Abstract
Engineered protein switches have a large dynamic range, high specificity for the activating ligand, and a modular architecture, and have been explored for a wide range of applications including biosensors and therapeutics. The ability to externally control switch function is important in extending applications for protein switches. We recently demonstrated that the on/off state could be controlled by the redox state of disulfide bonds introduced into the switches at select locations. Here, we demonstrate that an electrochemical signal can be used as an exogenous input to control switch function via reduction of the engineered disulfide bonds. This study suggests that disulfide-containing protein switch is a potentially useful platform for bioelectronic sensors with remote control of the sensing ability.
Keywords: allostery, disulfide bond, electrochemical reduction, protein engineering, protein switch
Introduction
In nature, cells have evolved to develop highly sensitive chemo-sensing capability through natural biomolecular switches. Inspired by these natural biosensors, there has been much effort in developing protein regulation through controllable inputs such as exogenous molecules, target-specific environmental variables, or endogenous biomarkers (Ostermeier, 2009; Vallee-Belisle and Plaxco, 2010). Engineered proteins, known as protein switches, have recently received attention for their potential applications as novel therapeutics and biomolecular sensors (Ostermeier, 2009; Stratton and Loh, 2011; Vallee-Belisle and Plaxco, 2010).
The cellular responses to oxidative conditions often involve redox-controlled molecular switches based on disulfide bond formation (Hogg, 2003; Nagahara, 2011). Inspired by these natural molecular switches, several engineered redox-controlled biosensors have been developed (Arias-Barreiro et al., 2010; Ostergaard et al., 2001). At the same time, redox-sensing has been integrated into nanostructured biosensors with electrochemical signals as readout (Huang and Mason, 2009; Lu et al., 2013). In addition, electrochemical methods have been used for disulfide bond reduction as a means to control and characterize protein structure and function (Cayot et al., 2002; Honeychurch, 1997; Zhu et al., 2001). Based on this previous work, we hypothesized that an electrochemical signal could be used to externally control the on/off state of protein switches by modulating the oxidation state of an introduced disulfide bond.
Here we use RG13 as a model protein switch to demonstrate the concept. RG13 is a fusion protein of maltose binding protein (MBP) and TEM1 β-lactamase (BLA) (Guntas et al., 2004). RG13’s β-lactamase activity increases in the presence of maltose (Supplementary Figure 1A) (Guntas et al., 2004). Evidence indicates that this activity increase is associated with a large hinge-bending conformational change in the MBP domain of RG13 from the open to the closed state like that observed in MBP (Kim and Ostermeier, 2006; Shilton et al., 1996; Wright et al., 2009). RG13 retains switching activity when tethered to a solid surface (Zayats et al., 2011).
Recently, we integrated redox-control into RG13 through the incorporation of disulfide bonds designed to lock the MBP domain in either the open and closed states (Choi and Ostermeier, 2014). We created switches with three different redox-mediated functionalities (Fig. 1 and Supplementary Table SI). We designed the engineered disulfide bond in RG13-AND2 to keep the MBP domain in an open conformation (Fig. 1B). Thus, RG13-AND2 requires pre-activation by a reducing agent such as tris(2-carboxyethyl) phosphine (TCEP) or dithiothreitol (DTT) in order to be activated by maltose (Supplementary Figure S1B). To create a switch that could be solely activated by a reducing agent, we introduced two mutations into the hinge region of the MBP domain of RG13-AND2 that are known to shift the equilibrium of MBP towards the closed state (Telmer and Shilton, 2003) (Fig. 1C). The resulting switch, RG13-YES, could be activated by disulfide bond reduction (Supplementary Figure S1C). Lastly, we designed the disulfide bond of RG13-ORN2 to hold the MBP domain in its closed state, even in the absence of maltose, such that the switch would be activated by disulfide formation (Fig. 1D). Thus RG13-ORN2’s catalytic activity decreases in the presence of reducing agent, although full deactivation was not achieved (Supplementary Figure S1D). RG13-YES and RG13-ORN2 can be viewed as opposite functionalities, with RG13-YES being activated by the reducing agent and R13-ORN2 being deactivated by the reducing agent.
