
- 12.Simpson GG. 1944. Tempo and mode in evolution. New York, NY: ColumbiaUniversity Press. [Google Scholar]
- 13.Svensson E, Calsbeek R. 2012. The adaptive landscape in evolutionary biology. Oxford, UK: Oxford University Press. [Google Scholar]
- 14.Schluter D, Grant PR. 1984. Determinants of morphological patterns in communities of Darwin's finches. Am. Nat. 123, 175–196. ( 10.1086/284196) [DOI] [Google Scholar]
- 15.Benkman CW. 2003. Divergent selection drives the adaptive radiation of crossbills. Evolution 57, 1176–1181. ( 10.1111/j.0014-3820.2003.tb00326.x) [DOI] [PubMed] [Google Scholar]
- 16.Phillips PC, Arnold SJ. 1989. Visualizing multivariate selection. Evolution 43, 1209–1222. ( 10.2307/2409357) [DOI] [PubMed] [Google Scholar]
- 17.Martin CH, Wainwright PC. 2013. Multiple fitness peaks on the adaptive landscape drive adaptive radiation in the wild. Science 339, 208–211. ( 10.1126/science.1227710) [DOI] [PubMed] [Google Scholar]
- 18.Turner BJ, Duvernell DD, Bunt TH, Barton MG. 2008. Reproductive isolation among endemic pupfishes (Cyprinodon) on San Salvador Island, Bahamas: microsatellite evidence. Biol. J. Linn. Soc. 95, 566–582. ( 10.1111/j.1095-8312.2008.01079.x) [DOI] [Google Scholar]
- 19.Herder F, Schwarzer J, Pfaender J, Hadiaty RK, Schliewen UK. 2006. Preliminary checklist of sailfin silversides (Pisces: Telmatherinidae) in the Malili Lakes of Sulawesi (Indonesia), with a synopsis of systematics and threats. Verh. Ges. Ichthyol. 5, 139–163. [Google Scholar]
- 20.Vaillant JJ, Haffner GD, Cristescu ME. 2011. The ancient lakes of Indonesia: towards integrated research on speciation. Integr. Comp. Biol. 51, 634–643. ( 10.1093/icb/icr101) [DOI] [PubMed] [Google Scholar]
- 21.Cerwenka AF, Wedekind JD, Hadiaty RK, Schliewen UK, Herder F. 2012. Alternative egg-feeding tactics in Telmatherina sarasinorum, a trophic specialist of Lake Matano's evolving sailfin silversides fish radiation. Hydrobiologia 693, 131–139. ( 10.1007/s10750-012-1099-8) [DOI] [Google Scholar]
- 22.Gray SM, McKinnon JS, Tantu FY, Dill LM. 2008. Sneaky egg-eating in Telmatherina sarasinorum, an endemic fish from Sulawesi. J. Fish. Biol. 73, 728–731. ( 10.1111/j.1095-8649.2008.01949.x) [DOI] [Google Scholar]
- 23.Pfaender J, Schliewen UK, Herder F. 2010. Phenotypic traits meet patterns of resource use in the radiation of ‘sharpfin’ sailfin silverside fish in Lake Matano. Evol. Ecol. 24, 957–974. ( 10.1007/s10682-009-9332-2) [DOI] [Google Scholar]
- 24.Herder F, Nolte AW, Pfaender J, Schwarzer J, Hadiaty RK, Schliewen UK. 2006. Adaptive radiation and hybridization in Wallace's Dreamponds: evidence from sailfin silversides in the Malili Lakes of Sulawesi. Proc. R. Soc. B 275, 2178–2195. ( 10.1098/rspb.2006.3558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herder F, Pfaender J, Schliewen UK. 2008. Adaptive sympatric speciation of polychromatic ‘roundfin’ sailfin silverside fish in Lake Matano (Sulawesi). Evolution 62, 2178–2195. ( 10.1111/j.1558-5646.2008.00447.x) [DOI] [PubMed] [Google Scholar]
- 26.Schwarzer J, Herder F, Misof B, Hadiaty RK, Schliewen UK. 2008. Gene flow at the margin of Lake Matano's adaptive sailfin silverside radiation: Telmatherinidae of River Petea in Sulawesi. Hydrobiologia 615, 201–213. ( 10.1007/s10750-008-9561-3) [DOI] [Google Scholar]
- 27.Gray SM, McKinnon JS. 2006. A comparative description of mating behaviour in the endemic telmatherinid fishes of Sulawesi's Malili Lakes. Environ. Biol. Fishes 75, 471–482. ( 10.1007/s10641-006-0037-x) [DOI] [Google Scholar]
- 28.Stelbrink B, Stöger I, Hadiaty RK, Schliewen UK, Herder F. 2014. Age estimates for an adaptive lake fish radiation, its mitochondrial introgression, and an unexpected sister group: Sailfin silversides of the Malili Lakes system in Sulawesi. BMC Evol. Biol. 14, 94 ( 10.1186/1471-2148-14-94) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zelditch M, Swiderski D, Sheets DH, Fink W. 2004. Geometric morphometrics for biologists: a primer. London, UK: Academic Press. [Google Scholar]
- 30.Rohlf FJ. 2006. tpsDig2. See http://tpsdig2.software.informer.com.
