Abstract
During bacterial chemotaxis, transmembrane chemoreceptor arrays regulate autophosphorylation of the dimeric, histidine-kinase CheA. The five domains of CheA (P1-P5) each play a specific role in coupling receptor stimulation to CheA activity. Biochemical and x-ray scattering studies of thermostable CheA from Thermotoga maritima find that the His-containing substrate domain (P1) is sequestered by interactions that depend upon P1 of the adjacent subunit. Non-hydrolyzable ATP analogs (but not ATP nor ADP) release P1 from the protein core (domains P3P4P5) and increase its mobility. Detachment of both P1 domains, or removal of one within a dimer, increases net autophosphorylation substantially at physiological temperature (55°C). However, nearly all activity is lost without the dimerization domain (P3). The linker length between P1 and P3 dictates inter-subunit (trans) versus intra-subunit (cis) autophosphorylation; with the trans reaction requiring a minimum length of 47 residues. A new crystal structure of the most active dimerization-plus-kinase unit (P3P4) reveals trans-directing interactions between the tether connecting P3 to P2-P1 and the adjacent ATP-binding (P4) domain. The orientation of P4 relative to P3 in the P3P4 structure supports a planar CheA conformation that is required by membrane array models, and suggests that the ATP-lid of CheA may be poised to interact with receptors and coupling proteins. Collectively, these data suggest that the P1 domains are restrained in the off-state as a result of cross-subunit interactions. Perturbations at the nucleotide-binding pocket increase P1 mobility and access of the substrate His to P4-bound ATP.
Keywords: kinase, dynamics, cooperativity, regulation, phosphorylation
Introduction
Kinases are essential sensors and transducers of cellular signals [1–4]. Regulation of their enzymatic activity often depends upon large-scale domain motions [1,5]. CheA is a multi-domain dimeric histidine kinase that acts as the primary enzyme in the bacterial chemotaxis signal transduction pathway [6,7]. During chemotaxis, signals propagate through membrane-incorporated chemoreceptor arrays composed of methyl-accepting chemotaxis proteins (MCPs), histidine kinases (CheA), and coupling proteins (CheW). Through complex interactions these proteins initiate an intracellular phospho-relay that ultimately regulates flagella rotational switching [8–10]. Central issues in understanding the mechanism of chemotaxis signaling concern how CheA autophosphorylation is regulated, and specifically, what conformational transitions convert the kinase from an inactive to an active form. Many biochemical and structural studies have been conducted to elucidate the organization of the receptor arrays, as well as identify the specific interactions between the MCPs, CheA, and CheW in the extended lattice [11–15]. Electron cryo-tomography (ECT) images [13,14,16,17] of the receptor arrays within intact cells reveal a hexagonal arrangement of proteins with features supported by the crystal structure of a CheA/CheW ring in complex with a Tm14 MCP dimer [18]. Previously determined Thermotoga maritima CheA structures [18–24] provide building blocks to understand the CheA domain arrangements within the lattice. However, domain positions in the most complete structure of CheA to date (comprising the core P3P4P5 domains or Δ289) [20] must be altered about the linker joining the P3 and P4 domains to fit the constraints of the lattice [12].
Each CheA subunit contains five distinct domains (P1-P5), which have discrete functions and display variable mobility [25]. The P1 domain contains the substrate histidine residue (His45 in T. maritima or His48 in E. coli), the P2 domain docks the response regulator CheY for phosphotransfer from P1, the P3 domain dimerizes the two subunits, the P4 domain acts as the ATP-binding kinase module and the P5 domain couples CheA to CheW and chemoreceptors [6,26–28]. CheA differs from sensor histidine kinases in several ways: CheA does not contain a transmembrane domain, relying instead on P5 and CheW for interaction with transmembrane components, it has the phosphorylatable His residue on a separate domain (P1) instead of the dimerization domain (P3), and it utilizes a separate docking domain (P2) for CheY. P2 is not necessary for phosphotransfer to the response regulator CheY per se [29] but variants lacking the P2 domain (ΔP2) exhibit a reduced phosphotransfer rate relative to full-length CheA (CheAFL) and support a lower extent of chemotaxis [29,30]. The linkers between the CheA domains have also been shown to play important roles in CheA activity [31,32]. For example, the P3-to-P4 linker influences the dynamical properties and kinase activity of the P4 domain [32,33]. NMR structures of E. coli and T. maritima CheA P1 assign the 20 linker residues C-terminal to the P1 4-helix bundle as an additional α-helix that runs along the other four helices [34] [24]. This additional helix constrains the P1 domain, and may restrict the movement and spatial orientation of activated CheA [24,34]. Notably, E. coli CheA autophosphorylates in trans, with one subunit phosphorylating the other [35,36].
Structural determinants for ATP binding and P1 phosphorylation are well defined within the P4 [19] and P1 [37] domains, although the manner and regulation of their interaction remains unclear. Nonetheless, recent work has elucidated possible contact points between these domains with one specific interaction identified between E. coli CheA Glu38 and Lys346 [38–42]. Additional studies indicate directional steering of the P1 domain toward the trans P4 subunit [36] and allosteric behavior of the two kinase active sites [42, 43]. Negative cooperativity for nucleotide binding [44], which implies interaction between the P4 domains, is not easily reconciled with the large P4 separation found in the crystal structure of P3P4P5 [19]. Domain interactions within the full-length kinase may also limit motions of the substrate domains important for phosphorylation. Indeed, CryoEM images of native arrays in different activity states suggest that the P1 and P2 domains are more ordered in the inhibited state [11].
Herein, we study the consequence of covalent attachment of the P1 substrate domain to the CheA kinase module using the structurally characterized CheA from T. maritima as our primary subject of investigation. The minimal linker length for trans autophosphorylation is defined; surprisingly, shorter linkers switch to cis phosphorylation and impart less activity. Small-angle x-ray scattering (SAXS) reveals that binding of non-hydrolyzable analogs of ATP release the P1 domains from a constrained environment. Targeted disulfide cross-linking between the P1 and P4 reactive centers correlates kinase activity with access of P1 to the P4 ATP pocket. Severing the P1 domains from the kinase core (P3P4P5) activates autophosphorylation at higher temperatures, as does the formation of heterodimers with only one P1 module. P4 alone has nearly no reactivity towards P1, but addition of the P3 dimerization greatly enhances kinase activity. The structure of the minimal active fragment, P3P4, exhibits a planar arrangement of the domains, consistent with the CheA conformation required by the receptor arrays. Coupling crystallographic and biochemical data, we generate an updated working model of T. maritima CheA within the context of the hexagonal receptor lattice that includes a productive association between P1 and P4. Overall, these data underscore the importance of specific interactions between CheA subunits that control accessibility of the histidine substrate domain to the ATP-binding kinase module. Rearrangement about inter-domain linkers appears critical for functional assembly of CheA into the receptor arrays.
Results
Generation of ΔP2 variants
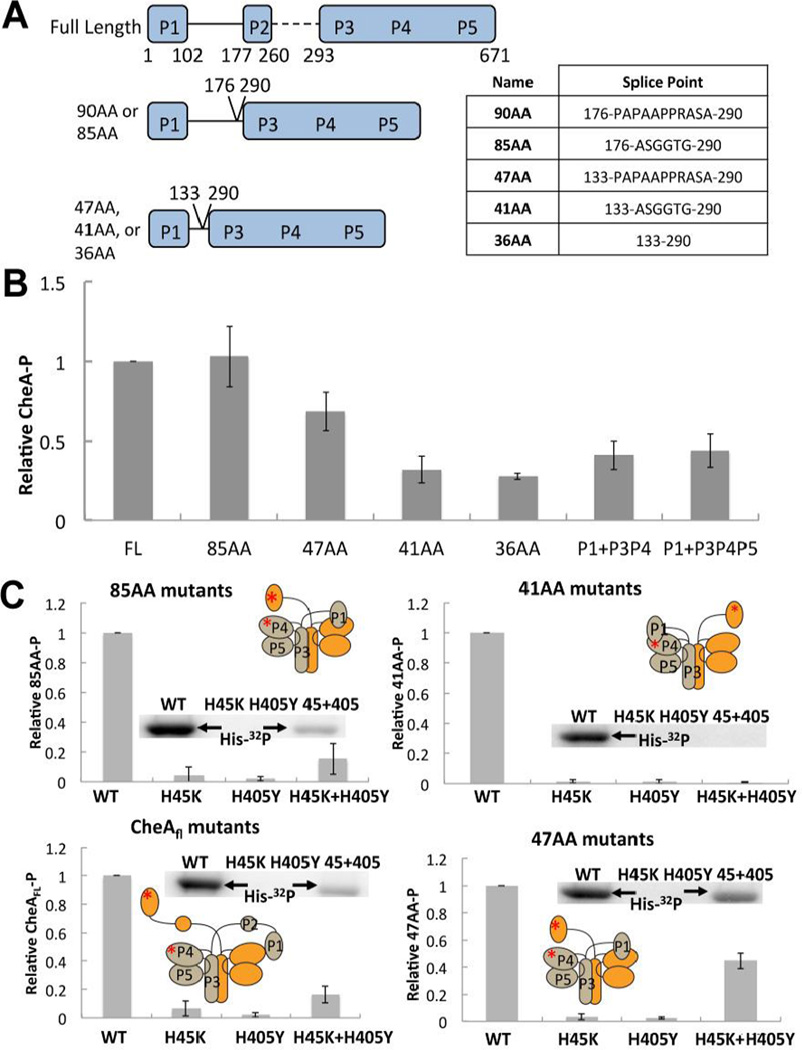
To generate CheA variants with altered linkages between the kinase core (P3P4P5, also known as Δ289) and the substrate P1 domain, P1 was fused to P3P4P5 with linkers that retain various sections of the native P1-P2 connection along with several short non-native sequences added to adjust length and flexibility (Fig. 1A). The ΔP2 variant nomenclature was assigned according to the number of residues that connect P3 residue 293 to P1 residue 100, the latter of which ends the fourth helix of the P1 domain of T. maritima CheA (Fig. 1A). The longest variant (85AA) contains all but the last residue (K177) of the native P1-P2 linker. The shorter variants (36AA, 41AA, and 47AA) retain the 33 residues C-terminal to the fourth P1 helix. The 41AA and 47AA variants also include a 5–14 residue spacer to facilitate linker flexibility.
Figure 1. ΔP2 variants and their autophosphorylation activity.
(A) Cartoon depiction of ΔP2 variants of Thermotoga maritima CheA with domain boundaries and splice points indicated, nomenclature is based on the linker length (see text). Table at right shows residues inserted between splice points marked on left. (B) Relative autophosphorylation activity of T. maritima CheAFL, ΔP2 variants, or separated domains. Levels of P1-P were determined after 6 min of exposure to γ-32P-ATP at 25 °C. All proteins are at 10 µM subunit concentration. (C) Rescue of autophosphorylation activity of CheAFL, 85AA, 47AA, and 41AA active site mutants (P1:H45K or P4:H405Y) by subunit exchange. A phosphorimage of the gel assays shows P1-P production. Autophosphorylation is only possible in the trans subunit reaction of a heterodimer that contains a WT P1 subunit and a WT P4 subunit (45 + 405) A cartoon depiction of trans or cis autophosphorylation for each variant is inset with a red asterisk depicting the point mutations that abrogate function of the P1 and P4 domains in the mixed dimer. All proteins are at a subunit concentration of 10 µM and were heated overnight at 55 °C to allow for subunit exchange prior to γ-32P-ATP being added for 6 minutes at 25 °C.
