Abstract
Prior studies identified allosteric information pathways connecting functional centers in the large ribosomal subunit to the decoding center in the small subunit through the B1a and B1b/c intersubunit bridges in yeast. In prokaryotes a single SSU protein, uS13, partners with H38 (the A-site finger) and uL5 to form the B1a and B1b/c bridges respectively. In eukaryotes, the SSU component was split into 2 separate proteins during the course of evolution. One, also known as uS13, participates in B1b/c bridge with uL5 in eukaryotes. The other, called uS19 is the SSU partner in the B1a bridge with H38. Here, polyalanine mutants of uS19 involved in the uS19/uS13 and the uS19/H38 interfaces were used to elucidate the important amino acid residues involved in these intersubunit communication pathways. Two key clusters of amino acids were identified: one located at the junction between uS19 and uS13, and a second that appears to interact with the distal tip of H38. Biochemical analyses reveal that these mutations shift the ribosomal rotational equilibrium toward the unrotated state, increasing ribosomal affinity for tRNAs in the P-site and for ternary complex in the A-site, and inhibit binding of the translocase, eEF2. These defects in turn affect specific aspects of translational fidelity. These findings suggest that uS19 plays a critical role as a conduit of information exchange between the large and small ribosomal subunits directly through the B1a, and indirectly through the B1b/c bridges.
Keywords: intersubunit bridges, ribosome, yeast, translational fidelity, uS19
Introduction
Conversion of the genetic information encoded in mRNAs into proteins is carried out by large ribonucleoprotein particles called ribosomes. Yeast ribosomes are 3.6 MDa structures composed of 79 ribosomal proteins and 4 rRNA molecules. The 40S small ribosomal subunit (SSU) directs the bulk of translational initiation, and contains the mRNA decoding center (DC). The large 60S subunit (LSU) harbors several functional centers, most notably the peptidyltransferase center (PTC) which is the catalytic site of polypeptide synthesis, 3 tRNA binding pockets, and the GTPase associated center. During the elongation stage of translation, the large and small subunits work in concert. With each cycle of amino acid incorporation into the nascent polypeptide chain, the 2 subunits rotate with respect to one another by ~8 Å.1-4 Ribosomes at the 2 extreme stages of this process are known as rotated and unrotated.5 In translating ribosomes, the 2 subunits are connected by highly conserved interactions called intersubunit bridges. The roles of these bridges include providing structural stability and transmitting information between distal functional centers of the ribosome.6-8 The core intersubunit bridges are highly conserved in all domains of life. In particular, the B1 family of bridges (divided into B1a and B1b/c) link the head of the SSU with the central protuberance of the LSU; these structures undergo the largest degree of intersubunit rotation during the elongation cycle.9,10 While these bridges are highly conserved, the identities of the interacting partners differ between prokaryotes compared to eukaryotes. In the prokaryotes, a single SSU protein, uS13, partners with Helix 38 of the large ribosomal subunit (abbreviated as H38, and also referred to as the A-site finger or ASL) and uL5 to form the B1a and B1b/c bridges respectively. In eukaryotes, it appears that these responsibilities were re-assigned to 2 separate proteins during the course of evolution. One of these proteins, also known as uS13 11 [also called S18 12], only participates in bridge B1b/c with uL5 in eukaryotes. The other, called uS19 11 [as also called S1512] is the SSU partner in the B1a bridge with H38. High resolution crystal structures of yeast ribosomes reveal that uS13 and uS19 interact extensively with one another7 (Fig. 1).
Figure 1.
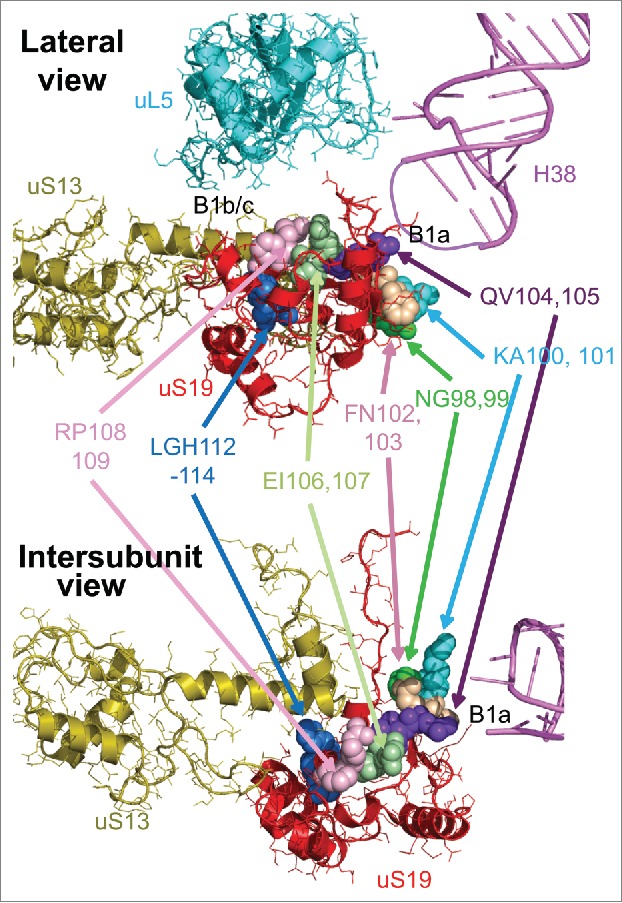
Two views of uS19 and its neighbors. Lateral (top, looking toward the SSU “head”), and intersubunit views of uS19 (red) intertwined with uS13 (olive). uL5 (cyan) and the B1b/c intersubunit bridge, and H38 (pink) and the B1a intersubunit bridges are indicated. The thinner purple line represents the approximate location of the distal tip of H38, which is not resolved in the original atomic resolution structure.41 Groups of amino acid residues of uS19 mutated in this study are labeled and color coded.
