SUMMARY
Mutations of the FUS gene were first reported to cause amyotrophic lateral sclerosis (ALS). Subsequent studies confirmed the role of mutations in ALS and also implicated them in frontotemporal dementia (FTD). Recently, through Next-Generation Exome sequencing approaches a mutation resulting in a substitution (p.Q290X) in the nuclear export domain of the FUS protein was nominated as a cause of autosomal dominant essential tremor (ET) in a large kindred. In addition, recent reports suggest a possible role for TDP-43 mutations in parkinsonism; TDP-43 is another RNA-binding protein implicated in ALS. Given these findings we investigated the role of FUS variants in Parkinson’s disease (PD). We sequenced specific regions of the gene encoding three functional domains of the FUS protein in 702 patients with PD. Our sequencing study did not identify any novel non-synonymous variant that would appear to affect the subjects’ susceptibility to Parkinson’s disease. These findings and previous studies have shown that variants within the FUS gene are not a common cause of PD or ET, in comparison to their role in ALS.
Keywords: Movement disorders, Genetics, FUS, Amyotrophic lateral sclerosis, Essential tremor, Parkinson’s disease
1. Introduction
The fused in sarcoma gene (FUS) was first identified as a fusion oncogene in human liposarcoma and encodes a ubiquitously expressed RNA/DNA-binding protein [1,2]. FUS plays a role in RNA transport between the nucleus and the cytoplasm; it is also reported to bind DNA and to be involved in DNA repair and transcriptional regulation [3]. FUS function appears to be closely related to TDP-43 which is also known to be important in neurodegeneration [3]. Genetic variation of the FUS and TDP-43 genes has been linked to several neurodegenerative diseases.
Mutations in FUS were first reported to cause amyotrophic lateral sclerosis (ALS) accounting for up to 4% of familial cases [4,5]. Further studies identified a substitution in FUS p.M254V in a patient suffering from frontotemporal dementia (FTD) [6]. The overlapping clinical, pathologic and genetic factors in ALS and FTD, and the presence of FUS pathology in a subset of cases with either disorder, supported pathogenicity. ALS/FTD-related FUS mutations are suspected to cause FUS protein mislocalization to the cytoplasm and result in protein inclusions although the underlying mechanisms are not fully understood. Recently, a missense mutation (p.Q290X) was nominated as the cause of autosomal dominant essential tremor (ET) in a large Canadian pedigree [7]. Subsequent ET studies have yet to identify the p.Q290X variant, and although a number of other rare variants have been identified the pathogenicity of FUS in ET remains to be determined. It is noteworthy, the p.Q290X substitution occurs in the nuclear export sequence (NES), unlike the FUS ALS/FTD mutations, and therefore may act through a different pathomechanism.
We recently reported the presence of a TDP-43 mutation in a patient with clinical Parkinson’s disease (PD) [8]. Parkinsonism has also been reported in the phenotype of ALS/FTD TDP-43 mutation carriers [9]. Given the evidence for the role for TDP-43 and FUS mutations in these movement and cognitive disorders, we investigated the role of FUS variants in PD. The present study focused on functional domains that harbor mutations related to neurodegeneration: (1) the glycine-rich domain, (2) the nuclear export signal (NES) and (3) the nuclear localization signal (NLS).
2. Methods
The combined patient–control series included 702 patients with clinical diagnosis of PD. We sequenced 372 late-onset sporadic patients, 211 late-onset patients with family history of PD, 69 patients with early-onset sporadic PD and 50 patients with early-onset familial PD. Key demographic and clinical data are summarized in Table 1. All subjects are unrelated, non-Hispanic Caucasians of mixed European ancestry, recruited at Mayo Clinic Florida. Average age was 77±11 years (range 35–103) for cases and average age at onset was 62±13 (16–94). Bi-directional sequencing was performed as previously described [10]. Primer and amplification conditions are available upon request. Call rate for sequencing was ≥99%. We sequenced exons 5 to 8 which included the glycine-rich domain (amino acids 166–267), exon 9 including the NES (amino acids 289–298) and exon 14 and 15 encompassing the predicted NLS (amino acids 495–526) [6] (see Fig. 1). The ethical review boards at the Mayo Clinic approved the study, and all participants provided informed consent.