- 31.Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 9. [Google Scholar]
- 32.Rasband WS. 1997–2009 ImageJ. See http://rsb.info.nih.gov/ij/ .
- 33.Ginter CC, DeWitt TJ, Fish FE, Marshall CD. 2012. Fused traditional and geometric morphometrics demonstrate pinniped whisker diversity. PLoS ONE 7, 1–10. ( 10.1371/journal.pone.0034481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. ( 10.1093/bioinformatics/btn129) [DOI] [PubMed] [Google Scholar]
- 35.R Development Core Team. 2012. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 36.Charrad M, Ghazzali N, Boiteau V, Niknafs A. 2014. NbClust: an R package for determining the relevant number of clusters in a data set. J. Stat. Softw. 61, 6.
- 37.Charrad M, Ghazzali N, Boiteau V, Niknafs A. 2014. Package ‘NbClust’. J. Stat. Soft 61, 1–36. [Google Scholar]
- 38.Pfaender J, Miesen FW, Hadiaty RK, Herder F. 2011. Adaptive speciation and sexual dimorphism contribute to diversity in form and function in the adaptive radiation of Lake Matanos's sympatric roundfin sailfin silversides. J. Evol. Biol. 24, 2329–2345. ( 10.1111/j.1420-9101.2011.02357.x) [DOI] [PubMed] [Google Scholar]
- 39.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. ( 10.1111/j.1654-1103.2003.tb02228.x) [DOI] [Google Scholar]
- 40.Vos P, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. ( 10.1093/nar/23.21.4407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Excoffier LAUR, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567. ( 10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 43.Joost S, Kalbermatten M, Bonin A. 2008. Spatial analysis method (SAM): a software tool combining molecular and environmental data to identify candidate loci for selection. Mol. Ecol. Res. 8, 957–960. ( 10.1111/j.1755-0998.2008.02162.x) [DOI] [PubMed] [Google Scholar]
- 44.Joost S, Kalbermatten M, Bezault E, Seehausen O. 2012. Use of qualitative environmental and phenotypic variables in the context of allele distribution models: detecting signatures of selection in the genome of Lake Victoria Cichlids. Methods Mol. Biol. 888, 295–314. ( 10.1007/978-1-61779-870-2_17) [DOI] [PubMed] [Google Scholar]
- 45.Carvajal-Rodriguez A, de Una-Alvarez J, Rolan-Alvarez E. 2009. A new multitest correction (SGoF) that increases its statistical power when increasing the number of tests. BMC Bioinf. 10, 209 ( 10.1186/1471-2105-10-209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah VB, McRae BH.2008. Circuitscape: a tool for landscape ecology. In Proc. of the 7th Python in Science Conference, pp. 62–66. See http://conference.scipy.org/proceedings/scipy2008 .