Activity of ΔP2 variants compared to CheAFL
CheA autophosphorylation activity generally decreases as the linker length in the ΔP2 variants shortens. ΔP2 variants with long linkers (90 AA and 85AA) undergo similar degrees of autophosphorylation compared to T. maritima full-length CheA at 25 °C (CheAFL; Fig. 1B). The ΔP2 variant with the longest linker (90AA) was indistinguishable from WT and not considered further. In contrast, the 41AA, 36AA variants show considerably reduced autophosphorylation. All of the ΔP2 variants retain the ability to then transfer phosphate from the P1 to T. maritima CheY (Fig. S1). This behavior is similar to that of a ΔP2 variant of E. coli CheA that is also competent for phosphotransfer to CheY [35,45–48] The fifth helix of the P1 domain [34] may substantially constrain P1 motion in the shorter constructs. Separation of the P1 domain from either P3P4 or P3P4P5 reduces autophosphorylation by ~50% at 25 °C (with all species at subunit concentrations of 10 µM). The change from an intramolecular to an intermolecular interaction should reduce phosphotransfer activity by orders of magnitude provided the unit concentrations are substantially below the binding constant for P1 (See analysis in the SI). Attempts were made to measure the binding constant between P1 and P3P4 at 25 °C by isothermal calorimetry and the Michaelis constant by enzyme kinetics [47–49]; in both cases the binding was too weak (> 250 µM) to provide reliable values.
CheA heterodimers to test for trans phosphorylation
Temperature dependent subunit exchange of T. maritima CheA allows for heterodimers of unequal subunits to be formed and trapped [49]. Prolonged incubation at 55 °C of T. maritima CheAFL, with CheA P3P4P5 produces heterodimers (with only one P1-P2 unit) that can be resolved on an analytical size-exclusion column at 4 °C (Fig. S2) [49]. To test for trans autophosphorylation, a CheAFL variant harboring a P4 mutation that abrogates kinase activity (H405Y) was exchanged with a CheAFL variant that lacks the substrate His (H45K) [36,50]. Thus, autophosphorylation is possible only when the subunit with a functional P4 domain trans-phosphorylates a functional P1 domain of the opposing subunit.
Based on the results from analytical size-exclusion chromatography (SEC) with CheAFL and P3P4P5, a statistical 1:2:1 ratio for H45K2:(H45K:H405Y):H405Y2 is expected after exchange, and hence the mixed sample will only have at most 50% activity (Fig. S2). In this system, CheAFL, 85AA, and 47AA dimers with complimentary active site defects regained activity upon heat-exchange (Fig. 1C). In contrast, the 41AA (Fig. 1C) and 36AA proteins (not shown) with active-site defects did not show any recovery of autophosphorylation when exchanged, despite unaltered 41AA and 36AA undergoing autophosphorylation as homodimers (Fig. 1B). Thus, the variants with the shortest P1-P2 linkers likely autophosphorylate in cis.
Conformational properties of P1 within CheAFL depend on nucleotide binding state
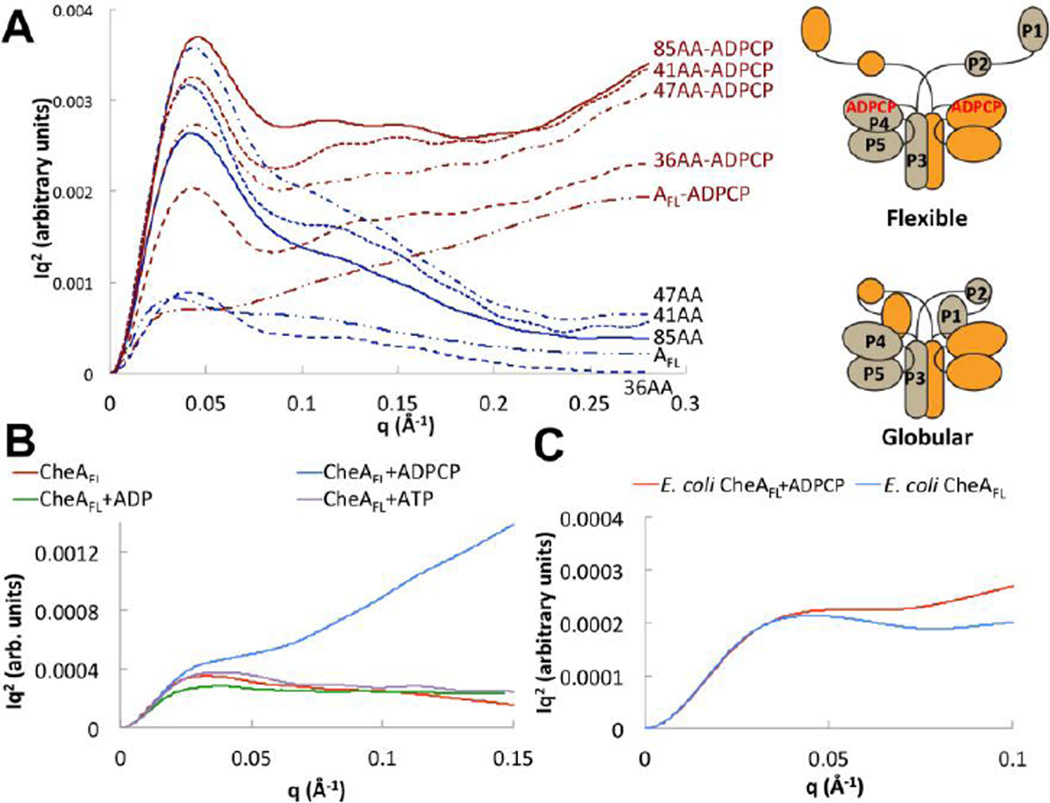
SAXS was used to evaluate the conformational properties of both native and variant CheA in solution. For all species studied, scattering data collected at several concentrations showed no evidence of aggregation (Fig. S3A) and intensity ratios between different concentrations at high scattering angle showed no evidence of buffer subtraction artifacts. The shape of Kratky plots (I(q)q2 vs. q, where q represents the momentum transfer: ), reports on the overall conformational properties of a polymer chain. For rod-like particles, I(q)q2 increases with positive slope; for random Gaussian chains, I(q)q2 plateaus at large values of q, and for globular particles I(q)q2 curves down toward baseline [51,52]. P3P4P5 has a globular shape in solution that is unaffected by addition of the non-hydrolyzable ATP analog ADPCP or ADP (Fig. S3B). CheAFL and the ΔP2 variants also appeared largely ordered and compact either in the absence of nucleotide (Fig. 2A). However, Kratky plots of the CheAFL and the ΔP2 variants displayed a dramatic change in conformation indicative of disordered Gaussian chains or flexible rods when ADPCP was added (Fig. 2A). This change in flexibility was not seen for CheAFL with ADP or ATP (Fig. 2B). These trends were observed for all of the T. maritima ΔP2 variants as well as E. coli CheA, although they were more pronounced for the T. maritima enzymes (Fig 2C). A residue substitution in the ATP binding pocket (D449A) that prevents nucleotide binding (Fig. S4A) prevented any change in SAXS profile upon addition of ADPCP for both CheAFL or 41AA (Fig S4B). Normalized Kratky plots [53] made dimensionless by factoring in VC (the volume per correlation length [54]), also showed that P1-containing proteins have a much higher surface-to-volume ratio when ADPCP is present (Fig. S5). Changes in the Porod exponents [55] also indicate much greater flexibility with the non-hydrolyzable nucleotide (Table 1). CD spectra indicate that overall secondary structure of the protein is not substantially altered by ADPCP (Fig. S6). Thermal melts of CheAFL and the ΔP2 variants with ADPCP confirmed that the enzymes remain highly stable and structured over a period of days (data not shown). Thus, mobility of the P1 domain in both CheAFL and the ΔP2 variants increases substantially upon addition of non-hydrolyzable ATP, but not ATP nor ADP.
Figure 2. Small-angle x-ray scattering indicates two CheA conformational states that depend on P1 and nucleotide.
(A) Kratky plots (I(q)q2 vs. q) of CheAFL and ΔP2 variants (3 mg/mL) with (red) and without (blue) ADPCP. A cartoon depicts the possible conformational transition suggested by the SAXS data. P3P4P5 shows no change in conformation with ADPCP (Figure S3). (B) Kratky plot of T. maritima CheAFL (3 mg/mL) without (red) and with ADPCP (blue), ADP (green) and ATP (purple) all at 5 mM. Only ADPCP increases CheA flexibility. (C) Kratky plot of E. coli CheAFL (5 mg/mL) without (blue) and with (red) 5 mM ADPCP. E. coli CheA also shows increased flexibility with ADPCP. SAXS curves are shown after smoothing and regularization. See Table 1 for SAXS parameters and SI Fig. 5 for dimensionless Kratky analysis of unsmoothed data.
Table 1.
SAXS Parameters for Full-length CheA and Select Variants Bound to Nucleotides
| Protein | Nucleotide | Rg (Å) | dmax (Å) | ravg (Å) | Px |
|---|---|---|---|---|---|
| CheAFL | none | 72.7 | 170 | 74.1 | 2.3 |
| ADP | 73.1 | 170 | 76.4 | 2.4 | |
| ATP | 73.8 | 170 | 74.3 | 2.2 | |
| ADPCP | 57.7 | 170 | 81.0 | 0.9 | |
| P3P4P5 | none | 54.4 | 130 | 61.9 | 2.8 |
| ADP | 50.2 | 130 | 59.3 | 2.7 | |
| ADPCP | 50.2 | 130 | 59.8 | 2.7 | |
| CheAFL D449A | none | 60.3 | 170 | 77.2 | 2.5 |
| ADPCP | 64.0 | 170 | 76.5 | 2.4 |
Rg represents the radius of gyration determined by Guinier analysis
dmax represents the maximum distance constraint in the pairwise distance distribution or P(r) generation.
ravg is the average radius from the P(r) distribution
Px is the Porod exponent; larger values represent more compact particles.
Above parameters were determined with Scatter: (biosis.net)
Cysteine cross-linking as a probe of P1-P4 domain interactions
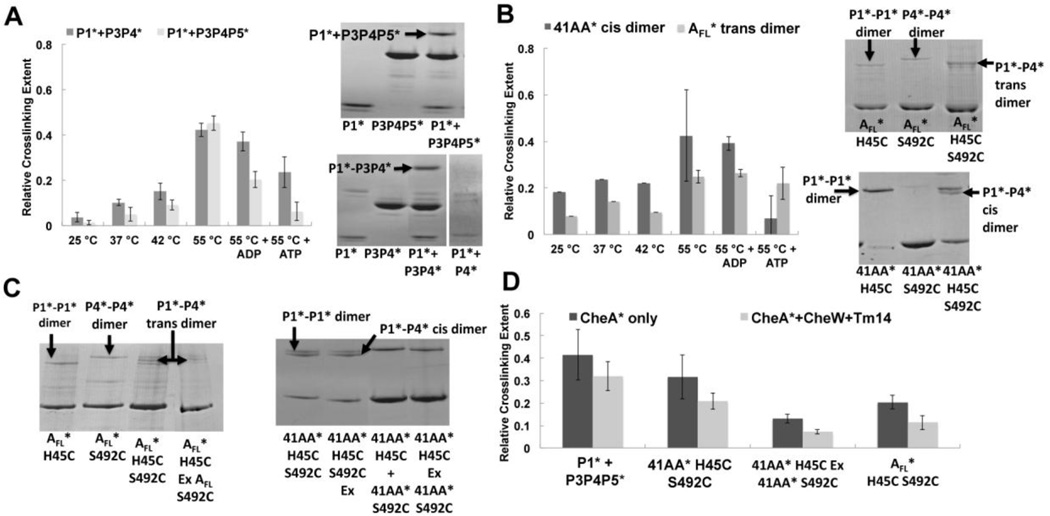
A cross-linking assay was devised to detect the association of the kinase (P4) and substrate (P1) domain within T. maritima CheA. Cysteine substitutions (Table 2) were generated in the P4 domain (S492C, P4*) and at the P1 substrate His (H45C, P1*) of an otherwise Cys-less CheA variant. Ser492 resides on the ATP-lid, near to the Mg2+ ion that coordinates the γ-phosphate of ATP in the P4 active site [19]. At 55 °C (but not 25 °C) P1*-P4* cross-linking produces a higher MW species resolvable on SDS-PAGE for CheAFL, 41AA and the P1-severed variants P3P4 and P3P4P5 (Fig. 3). Of the ΔP2 variants, 41AA was chosen for investigation because it was the longest variant for which exchange of subunits with complimentary active site defects did not rescue activity (Fig. 1). Disulfide cross-linking was observed between P1* and P3P4P5* and between P1* and P3P4*, but not between P1* and P4*, even after extended incubation (Fig. 3A). Notably, P1*-P4* cross-linking increases dramatically in the presence of the P3 dimerization domain (Fig. 3A).