Table 1.
PD patients’ demographicsa
| PD phenotype | Age | Age at onset | Gender (count)
|
|
|---|---|---|---|---|
| M | F | |||
| Late onset | ||||
| Sporadic (n = 372) | 81±8 (57–103) | 68±8 (51–94) | 236 | 131 |
| Familial (n = 211) | 79±8 (61–96) | 66±8 (51–83) | 134 | 77 |
| Early onset | ||||
| Familial (n = 50) | 62±12 (40–94) | 42±6 (29–50) | 30 | 20 |
| Sporadic (n = 69) | 61±10 (34–94) | 42±8 (16–50) | 45 | 29 |
| Total (n = 702) | 77±11 (34–103) | 62±13 (16–94) | 445 | 257 |
The sample mean±SD (minimum–maximum) is given for age and age at onset.
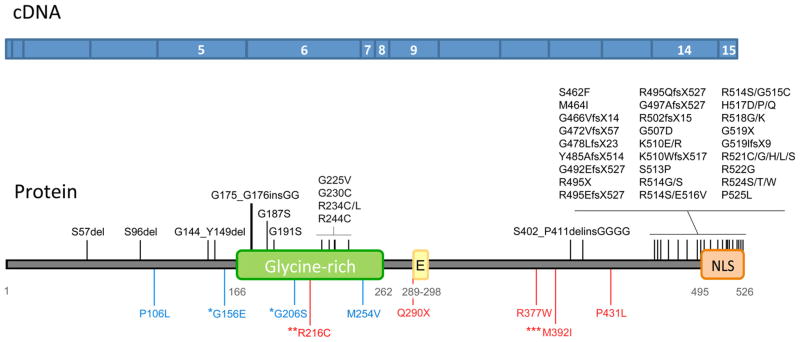
Fig. 1.
Protein domains and exons targeted for sequencing. The upper panel in blue represents the coding sequence of the FUS gene with the sequenced exons identified with white numbers. The lower panel represents the protein with the studied functional domain. Mutations shown in black were identified in patients with amyotrophic lateral sclerosis (ALS), mutations in blue were identified in patients with frontotemporal dementia (FTD), and mutations in red were identified in patients with essential tremor (ET). *G156 and *G206S were found in FTD/ALS. **R216C was identified in ALS, ET and one control. ***M392I is associated with an increased risk of developing ET. E, Nuclear export signal; NLS, nuclear localization signal.
3. Results
Sequencing of our US series of patients with PD (n = 702) for the FUS exons 5–9 and 14–15 identified a total of eight variants; six of the variants were within the exonic regions and two were within intron 5 (see Table 2). All the coding variants identified were synonymous variants. Of these variants three were located in exon 6 (p.G225G, p.G228G, and p.G229G), one in exon 15 within the NLS region (R522R) and two were located upstream of the NLS region in exon 14 (p.G482G, p.R487R). All the coding variants were databased within the NCBI dbSNP dataset except p.R487R which was novel. All the variants detected were rare, being observed only once or twice in 1404 chromosomes examined, and either rarely present or absent in the public exome variant server database (http://evs.gs.washington.edu/EVS); see Table 2.
Table 2.
FUS variants identified in patients with PD
| Position on Chr16 (GRCh37) | Location | rs number | Base change | AA | MAF (%) | MAF in EVS (%) |
|---|---|---|---|---|---|---|
| 31196245 | intron 5 | T>A | 0.07 | |||
| 31196255 | intron 5 | rs73530287 | C>T | 0.07 | 0.01 | |
| 31196411 | exon 6 | rs140003720 | C>T | G225G | 0.07 | 0.05 |
| 31196420 | exon 6 | rs151073460 | C>T | G228G | 0.07 | |
| 31196423 | exon 6 | rs140994262 | T>C | G229G | 0.14 | |
| 31202336 | exon 14 | rs112061837 | C>T | G482G | 0.07 | |
| 31202351 | exon 14 | C>T | R487R | 0.07 | ||
| 31202744 | exon 15 | rs138901914 | G>A | R522R | 0.14 | 0.2 |
AA, amino acid (transcript variant 1, NM_004960); MAF, minor allele frequency; EVS, Exome Variant Server – Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA, http://evs.gs.washington.edu/EVS/ (accessed July 2013).