The essential 142 amino acid eukaryotic uS19 also plays an integral role in small subunit biogenesis: while depletion of uS19 permits exit of 20S pre-rRNA molecules from the nucleolus, they are retained in the nucleus, demonstrating that assembly of uS19 into SSU precursors is required to render 43S particles competent for export from the nucleus to the cytoplasm.13 In mature ribosomes, crosslinking studies show that the unstructured C-terminal tail of eukaryotic uS19 reaches the ribosomal DC.14 Its LSU partner, H38 is a large structure that interacts with numerous functional centers in the LSU including the D-loop of aminoacyl tRNA (aa-tRNA), the PTC and GTPase associated center.8 During translation elongation, the B1a bridge is intact when ribosomes are in the unrotated state. However, when ribosomes become fully rotated, H38 and uS19 separate by 10 Å, breaking the B1a bridge. The 17kDa yeast ribosomal protein uS13 is encoded by 2 paralogous genes, RPS18A and RPS18B which are identical with the exception of silent mutations in their DNA sequences. Studies performed in bacteria suggest that prokaryotic uS13 plays an important role in ribosome function including subunit joining during initiation, and maintaining the pretranslocation state during translation. In yeast, uS13 is required for processing of 35S pre-rRNA at the A2 site.13 In mature ribosomes, while the B1b/c intersubunit bridge between uS13 and uL5 remains intact through ribosome rotation, the specific interactions between the two proteins change.6,15
uS19 and uS13 have been indirectly implicated in transmitting information between functional centers on the 2 subunits. For example, reverse genetics studies of yeast H38 revealed that information pertaining to aa-tRNA selection in the SSU DC is transmitted from uS19 across the B1a intersubunit bridge to H38, from which it is distributed to functional centers in the LSU.8 Similarly, analyses of uL5 (also called L1111) mutants identified a network of allosteric information exchange spanning the length of the LSU tRNA binding cleft, through uL5, across the B1b/c bridge to the DC.6,16 While uS13 and uS19 are different proteins, they physically interact with one another 7,13 (Fig. 1). Based on prior observations and the structural data, we suggest that they work in concert as part of a larger allosteric information exchange network that enables coordination of the spatially distinct functional centers of the ribosome. To examine this, the current study employed a series of uS19 mutants located at the uS13/uS19 and uS19/H38 interfaces. Polyalanine scanning mutagenesis revealed that uS19 residues L112-H114, located at the uS13/uS19 interface, are required to maintain normal rates of Ty1-directed programmed +1 ribosomal frameshifting (+1 PRF), the ability of ribosomes to distinguish between cognate and near-cognate tRNAs, and the ability of initiating ribosomes to distinguish between a bona fide AUG start codon, as compared to a UUG codon. Mutation of residues uS19 Q104V105, which are the closest amino acids to H38, inhibited +1 PRF. Biochemical analyses revealed that both mutants reduced the fraction of translating polyribosomes relative to isogenic wild type cells, shifted ribosomal rotational equilibrium toward the unrotated state, promoted increased affinity for elongation ternary complex in the ribosomal A-site and for peptidyl-tRNA in the P-site, and decreased affinity for the translocase eukaryotic elongation factor 2 (eEF2). These findings support the hypothesis that uS19 functions as an important conduit of information between the large and small subunits, perhaps directly through its interaction with the B1a bridge, and indirectly via its interaction with uS13, through the B1b/c intersubunit bridge.
Materials and Methods
Strains, plasmids, and media
The gene encoding uS19, RPS15, and its native 5’ and 3’ untranslated regions (UTRs) were amplified from S. cerevisiae genomic DNA. This PCR product was cloned into pRS313, a HIS3-based low-copy expression vector. Mutations in the open reading frame of pRPS15-HIS3 were generated by oligonucleotide site-directed mutagenesis using the Quikchange XL site-directed mutagenesis kit (Stratagene, Madison, WI, USA)(Table S1). Plasmids were amplified in the DH5α strain of E. coli. Mutations were introduced into the RPS15 knockout yeast strain JD1346 [his3Δ1; leu2Δ0; ura3Δ0; YOL040c::KanMX4; pFL38(Pgal::RPS15; URA3)] using a standard lithium acetate yeast transformation protocol 17 and subsequent 5-FOA plasmid shuffle.18 Strains generated in this study are shown in Table S2.
Genetic Assays
Standard 10-fold dilution spot assays were used as a qualitative measure of sensitivity to temperature and small molecule translational inhibitors. Dilution spot assays were performed as previously described.16 Briefly, yeast were grown in synthetic histidine deletion media (-His) to logarithmic growth phase. Serial dilutions were made from 105 to 1 colony-forming unit (CFU) per 2.2 µl and spotted on –His agar media. For pharmacogenetic assays, 10 µg/ml of anisomycin and 45 µg/ml of paromomycin were added to –His agar media and the plates were incubated at 20ºC for 3-5 days. To assay for temperature sensitivity, the plates were incubated in cold (20ºC), optimal growth temperature for yeast (30ºC) and elevated temperature (37ºC). The killer maintenance assay is a qualitative assay of translational fidelity and was also performed as previously described.19 Briefly, cells were replica plated on 4.7 MB plates seeded with a lawn of killer sensitive (K−) cells and plates were incubated at 20°C for 2-3 days. Killer phenotype was detected as a zone of growth inhibition around the patch of test cells. Mutants were scored based on their ability to maintain killer relative to wild-type.
