Abstract
Withdrawal of nutrients triggers an exit from the cell division cycle, the induction of autophagy, and eventually the activation of cell death pathways. The relation, if any, among these events is not well characterized. We found that starved mouse embryonic fibroblasts lacking the essential autophagy gene product Atg7 failed to undergo cell cycle arrest. Independent of its E1-like enzymatic activity, Atg7 could bind to the tumor suppressor p53 to regulate the transcription of the gene encoding the cell cycle inhibitor p21CDKN1A. With prolonged metabolic stress, the absence of Atg7 resulted in augmented DNA damage with increased p53-dependent apoptosis. Inhibition of the DNA damage response by deletion of the protein kinase Chk2 partially rescued postnatal lethality in Atg7−/− mice. Thus, when nutrients are limited, Atg7 regulates p53-dependent cell cycle and cell death pathways.
Cell cycle progression and autophagic flux are both sensitive to nutrient availability. Furthermore, with prolonged nutrient deprivation, cell death pathways and autophagy are simultaneously activated (1). However, our understanding of how autophagy intersects with cell cycle progression or apoptotic cell death is incomplete. We demonstrate that in the setting of low nutrients, cells lacking Atg7 have impaired p53-mediated cell cycle arrest, whereas with continued metabolic stress, cells and tissues lacking Atg7 display increased p53-mediated cell death.
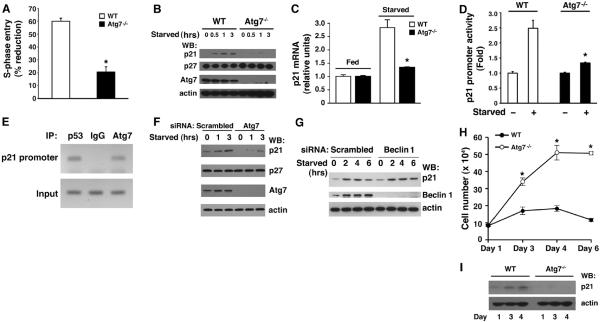
To address whether cell cycle progression and autophagy are linked, we examined the effects of acute nutrient withdrawal on subsequent S-phase entry in either wild-type or Atg7−/− mouse embryonic fibroblasts (MEFs). In wild-type MEFs, S-phase entry, as assessed by bromodeoxyuridine (BrdU) incorporation, decreased by about 60% in the first 3 hours after acute withdrawal of serum and amino acids (Fig. 1A and fig. S1). In contrast, only about 20% of Atg7−/− MEFs successfully exited the cell cycle under the same conditions (P < 0.001, n = 4). The withdrawal from the cell cycle under starved conditions is often mediated by accumulation of cyclin-dependent kinase inhibitors (CKIs) such as p27CDKN1B or p21CDKN1A (2–7). Although early-passage wild-type MEFs rapidly accumulated p21CDKN1A protein after being shifted to starvation media, this response was largely absent in Atg7−/− MEFs (Fig. 1B). In contrast, the abundance of p27CDKN1B was not appreciably different between the two cell types. Similarly, metabolic stress induced the accumulation of p21CDKN1A mRNA in wild-type but not Atg7−/− MEFs (Fig. 1C). Consistent with previous observations (8), the starvation-induced increase in p21CDKN1A expression was largely absent in human or mouse cells lacking p53 (fig. S2). Transcription from a p21CDKN1A promoter linked to a luciferase reporter was increased in wild-type but not Atg7−/− MEFs deprived of nutrients (Fig. 1D). Chromatin immunoprecipitation (ChIP) analysis demonstrated that under starved conditions, endogenous Atg7, along with p53, was present at the p21 promoter (Fig. 1E).
Fig. 1.
Requirement of Atg7 for p21CDKN1A expression and for cell cycle arrest. (A) Percentage reduction in S-phase entry as measured by BrdU incorporation during starved versus fed conditions for early-passage primary wild-type (WT) or Atg7−/− MEFs. Shown is one representative experiment performed in triplicate with greater than 200 individual cells assessed per condition. (B) Protein immunoblot assessment of p21CDKN1A and p27CDKN1B expression in WT and Atg7−/− MEFs after withdrawal of nutrients. Actin is used as a loading control. (C) Abundance of p21CDKN1A mRNA after starvation in WT and Atg7−/− MEFs. (D) Activity of the p21CDKN1A promoter containing a p53 binding element in WT or Atg7−/− MEFs under fed and starved conditions. (E) ChIP of the p21CDKN1A promoter prepared from starved HCT116 cells with antibodies to endogenous p53, Atg7, or an irrelevant immunoglobulin G (IgG) control. (F) Starvation-induced p21CDKN1A protein expression in the human HCT116 cell line after transfection with a scrambled control siRNA or after depletion of Atg7. (G) p21CDKN1A protein after depletion of Beclin 1 (Atg6). (H) Confluent density achieved by either WT or Atg7−/− primary MEFs seeded initially at a high but equal density. (I) Corresponding expression of p21CDKN1A as a function of confluent density in WT or Atg7−/− MEFs. *P < 0.01 (Student t test between WT and Atg7−/− MEFs), n ≥ 3.
