Abstract
Objective
Increasing evidence supports the role of the kidney as a viral reservoir for HIV-1. In vitro co-cultivation of HIV-infected T cells with renal epithelial cells results in virus transfer to the latter, while cell free virus infection of these cells is very inefficient. In this study we further characterized the fate of HIV-1 after it is internalized in renal epithelial cells.
Methods
Primary or immortalized CD4+ cells were infected with a GFP-expressing replication competent HIV-1. HIV-1 transfer from T cells to epithelial cells was carried out in a co-culture system and evaluated by FACS analysis. HIV-1 integration in renal epithelial cells was evaluated by Alu-PCR and the production of infectious particles was assessed by p24-ELISA and TZM-bl assay. HIV-infected renal cells were used as donor cells in a co-culture system to evaluate their ability to transfer the virus back to T cells.
Results
Renal epithelial cells become productively infected by HIV-1 and multiple copies of HIV-1 can be transferred from infected T cells to renal epithelial cells. Two separate cells populations were identified among infected renal cells based on the reporter gene GFP expression level (low vs high), with only the high showing sensitivity to AZT and Ritonavir. Co-cultivation of HIV-1 infected renal cells with non-infected T cells resulted in HIV-1 transmission to T cells, supporting bidirectional exchange of virus between T cells and kidney-derived cells.
Conclusions
These results support the kidney as a potential reservoir where virus is exchanged between interstitial T cells and renal tubule epithelial cells.
Keywords: HIV-1, Kidney, Cell to cell contact, Productive infection, Reservoir
INTRODUCTION
Direct HIV-1 infection and gene expression in renal tubular and glomerular epithelial cells is associated with the development of HIV-associated nephropathy (HIVAN) [1]. HIV-1 envelope sequences amplified from laser-capture microdissected renal tubules from HIVAN biopsies are distinct from those simultaneously derived from peripheral blood mononuclear cells (PBMCs), indicating that the kidney is a unique compartment for viral replication [2,3,4]. Furthermore, persistence of viral DNA and RNA in renal epithelial cells following treatment with antiretroviral therapy raises the concern that the kidney can serve as a long-term reservoir for the virus. Interstitial infiltrating lymphocytes with viral nucleic acid are a component of the pathology in HIVAN biopsies suggesting a role for T cells in HIV-1 spread within the tissue [2]. Several reports indicated that direct cell-to-cell contact between infected and non-infected cells can mediate transfer of HIV-1 to recipient cells with much greater efficiency (100 to 1000 times) than direct exposure of target cells to cell-free virus [5,6,7]. Renal epithelial cells must use an alternative mechanism to facilitate HIV-1 entry as some evidence suggests that they lack expression of CD4 receptor. The presence of known HIV-1 receptors was reported in only one study [8], but others failed to confirm these data [9,10]. While polysaccharides, such as dextran-sulfate and Iota-carrageenan, block transfer of virus from T cells to renal tubule epithelial cells, the virus and host cell molecules involved remain unknown [11]. It has been shown that the interaction of HIV-1 with the C-type lectin receptor DEC-205 results in cell-free virus internalization in the immortalized renal epithelial cell line HK2, but without mediating productive infection [12]. Heparan-sulfate proteoglycans (HSPGs) have been suggested to be alternate receptors for HIV-1 entry into CD4 negative cells, enabling HIV-1 entry in an envelope independent manner [11,13,14,15]. Despite strong evidence of intra-renal infection, little is known regarding the virus life cycle within the kidney and within renal epithelial cells. Here we show that after cell-to-cell contact between infected T cells and uninfected renal epithelial cells, HIV-1 is efficiently transferred to renal epithelial cells establishing a productive infection. Infected renal epithelial cells can transfer the virus back to T cells after cell-to-cell contact, delineating an intra-renal mechanism for virus propagation and evolution.
MATERIALS AND METHODS
Primary cells and cell lines
CD4+ cells positively isolated from healthy donor PBMCs by magnetic bead separation (Miltenyi-Biotec) were activated for 24 hours with 3μg/ml Phytohaemagglutinin (Roche) in presence of IL-2 (25U/ml). Primary human renal cortical epithelial cells (HRCEpCs) were obtained from Promocell. Healthy donor PBMCs were purchased from Gulf coast regional blood center (Houston, TX). The HK2 and the 293T Lenti-X cell lines were obtained from American Type Culture Collection (ATCC). The CD4+ T cell lines, CEM and CEM-SS, were kindly provided by Dr. Hal P. Bogerd (Duke University, NC).