Quantitative translational fidelity assays
Bicistronic luminescent reporter assays were employed to quantitatively probe programmed +1 ribosomal frameshifting (+1 PRF), -1 PRF, suppression of the nonsense UAA codon, and missense suppression.20,21 In the missense reporter, the AGC serine codon at position 218 in the firefly luciferase catalytic site is replaced with an AGA arginine codon. The dual luciferase assays were performed and data was analyzed as previously described22 using a Dual Luciferase Reporter Assay Kit (Promega, Madison, WI) and a Promega GloMax→-Multi Detection System Luminometer.
To monitor translation initiation at non-AUG start codons, assays of β-galactosidase activity in whole-cell extracts from strains containing the HIS4AUG-lacZ (p367) or his4UUG-lacZ (p391) reporter plasmids, were performed as previously described.23 Transformants were inoculated overnight in minimal media and grown to mid-log phase. OD595 values were recorded, 2 mL of each culture were sedimented by centrifugation, and cells were resuspended in 400µL of Z buffer (8.05g Na2HPO4•7H2O, 2.75g NaH2PO4•H2O, 0.375g KCl, 0.123g MgSO4•7H2O, and 1.35 mL β-mercaptoethanol in 500 mL). 150µL CH3Cl3 and 100µL of 0.1% SDS were added, the suspension was vortexed for 10 seconds and incubated at 28˚C for 15 minutes. O-nitrophenyl β-D-galactoside (ONPG) hydrolysis was measured as previously described.23 Briefly, 0.2mL of a 4 mg/mL solution of ONPG was added to samples. The reactions were quenched with 0.5 mL of Na2CO3 and cell debris was sedimented by centrifugation. OD420 was measured for each sample and ONPG hydrolyzing activity was determined. Assays were normalized to the OD600 of the culture and to the assay time. Three experimental replicates were performed, each in triplicate.
Ribosome purification and sucrose gradient analyses
Ribosomes were isolated from mutant yeast strains for in vitro ligand binding assays following a previously described protocol.24 Cultures were grown in 5 ml of YPAD media and inoculated in 500 ml of YPAD overnight in a 30°C shaker and grown to mid-log phase. Cells were incubated at 4°C for 1 hour to allow ribosomes to dissociate from mRNA transcripts, but remain intact. Cells were harvested by centrifugation in a ThermoScientific™ Sorvall™ Legend RT Plus Centrifuge and washed twice with ribosome purification binding buffer [20 mM HEPES-KOH pH 7.6, 60 mM NH4Cl, 5 mM Mg(CH3COO)2, 2 mM DTT]. Cells were resuspended in 1 ml of binding buffer per gram of cells and lysed in 2 ml screw cap vials half-filled with glass beads using a minibead beater. Lysates were clarified by centrifugation in a Beckman Coulter Optima™ XE-90 Ultracentifuge at 30,000g using an SW-41Ti rotor at 4ºC for 30 minutes. The supernatant was separated from the pelleted cell debris and ribosomes were isolated by affinity purification using Sulfolink beads (Pierce, Rockford, IL). The Sulfolink beads were first incubated on ice after addition of supernatant for 30 minutes. The column was centrifuged at 1000 rpm momentarily followed by 2 washes with binding buffer. Ribosomes were eluted from the resin in 8 ml of elution buffer [20 mM HEPES-KOH pH 7.6, 60 mM NH4Cl, 500 mM KCl, 10 mM Mg(CH3COO)2, 2 mM DTT, 0.5 mg/ml heparin]. Eluted ribosomes were treated with 1mM each puromycin and GTP for 30 minutes at 30ºC to ensure removal of endogenous peptidyl tRNA, mRNA, and soluble factors. Ribosome solution was layered atop 22 ml of glycerol cushion buffer (50 mM HEPES-KOH pH 7.6, 10 mM Mg(CH3COO)2, 60 mM NH4Cl, 500 mM KCl, 2 mM DTT, 25% glycerol) and pelleted by centrifugation overnight at 100,000g at 4ºC in a SW-32Ti rotor. Pellets were washed in 1 ml of storage buffer (50 mM HEPES-KOH pH 7.6, 5 mM Mg(CH3COO)2, 50 mM NH4Cl, 1 mM DTT, 25% glycerol) and resuspended in 200-400 µl of storage buffer. Ribosome concentrations were determined using a spectrophotometer (1 ODU at 260 nm = 20 pmoles of ribosomes). The ribosomes were stored at -80ºC in 500 picomole aliquots until needed for ligand binding assays. Lysates of cycloheximide arrested yeast cells were sedimented through 7% - 47% sucrose gradients and polysome profiles were determined by monitoring A254 nm as previously described.25
Ligand binding assays
6X-His-tagged eukaryotic elongation factor 2 (eEF2) was purified from Saccharomyces cerevisiae strain TKY675. Cells were grown overnight in 5 ml of YPAD and cultures were inoculated in 500 ml the following night. Once cell growth reached logarithmic phase (OD595 = 0.6-1), cells were harvested and washed with ultrapure water. Cells were resuspended in 1 ml of binding buffer (20 mM Sodium Phosphate Buffer pH 7.4, 500 mM NaCl, 20 mM Imidazole) per 1 gram of cells and lysed in a mini beadbeater using glass beads. The resulting lysate was centrifuged in an ultracentrifuge with an SW-32Ti fixed angle rotor at 32,000 RPM for 3 hours at 4°C. The supernatant was promptly removed from the cell debris and loaded onto Ni-NTA agarose affinity columns. Following purification, residual imidazole was removed by buffer exchange (100 mM Tris-HCl pH 7.5, 5 mM EDTA). The protein was flash-frozen in liquid N2 and stored in 50 µl aliquots in a -80°C freezer.