Figure 1.
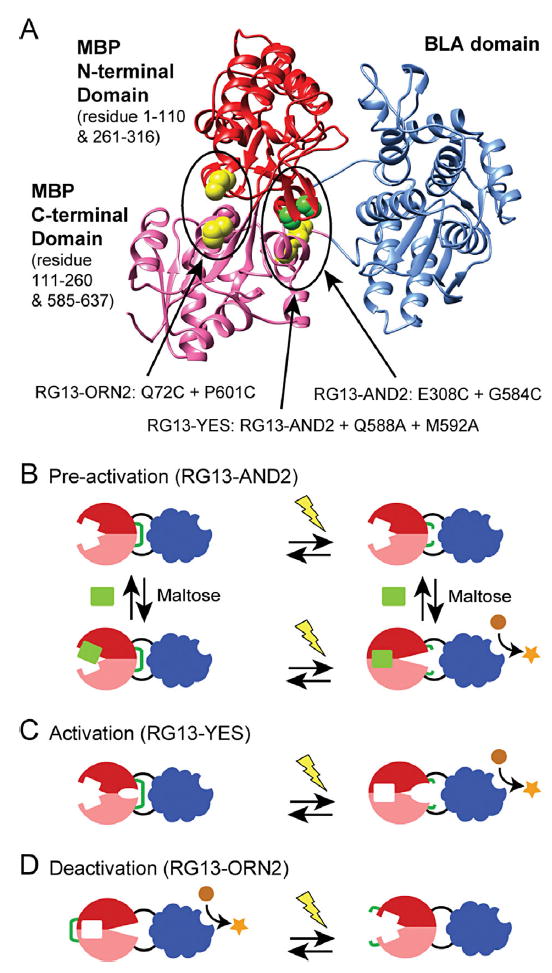
RG13 disulfide variants for electrochemical activation. A: The structure model of RG13 (PDB ID: 4DXC) with the sites of mutation to cysteine indicated in green and yellow spheres. The three proposed electrochemical control mechanisms of protein switches with conformational states regulated by disulfide bond reduction and maltose binding: (B) pre-activation—a disulfide bond (green line) introduced in the hinge region holds the MBP domain in an open conformation. Activation of enzyme activity requires both maltose and reduction of the disulfide bond, (C) activation—in addition to a disulfide in the hinge region holding the MBP domain in the open conformation, mutations in the hinge destabilize the open conformation such that reduction is sufficient to activate enzyme activity, and (D) deactivation—a disulfide bond (green line) is introduced to hold the MBP domain in the closed state resulting in an active enzyme domain. The switch is in an off state if the disulfide is reduced.
Here, we demonstrate the electrochemically-activated switching behavior of these engineered switches (Fig. 2A). We reduced the disulfide bonds in the switches in a conventional three-electrode cell with a gold working electrode by sweeping the potential from 0 to −0.8 V (vs. Ag/AgCl 3 M NaCl) (Fig. 2B, C). The disulfide bonds were designed to be sterically inaccessible to a surface, hence reaction of the disulfides with the gold surface is unlikely. The current-voltage curve for the RG13-AA switch, which is a variant of RG13 with two native cysteines in the BLA domain mutated to alanine, was indistinguishable from buffer alone (Fig. 2B). In contrast, reduction of the single disulfide bond in the RG13-AND2 switch resulted in a significant increase in current over the potential range from −0.3 V to −0.8 V (Fig. 2B), corresponding to the potential range for disulfide bond reduction in bovine serum albumin (Zhu et al., 2001). The RG13-ORN2 switch showed a similar increase in current to the RG13-AND2 switch, whereas the disulfide bond reduction current for the RG13-YES switch was somewhat smaller (Fig. 2C). These results were reproduced in three independent experiments. This difference may be due to the different environments around the disulfide bonds since RG13-YES has additional mutations near the engineered disulfide bond compared to RG13-AND2, while RG13-ORN2 has a disulfide bond on the opposite side of the MBP domain with increased solvent exposure compared to RG13-AND2 (Fig. 1A). No change in current from the background was observed on subsequent scans indicating irreversible reduction of the disulfide bonds in the protein switches (Supplementary Figure S2A, B). We speculate that the post-reduction conformation of the protein may be such that disulfide bond formation is no longer favorable, much like what was observed in the irreversible electrochemical reduction of BSA (Zhu et al., 2001).