Virus preparation and T cells infection
293T Lenti-X cells were transiently transfected with 12 μg of the viral construct pNL-GI [16], using the PEI transfection kit from Polyplus-Transfection SA following the manufacturer’s instructions. To generate viral particles pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G), 293T Lenti-X were transfected with 10μg of pNL-GI and 2μg of pHCMV-VSV-G [17]. Transduction efficiency for each virus stock was assessed on infected CEM T cells by flow cytometry. CEM T cells or Phytohaemagglutinin-activated primary CD4+ cells were inoculated with 1 MOI of HIV-1, and spinoculated for 1 hour at 1500rpm at 25°C to improve transduction efficiency. 24h post infection, infected donor cells were washed with PBS and cultured for additional 48 hours before the co-culture. Donor cells were washed again with PBS before co-culture with target cells.
RTE- T cells co-culture and drugs treatments
Target renal epithelial cells were labeled with 2μM of CMTMR fluorescent dye (Molecular Probes) at 37°C for 30 minutes, as described in manufacturer’s instructions. Labeled cells were washed with PBS, cultured overnight, and rinsed again with PBS before co-culture with donor T cells. Infected donor T cells were added in a 4:1 ratio to target cells and co-cultured overnight. The co-culture was terminated by removal of donor T cells followed by four PBS washes. At 48h post co-culture, epithelial cells were analyzed by flow cytometry (FACS-Calibur, FACS-CantoII; BD Biosciences). Renal epithelial cells double positive for CMTMR and GFP were flow sorted and were either re-plated for subsequent analyses or pelleted for genomic DNA extraction. For the inhibition studies, target renal epithelial cells were pretreated with the azidothymidine (AZT) at 37°C for 30 to 45 minutes, followed by co-culture at 37°C overnight still in the presence of the drugs. In the experiments using the HIV protease inhibitor Ritonavir, donor T cells were treated with the drug before and during virus infection and throughout the co-culture with renal epithelial cells. After the removal of the donor T cells, the drugs were added again with fresh medium.
p24 ELISA and TZM-bl infectivity assay
Supernatants from flow sorted GFP positive HK2 were collected at different time points to evaluate the presence of p24 by ELISA (PerkinElmer). For TZM-bl infectivity assay, serial dilutions of the cell supernatants were performed in quadruplicate wells. Cells containing 10 μg/ml of DEAE-Dextran were added to each well, incubated at 37 °C for 48-hours. Culture medium was then removed and a luciferase reporter gene assay system reagent (Britelite; PerkinElmer) was added to each well. Infectivity was measured as relative luminescence units (RLU) compared to virus control wells after subtraction of background RLU (wells with RLU<2.5 times background are considered negative for the calculation).
Molecular characterization of HIV-1 DNA in RTE cells
Genomic DNA was extracted using QiAmp DNA micro-kit (Qiagen) from flow sorted GFP positive HK-2 cells pelleted either soon after sorting or after being cultured for one week post-sorting. PCR reactions were performed as described in table 1. Intermediates of reverse transcription were detected using a previously described PCR approach [18,19]. The initiation of reverse transcription was detected with primers M667 and AA55M (LTR). Primers specific for middle and late reverse transcription products were detected with primer pairs M667/BB30 and M667/M661 respectively. Integrated HIV-1 DNA was detected by nested PCR to increase sensitivity and fidelity. Standard curves specific for each PCR amplification were used for DNA quantification. Briefly, the standard curve for total HIV-1 DNA PCR and 2-LTR circles were generated using the previously described plasmids pNL-GI [11,16] and p2-LTR-luc [19], respectively. For integrated HIV-1 DNA quantification we constructed a puromycin resistant molecular clone of HIV-1, NL-Puro, to stably transduce the HK2 cell line (HK2/NL-Puro). PCR products were separated by electrophoresis on 2% agarose gels stained with ethidium bromide. The reference gene β-actin was used as loading control.
Table 1.