Aminoacylated and acetylated aminoacylated tRNAs ([14C]Phe-tRNAPhe and N-Ac-[14C]Phe-tRNAPhe, respectively) were synthesized from CCA-repaired phenylalanyl tRNAs and purified by high performance liquid chromatography (HPLC) as previously described.26 For A-site ligand binding assay, 100 picomoles of purified ribosomes were incubated for 30 minutes at 30ºC with polyuridylic acid (Poly U), cold phenylalanyl tRNA, and 4X Binding Buffer [80 mM Tris-HCl pH 7.4, 160 mM NH4Cl, 15 mM Mg(CH3COO)2, 2 mM spermidine, 0.5 mM spermine, 6 mM β-mercaptoethanol] to a total volume of 300 µl. Ternary complex was formed by combining 512 picomoles of [14C]Phe-tRNAPhe, GTP, soluble protein factors, and 4X Binding Buffer to a final volume of 270 µl and incubated for 30 minutes at 30ºC. Eight two-fold serial dilutions of ternary complex mix were prepared ranging from 128 picomoles to 1 picomole concentrations. Fifteen µl of the ribosome mix was added to each tube and the ternary complex + ribosome mix was incubated for 30 minutes at 30ºC. The solutions were then poured onto moistened Millipore HA 0.45 micron nitrocellulose filters and incorporated radioactivity was determined by scintillation counting.
For P-site ligand binding, 100 picomoles of purified ribosomes were incubated for 10 minutes at 30ºC with polyuridylic acid (Poly U), and 4X Binding Buffer [80 mM Tris-HCl pH 7.4, 160 mM NH4Cl, 11 mM Mg(CH3COO)2, 2 mM spermidine, 0.5 mM spermine, 6 mM β-mercaptoethanol] to a total volume of 300 µl. tRNA mix was formed by combining 512 picomoles of N-Ac-[14C]Phe-tRNAPhe, and 4X Binding Buffer to a final volume of 270 µl and incubating for 10 minutes at 30ºC. Eight two-fold serial dilutions of tRNA mix were prepared ranging in concentration from 128 picomoles to 1 picomole. 15 µl of the ribosome mix was added to each tube and the tRNA + ribosome mix was incubated for 30 minutes at 30ºC. The solutions were then poured onto moistened Millipore HA 0.45 micron nitrocellulose filters and incorporated radioactivity was determined by scintillation counting. Elongation factor 2 (eEF2) binding was performed as previously described.27 Binding data were analyzed and KD values were calculated via single binding site with ligand depletion models using GraphPad Prism software.
rRNA chemical modification and analysis
For chemical modification of rRNA, 50 picomoles of ribosomes were first reactivated with 50 µl of 2X SHAPE buffer [160 mM HEPES-KOH pH 7.5, 100 mM NaCl, 5 mM Mg(CH3COO)2, 6 mM β-mercaptoethanol] at 30ºC for 10 minutes. For the SHAPE reaction, 75 µl of 1X SHAPE buffer and 12.5 µl of either DMSO (negative control) or 1-methyl-7-nitroisatoic anhydride (1M7) was added to 50 µl of reactivated ribosomes. The mixture was incubated for 10 minutes at 30ºC and ethanol precipitated overnight. The modified rRNA was then purified using an RNAqueous micro kit (LifeTechnologies, Grand Island, NY) and used in primer extension reactions. Primer extension was performed as previously described and 10 µl samples in Hi-Di Formamide were sent to Genewiz (Frederick, MD) for capillary sequencing. Data were analyzed using ShapeFinder.28
Results
A polyalanine screen reveals inviable uS19-uS13 interface mutants
Using available cryo-EM 29 and X-ray crystal 7 structures of yeast ribosomes as a guide, a region of 19 residues of uS19 was identified as potentially interacting with uS13 and H38. Initially, oligonucleotide site-directed mutagenesis was used to mutate 3 stretches of 6 or 7 amino acids in this region to polyalanine (uS19 residues 98 – 114) (Table S3). All three mutants were inviable as the sole form of uS19. Consequently, 8 additional mutants were synthesized with stretches of 2 or 3 alanines, only one of which (EM110-111AA) was inviable as the sole form of uS19 (Table S3). The seven viable mutants mapped onto the structure of uS19 are shown in Figure 1.
Genetic evaluations reveal that the QV105-105AA and LGH112-114AAA mutants of uS19 affect cell growth and translational fidelity
Mutants were assayed using standard 10-fold dilution spot assays at low temperature (20ºC), optimal temperature for yeast growth (30ºC), and high temperature (37ºC) (Fig. 2A). At 30ºC, all mutants grew comparably to wild-type except for LGH112-114AAA, which exhibited reduced growth, and the EI106-107AA mutant, which exhibited a milder growth defect. At 37ºC, these 2 mutants were not viable, and additional mutants, i.e. FN102-103AA, QV104-105AA and RP108-109AA conferred slightly reduced growth phenotypes. Cold conditions (20ºC) did not appear to exacerbate any of the growth characteristics of any of the mutants, nor did the small molecule translation inhibitors anisomycin or paromomycin (not shown).
Figure 2.