Figure 2.
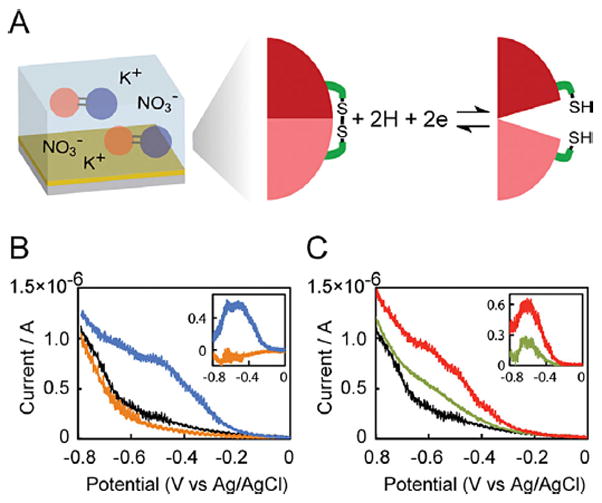
Electrochemical reduction. A: A schematic diagram of electrochemical reduction of the protein switch. (B) Current - voltage curves for electrochemical reduction of buffer (black), RG13-AA (orange) and RG13-AND2 (blue). Inset: Current–voltage curves of net current after baseline (buffer) subtraction of RG13-AA (orange) and RG13-AND2 (blue). (C) Current–voltage curves for electrochemical reduction of buffer (black), RG13-YES (green) and RG13-ORN2 (red). Inset: Current–voltage curves of net current after baseline (buffer) subtraction of RG13-YES (green) and RG13-ORN2 (red). The proteins (0.5 mg mL−1) were in a buffer containing 25 mM phosphate and 200 mM KNO3 (pH 7.5). The scan rate was 50 mV s−1, and the electrode area was 0.02 cm2.
We confirmed that electrochemical reduction resulted in the expected modulation of the switching activity. We measured the enzymatic activity of the switches by the initial rates of nitrocefin hydrolysis before and after reduction. In addition, we measured the catalytic activity of RG13-AA and RG13-AND2 in the presence and absence of maltose both before and after electrochemical reduction. We applied a potential of −0.7 V for 10 min for all switches. RG13-AA, a version of RG13 lacking any cysteine residues, showed no effect of electrochemical treatment on maltose-regulated switching behavior (Fig. 3A and 3B) illustrating that neither catalytic activity nor switch activity is inherently influenced by the conditions used for electrochemical reduction. For RG13-AND2, we observed significant maltose-regulated switching behavior only after electrochemical reduction. The maltose-dependent fold-increase in catalytic activity of RG13-AND2 upon electrochemical reduction was about a third as large as that observed with RG13-AA (Supplementary Table SII), suggesting that electrochemical reduction was less efficient than chemical reduction. Electrochemical reduction of RG13-YES and RG13-ORN2 resulted in ~2.5-fold activation and ~2-fold deactivation of catalytic activity, respectively (Fig. 4 and Supplementary Table SII), which is the same trend observed with chemical reduction of these switches (Supplementary Figure S1C, D). The amount of activated switches as estimated by the charge passed during electrochemical reduction was 2.6% of the amount required to account for the observed enzyme activity. However, the efficiency of electrochemical reduction of protein disulfide bond is difficult to quantify by electrochemical means as previously described (Kwee, 1976).