Table 2.
List of Cys-modified variants and their molecular weights
| Proteins | Mutants* | MW (subunit) |
|---|---|---|
| P1* | H45C | 14.7 |
| P4* | S492C | 20 |
| P3P4* | S492C | 27.1 |
| P3P4P5* | S492C | 42.5 |
| 41AA* | H45C (P1*), S492C (P4*), H45C, S492C (P1*P4*) | 53 |
| CheAFL* | H45C (P1*), S492C (P4*), H45C, S492C (P1*,P4*) | 74 |
Figure 3. Disulfide cross-linking to probe interactions between P1 and P4.
(A) Disulfide cross-linking between P1*(H45C) to P4*(S492C) for P1* and either P3P4* or P3P4P5* (stars denote Cys-modified proteins). Left: Quantification of P1*-P4* cross-linked band on SDS-PAGE for P1* (12 µM) reacting with P3P4* or P3P4P5* (24 µM) at the indicated temperatures for 1 hour. Addition of 2mM ADP or ATP reduces P1*-P4* cross-linking, with ATP having the greater effect. Right: SDS-PAGE of P1* cross-linked to either P3P4P5* or P3P4* after 1 hour at 55 °C. P1* and P4* give no cross-linking under identical conditions. (B) Disulfide cross-linking for CheAFL* (AFL*) and 41AA containing the H45C and S492C Cys substitutions Left: Comparison of trans P1*-P4* cross-linking within the CheAFL (P1*P4*) homodimer to cis cross-linking within the 41AA* (P1*P4*) homodimer. ATP reduces cross-linking with 41AA, but not with AFL. Right: SDS-PAGE of CheAFL* and 41AA* (12 µM) cross-linked dimers after 1 hour at 55 °C: P1* mutant homodimers form only P1*-P1* bands, P4* mutants form only P4*-P4* bands, whereas P1*P4* double mutants form all three cross-link types: P1*-P1*, P1*-P4* and P4*-P4*. P4*-P4* crosslinks appear reduced in the double mutants, which favor P1*-P4* cross-linking. (C) Exchange of P1* and P4* homodimers at 55 °C overnight (Ex) rescues P1*-P4* cross-linking in CheAFL* but not in 41AA*. Left: SDS-PAGE of CheAFL* (6 µM) + CheW (6 µM) cross-linking at 55 °C for 1 hour; the exchanged (Ex) P1* and P4* homodimers produce the same banding pattern as the double mutant (P1*P4* = H45C,S492C) for CheAFL*. Right: SDS-PAGE of P1* and P4* exchange for 41AA* (6 µM), which fails to form the P1*-P4* cross-link upon mixing of the single mutants (third lane) or after many hours of exchange at 55 °C (last lane). Lane 1 and Lane 2 show that overnight subunit exchange at 55 °C does not affect cross-linking of the double mutant. Cross-linking was performed by adding 5 mM Cu(II)phen3 at 55 °C for 1 hr whether or not preceded by an exchange period. (D) Quantified band intensities of the cross-linked CheA* (6 µM) with and without CheW (6 µM) and Tm14 (18 µM) at 55 °C for 1 hour. Tm14 and CheW inhibit P1*-P4* cross-linking in all cases.
Cross-linking of CheAFL* (P1*P4*) generated multiple bands on SDS-PAGE, indicative of P1*-to-P1*, P4*-to-P4* and P1*-to-P4* disulfide-linked species (Fig. 3B,C). Positions of the P1*-to-P1* and P4*-to-P4* bands were confirmed in variants that only harbored the P1* or P4* cysteine substitutions, respectively (Fig. 3A,B). For CheAFL*, the P1*-to-P4* cross-linked species runs between that of P1*-to-P1* and P4*-to-P4* on SDS-PAGE (Fig. 3B,C). Surprisingly, the cross-linking efficiency for P1*-to-P4* in CheAFL* and 41AA* was significantly less than when P1* was supplied as a separate protein (Fig. 3A,B and D). For the single Cys mutants, 41AA* forms P1*-to-P1* cross-links, but not P4*-to-P4* crosslinks (Fig. 3B). In the 41AA* double Cys mutant (P1*P4*), the P1*-to-P4* cross-linked band runs below the P1*-to-P1* band and, importantly, is not present when 41AA* homodimers with single mutations (P1*-P1* and P4*-P4*) are exchanged to produce heterodimers (P1*P4-P1P4*; Fig. 3C). This behavior contrasts with that of CheAFL, which produces the same banding pattern on subunit exchange of the single Cys mutants, as does the double mutant (Fig. 3C). The inability of subunit exchange (which depends primarily on the P3 domain) to rescue P1*P4* cross-linking of the single mutants is consistent with a cis interaction, which is also consistent with the autophosphorylation data (Fig. 1). Note that the 41AA* cis dimer band runs on SDS-PAGE at twice the subunit MW, which is unusual for intra-subunit cross-linking. Nonetheless, we have found that some thermostable non-covalent dimers are stable in SDS. To further confirm that the P1*-to-P4* band derives from a cis-crosslink we exchanged the 41AA* double mutants with excess 41AA or P3P4 subunits that did not contain engineered cysteine residues. SEC experiments demonstrated that the 41AA variant does indeed undergo such exchange reactions (Fig. S2B). However, no reduction in cross-linking was found even with a 3-fold molar excess of the native subunits during the exchange processes (Fig. S7), which again indicates that P1*-to-P4* cross-linking occurs within a subunit for 41AA*.
The cytosolic MCP Tm14 deactivates CheA autophosphorylation [56]. Tm14 (residues 41–254) was used to probe the effect of inhibitory receptors on the P1*-P4* interaction (Fig. 3D). Deactivation by Tm14 also requires CheW. For all CheA species, addition of CheW and deactivating MCP (Tm14) reduced disulfide formation between P1* and P4* in CheAFL*, 41AA* and for P3P4P5*. ATP, and to a lesser extent ADP, affected cross-linking of the various species (with perhaps the exception of CheAFL*), likely because the nucleotide phosphates interfere with approach of the H45C residue to S492C.
T. maritima CheA autophosphorylation kinetics
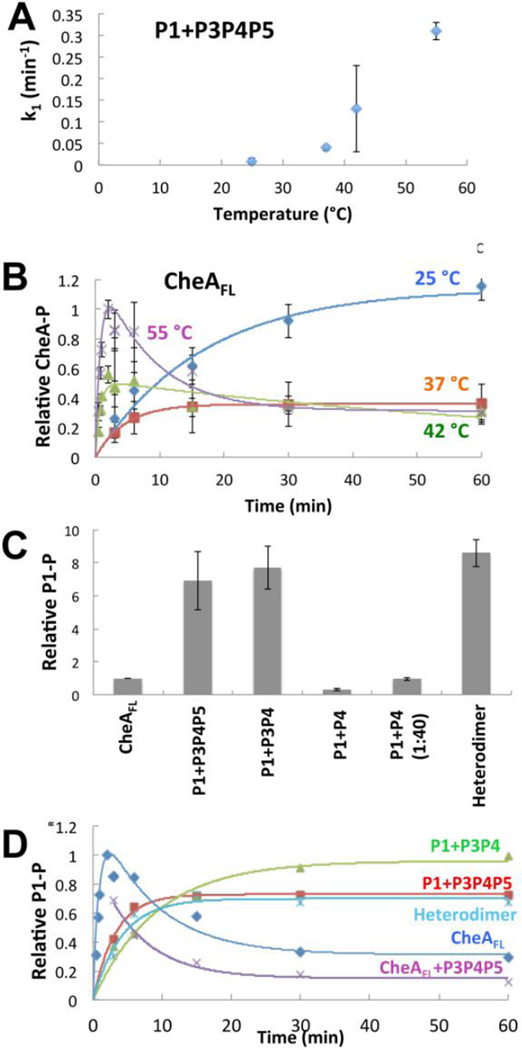
P3P4P5 or P3P4 phosphorylated free P1 (P1-P) by first order kinetics, with a direct relationship between rate constant and temperature (Fig. 4A, Table 3). In contrast, the progress curve for CheAFL autophosphorylation became biphasic at 42 °C; and this effect intensified as temperature increased toward the physiological level for T. maritima (Fig. 4B). At 55 °C, P1 phosphorylation increased rapidly for the first two minutes, peaked, and then diminished at times extending to 60 min. Phosphorylated P1 is stable at 55 °C for hours [37], and thus these data indicate a dephoshorylation activity that increases with time and temperature. Such an activity could be due to the reverse reaction between P1-P and P4-bound ADP [45,57,58], if ADP levels were to increase with time. Indeed, chasing CheAFL with excess ADP after 6 min of autophosphorylation depletes P1-P for all species studied (Fig. S8A). However, with ATP in excess, P4-bound ADP will exchange rapidly with ATP and prevent the back reaction (kd for ADP ~ 1 sec−1 at 4 °C [59]). Furthermore, we detected no appreciable increase in ADP concentrations at 55 °C after one hour (by thin layer chromatography [60] and by detecting ADP with an enzyme-coupled spectrophotometric method (Fig. S8B) [61–63]). The loss of P1-P at 55 °C for CheAFL correlates with a release of inorganic phosphate from P1 that is not observed at lower temperatures; thus reduced P1-P results from an increase in phosphatase activity within CheAFL (Fig. S8B), Remarkably, if a heterodimer is formed between CheAFL and P3P4P5, there is no loss of P1-P at longer times and the reaction proceeds with first order kinetics (Fig. 4C, 4D). In contrast, simply adding P3P4P5 to CheAFL without subunit heat exchange does not eliminate phosphatase activity (Fig. 4D). Importantly, at 55 °C, the separated domains, P1 + P3P4 and P1 + P3P4P5 produced sustained levels of P1-P at long times (Fig. 4C, 4D). These amounts are much greater than those seen for CheAFL, under the same conditions (Fig. 4C), but similar to the peak values of CheAFL at short times (Fig. 4D). Thus, for T. maritima CheAFL, the presence of P1 in both subunits, but not one, destabilizes P1-P at increased temperature. (Note that the relative activity of the separated domains is greater at 55 °C than at 25 °C (Fig. 1B).) The kinetic progress curves can be described by a multi-state process in which phosphorylated CheA (P1-P) converts to an un-phosphorylated form (P1#) that differs from the starting species and can only re-phosphorylate slowly (P1 → P1-P ←→ P1 #). As indicated by this kinetic model, P1-P will reach an equilibrium level at long time that is less than the peak value (Fig. 4B). Slower net autophosphorylation by P1# could result from an increased intrinsic phosphatase activity. Importantly, the P4 domain alone shows nearly no ability to phosphorylate P1 (Fig. 4C, Table 3). Thus, the P3P4 dimer is the minimal unit required to achieve high, sustained levels of P1-P.
Figure 4. Autophosphorylation activity for T. maritima CheAFL and separated domains.
(A) Rate constants for accumulation of P1-P by P1+P3P4P5 as a function of temperature. Higher temperatures increase P1 phosphorylation rates, which fit well to first order kinetics. (B) Time courses for autophosphorylation of CheAFL as a function of temperature. At 42 °C CheAFL begins to show biphasic behavior in which P1-P peaks at early times and then diminishes to a lower constant level. This behavior accentuates at 55 °C. All proteins are at 10 µM. Data were fit to first order kinetics for low temperatures and to the kinetic described in the Materials and Methods for 42 °C and 55 °C. (C) Production of phosphorylated CheAFL and separated domains (all at 10 µM) after 60 min at 55 °C. Separation of P1 from P4 increases activity, but only when the P3 domain is fused to P4. Even in high molar excess (40:1) P4 only weakly phosphorylates P1. In contrast to CheAFL, CheAFL:P3P4P5 heterodimers (Heterodimer) containing only one P1 domain show efficient P1 phosphorylation. (D) Time courses for P1-P formation for CheA (10 µM) and separated domains at 55 °C. The separated domains and the CheAFL:P3P4P5 heterodimer accumulate P1-P, whereas P1-P diminishes over time with CheAFL with or without added (but not exchanged) P3P4P5. Error bars of similar magnitude as shown in (A), but not shown for clarity. Each point is an average from 3–5 individual experiments.