4. Discussion
FUS mutations are an established cause of ALS. A number of specific variants have also been suggested to cause FTD and ET. These studies would implicate FUS in a myriad of movement disorders. In addition, FUS protein pathology is observed in a subset of FTD patients. The overlapping genetic and functional roles of mutations in FUS and TDP-43 nominate these two genes and altered proteins as determinants of specific subtypes of neurodegeneration that may present as alternative clinical syndromes. A role for TDP-43 has been suggested in parkinsonism, and based on these results, we set out to determine if genetic variation at the FUS locus may also play a role in PD.
Interestingly, the FUS gene is located on chromosome 16 within one megabase of a PD genome-wide association study nominated locus surrounding variant rs4889603 [11]. Correlation between rs4889603 and the HapMap SNPs in FUS is however low, r2 < 0.4 (tested using SNAP [12]). The region includes 48 genes but STX1B has been nominated as the putative causal gene. The results of the sequencing study in our series of PD patients would suggest that variants in the FUS glycine-rich region, NES or NLS are not a common cause of parkinsonism. It is possible that variants outside of the coding region of FUS which influence splicing or expression level may still have an effect on disease risk; additional studies will have to be performed to assess this possibility.
It remains a possibility that FUS mutations may play a role in a specific subset of patients with parkinsonism and it is becoming evident that a number of genes can harbor mutations that present with overlapping clinical features. The identification of the FUS p.Q290X nonsense mutation in a large family with ET occurred from the unbiased approach of whole-exome sequencing. As next-generation sequencing approaches become more widely accessible and cost-affordable we may find an increase in the identification of pathogenic mutations in neurodegenerative-related genes for clinical phenotypes or signs that would not typically be expected. These findings may provide important insights within the setting of clinical diagnosis, prognosis and potential therapeutic applications.
Acknowledgments
We would like to thank all those who have contributed to our research, particularly the patients and families who donated DNA samples for this work. We also wish to thank Margaret A. McKinney, medical illustrator, for her help in creating Figure 1.
This work is supported by a Morris K. Udall Parkinson’s Disease Research Center of Excellence (NINDS P50 #NS072187), NINDS R01 #NS078086 and a gift from Carl Edward Bolch, Jr. and Susan Bass Bolch.
Footnotes
Conflict of interests
The authors have no financial or other conflicts of interest to declare.
References
- 1.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–4. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 2.Rabbitts TH, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–80. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 3.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904–19. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 5.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Langenhove T, van der Zee J, Sleegers K, Engelborghs S, Vandenberghe R, Gijselinck I, et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. 2010;74:366–71. doi: 10.1212/WNL.0b013e3181ccc732. [DOI] [PubMed] [Google Scholar]
- 7.Merner ND, Girard SL, Catoire H, Bourassa CV, Belzil VV, Riviere JB, et al. Exome sequencing identifies FUS mutations as a cause of essential tremor. Am J Hum Genet. 2012;91:313–9. doi: 10.1016/j.ajhg.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rayaprolu S, Fujioka S, Traynor S, Soto-Ortolaza AI, Petrucelli L, Dickson DW, et al. TARDBP mutations in Parkinson’s disease. Parkinsonism Relat Disord. 2013;19:312–5. doi: 10.1016/j.parkreldis.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quadri M, Cossu G, Saddi V, Simons EJ, Murgia D, Melis M, et al. Broadening the phenotype of TARDBP mutations: the TARDBP Ala382Thr mutation and Parkinson’s disease in Sardinia. Neurogenetics. 2011;12:203–9. doi: 10.1007/s10048-011-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labbé C, Soto-Ortolaza AI, Rayaprolu S, Harriott AM, Strongosky AJ, Uitti RJ, et al. Investigating the role of FUS exonic variants in essential tremor. Parkinsonism Relat Disord. 2013;19:755–7. doi: 10.1016/j.parkreldis.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Parkinson’s Disease Genomics Consortium (IPDGC); Wellcome Trust Case Control Consortium 2 (WTCCC2) A two-stage meta-analysis identifies several new loci for Parkinson’s disease. PLoS Genet. 2011;7:e1002142. doi: 10.1371/journal.pgen.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]