Similar analysis in human cells in which Atg7 expression was decreased with small interfering RNA (siRNA) confirmed the impaired expression of p21CDKN1A in nutrient-deprived cells (Fig. 1F). In contrast, depletion of Beclin 1 (Atg6) had no effect on the starvation-induced increase in either p21CDKN1A protein or mRNA expression (Fig. 1G and fig. S3). A similar analysis in Atg5−/− MEFs again revealed no alteration in expression of p21CDKN1A (fig. S4) or other cell cycle parameters (9). Thus, the observed defects in cell cycle arrest and the lack of p21CDKN1A accumulation after nutrient withdrawal appear to be specific for Atg7. This defect was not confined to nutrient withdrawal, as Atg7-deficient cells also appeared to have impaired confluence-dependent growth arrest (Fig. 1, H and I).
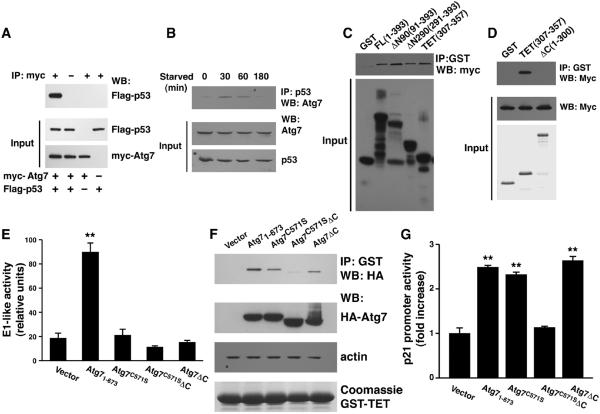
We used epitope-tagged proteins to demonstrate that p53 and Atg7 are present in a single complex (Fig. 2A). Analysis of endogenous proteins revealed a similar interaction that was enhanced after nutrient withdrawal (Fig. 2B). Complexes containing Atg7 and p53 complex were evident in both the cytosol and the nucleus (fig. S5). Using p53 glutathione S-transferase (GST) constructs, we showed that the region corresponding to the p53 tetramerization (TET) domain mediated the interaction with Atg7 (Fig. 2, C and D, and fig. S6) and that Atg7 could promote the formation of p53 tetramers (fig. S7).
Fig. 2.
Interaction of Atg7 with p53. (A) Protein-protein interaction in transfected HCT116 cells assessed by protein immunoblot (WB) analysis with myc-tagged Atg7 and Flag-tagged p53. Immunoprecipitation (IP) was performed using an antibody to the myc epitope or, where indicated (−), an irrelevant IgG iso-type control serum. IP was performed using 2 mg of protein lysate; the input represents 50 μg of lysate. (B) Interaction between endogenous Atg7 and p53 in HCT116 cells under fed conditions (t=0) and after withdrawal of nutrients. (C) In vitro interaction between full-length myc-tagged Atg7 and various GST full-length (FL) or truncation mutants of p53. (D) A GST-p53 construct (ΔC) consisting of the first 300 amino acids of p53 does not bind Atg7. (E) Autophagy is stimulated by full-length Atg7 (1–673), whereas constructs lacking either the active cysteine (Cys571), the C terminus (ΔC), or both of these alterations lack E1-like enzymatic activity. A minimum of 300 cells per sample were counted using quadruplicate samples per construct (**P < 0.01, full-length Atg7 compared to other conditions). (F) In vitro binding of hemagglutinin (HA)–tagged Atg7 constructs to the GST-p53 tetramerization domain. (G) p21CDKN1A promoter activity in fed Atg7−/− MEFs transfected with WT Atg7 or Atg7 constructs with and without E1-like enzymatic activity (n = 4 per condition, **P < 0.01 compared to vector control).