Table 3.
Kinetic parameters for CheA autophosphorylation.
| Components | A0 (CheA-P formation) |
k1 (min−1) | k2 (min−1) | k1A0 | R2 |
|---|---|---|---|---|---|
| P1+P3P4P5 25 °C | 1.3 ± 0.08 | 0.009 ± 0.007 | - | 0.011 | 0.98 |
| P1+P3P4P5 37 °C | 0.54 ± 0.04 | 0.040 ± 0.006 | - | 0.022 | 0.99 |
| P1+P3P4P5 42 °C | 0.33 ± 0.08 | 0.13 ± 0.10 | - | 0.043 | 0.60 |
| P1+P3P4P5 55 °C | 0.73 ± 0.01 | 0.31 ± 0.02 | - | 0.23 | 0.99 |
| CheAFL 25 °C | 1.14 ± 0.09 | 0.06 ± 0.01 | - | 0.068 | 0.97 |
| CheAFL 37 °C | 0.358 ± 0.001 | 0.216 ± 0.004 | - | 0.0773 | 0.99 |
| CheAFL 42 °C | 0.5 ± 0.1 | 1.3 ± 0.9 | 0.01 ± 0.009 | 0.65 | 0.82 |
| CheAFL 55 °C | 1.0 ± 0.3 | 1.2 ± 0.7 | 0.03 ± 0.02 | 1.2 | 0.86 |
| P1+P3P4 55 °C | 0.96 ± 0.05 | 0.11 ± 0.02 | - | 0.11 | 0.97 |
| CheAFL+P3P4P5 55 °C | 0.52 ± 0.05 | 0.15 ± 0.04 | - | 0.078 | 0.97 |
| Mixed CheAFL+P3P4P5 55 °C | 0.70 ± 0.03 | 0.24 ± 0.04 | - | 0.17 | 0.97 |
| CheAFL+ADP 55 °C | 0.510 ± 0.003 | 0.66 ± 0.02 | - | 0.335 | 0.99 |
| P1+P3P4P5+ADP 55 °C | 0.52 ± 0.05 | 0.24 ± 0.06 | - | 0.12 | 0.96 |
Data were fit to the first order rate equation (Pt = A0(1−e−k1t)), except for the biphasic behavior of CheAFL at 42 °C and 55 °C; in these case k2 represents the rate constant for P1-P decay to P1#
Crystallographic structure of CheA P3P4
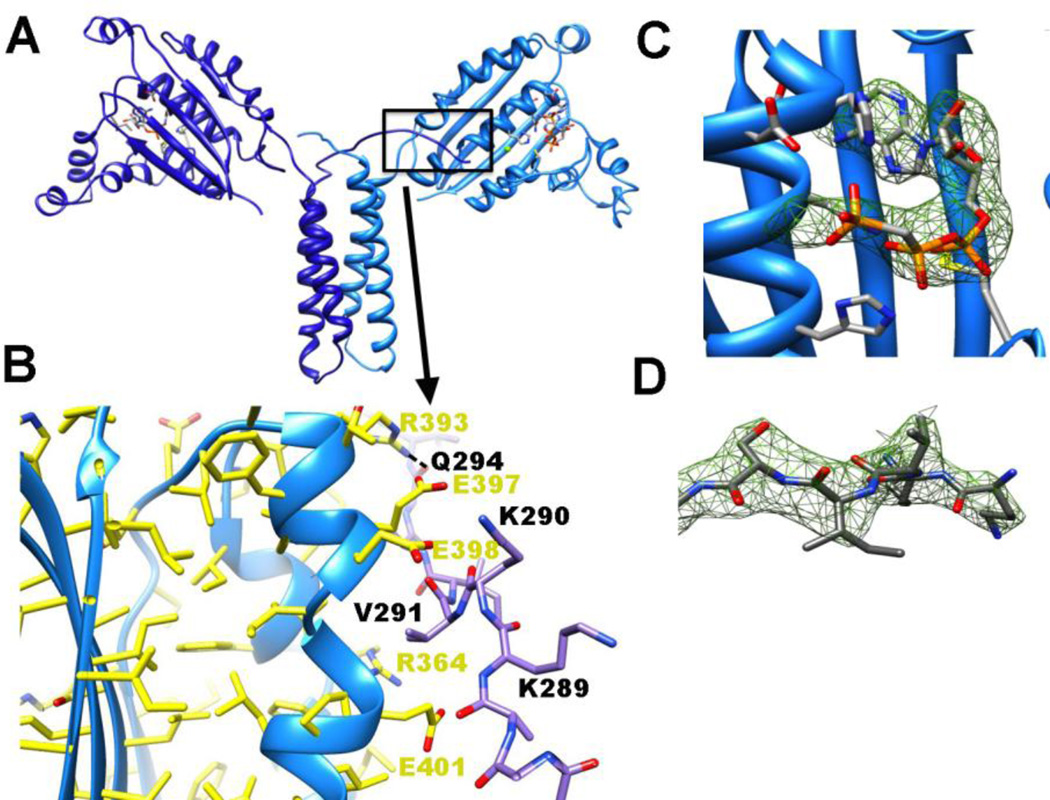
The crystal structure of the T. maritima CheA P3P4 domains bound to ADPCP were determined to 3.0 Å resolution (Fig. 5). The P3 domain is composed of two α-helices that form a coiled-coil dimer. The P4 domain has a characteristic Bergerat ATP-binding fold, common for the GHKL family of ATPases [2]. Unlike the previously determined P3P4P5 structure (CheA Δ289 [20]) the P4 domains reside in a narrow plane with their β-sheets roughly aligned to the helical axis of the P3 domain and their ATP pockets face 180° from one another (Fig. 5A). In contrast, the P4 domains of the P3P4P5 dimer have aroughly perpendicular juxtaposition [20]. The unit cell and space group of the P3P4 crystals differ from those of the P3P4P5 crystals (Table 4) owing to the absence of the P5 domains, which form intermolecular lattice contacts in the latter [20].
Figure 5. P3P4 crystal structure.
(A) 3.0 Å resolution of P3P4 (light and dark blue ribbons denote the subunits) with ADPCP (orange bonds) bound in P4. The P3 and P4 domains roughly form a plane and residues 289–293 in the N-terminal linker project toward the ATP-binding pocket of the opposing subunit. (B) The N-terminal P3 linker of one subunit (dark blue bonds) interacts with the P4 domain of another (light blue ribbons and yellow bonds) by binding along side a break in helix α4. Hydrogen bonds are formed between the side chains and main chains of residues on each subunit and favorable electrostatics between negatively charged residues on P4 (Glu297, Glu298, and Glu401) and positively charged residues on the linker (Lys289, Lys290) may stabilize the contact. (C,D) Omit electron density (green mesh) for ADPCP within the ATP binding pocket (C) and (D) residues 289–293 from the C-terminal end of the P2-P3 linker (both shown at 3 σ).
Table 4.
Data collection and refinement statistics for the P3P4 structure.
| Diffraction Data Statistics | |
| Space Group | P212121 |
| Unit Cell: | |
| Length (Å) | Angle (°) |
| a = 66.4 | α = 90 |
| b = 131.4 | β = 90 |
| c = 147.0 | γ =90 |
| Unique Reflections | 57338 |
| Resolution (Å) | 49.2 – 3.0 (3.08 – 3.0)* |
| Redundancy | 5.2 (5.4) |
| Completeness (%) | 98 (97.4) |
| I/σ | 23.1 (8.4) |
| †Rpim (3.0 Å) | 0.075 (0.659) |
| CC ½ (3.0 Å) | 0.511 |
| Refinement Metrics | |
| ††Rwork | 0.242 (0.351) |
| Rfree | 0.282 (0.395) |
| No. atoms | 4030 |
| RMSD from ideal bond lengths (Å) | 0.003 |
| RMSD from ideal bond angles (Å) | 0.8 |
| B-factors (Å2) | |
| Main chain | 48.1 |
| Side Chain | 56.7 |
| Total | 53.0 |
| Ramachandran Favored (% residues) | 95 |
| Ramachandran Outliers (% residues) | 0.8 |
* Highest resolution range for compiling statistics.
, where Ij = the intensity of the jth observation of reflection i, <II> = the average intensity of reflection i and Ni = redundancy of reflection i.
Comparison of P3P4 to the structure of P3P4P5
Superimpositions of the kinase domain from either P3P4P5 [20] or P4 [19] onto the P3P4 structure show little difference in the main chain of the kinase domain (Fig. S9). Electron density in the binding pocket reveals the presence of ADPCP in a similar conformation as that found in the P4 crystal structure (Fig. 5C) [19]. However, unlike the ATP-bound structure of the isolated P4 domain, a portion of the ATP lid (493–503) [12] is poorly ordered in P3P4 and its precise structure is difficult to discern. Electron density corresponding to the C-terminal P3 linker (residues 289–292) remains well defined in one subunit (Fig. 5B,D) [20]. These residues project toward the ATP-lid and nucleotide pocket of the P4 domain of the opposing subunit where they contact the P4 α1 and α2 helices (Fig. 5B). In particular, Val291 packs into a cusp of the P4 α2 helix that results from Glu387 failing to provide a main chain hydrogen bond in helical register (Fig. 5B). Ser293 hydrogen bonds to the main-chain of Phe360 on the opposing subunit, whereas the carbonyl of Gln294 receives a hydrogen bond from Arg393. Complementary electrostatic interactions between Lys290 and Glu398 may also stabilize the contact (Fig. 5B). The two Arg297 residues project to solvent from the top of the P3 domain and appear to support the trans directionality of the P2-P3 linker. Thus, the conformation of the N-terminal linker visible in the P3P4 structure further supports trans autophosphorylation by T. maritima CheA. The N-terminal linker of P3 may help stabilize the interaction of P1 to P4, and therefore enhance autophosphorylation.
Modeling the CheA:CheW complex on the P3P4 conformation
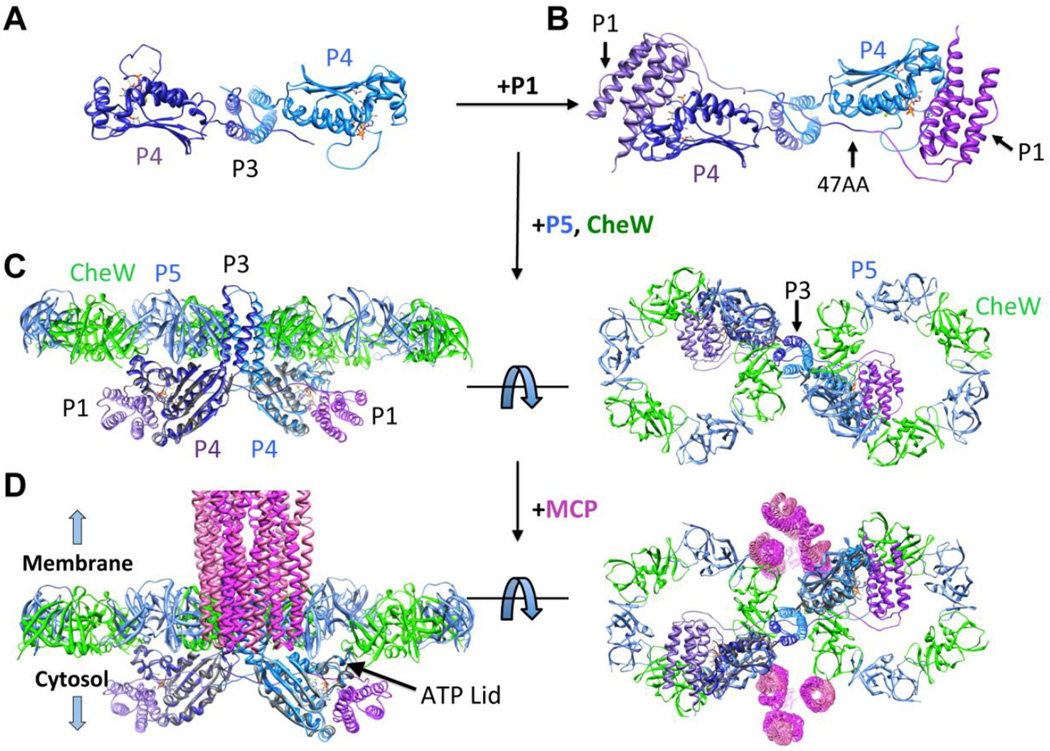
In constructing the receptor:kinase array models from the original P3P4P5 structure, the P3P4 linker conformation was adjusted so that the P5 and CheW units could form planar rings [12]. Fortuitously, the P4 orientation relative to P3 in the new P3P4 structure produces this very juxtaposition (Figs 5 and 6). Superposition of the P3 domains from the two structures reveals that the P4 domains in P3P4 lie in the same plane, largely because the P3-to-P4 linker residues Arg354 and Met355 change conformation compared to the P3P4P5 structure. To generate a model of CheAFL bound to CheW based on the P3P4 conformation, the P5 module from the T. maritima P3P4P5 (Δ289) was matched with the P3P4 structure by superimposing the respective P4 domains [20]. The resulting P5 domain positions do not clash with the P3 domain and sit roughly perpendicular to the P4 positions. P5 and CheW [18] were then superimposed from their complex structure based on an alignment of the respective P5 modules; the resulting CheW units were also free of any steric clashes (Fig. 6). To further develop the CheA model, the P1 domain [34] was docked in a conformation conducive for autophosphorylation based on the following constraints: the minimum linker length for trans autophosphorylation (47AA), the required distance for an interaction between His45 and the γ-phosphate of ATP, the distance for effective cross-linking between residues 45 and 492, information from E. coli CheA mutants that disrupt interactions between P1 and P4 [40], and knowledge of sites that cross-link when changed to cysteine residues [39] (Fig. S10). A majority of the linker was modeled as a 5th α-helix as found in solution structures of the P1 domain [34]. The ordered connection N-terminal of P3 (Fig. 5) constrains P1 positioning (Fig. 6B) when the linker is shortened to the minimal length that allows for trans interaction to P4 (47AA).