Figure 3.
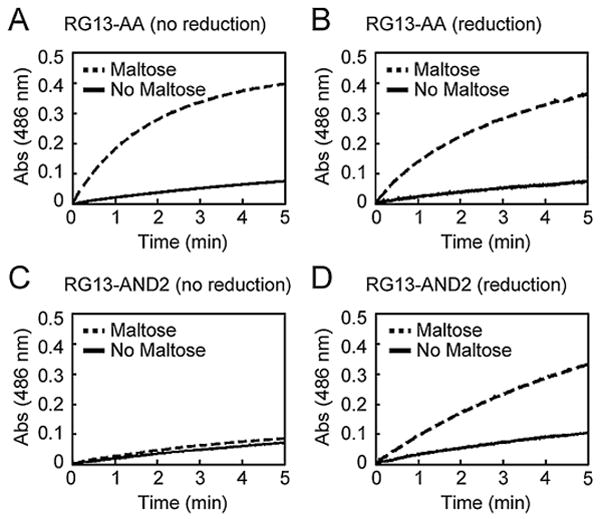
The effect of electrochemical reduction on enzymatic activity Enzymatic activity in the absence and presence of maltose was measured using the colorimetric β-lactamase substrate nitrocefin by monitoring the absorbance at 486 nm as a function of time. The catalytic activity of RG13-AA (A) before and (B) after electrochemical reduction did not differ significantly. In contrast, the catalytic activity of RG13-AND2 (C) before and (D) after electrochemical reduction showed that electrochemical reduction enabled maltose-activation of catalytic activity.
Figure 4.
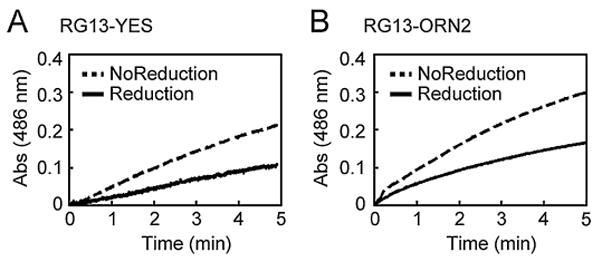
The effect of electrochemical reduction on enzymatic activity. Enzymatic activity of (A) RG13-YES and (B) RG13-ORN2 was measured before and after electrochemical reduction using the colorimetric β-lactamase substrate nitrocefin by monitoring the absorbance at 486 nm as a function of time.
These results demonstrate the potential for electrochemical control of protein switches. We demonstrated that all three types of redox-mediated switch mechanisms (pre-activation, activation, and deactivation) could be implemented via electrochemical input. We also demonstrated that the maltose-induced allosteric switching property of RG13-AND2 could be activated by electrochemical reduction of the engineered disulfide bond. Thus, RG13-AND2 switching activity is controlled at two input levels: an electrochemical signal as a pre-activation control and effector binding as an activation control input. The catalytic activities of RG13-YES and RG13-ORN2 were controlled solely by electrochemical input. The current results support the hypothesis of controlling protein-switching activity via electrochemical means, and this study opens up the possibility of regulating the activity of protein by manipulating allosteric behavior with electrochemical signals.
Methods
All chemicals used were purchased from Sigma (St. Louis, MO) unless otherwise noted. Nitrocefin was purchased from TOKU-E (Bellingham, WA). Bovine Serum Albumin (BSA) was purchased from New England Biolabs (Ipswich, MA). BL21 competent cells purchased from Agilent Technologies (Santa Clara, CA). pDIMC8-RG13-AND2, -YES, and -ORN2, plasmids that contain the RG13 disulfide variant genes, were previously described (Choi and Ostermeier, 2014). Electrochemical cell kit was purchased from BASi (West Lafayette, IN). The potentiometer was from purchased from Princeton Applied Research (Oak Ridge, TN).