In yeast Atg7, a specific C-terminal active-site cysteine residue and the C-terminal 40 amino acids are required for full enzymatic activity (10). We therefore analyzed the biological activity of mammalian Atg7 mutants containing a serine residue at the active-site cysteine (Atg7C571S), a deletion of the C terminus (Atg7ΔC), or a combination of the two (Atg7C571SΔC). Increased Atg7 expression can stimulate autophagic flux (11, 12). The overexpression of wild-type Atg7 but not the mutant proteins caused an increase in the conversion of light chain 3 form 1 (LC3-I) to LC3-II. This posttranslational modification of LC3 is indicative of the E1-like ubiquitinconjugating activity of Atg7 (Fig. 2E and figs. S8 and S9). However, full-length Atg7, as well as the enzymatically deficient Atg7C571S and Atg7ΔC constructs, all appeared to bind to GST-p53 (Fig. 2F). This interaction was lost with the combined Atg7C571SΔC construct., When assayed in Atg7−/− MEFs, the combined Atg7C571SΔC mutant failed to stimulate transcription of the p21CDKN1A gene (Fig. 2G), consistent with the absence of an in vitro interaction. In contrast, the autophagy-deficient Atg7C571S and Atg7ΔC constructs were indistinguishable from wild-type Atg7 in stimulating transcription at the p21CDKN1A promoter. As such, the ability of Atg7 to bind p53 and regulate transcription of p21CDKN1A appears separable from its E1-like enzymatic function.
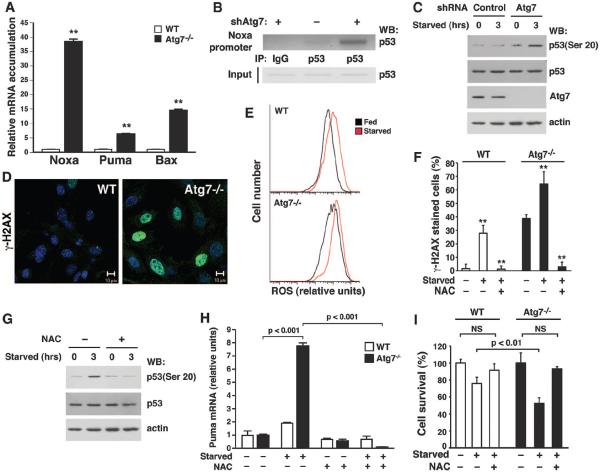
Taken together, our data indicate that under conditions of metabolic stress, Atg7 is required for the p53-dependent expression of p21CDKN1A. Analysis of several other known p53-regulated genes revealed no significant differences in expression between wild-type and Atg7−/− MEFs (fig. S10). In contrast, analysis of a subset of p53-regulated proapoptotic genes—most notably Noxa, Bax, and Puma—demonstrated increased expression of mRNA and protein in starved Atg7-deficient cells (Fig. 3A and fig. S11A). ChIP analysis revealed that Atg7-deficient cells deprived of nutrients had increased p53 binding at promoters of proapoptotic target genes (Fig. 3B and figs. S11B and S12). Thus, the absence of Atg7 reciprocally regulates p53-dependent cell cycle and cell death pathways.
Fig. 3.
The absence of Atg7 increases mitochondrial ROS, augments DNA damage, and activates p53. (A) Relative mRNA expression in starved WT or Atg7−/− MEFs for a panel of proapoptotic p53-responsive genes (**P < 0.01 compared to WT MEFs). (B) ChIP assay 3 hours after the induction of nutrient stress, demonstrating increased p53 binding to the Noxa promoter in HCT116 cells depleted of Atg7. (C) Ser20 phosphorylation in control or Atg7-depleted cells. (D) Images of DNA damage in WT and Atg7−/− MEFs as assessed by γ-H2AX staining. (E) Fluorescence-activated cell sorter analysis of WT and Atg7−/− MEFs under fed (black) or starved (red) conditions, using the redox-dependent fluorophore dichlorodihydrofluorescein diacetate (DCFDA). A rightward shift indicates higher intracellular ROS levels. (F) γ-H2AX foci formation in fed or starved WT and Atg7−/− MEFs. Cells were treated as indicated with the antioxidant NAC (**P < 0.01 compared to starved cells not treated with NAC). (G) NAC treatment inhibits starvation-induced p53 Ser20 phosphorylation in Atg7-depleted cells. (H) Puma expression in WT and Atg7−/− MEFs under fed (−) or starved (+) conditions, in the presence or absence of the cell permeant antioxidant N-acetylcysteine (NAC). (I) Cell death during fed or starved conditions in WT or Atg7−/− MEFs in the presence or absence of NAC. Unless specified, all results are from at least triplicate determinations.