Figure 6. P3P4 crystal structure and model.
(A) Structure of P3P4 (each subunit colored as light and dark ribbons) obtained with ADPCP (orange bonds) present in the nucleotide-binding pocket. View is perpendicular form Fig. 5A. (B) Model of P1 (light and dark purple ribbons) near the P4 ATP-binding pocket in 47AA. (C) CheA P3P4P5 structure based on the P3P4 conformation with CheW/P5 ring overlay [18]. (D) Model of CheA within the array. Trimers of receptor dimers (magenta) [85] located at the vertices of the hexagonal P5/CheW rings are shown from the side and below. The P3P4 structure predicts that the ATP-lids of the P4 domains will be oriented up toward the P5-CheW layer.
Receptor array model
The new P3P4 structure has the appropriate juxtaposition of the P3 and P4 domains to form the planar receptor array lattice (Fig. 6), thus demonstrating that the P3-P4 linker can achieve a conformation that was previously hypothesized [12]. The new CheA:CheW model readily superimposed onto the hexagonal P5/CheW ring structure that was previously determined [32] (Fig. 6D). In contrast, the P3P3P5 [20] conformation generates P5-CheW rings that are oriented perpendicular to each other and hence not compatible with the flat membrane arrays [14] (Fig. S11). The MCP trimer-of-dimers resides at the vertices of the hexagon and interact directly with CheW and P5 based upon the location of the MCP Tm14 in the ternary crystal structures [18]. Overall, the planarity of the CheA core allows the CheW/P5 rings to extend into a hexagonal two-dimensional lattice. The P4 domains reside below, in a position that can accommodate their interaction with P1 (Fig. 6C,D). Interestingly, the model predicts that the ATP-lid of the kinase domain will be oriented toward the P5-CheW rings (Fig. 6D). This region of P4 participates in ATP binding and the recognition of interaction partners in other GHKL ATPases [2,25]. The model suggests that the CheW-P5 ring structure formed with receptors may modulate interactions of the ATP lid with nucleotide or P1 or both.
Discussion
Tethering P1 to its kinase module
E. coli CheA has been shown to undergo trans autophosphorylation [35,36,46,48] whereas other sensor kinases in the GHKL family show either cis or trans phosphotransfer [1,2,64]. T. maritima CheAFL autophosphorylates in trans, but can switch to cis, depending on the P1-to-P3 linker length. Trans autophosphorylation in 47AA and longer ΔP2 variants was demonstrated by the ability of mixed mutant dimers to regain activity when an active kinase domain (P4) of one subunit complements an active substrate domain (P1) on the other. Although both 41AA and 36AA are capable of some autophosphorylation, their mixed mutant dimers are inactive (Fig. 1C), thus indicating that these variants undergo cis phosphorylation exclusively. The reasons for reduced activity in the shorter variants may derive from restraints imposed by the shorter linkers, and in the case of 41AA and 36AA, cis-autophosphorylation. Trans subunit cross-linking between P1 and P4 is also not observed for 41AA*. Not surprisingly, 41AA is less active than CheAFL (Fig. 1B) with cis phosphorylation producing only ~50% of P1-P compared to the trans event. The inclusion of only six amino acids (41AA to 47AA) improves the flexibility and reach of the linker to enable trans autophosphorylation and an activity level similar to that of CheAFL (Fig. 1C). The affinity of the isolated P1 domain for P3P4 is low at 25 °C (Kd > 250 µM), yet at near physiological temperature P3P4 phosphorylates P1 with an efficiency that exceeds CheAFL, despite a ~1000 fold effective concentration increase in substrate for the latter (see SI). The cross-linking experiments also show that P1 to P3P4 interactions increase in the separated domains. Additionally, the ΔP2 variants are less active than expected based on the tethering linker length (See SI). Thus, it appears that in CheAFL and the ΔP2 variants, P1 is constrained from fully accessing the P4 domain.
Conformational properties of CheA P1-P2
SAXS data provide evidence for a conformation of CheAFL that has limited mobility of P1 and P2. Crystallography indicates that the P3P4P5 domains of CheAFL compose an ordered globular core of the enzyme [20] and solution SAXS of P3P4P5 support this view (Fig. S3–S5). Similarly, T. maritima and E. coli CheAFL, as well as the ΔP2 variants, were found to be relatively compact in most states (Fig. 2B, C, S5) [65]. Thus, the P1 domains are held to the core in manner that could constrain productive interactions with the P4 active site. However, upon addition of ADPCP CheAFL and the ΔP2 variants show enhanced flexibility (Fig. 2A). In contrast, P3P4P5 undergoes little to no changes upon addition of nucleotide (Fig. S3B). CheAFL has a more drastic conformational change than the ΔP2 variants presumably owing to presence of the P2 domain, which likely exhibits motion independent from the CheA core. Thus, ADPCP converts an ordered P1-P5 unit to a species with a smaller compact core (P3P4P5), and a conformationally flexible region (P1P2). It is surprising that ADPCP, but not ATP nor ADP, triggers P1 release. Either the ADPCP γ-phosphate provides a structural perturbation that the γ-phosphate of ATP does not, or the inability to transfer phosphate from ADPCP favors an otherwise unstable conformational state. An inhibitory site for P1 on P4 found in NMR experiments [66] may participate in the restraint of P1 and could be sensitive to ADPCP binding, thus releasing P1 upon ADPCP addition. Interactions that constrain P1 mobility within the CheA dimer may also explain subunit interference and negative cooperativity effects observed in the E. coli enzyme [31,32,43,44].
Interactions between P1 and P4
Targeted disulfide cross-linking experiments probe the proximity of the P1* substrate site to the P4* ATP-γ-phosphate position (Fig. 3A,B). The specificity of this contact was confirmed by the ability of ATP (and ADP) to diminish cross-linking in some cases. Compared to CheAFL*, interactions between P1* and P4* increase when P1* is separated from P3P4* or P3P4P5*. Furthermore, there is virtually no interaction between the isolated P1* domain and P4*, which is consistent with the inability of P4 to phosphorylate isolated P1 to any appreciable extent. Thus, the P3 dimerization domain either facilitates interactions between P1 and P4 or influences the structural properties of P4 [31]. Furthermore, promotion of P1-to-P4 interactions by P3 may allow for allosteric coupling between subunits within the assembled core ternary complex [67].
Comparing the 41AA* and CheAFL* cross-linked dimers provides insight into interactions between the P1 and P4 domains (Fig. 3B). When P4* Cys variants of CheAFL* exchange with P1* Cys variants of CheAFL* all cross-linked dimers form: P1*-to-P1*, P1*-to-P4*, and P4*-to-P4*. In contrast, the mixture of the 41AA* single mutants forms only the P1*-to-P1* dimer (Fig. 3C). Thus, 41AA lacks inter-subunit P1-P4 contacts, consistent with this variant being incapable of trans autophosphorylation. Note that the mutations differ in phosphorylation and cross-linking studies and are unlikely to influence subunit exchange as the sites are far removed from P3. The ability of the soluble Tm14 MCP [56] to decrease P1*-to-P4* cross-linking suggests that inhibitory receptors may limit association of the P1 and P4 domains (Fig. 3D). Importantly, the 45 and 492 Cys reporter sites may serve as effective in vivo probes to monitor P1-P4 interactions within native transmembrane receptors arrays.
Subunit effects on CheA activity
T. maritima grows at temperatures between 55–90 °C [68] and, not surprisingly, initial CheAFL autophosphorylation rates increase with temperature [69]. The separated domains also show temperature dependent activity (Fig. 4A); at higher temperatures and longer times they produce more P1-P than CheAFL. In contrast to the separated domains, CheAFL gives biphasic kinetics as temperature increases. The buildup and decay of P1-P indicates that a phosphatase activity increases with time and temperature. This activity does not result from increasing ADP/ATP ratios at elevated temperatures, which would accelerate the reverse reaction between phosphorylated histidine and P4-bound ADP. Modeling the progress curve for CheAFL autophosphorylation at 55 °C suggests that P1-P converts to an un-phosphorylated form that differs from the starting enzyme and can only re-phosphorylate relatively slowly. Interestingly, neither separated domains nor CheAFL:P3P4P5 heterodimers that contain only one P1 domain show P1-P depletion. Thus, the mechanism underlying this behavior depends upon the P1 domain in the adjacent subunit, but is otherwise unclear. However, we note that the T. maritima P3 dimer dissociates on the same timescale as the second kinetic process associated with loss of P1-P (k~0.07 min−1 at 55 °C) [69] and thus subunit dissociation could potentially play a role in P1-P destabilization. The P1 domain of E. coli CheA is known to enhance dimer stability [70], and perhaps phosphorylation can alter this stability. Overall, the kinetic data support communication between the two P1 domains within the CheA dimer, which may arise from their direct interaction.
The P1 domains within a dimer are inaccessible to kinase domains from other CheA molecules in solution. Although P3P4 or P3P4P5 produce sustained levels of P1-P, addition of P3P4P5 to CheAFL does not increase formation of P-CheAFL. However, when the CheAFL subunits are exchanged with those of P3P4P5, autophosphorylation levels are similar to those of the separated domains. Thus, the two P1 domains within the CheA dimer may act to sequester or facilitate dephosphorylation of one another. Removal of one P1 in the CheAFL:P3P4P5 heterodimer relieves this inhibition, freeing the remaining P1 domain to access the trans P4 domain. Recent cryoEM studies supported by proteolytic protection assays also found evidence for greater ordering of the P1-P2 domains in the inhibited state of the kinase [11]. Thus, intersubunit contacts may facilitate domain interactions that limit P1 mobility and destabilize its phosphorylated form.
Notably, the sensor histidine kinase PhoQ phosphorylates the response regulator (RR) PhoP with a burst phase [71]. Phosphorylated PhoP increases rapidly and then diminishes owing to the ability of kinase-bound ADP to enhance PhoQ phosphatase activity. However, unlike the CheA phosphatase activity studied here, the PhoQ phosphatase does not act on the intermediate phospho-His and it depends on ADP formation. Although manifested differently, the intrinsic phosphatase activity of Thermotoga CheA may also act in negative feedback to diminish net phosphotransfer.