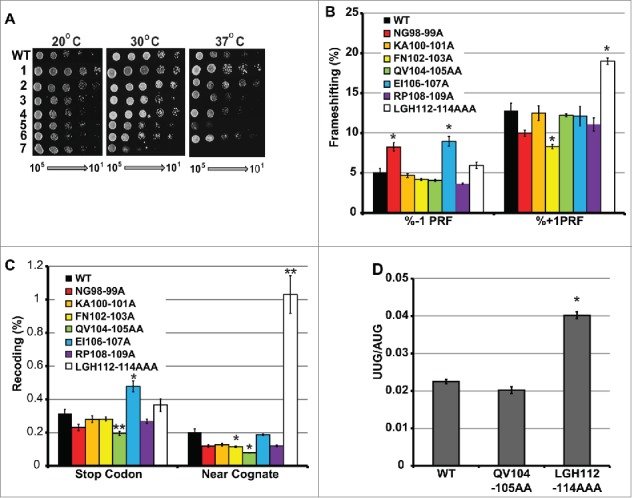
Effects of uS19 mutants on cell growth and translational fidelity. (A) Growth assays. Serial dilutions (105 – 101 CFU) of yeast cells expressing either wild-type or mutant forms of uS19 were transformed spotted onto rich medium and incubated at the indicated temperatures. Viable mutants of amino acid residues uS19 mutated to alanine are numbered as follows. 1: NG98-99AA. 2: KA100-101AA. 3: FN102-103AA. QV104-105AA. 5: EI106-107AA. 6: RP108-109AA. 7: LGH112-114AAA. (B – C)Translational recoding assays. Isogenic cells expressing wild-type uS19 or the indicated mutants were transformed with dual luciferase reporter and in-frame control plasmids and rates of translational recoding were determined. Rates of L-A virus directed -1 PRF, and Ty1 directed +1 PRF. Nonsense codon suppression construct consists of an in-frame stop codon between the 2 ORFs. Missense suppression rates were determined by incorporation of cognate arginine (AGA) in place of near cognate serine (AGC) at position 248 in the firefly ORF. (D) Start codon recognition assay. The ability to initiate translation at a UUG codon was monitored as a ratio of β-galactosidase activities generated from reporters harboring UUG or bona fide AUG codons. Error bars indicate SEM. *P < 0.05, **P < 0.01.
To further characterize the mutants, a series of bicistronic luminescence reporter assays were employed to quantitatively measure different aspects of translational fidelity. Four reporter plasmids were used to assay for -1 PRF, +1 PRF, stop codon recognition (suppression of a UAA nonsense codon),20 and aa-tRNA selection (suppression of a near-cognate missense codon).21 Mutant strains were transformed separately with each of the 4 test constructs as well as an in-frame control reporter, and translational fidelity was measured in terms of percentages of in-frame controls. None of the mutants promoted significant effects on L-A virus directed -1 PRF, with the exception of NG98-99AA and EI106-107AA, which conferred approximately two-fold increases (Fig. 2B). However, FN102-103AA caused a significant decrease in Ty1 directed +1 PRF, and LGH112-114AAA had a stimulatory effect. UAA stop codon decoding was enhanced by the QV104-105AA mutant, and was decreased by EI106-107AA (Fig. 2C). Decoding of a near-cognate missense codon was enhanced by 3 of the mutations (FN102-103AA, QV104-105AA, and RP108-109AA), and was stimulated approximately 5-fold in cells expressing the LGH112-114AAA form of uS19 (Fig. 2C). Initiation at a UUG codon was enhanced by approximately two-fold by the uS19 LGH112-114AAA mutant (Fig. 2D).
The uS19 LGH112-114AAA mutant promotes opposing effects on binding of A-site substrates and increased affinity for tRNAs to the ribosomal P-site
In light of the genetic analyses, further biochemical studies focused on the 2 mutants with the most pronounced phenotypes: QV104-105AA and LGH112-114AAA. Ribosomes isolated from isogenic wild type and mutant cells were assayed with regard to their effects on the steady state binding of one acetylated-Phe-tRNAPhe (Ac-aa-tRNA, a P-site substrate), and two A-site substrates: ternary complex (TC) and eEF2. In general, the biochemical profiles of both mutants were similar to one another. Consistent with the growth phenotypes the magnitude of the defects conferred by the LGH112-114AAA mutant were greater than those promoted by QV104-105AA. Ribosomes isolated from cells expressing the LGH112-114AAA mutant exhibited an ~two-fold decrease in KD for Ac-aa-tRNA (Fig. 3A-D), an ~three-fold decrease in KD for ternary complex (Fig. 3B-D) and a nearly three-fold increase in KD for the translocase eEF2 (Fig. 3C-D).
Figure 3.
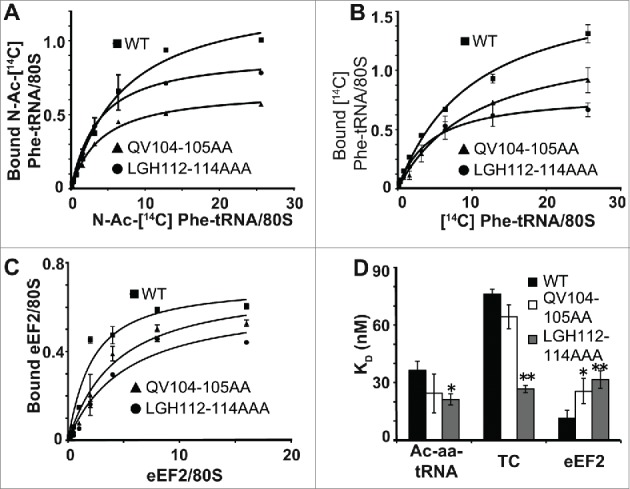
The uS19 mutants promote increased affinity for TC and decreased affinity for the eEF2 translocase binding. (A) P-site tRNA binding. Salt-washed ribosomes were incubated for 10 minutes at 30ºC with two-fold dilutions of [14C]-ac-aa-tRNA and polyuridylic acid (Poly U). (B) A-site tRNA binding. Salt-washed ribosomes were incubated for 10 minutes at 30ºC with two-fold dilutions of pre-incubated ternary complex ([14C]-aa-tRNA•eEIF1A•GTP) and poly U. (C) eEF2 binding. Salt-washed ribosomes were incubated for 30 minutes at 30ºC with two-fold dilutions of purified eEF2, diphtheria toxin, and [14C]-NAD. In all 3 cases, binding was determined using liquid scintillation counting. (D) KD values calculated using ligand depletion model in GraphPad Prism. *P < 0.05, **P < 0.01.