Expression and Purification of RG13 Variants
Proteins were expressed and purified from BL21 cells grown in M9 minimal media, as previously described (Choi and Ostermeier, 2014). For production of each protein, 1 L of minimal media was inoculated with 10 mL overnight culture of BL21 cells harboring the pDIMC8 plasmid that encoded the proteins and shaken at 37°C until the OD600 reached 0.8. The culture was induced with 1 mM IPTG and incubated at 25°C for another 48 h. After expression, the cells were harvested, resuspended and lysed using a French press. The soluble proteins were purified using the HisTrap and size-exclusion column in the AKTA FPLC purifier system from GE Healthcare Life Sciences. Proteins were dialyzed against 5 L of a dialysis buffer (25 mM phosphate and 200 mM KNO3, pH 7.5). Approximately 5 mg of purified proteins were obtained with >95% purity, estimated by coomassie blue staining of SDS-PAGE gels.
Electrochemical Reduction of Disulfide Bond in Engineered RG13 Disulfide Variants
The electrochemical experiments were carried out in three-electrode electrochemical cell, MF-2040 Low volume cell from BASi with disk gold electrode with a surface area of 0.02 cm2 (MF-2014, BASi) as a working electrode, Ag/AgCl/3M NaCl as a reference electrode (MF-2052, BASi) and Pt wire as an auxiliary electrode (MW-4130, BASi). A low volume sample cell (MF-2031, BASi) was used during experiment. The working electrode was cleaned based on the standard procedure provided by BASi prior to each measurement.
The proteins were in a solution containing 25 mM phosphate (pH 7.5) and 200 mM KNO3 as supporting electrolyte with the final concentration of 0.5 mg mL−1. Before conducting electrochemical reduction, the dissolved oxygen was eliminated by purging with nitrogen into the solution for 2–5 min. The cell was continuously blanketed with nitrogen during the measurements. The linear sweep voltammetry measurements were carried out at the potential range from 0 V to −0.8 V at scan rate of 50 mV s−1, and the chronoamperometric experiments were carried out by applying the electrochemical potential of −0.7 V for 10 min with VersaSTAT 3 potentiostat from Princeton Applied Research. The voltammograms were recorded and analyzed using VersaStudio software package.
Nitrocefin Hydrolysis Enzymatic Assay
Enzymatic assays were performed as previously described (Choi and Ostermeier, 2014). Proteins were added in 100 mM sodium phosphate buffer, pH 7.5 to the final concentration of 1–5 nM, and incubated either in the absence and presence of 10 mM maltose for 30 min at 25°C. Nitrocefin was added to the final concentration of 50 μM. The absorbance at the wavelength of 486 nm was recorded by using a Varian Cary 50 UV-Visible Spectrophotometer (Agilent Technologies) and the initial rate of nitrocefin hydrolysis reaction was determined by Varian Cary WinUV software package.
Supplementary Material
Acknowledgments
This work was supported by the Defence Threat Reduction Agency [HDTRA1-09-1-0016] and the National Institute of General Medicine at the National Institutes of Health [R01 GM066972].