The importance of P3 for directing P1
P3P4 is the minimal unit required for stable P1 phosphorylation. Surprisingly, when separated from P1, the P4 domain alone has very low kinase activity even when P4 is present in large excess (Fig. 4). The P3 dependence of P4 activity could owe to several factors: influences of the P3-P4 linker on the structure and dynamics of the P4 domain [31], changes to the spatial orientation of the kinase domains when P3 is present [32], direct interaction of P3 with P1, or the ability of one kinase domain to facilitate phosphorylation by the other. The structure of the P3P4 unit reveals that the P4 domains are far separated from one another in space, and thus rather large domain motions would be required for one kinase domain to assist P1 binding or phosphorylation by the other. Alternatively, the P3-P4 linker does assume a new conformation in the P3P4 structure, which could affect the structure and dynamics of the P4 ATP pocket. There appears to be no significant conformational changes in the ATP pocket when comparing the crystal structures of P4 [19] and P3P4 (Fig. S9); however, the ATP-lid is not fully engaged with ADPCP in the P3P4 structure and the 3.0 Å resolution may prevent detection of small structural changes important for catalysis. The P3 domain does direct toward the trans subunit the N-terminal linker (Fig. 5), which in itself could contribute to interactions with P1.
The crystal structure of a sensor kinase in complex with ATP (3DGE) [72] has the His-containing dimerization domain closely associated with the kinase domain. Although the dimerization domains of sensor kinases assume a 4-helix bundle structure like P1, the 3DGE structure is not compatible with P1 binding to P4. Superposition of the respective kinase domains reveals that the much larger ATP-lid of CheA P4 clashes with the position occupied by the substrate domain of the sensor kinase (Fig. S12). Thus, in comparison to sensor kinases, CheA likely uses a unique mode of recognition for its His-containing substrate domain, such as the one proposed here.
The P3P4 conformation is readily elaborated into a P3P4P5:CheW structure by superposition of the P4 domain with CheA structures that also contain P5 and CheW. The P3P4 linker sets the P5 and CheW units in conformations that allow ring arrangements supported by other crystallographic and cyroEM data [11–14]. The addition of the P1 domain was modeled without steric clash under constraints of the minimal 47AA linker length, proximity requirements for phosphotransfer between His45 and ATP [19], and results from cross-linking [36] and mutagenesis [40]. The P3P4 structure allows for a planar arrangement of the P5:CheW rings and hence this conformation may anchor CheA to a 2-dimensional hexagonal receptor lattice. With the addition of a trimer-of-receptor dimers the array approaches what would be expected from CryoEM maps (Fig. 6) [14,73]. In this context, the inactive form of the CheAFL likely involves interactions between P1 domains that prevent productive trans associations with P4.
Summary
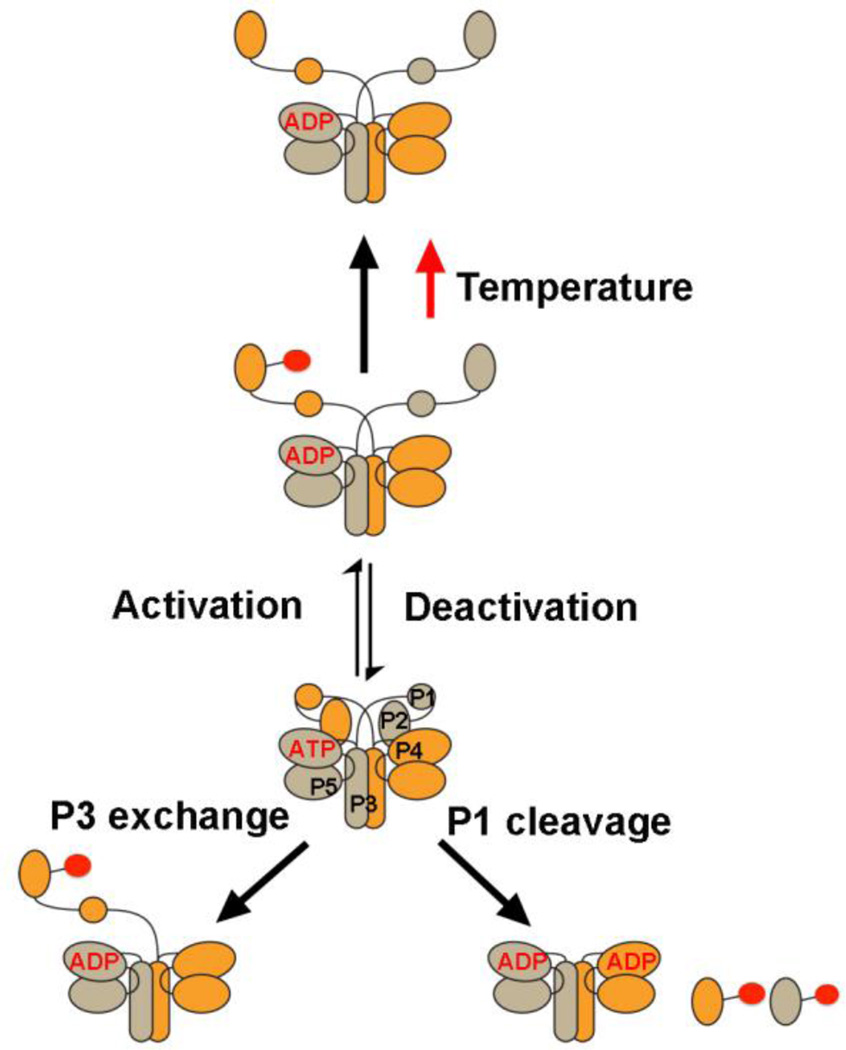
Collectively, the data support an inhibited state of CheA in which sequestered P1 domains cannot access P4 due to interactions that involve P1 of the adjacent subunit (Fig. 7). Surprisingly, the adjacent P1 domain not only affects accessibility of its neighbor domain, but also the stability of its phosphorylated form. Changes to the nucleotide-binding pocket in the vicinity of the ATP-γ-phosphate relieve this inhibition, suggesting that the P1 domains and two P4 domains are all closely associated in the inhibited state. In the activated form of the enzyme, P1 projects toward the trans subunit for interaction with P4 through interactions made by the linker N-terminal to P3. The P3 domain plays a key role in productive interactions between P1 and P4. P4 domain orientations in the receptor arrays based on the P3P4 structure suggest that conformational changes may be propagated between the P5-CheW layer and the ATP-lid of the P4 domain, which may then alter P4 interactions with ATP or the P1 domains.
Figure 7. CheA conformational states.
The P1 substrate domain is constrained within the core CheA kinase. Activation by MCPs causes release of P1 allowing access to P4 of the adjacent subunit. Perturbations close to the ATP binding pocket trigger release. Detachment of P1 from the kinase core stimulates phosphotransfer, as does removal of one P1 domain from a CheAFL dimer. At physiological temperatures, P1-P is destabilized in a manner that also depends on the P1 domain of the adjacent subunit.
Materials and Methods
Generation of CheA variants and protein expression
To generate the ΔP2 variants, the P1 and P3P4P5 regions of T. maritima CheA were covalently joined via a linker of varying size primarily composed of native P1-P2 linker residues (Fig. 1A) as well as several non-native residues added to achieve the desired length. The constructs were PCR cloned into the pET28a expression vector between the NdeI and BamHI restriction sites of the multiple-cloning region. Other CheA domain truncations were similarly produced. Site-directed mutations were introduced via Quikchange mutagenesis (Agilent Technologies). Two endogenous cysteine residues (Cys63 and Cys208) were substituted to serine to provide a Cys-less background for targeted cross-linking studies. Nomenclature of the ΔP2 variants was assigned according to the linker lengths between the P1 and P3 domains. Proteins contained a His6 affinity tag and were purified to homogeneity according to protocols described previously [57]. Briefly, E. coli BL-21 DE3 cells transformed with the pET28a vectors were grown in the presence of kanamycin to an optical density of A 600 = 0.6 prior to induction with 146 µM IPTG, followed by growth for overnight at 25 °C. Cells were harvested and sonicated. Prior to sonication, 10 mM PMSF was added to lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM imidazole, 10% glycerol) to limit proteolysis. After centrifugation (1 hour at 22k rpm) the lysate was run over Ni2+-NTA affinity resin and washed with buffer containing 20 mM imidazole, 50 mM Tris [pH 7.5], 150 mM NaCl, 10% glycerol, followed by elution with the same buffer containing 200 mM imidazole. The sample was then purified via a Sephadex 200 size-exclusion chromatography (SEC) prep column (GE) run with GF buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 10% glycerol).
Autophosphorylation Activity
T. maritima CheAFL, ΔP2 variants, or isolated P1, P3P4, and P3P3P5 domains were prepared in 10 µM in TKEDM buffer (50 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 0.5 mM EDTA, 50 mM Tris [pH 7.5]) and made up to a total volume of 23 µL. After 12 hours of incubation at 4 °C, 2 µL of a solution containing 2.3 mM cold ATP and 3–8 µL of [γ-32P] ATP (3000 Ci/mmol, 10 µCi/µL, Perkin Elmer) was added to the samples for 10 seconds-to-60 minutes at the indicated temperatures. Reactions were then quenched with 25 µL of 3×SDS buffer (Life Technologies) containing 50 mM EDTA pH 8.0 and 30 µL of the quenched samples were subjected to gel electrophoresis on a gradient 4–20% Tris-glycine SDS-PAGE gel. The gel was stained with Coomassie blue, destained with water, and dried with a GelAir dryer. The dried gel was placed in a cassette for at least 24 hours, imaged with Storm phosphoimager (GE Healthcare), and then analyzed with ImageJ. The first order kinetics time courses were fit to Pt = A0(1−e−k1t), where Pt = CheA-P formed at time t, A0 = the plateau value of CheA-P and k1 = the first order rate constant. For non-first order behavior kinetics, data were simulated under the model A → B ←→ C, where A = unphosphorylated P1, B = P1-P, and C = a form of unphosphorylated CheA different from A (P1#). After reasonable values were established by simulation, rate constants were then optimized by non-linear least square fitting during numerical integration with Tenua 2.1 (bililite.com/tenua/).
Phosphotransfer to CheY
Solutions of 5 µM (subunit concentration) T. maritima CheA (or ΔP2) and 20 µM T. maritima CheY were prepared in TKEDM buffer to a total volume of 23 µL and incubated for 12 hours at 4 °C. 32P-γ-ATP solutions were added to the samples to initiate the reactions as described above and the assays were processed similarly.
Assessment of trans vs. cis autophosphorylation
Solutions of 5 or 10 µM (per subunit) T. maritima CheA with or without the H45K and H405Y substitutions in the context of full-length and ΔP2 variants were prepared overnight in TKEDM buffer to a total volume of 25 µL. Prior to autophosphorylation assays the samples were heated for 3–18 hours at 55 °C to allow subunit exchange. Autophosphorylation is only rescued by the trans reaction between one subunit with a native P1 subunit (but defective P4, H405Y) and a native P4 subunit (but defective P1, H45K).
Heterodimer formation and analysis
Solutions containing 35 µM (subunit concentration) Δ449A T. maritima CheAFL and 35 µM T. maritima CheA P3P4P5 were incubated in TKEDM buffer to a total volume of 500 µL. Samples were heated to 55 °C for 0–18 hours prior to injection onto an analytical 200 Sepharose SEC (GE) for separation.
Affinity of isolated P1 for P3P4
Michealis constants of P1 for P3P4 were investigated at 25 °C and 55 °C by monitoring the initial rates of autophosphorylation through the production of ADP, as detected by an enzyme coupled assay [61–63]. Production of ADP by CheA was determined to be rate limiting under the conditions tested. Initial rates were determined by fitting ADP production to a first order expression P = A0e−kt and setting v = kA0. Data were fit to the Michealis expression where Vmax is the maximum initial rate at substrate saturation, but little saturation was observed at high concentrations of P1 (> 200 µM).