In other contexts such as exogenous γ-irradiation, activation of p53-dependent apoptosis occurs in conjunction with DNA damage and the phosphorylation of p53 on specific N-terminal residues such as Ser20 (13). When deprived of nutrients, cells lacking Atg7 had increased phosphorylation of p53 at Ser20 (Fig. 3C) along with other parameters indicating increased activation of the DNA damage response pathway (Fig. 3D). Cells deficient in other essential autophagy gene products including Beclin 1 and Atg5 also exhibit increased activation of the DNA damage response (14–16), along with an increase in reactive oxygen species (ROS) (14, 17). We similarly found that Atg7−/− MEFs had higher levels of ROS under basal and starved conditions (Fig. 3E) that were reduced by addition of the cell-permeant antioxidant N-acetylcysteine (NAC) or a mitochondrial uncoupler (fig. S13).
Treatment of wild-type or Atg7−/− MEFs with NAC reduced the number of cells with DNA damage foci and starvation-induced phosphorylation of p53 Ser20 (Fig. 3, F and G). Nutrient depletion–induced expression of Puma and other proapoptotic genes was also abrogated by antioxidant treatment of wild-type or Atg7−/− MEFs (Fig. 3H and fig. S14). Underscoring the difference between the p53-mediated growth arrest and p53-mediated cell death pathways, NAC treatment had no effect on p53-dependent expression of p21CDKN1A during metabolic stress (fig. S15). Finally, consistent with the augmented DNA damage and increased activation of proapoptotic genes, Atg7−/− MEFs were significantly more sensitive to starvation (fig. S16). Nonetheless, treatment with NAC largely protected these cells from starvation-induced cell death (Fig. 3I).
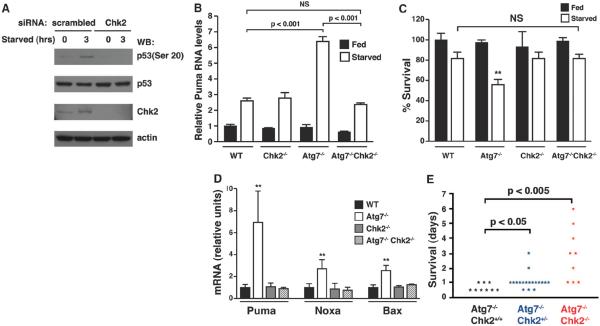
After DNA damage induced by γ-irradiation, activation of Chk2 activation is required for p53 Ser20 phosphorylation and subsequent p53-dependent cell death (18). In certain genetic models such as the absence of the Polycomb gene product Bmi-1, a rise in mitochondrial oxidants is sufficient to induce DNA damage and Chk2 activation (19). Thus, we analyzed the effect of depletion of Chk2 on p53 Ser20 phosphorylation induced by metabolic stress. In Atg7-deficient cells, depletion of Chk2 reduced starvation-induced phosphorylation of p53 Ser20 (Fig. 4A and fig. S17). The increase in proapoptotic gene expression observed in Atg7-deficient cells was not observed in Atg7−/−Chk2−/− MEFs (Fig. 4B and figs. S18 and S19). Similarly, the reduced viability observed in starved Atg7−/− MEFs was also absent in Atg7−/−Chk2−/− MEFs (Fig. 4C).
Fig. 4.
Rescue of Atg7 deficiency after Chk2 deletion. (A) p53 Ser20 phosphorylation in HCT116 cells with stable short hairpin RNA depletion of Atg7 with and without additional siRNA-mediated depletion of Chk2. (B) Starvation-induced Puma expression in primary MEFs with the indicated genotype (n ≥ 3 per genotype per condition). (C) Cell viability of MEFs with the indicated genotype under fed or starved conditions (**P < 0.01 between starved WT and Atg7−/− cells, n = 4 per genotype per condition). (D) Expression of proapoptotic gene products in the liver of pups about 8 hours after birth (**P < 0.01, n = 9 determinations per genotype). (E) Survival of consecutive births of Atg7−/− pups with the indicated Chk2 status.