- 47.Thioulouse J, Chessel D, Doledec S, Olivier JM. 1997. ADE-4: a multivariate analysis and graphical display software. Stat. Comput. 7, 75–83. ( 10.1023/A:1018513530268) [DOI] [Google Scholar]
- 48.Bolger T, Connolly PL. 1989. The selection of suitable indices for the measurement and analysis of fish condition. J. Fish Biol. 34, 171–182. ( 10.1111/j.1095-8649.1989.tb03300.x) [DOI] [Google Scholar]
- 49.Bolnick DI. 2004. Can intraspecific competition drive disruptive selection? An experimental test in natural populations of sticklebacks. Evolution 58, 608–618. ( 10.1111/j.0014-3820.2004.tb01683.x) [DOI] [PubMed] [Google Scholar]
- 50.Sheridan MA. 1988. Lipid dynamics in fish: aspects of absorption, transportation, deposition and mobilization. Comp. Biochem. Physiol. B 90, 679–690. ( 10.1016/0305-0491(88)90322-7) [DOI] [PubMed] [Google Scholar]
- 51.Chellappa S, Huntingford FA, Strang RHC, Thomson RY. 1995. Condition factor and hepatosomatic index as estimates of energy status in male three-spined stickleback. J. Fish. Biol. 47, 775–787. ( 10.1111/j.1095-8649.1995.tb06002.x) [DOI] [Google Scholar]
- 52.Koops MA, Hutchings JA, McIntyre TM. 2004. Testing hypotheses about fecundity, body size and maternal condition in fishes. Fish Fish. 5, 120–130. ( 10.1111/j.1467-2979.2004.00149.x) [DOI] [Google Scholar]
- 53.Marshall CT, Yaragina NA, Lambert Y, Kjesbu OS. 1999. Total lipid energy as a proxy for total egg production by fish stocks. Nature 402, 288–290. ( 10.1038/46272) [DOI] [Google Scholar]
- 54.Blukacz EA, et al. 2010. Linking lake whitefish (Coregonus clupeaformis) condition with male gamete quality and quantity. J. Gt Lakes Res. 36, 78–83. ( 10.1016/j.jglr.2009.12.014) [DOI] [Google Scholar]
- 55.Burness G, Schulte-Hostedde AI, Montgomerie R. 2008. Body condition influences sperm energetics in lake whitefish (Coregonus clupeaformis). Can. J. Fish. Aquat. Sci. 65, 615–620. ( 10.1139/f07-188) [DOI] [Google Scholar]
- 56.Rätz HJ, Lloret J. 2003. Variation in fish condition between Atlantic cod (Gadus morhua) stocks, the effect on their productivity and management implications. Fish. Res. 60, 369–380. ( 10.1016/S0165-7836(02)00132-7) [DOI] [Google Scholar]
- 57.Wood S. 2006. Generalized additive models: an introduction with R. Boca Raton, FL: Chapman & Hall/CRC. [Google Scholar]
- 58.Wood SN. 2004. Stable and efficient multiple smoothing parameter estimation for generalized additive models. J. Am. Stat. Assoc. 99, 673–686. ( 10.1198/016214504000000980) [DOI] [Google Scholar]
- 59.Fields development team. 2006. Fields: tools for spatial data. Boulder, CO: National Center for Atmospheric Research. [Google Scholar]
- 60.Hijmans RJ, Etten J.2011. raster: geographic analysis and modeling with raster data. R package v. 1.8-39. See https://cran.r-rpoject.org/web/packages/raster/raster.pdf .