Phosphatase Activity
T. maritima CheAFL was prepared in 10 mM TKEDM buffer (50 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 0.5 mM EDTA, 5 mM Tris pH 7.5) to a total concentration of 40 µM and volume of 50 µl. At either 25°C or 55°C, 2.5 µl of 4 mM ATP was added to each reaction for 3 to 90 minutes. Reactions were then quenched with 100 µl of Biomol Green reagent (Enzo Life Sciences, BML-AK111). The reactions were initiated with ATP such that all samples were quenched simultaneously. Samples were then loaded into an optically transparent 96-well microtiter plate and measured for absorbance at 620 nm. The concentration of inorganic phosphate (Pi) present in each reaction was determined by A620nm of phosphate standards (80 µM-0 µM Pi). To ensure that the temperature sensitivity of Biomol Green did not affect the calculated values for [Pi], a set of phosphate standards was conducted at each temperature, and then used with the CheAFL samples of the corresponding temperature.
Disulfide cross-linking
Solutions of T. maritima CheA single site Cys variants P1* (H45C) and P4* (S492C) were incubated for 1 hour at 55°C in the presence of freshly prepared initiator 5 mM Cu(1,10 phenanthroline)32+, at concentrations of 6 µM and in a total volume of 10 µL. For some samples 1 mM ADP or ATP, along with 10 mM MgCl2 was also added. Similarly, some samples included the T. maritima Tm14 (residues 41–254) at a 1:3 ratio (CheA subunit:Tm14 dimer), with CheW added at the same concentration as the CheA subunit. Tm14 KCM (41–254) contains the adaptation region yet is more stable to heat in vitro than the full length Tm14. An optimal temperature (55 °C, near physiological [49]) was used to promote cross-linking, maximize activity, and avoid degradation of the nucleotide or the receptor. Disulfide cross-linking was quenched by the addition of 8 µL of 3×SDS sample buffer supplemented with 10 mM imidazole. The samples were electrophoresed on a 4–12% gradient Bis-Tris SDS PAGE gel in MES (or MOPS in the case of CheAFL) running buffer. The gel was stained with Coomassie blue, destained with an acetic acid/ethanol solution, and then dried overnight at room temperature. All bands were analyzed using ImageJ.
Small-angle x-ray scattering (SAXS)
For SAXS experiments the protein was exchanged into 50 mM HEPES pH 7.5, 150 mM NaCl, 2 mM DTT, and 5% glycerol the day of the experiment then prepared at three concentrations (6, 3, and 1.5 mg/mL). Other sets of samples were prepared with the same protein concentrations containing 5 mM α-β-methyleneadenosine 5’-triphosphate (ADPCP), ATP, and ADP. The samples were centrifuged at 13,000 rpm for 20 minutes at 4 °C, then kept in the sample tray at 4 °C prior to x-ray exposure. Data were collected at the F2 or G1 beam line of CHESS with a Pilatus 100K detector [74]. Buffer blanks that contained all components (including nucleotides) were collected under each condition and exposure for background subtraction. Each sample was exposed during continuous oscillation for 30 seconds for 10 frames. During exposures 30 µL of sample is continuously oscillated through the illuminated volume (0.125 µL) at 2–4 µL s−1, thereby reducing the absorbed dose by two orders of magnitude compared to static illumination [75]. Intensity ratios between the different protein concentrations as a function of q were examined to rule out artifacts from buffer subtraction at high scattering angle. RAW [76] and Primus [77] were used for data processing and generation of Guinier and Kratky plots. Molecular weight prediction was based upon a standard of 3 mg/mL of lysozyme (13 kDa). Envelope reconstructions were calculated using the ATSAS programs[78–80]. A total of ten independent models were generated for each data set and then averaged with Damaver into one envelope. Flexibility analysis was further conducted through the analysis of Porod-Debye and Porod-Kratky plots with ScÅtter ([81] Rambo R, bioisis.net).
Circular dichroism
For CD experiments the protein was exchanged into 10 mM sodium phosphate pH 7.2 and 150 mM NaCl the day of the experiment and concentrated to 0.5 mg/mL. For comparison, two sets of samples were generated, one of which contained 1 mM of ADPCP and one without. A subsequent data set was collected on the sample with nucleotide 24 hours later to check for any long-term changes. Samples were placed in a 0.1 cm quartz cell and analyzed with an AVIV Biomedical (model 202-01) spectropolarimeter. Data were collected at 1 nm intervals with 5 second averaging time at 25 °C from 260-200 nm. Degree of elipicity was plotted versus wavelength.
TNP-ATP binding assay by fluorescence enhancement
T. maritima CheAFL (2 µM subunit concentration) was placed in 2 mL of Fluoro buffer (25 mM Tris [pH 7.5], 0.5 mM Na2EDTA, 10% glycerol, 25 mM NaCl, 54.7 mM potassium glutamate, 20 mM MgCl2) at 25 °C. Stock 2,3-O-(2,4,6- trinitrophenyl) adenosine 5′-triphosphate (TNP-ATP) solutions were added in small increments (1–2 µL) over time to final concentrations of 0–50 µM. Emission spectra were collected over 530–600 nm (545 nm peak intensity) using a □ex of 520 nm with 4 nm slits on a Horiba Jobin Yvon Flourolog-3 fluorescence spectrophotometer equipped with a with 450 W Xe lamp, double excitation and emission monochromators, and a digital photon-counting multiplier. Three scans were collected for each addition.
Crystallization and data collection
The P3P4 domain structure of T. maritima CheA (4XIV) was determined to 3.0 Å resolution in space group P212121. The P3P4 crystals were obtained by vapor diffusion of 1 µL of protein (700 µM monomer and 1.3 µM ADPCP) mixed with 1 µL of reservoir solution. The reservoir contained 0.5 M ammonium sulfate, 0.1 M sodium citrate tribasic dihydrate pH 5.6, and 1.0 M lithium sulfate monohydrate at 4 °C. The crystal was flash cooled in 25% glycerol. Diffraction data were collected at the Cornell High Energy Synchrotron Source (CHESS) A1 beam line on an ADSC Quantum 210 CCD detector. Data were processed with HKL2000 [82]. Molecular replacement was accomplished by Phenix [83] with individual domains composed of P3 residues 293–353 and P4 residues 354–540 from the Protein Data Bank (PDB) model 1B3Q (LLG=329, TFZ=16). Model Refinement was carried out with Phenix[83] amidst model building with Coot [84].
Modeling of P1 and array complexes
Structural superpositions were generated using the supramolecular overlap function in Coot. [84] T. maritima P5 and CheW domains were taken from 1B3Q14, 4JPB [18,20], and P1 from 2LD6 [24]. Trimer-of-receptor dimers were modeled as in Briegel et al. [12]. Manual placement of P1 was based on constraints of linker length, requirements for phosphorylation, cross-linking data, and functional data as described in the text.
Supplementary Material
HIGHLIGHTS.
The chemotaxis histidine kinase CheA has on and off states that depend on conformational dynamics.
The substrate domain (P1) is sequestered in the off-state by interactions that involve the adjacent subunit.
Structural changes at the ATP pocket relieve P1 inhibition.
The structure of the minimal active unit containing the kinase and dimerization domain is consistent with receptor array integration and explains trans-subunit phosphorylation.
Acknowledgements
We also thank Cornell High Energy Synchrotron Source (CHESS) for access to data collection facilities. This work was also supported by a grant from the National Institutes of Health (R01 GM066775) to BRC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers:
The atomic coordinates of the structure have been deposited to the Protein Data Bank with the accession code 4XIV.
REFERENCES
- 1.Casino P, Miguel-Romero L, Marina A. Visualizing autophosphorylation in histidine kinases. Nat. Commun. 2014;5 doi: 10.1038/ncomms4258. [DOI] [PubMed] [Google Scholar]
- 2.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 3.Hoch JA, Silhavy TJ. Two-component signal transduction. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 4.Bhate MP, Molnar KS, Goulian M, DeGrado WF. Signal Transduction in Histidine Kinases: Insights from New Structures. Structure. 2015;23:981–994. doi: 10.1016/j.str.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional Characterization in Vitro of All Two-component Signal Transduction Systems from Escherichia coli. J. Biol. Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 6.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 7.Falke JJ, Piasta KN. Architecture and signal transduction mechanism of the bacterial chemosensory array: Progress, controversies, and challenges. Curr. Opin. Struct. Biol. 2014;29:85–94. doi: 10.1016/j.sbi.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aizawa S-I, Harwood CS, Kadner RJ. Signaling Components in Bacterial Locomotion and Sensory Reception. J. Bacteriol. 2000;182:1459–1471. doi: 10.1128/jb.182.6.1459-1471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blair DF. How Bacteria Sense and Swim. Annu. Rev. Microbiol. 1995;49:489–520. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 10.Scharf BE, Aldridge PD, Kirby JR, Crane BR. Upward mobility and alternative lifestyles: a report from the 10th biennial meeting on Bacterial Locomotion and Signal Transduction. Mol. Microbiol. 2009;73:5–19. doi: 10.1111/j.1365-2958.2009.06742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briegel A, Beeby M, Thanbichler M, Jensen GJ. Activated chemoreceptor arrays remain intact and hexagonally packed. Mol. Microbiol. 2011;82:748–757. doi: 10.1111/j.1365-2958.2011.07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, et al. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briegel A, Wong ML, Hodges HL, Oikonomou CM, Piasta KN, Harris MJ, et al. New Insights into Bacterial Chemoreceptor Array Structure and Assembly from Electron Cryotomography. Biochemistry. 2014;53:1575–1585. doi: 10.1021/bi5000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu TS, Le Novère N, Levin MD, Beavil AJ, Sutton BJ, Bray D. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 16.Weis RM, Hirai T, Chalah A, Kessel M, Peters PJ, Subramaniam S. Electron Microscopic Analysis of Membrane Assemblies Formed by the Bacterial Chemotaxis Receptor Tsr. J. Bacteriol. 2003;185:3636–3643. doi: 10.1128/JB.185.12.3636-3643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khursigara CM, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: Trimers of Receptor Dimers in a Partially Ordered Hexagonally Packed Array. J. Bacteriol. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Fleetwood AD, Bayas C, Bilwes AM, Ortega DR, Falke JJ, et al. The 3.2 Å Resolution Structure of a Receptor:CheA:CheW Signaling Complex Defines Overlapping Binding Sites and Key Residue Interactions within Bacterial Chemosensory Arrays. Biochemistry. 2013;52:3852–3865. doi: 10.1021/bi400383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI. Nucleotide binding by the histidine kinase CheA. Nat. Struct. Mol. Biol. 2001;8:353–360. doi: 10.1038/86243. [DOI] [PubMed] [Google Scholar]
- 20.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a Signal-Transducing Histidine Kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 21.Park S-Y, Beel BD, Simon MI, Bilwes AM, Crane BR. In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11646–11651. doi: 10.1073/pnas.0401038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park S-Y, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, et al. Reconstruction of the chemotaxis receptor-kinase assembly. Nat. Struct. Mol. Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 23.Quezada CM, Grǎdinaru C, Simon MI, Bilwes AM, Crane BR. Helical Shifts Generate Two Distinct Conformers in the Atomic Resolution Structure of the CheA Phosphotransferase Domain from Thermotoga maritima. J. Mol. Biol. 2004;341:1283–1294. doi: 10.1016/j.jmb.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 24.Vu A, Hamel DJ, Zhou H, Dahlquist FW. The structure and dynamic properties of the complete histidine phosphotransfer domain of the chemotaxis specific histidine autokinase CheA from Thermotoga maritima. J. Biomol. NMR. 2011;51:49–55. doi: 10.1007/s10858-011-9540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilwes AM, Park SY, Quezada CM, Simon MI, Crane BR. Structure and Function of CheA, the Histidine Kinase Central to Bacterial Chemotaxis. In: Inouye M, editor. Histidine Kinases Signal Transduct. San Diego, CA: Academic Press; 2003. pp. 48–74. [Google Scholar]