Mice lacking Atg7 are born with the expected Mendelian frequency but rapidly succumb in the first few hours of life (20). The basis for this lethality is incompletely understood (21). We wondered whether oxidative stress and the activation of the DNA damage response played a role. Metabolically active tissues of Atg7−/− mice demonstrated augmented expression of p53-dependent proapoptotic genes (Fig. 4D and fig. S20), but this was not observed in Atg7−/−Chk2−/− mice. Fewer than 5% of ATG7−/− pups survived more than 24 hours and none survived more than 48 hours. Deletion of one or both alleles of Chk2, however, significantly extended survival of Atg7-deficient animals (Fig. 4E). Long-term survival of double-knockout mice appeared to be limited by persistent neurological defects (fig. S21).
Our data demonstrate that Atg7 and p53 bind each other directly, that starvation increases this interaction, and that in the absence of Atg7, cells fail to properly induce p21CDKN1A expression. This cell cycle defect is specific for Atg7 and is not observed in Atg5- or Atg6-deficient cells, nor does it require the E1-like enzymatic activity of Atg7. In the absence of Atg7, prolonged metabolic stress leads to augmented p53 proapoptotic activity. The increase in p53 proapoptotic activity appears to be similarly dysregulated in Atg7−/− and Atg5−/− cells (figs. S22 and S23). The physiological importance of this shared ROS-Chk2-p53–dependent pathway is underscored by the rescue of Atg7−/− postnatal lethality by Chk2 deletion. Previous results suggest that p53 can regulate autophagy (22–25). Our observations suggest that an additional and reciprocal regulation also exists whereby Atg7 can regulate p53.
Supplementary Material
Acknowledgments
We thank M. Komatsu for providing the Atg7+/− mice. Supported by National Natural Science Foundation of China grants 81130042 and 31171323 (L.C.), NIH intramural funds, and a grant from the Ellison Medical Foundation (T.F.).
Footnotes
Supplementary Materials www.sciencemag.org/cgi/content/full/336/6078/225/DC1
References and Notes
- 1.Rabinowitz JD, White E. Science. 2010;330:1344. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. Cell. 1993;75:805. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 3.Delavaine L, La Thangue NB. Oncogene. 1999;18:5381. doi: 10.1038/sj.onc.1202923. [DOI] [PubMed] [Google Scholar]
- 4.Perucca P, et al. Cell Cycle. 2009;8:105. doi: 10.4161/cc.8.1.7507. [DOI] [PubMed] [Google Scholar]
- 5.Rivard N, L'Allemain G, Bartek J, Pouysségur J, Biol J. Chem. 1996;271:18337. doi: 10.1074/jbc.271.31.18337. [DOI] [PubMed] [Google Scholar]
- 6.Ladha MH, Lee KY, Upton TM, Reed MF, Ewen ME. Mol. Cell. Biol. 1998;18:6605. doi: 10.1128/mcb.18.11.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon YH, Jovanovic A, Serfas MS, Kiyokawa H, Tyner AL. J. Biol. Chem. 2002;277:41417. doi: 10.1074/jbc.M203388200. [DOI] [PubMed] [Google Scholar]
- 8.El-Deiry WS, et al. Cell. 1993;75:817. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 9.Valentin M, Yang E. Cell Cycle. 2008;7:2762. doi: 10.4161/cc.7.17.6595. [DOI] [PubMed] [Google Scholar]
- 10.Komatsu M, et al. J. Biol. Chem. 2001;276:9846. doi: 10.1074/jbc.M007737200. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Cell Metab. 2010;11:467. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pattison JS, Osinska H. J. Robbins, Circ. Res. 2011;109:151. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vazquez A, Bond EE, Levine AJ, Bond GL. Nat. Rev. Drug Discov. 2008;7:979. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 14.Mathew R, et al. Cell. 2009;137:1062. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathew R, et al. Genes Dev. 2007;21:1367. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karantza-Wadsworth V, et al. Genes Dev. 2007;21:1621. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tal MC, et al. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2770. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirao A, et al. Science. 2000;287:1824. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, et al. Nature. 2009;459:387. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu M, et al. J. Cell Biol. 2005;169:425. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiaffino S, Mammucari C, Sandri M. Autophagy. 2008;4:727. doi: 10.4161/auto.6143. [DOI] [PubMed] [Google Scholar]
- 22.Crighton D, et al. Cell. 2006;126:121. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 23.Feng Z, Zhang H, Levine AJ, Jin S. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8204. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amaravadi RK, et al. J. Clin. Invest. 2007;117:326. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tasdemir E, et al. Cell Cycle. 2008;7:3006. doi: 10.4161/cc.7.19.6702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.