Both the uS19 QV104-105AA and LGH112-114AAA mutants perturb the rotational equilibrium of ribosomes and reduce the fraction of polysomes
Recent studies have suggested a correlation between ribosome rotational status and affinity of ribosomes for elongation factors. For example, yeast mutants of uL16 were used to support a model in which unrotated ribosomes have higher affinity for ternary complex (eEF1A, aa-tRNA, and GTP) while rotated ribosomes have higher affinity for the translocase eEF2.30 In a second study, the same criteria were used identify two uL2 mutants that drive ribosomal rotational equilibrium toward the rotated state.31 Thus, the increased affinity for A-site substrate (ternary complex) and decreased affinity for the translocase, eEF2, is indicative of a shift in the ribosomal rotational equilibrium toward the unrotated state. As the ribosome transitions from the unrotated to the rotated state, a number of intersubunit bridges are rearranged. In particular, G913 of the 18S rRNA and A2207 of the 25S rRNA interact in unrotated ribosomes, but this interaction is broken in rotated ribosomes (Fig. 4A). Their interaction renders them inaccessible to chemical modification in unrotated ribosomes while their dissociation allows them to react with chemicals upon rotation. The reactivity of G913 was probed using kethoxal, a guanine specific chemical, while the reactivity of A2207 was probed using 1-methyl-7-nitroisatoic anhydride (1M7), and high-throughput Selective 2’-Hydroxyl Acylation Analyzed by Primer Extension (hSHAPE) 32 was utilized to monitor the reactivity of these bases in isogenic wild-type and uS19 QV104-105AA and LGH112-114AAA mutant ribosomes. Both 18S rRNA G913 and 25S rRNA A2207 exhibited decreased reactivity in both sets of mutant ribosomes (Fig. 4B-C), indicative of a shift in the ribosomal rotational equilibrium toward the unrotated state. Sucrose gradient fractionation of cycloheximide arrested ribosomes revealed an overall reduction of monosomes and polysomes in both mutants (Fig. 4D). Additionally, the QV104-105AA mutant displayed a strong accumulation of 80S vacant ribosomes.
Figure 4.
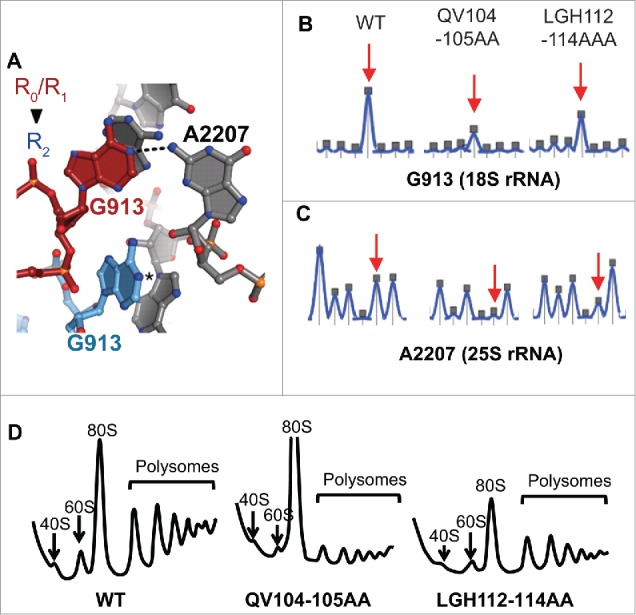
The uS19 mutants perturb ribosomal rotational equilibrium and decrease the fraction of polysomes. (A) In unrotated ribosomes, 18S base G913 (red) and 25S base A2207 (A702 and G1846 in E. coli respectively) interact while, in unrotated ribosomes, G913 is displaced (blue) rendering both bases solvent accessible. (B) Kethoxal probing of G913 (18S rRNA). (C) 1M7 probing of A2207 (25S rRNA). Images created in ShapeFinder.28 (D) A260 tracings of cycloheximide arrested polyribosomes separated through 7% - 47% sucrose gradients by ultracentrifugation. 40S, 60S, 80S and polysome fractions are indicated.
Discussion
While the ribosome is central to life on earth, over the course of evolution significant differences have arisen among the 3 kingdoms. For example, all 3 precursor rRNA molecules are derived from one primary transcript in prokaryotic ribosome biogenesis, while the 4 precursor rRNAs in eukaryotes are derived from 2 primary transcripts.33 Similarly, while ribosome biogenesis and translation occur in the same compartment in prokaryotic cells, eukaryotic ribosome biogenesis occurs in 3 subcellular compartments, and eukaryotic ribosomes are larger and more complex than their bacterial and archael counterparts.34 There are also significant mechanistic differences between the kingdoms. During the more complex eukaryotic translation initiation program the eukaryotic initiation factors eIF1 and eIF1A induce an “open conformation” of the 40S subunit to enable efficient 43S pre-initiation complex scanning along the mRNA, and upon AUG recognition, a major rearrangement occurs in which the head region interacts with the shoulder region clamping the incoming mRNA and locking the initiator tRNA onto the AUG codon.35 This rearrangement does not occur in prokaryotic translation initiation because the AUG codon is positioned in the P-site by base pairing interactions between the Shine-Dalgarno sequence on the mRNA and the anti-Shine Dalgarno sequence on the 16S rRNA.36-38 Head domain flexibility is also observed during the elongation cycle; as ribosomes rotate, the head region rotates 14° with respect to the rest of the SSU.39 Furthermore, mammalian ribosomes undergo an additional eukaryote-specific motion called “subunit rolling."10 These findings suggest that, eukaryotic ribosomes require a greater degree of structural complexity and intersubunit coordination than their prokaryotic counterparts. In prokaryotes, a single SSU protein, uS13, partners with both H38 (the A-site finger) and uL5 to form the B1a and B1b/c bridges respectively. In eukaryotes, it appears that the SSU side of these bridges was separated between 2 different proteins. Thus, while the eukaryotic uS13 retained its B1b/c bridge function, uS19 took on a new role as the SSU partner in the B1a bridge with H38. In this work, some of the consequences of this split were explored by monitoring the effects of uS19 mutants mapping to the interfaces that uS19 makes with uS13 and H38.