- 61.Albertson RC. 2008. Morphological divergence predicts habitat partitioning in a Lake Malawi cichlid species complex. Copeia 3, 689–698. ( 10.1643/CG-07-217) [DOI] [Google Scholar]
- 62.Wainwright PC, Richard BA. 1995. Predicting patterns of prey use from morphology of fishes. Environ. Biol. Fishes 44, 97–113. ( 10.1007/BF00005909) [DOI] [Google Scholar]
- 63.Webb PW. 1984. Body form, locomotion and foraging in aquatic vertebrates. Am. Zool. 24, 107–120. ( 10.1093/icb/24.1.107) [DOI] [Google Scholar]
- 64.Kottelat M. 1991. Sailfin silversides (Pisces: Telmatherinidae) of Lake Matano, Sulawesi, Indonesia, with descriptions of six new species. Ichthyol. Explor. Freshw. 1, 321–344. [Google Scholar]
- 65.Schluter D. 2009. Evidence for ecological speciation and its alternative. Science 323, 737–741. ( 10.1126/science.1160006) [DOI] [PubMed] [Google Scholar]
- 66.Amundsen P-A, Bøhn T, Vågå G. 2004. Gill raker morphology and feeding ecology of two sympatric morphs of European whitefish (Coregonus lavaretus). Ann. Zool. Fenn. 41, 291–300. [Google Scholar]
- 67.Nosil P, Harmon LJ, Seehausen O. 2009. Ecological explanations for (incomplete) speciation. Trends Ecol. Evol. 24, 145–156. ( 10.1016/j.tree.2008.10.011) [DOI] [PubMed] [Google Scholar]
- 68.Wu CI. 2001. The genetic view of the process of speciation. J. Evol. Biol. 14, 851–865. ( 10.1046/j.1420-9101.2001.00335.x) [DOI] [Google Scholar]
- 69.Seehausen O. 2004. Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207. ( 10.1016/j.tree.2004.01.003) [DOI] [PubMed] [Google Scholar]
- 70.Feder JL, Egan SP, Nosil P. 2012. The genomics of speciation-with-gene-flow. Trends Genet. 28, 342–350. ( 10.1016/j.tig.2012.03.009) [DOI] [PubMed] [Google Scholar]
- 71.Bolnick DI, Araújo MS. 2011. Partitioning the relative fitness effects of diet and trophic morphology in the threespine stickleback. Evol. Ecol. Res. 13, 439–459. [Google Scholar]
- 72.Roesch C, Lundsgaard-Hansen B, Vonlanthen P, Taverna A, Seehausen O. 2013. Experimental evidence for trait utility of gill raker number in adaptive radiation of a north temperate fish. J. Evol. Biol. 26, 1578–1587. ( 10.1111/jeb.12166) [DOI] [PubMed] [Google Scholar]
- 73.Ohlberger J, Brännström Â, Dieckmann U. 2013. Adaptive phenotypic diversification along a temperature-depth gradient. Am. Nat. 182, 359–373. ( 10.1086/671169) [DOI] [PubMed] [Google Scholar]
- 74.Bhat S, et al. 2014. Speciation reversal in European whitefish (Coregonus lavaretus (L.)) caused by competitor invasion. PLoS ONE 9, e91208 ( 10.1371/journal.pone.0091208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hudson AG, Vonlanthen P, Bezault E, Seehausen O. 2013. Genomic signatures of relaxed disruptive selection associated with speciation reversal in whitefish. BMC Evol. Biol. 13, 108 ( 10.1186/1471-2148-13-108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamilton WB. 1979. Tectonics of the Indonesian region. Washington, DC: US Govt. Print. Off. [Google Scholar]
- 77.Herder F, Schliewen UK, Geiger MF, Hadiaty RK, Gray SM, McKinnon JS, Walter RP, Pfaender J. 2012. Alien invasion in Wallace's dreamponds: records of the hybridogenic ‘flowerhorn’ cichlid in Lake Matano, with an annotated checklist of fish species introduced to the Malili Lakes system in Sulawesi. Aquat. Invasions 7, 521–535. ( 10.3391/ai.2012.7.4.009) [DOI] [Google Scholar]
- 78.Herder F, Schliewen UK. 2010. Beyond sympatric speciation: radiation of sailfin silverside fishes in the Malili Lakes (Sulawesi). In Evolution in action (ed. M Glaubrecht), pp. 465–483. Berlin, Germany: Springer. [Google Scholar]
- 79.Day JJ, Cotton JA, Barraclough TG. 2008. Tempo and mode of diversification of Lake Tanganyika cichlid fishes. PLoS ONE 3, e1730 ( 10.1371/journal.pone.0001730) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.