- 26.Adler J. Chemotaxis in Bacteria. Annu. Rev. Biochem. 1975;44:341–356. doi: 10.1146/annurev.bi.44.070175.002013. [DOI] [PubMed] [Google Scholar]
- 27.Koshland DE. Biochemistry of Sensing and Adaptation in a Simple Bacterial System. Annu. Rev. Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- 28.Falke JJ, Bass RB, L Butler S, Chervitz SA, Danielson MA. THE TWO-COMPONENT SIGNALING PATHWAY OF BACTERIAL CHEMOTAXIS: A Molecular View of Signal Transduction by Receptors, Kinases, and Adaptation Enzymes. Annu. Rev. Cell Dev. Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart RC, Jahreis K, Parkinson JS. Rapid Phosphotransfer to CheY from a CheA Protein Lacking the CheY-Binding Domain†. Biochemistry. 2000;39:13157–13165. doi: 10.1021/bi001100k. [DOI] [PubMed] [Google Scholar]
- 30.Jahreis K, Morrison TB, Garzón A, Parkinson JS. Chemotactic signaling by an Escherichia coli CheA mutant that lacks the binding domain for phosphoacceptor partners. J. Bacteriol. 2004;186:2664–2672. doi: 10.1128/JB.186.9.2664-2672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Vallurupalli P, Vu A, Lee K, Sun S, Bai W-J, et al. The Linker between the Dimerization and Catalytic Domains of the CheA Histidine Kinase Propagates Changes in Structure and Dynamics That Are Important for Enzymatic Activity. Biochemistry. 2014;53:855–861. doi: 10.1021/bi4012379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Wu C, Vu A, Shea J-E, Dahlquist FW. Computational and Experimental Analyses Reveal the Essential Roles of Interdomain Linkers in the Biological Function of Chemotaxis Histidine Kinase CheA. J. Am. Chem. Soc. 2012;134:16107–16110. doi: 10.1021/ja3056694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Vallurupalli P, Vu A, Lee K, Sun S, Bai W-J, et al. The Linker between the Dimerization and Catalytic Domains of the CheA Histidine Kinase Propagates Changes in Structure and Dynamics That Are Important for Enzymatic Activity. Biochemistry. 2014;53:855–861. doi: 10.1021/bi4012379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mo G, Zhou H, Kawamura T, Dahlquist FW. Solution Structure of a Complex of the Histidine Autokinase CheA with Its Substrate CheY. Biochemistry. 2012;51:3786–3798. doi: 10.1021/bi300147m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson RV, Bourret RB, Simon MI. Intermolecular complementation of the kinase activity of CheA. Mol. Microbiol. 1993;8:435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 36.L Gloor S, Falke JJ. Thermal Domain Motions of CheA Kinase in Solution: Disulfide Trapping Reveals the Motional Constraints Leading to Trans-autophosphorylation†. Biochemistry. 2009;48:3631–3644. doi: 10.1021/bi900033r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quezada CM, Hamel DJ, Grǎdinaru C, Bilwes AM, Dahlquist FW, Crane BR, et al. Structural and Chemical Requirements for Histidine Phosphorylation by the Chemotaxis Kinase CheA. J. Biol. Chem. 2005;280:30581–30585. doi: 10.1074/jbc.M505316200. [DOI] [PubMed] [Google Scholar]
- 38.Natale AM, Duplantis JL, Piasta KN, Falke JJ. Structure, Function, and On-Off Switching of a Core Unit Contact between CheA Kinase and CheW Adaptor Protein in the Bacterial Chemosensory Array: A Disulfide Mapping and Mutagenesis Study. Biochemistry. 2013;52:7753–7765. doi: 10.1021/bi401159k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Two-Compon. Signal. Syst. Pt B. 2007. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: Applications to soluble and transmembrane receptors of bacterial chemotaxis, in; pp. 25–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama S, Garzón A, Parkinson JS. Mutational Analysis of the P1 Phosphorylation Domain in Escherichia coli CheA, the Signaling Kinase for Chemotaxis. J. Bacteriol. 2014;196:257–264. doi: 10.1128/JB.01167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Xu Y, Shen J, Luo X, Chen J, Chen K, et al. Dynamic Mechanism for the Autophosphorylation of CheA Histidine Kinase: Molecular Dynamics Simulations. J. Am. Chem. Soc. 2005;127:11709–11719. doi: 10.1021/ja051199o. [DOI] [PubMed] [Google Scholar]
- 42.Shi T, Lu Y, Liu X, Chen Y, Jiang H, Zhang J. Mechanism for the Autophosphorylation of CheA Histidine Kinase: QM/MM Calculations. J. Phys. Chem. B. 2011;115:11895–11901. doi: 10.1021/jp203968d. [DOI] [PubMed] [Google Scholar]
- 43.Eaton AK, Stewart RC. The Two Active Sites of Thermotoga maritima CheA Dimers Bind ATP with Dramatically Different Affinities. Biochemistry. 2009;48:6412–6422. doi: 10.1021/bi900474g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levit M, Liu Y, Surette M, Stock J. Active Site Interference and Asymmetric Activation in the Chemotaxis Protein Histidine Kinase CheA. J. Biol. Chem. 1996;271:32057–32063. doi: 10.1074/jbc.271.50.32057. [DOI] [PubMed] [Google Scholar]
- 45.Borkovich KA, Simon MI. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 46.Ellefson DD, Weber U, Wolfe AJ. Genetic analysis of the catalytic domain of the chemotaxis-associated histidine kinase CheA. J. Bacteriol. 1997;179:825–830. doi: 10.1128/jb.179.3.825-830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hess JF, Bourret RB, Simon MI. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988;336:139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- 48.Wolfe AJ, Stewart RC. The short form of the CheA protein restores kinase activity and chemotactic ability to kinase-deficient mutants. Proc. Natl. Acad. Sci. 1993;90:1518–1522. doi: 10.1073/pnas.90.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park S-Y, Quezada CM, Bilwes AM, Crane BR. Subunit Exchange by CheA Histidine Kinases from the Mesophile Escherichia coli and the Thermophile Thermotoga maritima†. Biochemistry. 2004;43:2228–2240. doi: 10.1021/bi0352419. [DOI] [PubMed] [Google Scholar]
- 50.Hirschman A, Boukhvalova M, VanBruggen R, Wolfe AJ, Stewart RC. Active Site Mutations in CheA, the Signal-Transducing Protein Kinase of the Chemotaxis System in Escherichia coli†. Biochemistry. 2001;40:13876–13887. doi: 10.1021/bi0113622. [DOI] [PubMed] [Google Scholar]
- 51.Hammel M. Validation of macromolecular flexibility in solution by small-angle X-ray scattering (SAXS) Eur. Biophys. J. 2012;41:789–799. doi: 10.1007/s00249-012-0820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 53.Durand D, Vivès C, Cannella D, Pérez J, Pebay-Peyroula E, Vachette P, et al. NADPH oxidase activator p67phox behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 2010;169:45–53. doi: 10.1016/j.jsb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 54.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhatnagar J, Borbat PP, Pollard AM, Bilwes AM, Freed JH, Crane BR. Structure of the Ternary Complex Formed by a Chemotaxis Receptor Signaling Domain, the CheA Histidine Kinase, and the Coupling Protein CheW As Determined by Pulsed Dipolar ESR Spectroscopy. Biochemistry. 2010;49:3824–3841. doi: 10.1021/bi100055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ninfa EG, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 58.Tawa P, Stewart RC. Kinetics of CheA Autophosphorylation and Dephosphorylation Reactions. Biochemistry. 1994;33:7917–7924. doi: 10.1021/bi00191a019. [DOI] [PubMed] [Google Scholar]
- 59.Eaton AK, Stewart RC. Kinetics of ATP and TNP-ATP Binding to the Active Site of CheA from Thermotoga maritima. Biochemistry. 2010;49:5799–5809. doi: 10.1021/bi100721b. [DOI] [PubMed] [Google Scholar]
- 60.Bronnikov GE, Zakharov SD. Microquantitative determination of Pi-ATP and ADP-ATP exchange kinetics using thin-layer chromatography on silica gel. Anal. Biochem. 1983;131:69–74. doi: 10.1016/0003-2697(83)90136-7. [DOI] [PubMed] [Google Scholar]
- 61.Surette MG, Levit M, Liu Y, Lukat G, Ninfa EG, Ninfa A, et al. Dimerization Is Required for the Activity of the Protein Histidine Kinase CheA That Mediates Signal Transduction in Bacterial Chemotaxis. J. Biol. Chem. 1996;271:939–945. doi: 10.1074/jbc.271.2.939. [DOI] [PubMed] [Google Scholar]
- 62.Nørby JG. [11] Coupled assay of Na+,K+-ATPase activity. In: Fleischer BF Sidney., editor. Methods Enzymol. Academic Press; 1988. [accessed April 5, 2015]. pp. 116–119. http://www.sciencedirect.com/science/article/pii/0076687988560147. [DOI] [PubMed] [Google Scholar]
- 63.Shrout AL, Montefusco DJ, Weis RM. Template-Directed Assembly of Receptor Signaling Complexes†. Biochemistry. 2003;42:13379–13385. doi: 10.1021/bi0352769. [DOI] [PubMed] [Google Scholar]
- 64.Saul VV, de la Vega L, Milanovic M, Krüger M, Braun T, Fritz-Wolf K, et al. HIPK2 kinase activity depends on cis-autophosphorylation of its activation loop. J. Mol. Cell Biol. 2013;5:27–38. doi: 10.1093/jmcb/mjs053. [DOI] [PubMed] [Google Scholar]
- 65.Briegel A, Ames P, Gumbart JC, Oikonomou CM, Parkinson JS, Jensen GJ. The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signalling state. Mol. Microbiol. 2013;89:831–841. doi: 10.1111/mmi.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamel DJ, Zhou H, Starich MR, Byrd RA, Dahlquist FW. Chemical-Shift-Perturbation Mapping of the Phosphotransfer and Catalytic Domain Interaction in the Histidine Autokinase CheA from Thermotoga maritima†,‡. Biochemistry. 2006;45:9509–9517. doi: 10.1021/bi060798k. [DOI] [PubMed] [Google Scholar]
- 67.Li M, Hazelbauer GL. Selective allosteric coupling in core chemotaxis signaling complexes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15940–15945. doi: 10.1073/pnas.1415184111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huber R, Langworthy TA, König H, Thomm M, Woese CR, Sleytr UB, et al. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 1986;144:324–333. [Google Scholar]
- 69.Park S-Y, Quezada CM, Bilwes AM, Crane BR. Subunit Exchange by CheA Histidine Kinases from the Mesophile Escherichia coli and the Thermophile Thermotoga maritima†. Biochemistry. 2004;43:2228–2240. doi: 10.1021/bi0352419. [DOI] [PubMed] [Google Scholar]
- 70.Kott L, Braswell EH, Shrout AL, Weis RM. Distributed subunit interactions in CheA contribute to dimer stability: a sedimentation equilibrium study. Biochim. Biophys. Acta BBA - Proteins Proteomics. 2004;1696:131–140. doi: 10.1016/j.bbapap.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 71.Yeo W-S, Zwir I, Huang HV, Shin D, Kato A, Groisman EA. Intrinsic Negative Feedback Governs Activation Surge in Two-Component Regulatory Systems. Mol. Cell. 2012;45:409–421. doi: 10.1016/j.molcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Casino P, Rubio V, Marina A. Structural Insight into Partner Specificity and Phosphoryl Transfer in Two-Component Signal Transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 73.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc. Natl. Acad. Sci. 2012;109:E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skou S, Gillilan RE, Ando N. Synchrotron-based small-angle X-ray scattering of proteins in solution. Nat. Protoc. 2014;9:1727–1739. doi: 10.1038/nprot.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nielsen SS, Møller M, Gillilan RE. High-throughput biological small-angle X-ray scattering with a robotically loaded capillary cell. J. Appl. Crystallogr. 2012;45:213–223. doi: 10.1107/S0021889812000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nielsen SS, Toft KN, Snakenborg D, Jeppesen MG, Jacobsen JK, Vestergaard B, et al. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J. Appl. Crystallogr. 2009;42:959–964. [Google Scholar]
- 77.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 78.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001;34:33–41. [Google Scholar]
- 80.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rambo RP, Tainer JA. Super-Resolution in Solution X-Ray Scattering and Its Applications to Structural Systems Biology. Annu. Rev. Biophys. 2013;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- 82.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode, in. In: Charles J, Carter W, editors. Methods Enzymol. Academic Press; 1997. [accessed November 7, 2014]. pp. 307–326. http://www.sciencedirect.com/science/article/pii/S007668799776066X. [DOI] [PubMed] [Google Scholar]
- 83.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot . Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim KK, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.