The results of genetic analyses served to focus further analyses on 2 mutants: LGH112-114AAA and QV104-105AA. In a prior study using mutants of uL5, we identified a pathway of allosteric information flow through the B1b/c intersubunit bridge that connects the PTC in the LSU with the DC in the SSU6. uS19 LGH112-114AAA makes intimate contacts with uS13. We suggest that this tight junction is important for maintaining the integrity of this biophysical pathway. A different study using mutants of H38 identified another allosteric information pathway connecting the PTC with the DC through the B1a intersubunit bridge8 QV104-105AA appears to be the closest amino acids to H38, and we suggest that these are important partners on the uS19 side of this bridge. Consistent with those prior studies, mutants affecting the B1b/c bridge elicited stronger biochemical and phenotypic changes than mutants affecting the B1a bridge. Also, as previously observed in studies using mutants of uL1629 (also known as L10 11) and uL230 (also called L2 11), the propensity of both of the uS19 mutants to redistribute ribosomal rotational equilibrium toward the unrotated state is consistent with their observed biochemical defects, i.e., increased affinity for tRNAs in the A-and P-sites, and decreased affinity for the eEF2 translocase. The non-specific increases in the affinities for tRNAs in the P-site are consistent with uS19 LGH112-114AAA mutant cells stimulation of initiation at a UUG codon relative to a bona fide AUG initiation codon.23 While this would appear to argue against the observed increase in +1 PRF (because the P-site tRNA would be less inclined to slip), perhaps this is offset by the larger increase in affinity for A-site substrate, which may render these ribosomes more likely to accept an aa-tRNA in the +1 frame. Similarly, increased affinity for A-site substrate is also consistent with increased misreading of a near cognate codon. The enhanced decoding of UAA and near-cognate codons by the QV104-105AA mutant cannot be readily explained. Perhaps, the very strong effect of this mutant on ribosome rotation causes it to remain longer in the unrotated state, allowing more time for proofreading. The accumulation of 80S vacant ribosomes by the QV104-105AA mutant is suggestive of a defect in the Dom34/Hbs1 mediated recycling of vacant ribosomes.40 From a broader view, the ability to link the effects of mutants on ribosome structure and biochemistry with changes in translational fidelity and gene expression has implications for understanding and eventual treatment of diseases such as ribosomopathies, which are caused by mutations in the translational apparatus.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to thank Tom Dever and Terri Kinzy for the gifts of plasmids.
Supplemental Material
Supplemental data for this article can be accessed on the publisher’s website
Funding
AMB was partially supported by a Diversity Supplement to a grant from the NIH (3R01GM058859-10A1S3). This work was supported by grants from the National Institutes of Health to JDD (R01GM058859, R01HL119439, and R01GM117177).
References
- 1.Zhang W, Dunkle JA, Cate JHD. Structures of the ribosome in intermediate states of ratcheting. Science 2009; 325:1014-7; PMID:19696352; http://dx.doi.org/ 10.1126/science.1175275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Julián P, Konevega AL, Scheres SHW, Lázaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A 2008; 105:16924-7; http://dx.doi.org/ 10.1073/pnas.0809587105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 2000; 406:318-22; PMID:10917535; http://dx.doi.org/ 10.1038/35018597 [DOI] [PubMed] [Google Scholar]
- 4.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA 2007; 104:4881-5; http://dx.doi.org/ 10.1073/pnas.0700762104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank J. Intermediate states during mRNA-tRNA translocation. Curr Opin Struct Biol 2012; 22:778-85; PMID:22906732; http://dx.doi.org/ 10.1016/j.sbi.2012.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhodin MHJ, Dinman JD. An Extensive Network of Information Flow through the B1b/c Intersubunit Bridge of the Yeast Ribosome. PLoS One 2011; 6:e20048; PMID:21625514; http://dx.doi.org/ 10.1371/journal.pone.0020048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 2011; 334:1524-9; PMID:22096102; http://dx.doi.org/ 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- 8.Rakauskaite R, Dinman JD. An arc of unpaired “hinge bases” facilitates information exchange among functional centers of the ribosome. Mol Cell Biol 2006; 26:8992-9002; PMID:17000775; http://dx.doi.org/ 10.1128/MCB.01311-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ratje AH, Loerke J, Mikolajka A, Brünner M, Hildebrand PW, Starosta AL, Dönhöfer A, Connell SR, Fucini P, Mielke T, et al. . Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 2010; 468:713-6; PMID:21124459; http://dx.doi.org/ 10.1038/nature09547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budkevich TV, Giesebrecht J, Behrmann E, Loerke J, Ramrath DJ, Mielke T, Ismer J, Hildebrand PW, Tung CS, Nierhaus KH, et al. . Regulation of the Mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell 2014; 158:121-31; PMID:24995983; http://dx.doi.org/ 10.1016/j.cell.2014.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. . A new system for naming ribosomal proteins. Curr Opin Struct Biol 2014; 24:165-9; PMID:24524803; http://dx.doi.org/ 10.1016/j.sbi.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mager WH, Planta RJ, Ballesta JG, Lee JC, Mizuta K, Suzuki K, Warner JR, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res 1997; 25:4872-5; PMID:9396790; http://dx.doi.org/ 10.1093/nar/25.24.4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leger-Silvestre I, Milkereit P, Ferreira-Cerca S, Saveanu C, Rousselle JC, Choesmel V, Guinefoleau C, Gas N, Gleizes PE. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J 2004; 23:2336-47; PMID:15167894; http://dx.doi.org/ 10.1038/sj.emboj.7600252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khairulina J, Graifer D, Bulygin K, Ven’yaminova A, Frolova L, Karpova G. Eukaryote-specific motif of ribosomal protein S15 neighbors A site codon during elongation and termination of translation. Biochimie 2010; 92:820-5; PMID:20206660; http://dx.doi.org/ 10.1016/j.biochi.2010.02.031 [DOI] [PubMed] [Google Scholar]