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Arias-Barreiro CR, Okazaki K, Koutsaftis A, Inayat-Hussain SH, Tani A, Katsuhara M, Kimbara K, Mori IC. A bacterial biosensor for oxidative stress using the constitutively expressed redox-sensitive protein roG FP2. Sensors (Basel) 2010;10(7):6290–6306. doi: 10.3390/s100706290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayot P, Rosier H, Roullier Lc, Haertléc T, Tainturier G. Electrochemical modifications of proteins: Disulfide bonds reduction. Food Chem. 2002;77(3):309–315. [Google Scholar]
- Choi JH, Ostermeier M. Rational design of a fusion protein to exhibit disulfide-mediated logic gate behavior. ACS Synth Biol. 2014;4(4):400–406. doi: 10.1021/sb500254g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas G, Mitchell SF, Ostermeier M. A molecular switch created by in vitro recombination of nonhomologous genes. Chem Biol. 2004;11(11):1483–1487. doi: 10.1016/j.chembiol.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Hogg PJ. Disulfide bonds as switches for protein function. Trends Biochem Sci. 2003;28(4):210–214. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- Honeychurch MJ. The reduction of disulfide bonds in proteins at mercury electrodes. Bioelectrochemistry Bioenerg. 1997;44(1):13–21. [Google Scholar]
- Huang Y, Mason AJ. A redox-enzyme-based electrochemical biosensor with a CMOS integrated bipotentiostat. IEEE BioCAS 2009 Conf; 2009. pp. 29–32. [Google Scholar]
- Kim JR, Ostermeier M. Modulation of effector affinity by hinge region mutations also modulates switching activity in an engineered allosteric TEM1 beta-lactamase switch. Arch Biochem Biophys. 2006;446(1):44–51. doi: 10.1016/j.abb.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Kwee S. The electrochemical reduction of disulfide bonds in proteins. Bioelectrochemistry Bioenerg. 1976;3(2):264–271. [Google Scholar]
- Lu C-S, Wen P-C, Chang H-Y, Tseng FG. Nanoparticles-based electrochemical biosensor for single bacterium detection by redox signal amplification. 17th International Conference on Miniaturized Systems for Chemistry and Life Sciences; 2013. pp. 239–241. [Google Scholar]
- Nagahara N. Intermolecular disulfide bond to modulate protein function as a redox-sensing switch. Amino Acids. 2011;41(1):59–72. doi: 10.1007/s00726-010-0508-4. [DOI] [PubMed] [Google Scholar]
- Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: Engineering a redox switch in green fluorescent protein. EMBO J. 2001;20(21):5853–5862. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier M. Designing switchable enzymes. Curr Opin Struct Biol. 2009;19(4):442–448. doi: 10.1016/j.sbi.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilton BH, Shuman HA, Mowbray SL. Crystal structures and solution conformations of a dominant-negative mutant of Escherichia coli maltose-binding protein. J Mol Biol. 1996;264(2):364–376. doi: 10.1006/jmbi.1996.0646. [DOI] [PubMed] [Google Scholar]
- Stratton MM, Loh SN. Converting a protein into a switch for biosensing and functional regulation. Protein Sci. 2011;20(1):19–29. doi: 10.1002/pro.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telmer PG, Shilton BH. Insights into the conformational equilibria of maltose-binding protein by analysis of high affinity mutants. J Biol Chem. 2003;278(36):34555–34567. doi: 10.1074/jbc.M301004200. [DOI] [PubMed] [Google Scholar]
- Vallee-Belisle A, Plaxco KW. Structure-switching biosensors: inspired by Nature. Curr Opin Struct Biol. 2010;20(4):518–526. doi: 10.1016/j.sbi.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CM, Majumdar A, Tolman JR, Ostermeier M. NMR characterization of an engineered domain fusion between maltose binding protein and TEM1 beta-lactamase provides insight into its structure and allosteric mechanism. Proteins. 2009;78(6):1423–1430. doi: 10.1002/prot.22657. [DOI] [PubMed] [Google Scholar]
- Zayats M, Kanwar M, Ostermeier M, Searson PC. Surface-tethered protein switches. Chem Commun (Camb) 2011;47(12):3398–3400. doi: 10.1039/c1cc10146c. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Cheng G, Dong S. The electrochemically induced conformational transition of disulfides in bovine serum albumin studied by thin layer circular dichroism spectroelectrochemistry. Biophys Chem. 2001;90(1):1–8. doi: 10.1016/s0301-4622(00)00238-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