- 15.Mitra K, Frank J. Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct 2006; 35:299-317; PMID:16689638; http://dx.doi.org/ 10.1146/annurev.biophys.35.040405.101950 [DOI] [PubMed] [Google Scholar]
- 16.Rhodin MHJ, Dinman JD. A flexible loop in yeast ribosomal protein L11 coordinates P-site tRNA binding. Nucleic Acids Res 2010; 38:8377-89; PMID:20705654; http://dx.doi.org/ 10.1093/nar/gkq711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol 1983; 153:163-8; PMID:6336730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor Lab 1982; Cold Spring Harbor, NY [Google Scholar]
- 19.Dinman JD, Wickner RB. Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virology 1992; 66:3669-76; PMID:1583726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 2003; 9:1019-24; PMID:12869712; http://dx.doi.org/ 10.1261/rna.5930803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plant EP, Nguyen P, Russ JR, Pittman YR, Nguyen T, Quesinberry JT, Kinzy TG, Dinman JD. Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae. PLoS One 2007; 2:e517; PMID:17565370; http://dx.doi.org/ 10.1371/journal.pone.0000517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res 2004; 32:e160-70; PMID:15561995; http://dx.doi.org/ 10.1093/nar/gnh157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moehle CM, Hinnebusch AG. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. MolCell Biol 1991; 11:2723-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meskauskas A, Dinman JD. Ribosomal protein L3 functions as a “rocker switch” to aid in coordinating of large subunit-associated functions in eukaryotes and Archaea. Nucleic Acids Res 2008; 36:6175-86; PMID:18832371; http://dx.doi.org/ 10.1093/nar/gkn642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 1989; 58:857-67; PMID:2673535; http://dx.doi.org/ 10.1016/0092-8674(89)90938-0 [DOI] [PubMed] [Google Scholar]
- 26.Meskauskas A, Dinman JD. A molecular clamp ensures allosteric coordination of peptidyltransfer and ligand binding to the ribosomal A-site. Nucleic Acids Res 2010; 38:7800-13; PMID:20660012; http://dx.doi.org/ 10.1093/nar/gkq641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem 2006; 281:32639-48; PMID:16950777; http://dx.doi.org/ 10.1074/jbc.M607076200 [DOI] [PubMed] [Google Scholar]
- 28.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 2008; 14:1979-90; PMID:18772246; http://dx.doi.org/ 10.1261/rna.1166808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J. Comprehensive molecular structure of the eukaryotic ribosome. Structure 2009; 17:1591-604; PMID:20004163; http://dx.doi.org/ 10.1016/j.str.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulima SO, Gülay SP, Anjos M, Patchett S, Meskauskas A, Johnson AW, Dinman JD. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res 2014; 42:2049-63; PMID:24214990; http://dx.doi.org/ 10.1093/nar/gkt1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musalgaonkar S, Moomau CA, Dinman JD. Ribosomes in the balance: structural equilibrium ensures translational fidelity and proper gene expression. Nucleic Acids Res 2014; 42:13384-92; PMID:25389262; http://dx.doi.org/ 10.1093/nar/gku1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson KA, Merino EJ, Weeks KM. Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. NatProtoc 2006; 1:1610-6 [DOI] [PubMed] [Google Scholar]
- 33.Kaczanowska M, Rydén-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 2007; 71:477-94; PMID:17804668; http://dx.doi.org/ 10.1128/MMBR.00013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinman JD. The eukaryotic ribosome: current status and challenges. J Biol Chem [Internet] 2009; 284:11761-5; http://dx.doi.org/ 10.1074/jbc.R800074200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 2007; 26:41-50; PMID:17434125; http://dx.doi.org/ 10.1016/j.molcel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- 36.Nakamoto T. The initiation of eukaryotic and prokaryotic protein synthesis: a selective accessibility and multisubstrate enzyme reaction. Gene 2007; 403:1-5; PMID:17869453; http://dx.doi.org/ 10.1016/j.gene.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 37.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene 1999; 234:187-208; PMID:10395892; http://dx.doi.org/ 10.1016/S0378-1119(99)00210-3 [DOI] [PubMed] [Google Scholar]
- 38.Boni IV. [Diverse molecular mechanisms for translation initiation in prokaryotes]. Mol Biol (Mosk) 2006; 40:658-68; PMID:16913225; http://dx.doi.org/ 10.1134/S002689330604011X [DOI] [PubMed] [Google Scholar]
- 39.Dunkle JA, Cate JH. Ribosome Structure and Dynamics during Translocation and Termination. Annu Rev Biophys 2010; 39:227-44 [DOI] [PubMed] [Google Scholar]
- 40.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 2010; 330:369-72; PMID:20947765; http://dx.doi.org/ 10.1126/science.1192430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenner L, Melnikov S, de Loubresse NG, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. Crystal structure of the 80S yeast ribosome. Curr Opin Struct Biol 2012; 22:759-67; PMID:22884264; http://dx.doi.org/ 10.1016/j.sbi.2012.07.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