Primer sequences and PCR conditions
| Target | Primer | Sequences (5′-3′) | PCR conditions |
|---|---|---|---|
| Total HIV-1 DNA | M667/AA55M | GGC TAA CTA GGG AAC CCA CTG / GCT AGA GAT TTT CCA CAC TGA CTA A | 95°C 3 min - 95°C 30s - 60°C 30s- 72°C 30s - 72°C 5 min for 35 cycles |
| M667/BB301 | GGC TAA CTA GGG AAC CCA CTG/ CCC TGT TCG GGC GCC ACT G | 95°C 3 min - 95°C 30s - 60°C 30s- 72°C 30s - 72°C 5 min for 35 cycles | |
| M667/M661 | GGC TAA CTA GGG AAC CCA CTG / CCT GCG TCG AGA GAT CTC CTC TGG | 95°C 3 min - 95°C 30s - 60°C 30s- 72°C 30s - 72°C 5 min for 35 cycles | |
|
| |||
| Integrated HIV-1 DNA | |||
| 1st PCR | Nef Alu1 Alu2 |
ATG CTG CTT GTG CCT GGC TAG AAG C TCC CAG CTA CTG GGG GAG GCT GAG G GCC TCC CAA AGT GCT GGG ATT ACA G |
95°C 3 min - 95°C 30s - 60°C 30s- 72°C 3 min - 72°C 5 min for 18 cycles |
| Nested PCR | AA55M M667 |
GCT AGA GAT TTT CCA CAC TGA CTA A GGC TAA CTA GGG AAC CCA CTG |
95°C 5 min - 95°C 30s - 60°C 30s- 72°C 30s - 72°C 3 min for 25 cycles |
|
| |||
| 2-LTR circles | 9600 515c |
GCT TAA GCC TCA ATA AAG CTT GCC T GTG TGT AGT TCT GCT AAT CAG GGA A |
95°C 5 min - 95°C 30s - 60°C 30s- 72°C 30s - 72°C 10 min for 40 cycles |
|
| |||
| β-Actin | Actin F Actin R |
ATC TGG CAC CAC AAC AAT GAG CTG CG CGT CAT ACT CCT GGA TCC ACA TCT GC |
94°C 2 min - 94°C 30s - 55°C 30s- 72°C 1 min - 72°C 10 min for 25 cycles |
HIV-1 infected RTE cell–T cell co-culture
Flow sorted GFP positive HK2 or HRCEpC cells were co-cultured, at 4 or 6 days post-sorting, with uninfected CEM-SS T cells or primary CD4+ cells. Target T cells were labeled with 2μM of CMTMR, washed with PBS, cultured overnight, and rinsed again with PBS before overnight co-culture with infected renal epithelial cells. The co-culture was terminated by recovering the target T cells from the cell medium. At different time points post co-culture, GFP expression by recovered T cells was analyzed by flow cytometry or fluorescence microscopy (Nikon TE2000-E).
RESULTS
Cell-to-cell contact is necessary for HIV-1 transfer to renal epithelial cells
Prior work has demonstrated that co-cultivation of infected T cells with renal tubule epithelial cells (RTE) results in expression of viral RNA and proteins in RTE cells [11]. To determine the requirements for this transfer and to study the kinetics in RTEs, we used a previously described infectious molecular clone of HIV-1, NL-GI, which carries the green fluorescent protein (GFP) in place of nef and restores nef expression with an internal ribosome entry site (IRES) [11]. CEM T cells were incubated overnight with NL-GI viral particles to infect 60–80% of the cells. Forty-eight hours post infection, CEM T cells were co-cultured with HK2 renal epithelial cells for ~24 hours. Target epithelial cells were labeled with Cell Tracker orange CMTMR to distinguish from donor T cells. To show that cell-to-cell contact is necessary for HIV-1 transfer from infected T cells to renal epithelial cells, we used a transwell membrane (0.4μm pore-size) to separate the two cell populations. After ~24 hours co-culture, T cells were removed by extensive PBS washes and the adherent epithelial cells were incubated at 37°C for an additional 24 hours. GFP expression by HK2 cells was analyzed by flow cytometry at 48h post co-culture. In the presence of a transwell membrane between the two cell populations no HIV-1 infection of the renal epithelial cells was observed, while about 2.5% of HK2 cells expressed GFP after direct contact with infected T cells (data not shown). Furthermore, as previously observed [11], the incubation of HK2 with a large amount of cell-free virus (MOI-20) resulted in low to undetectable infection of epithelial cells (data not shown), confirming the need for cell contact for HIV-1 transfer from infected T cells to uninfected RTEs.
RTE cells support HIV-1 reverse transcription and integration
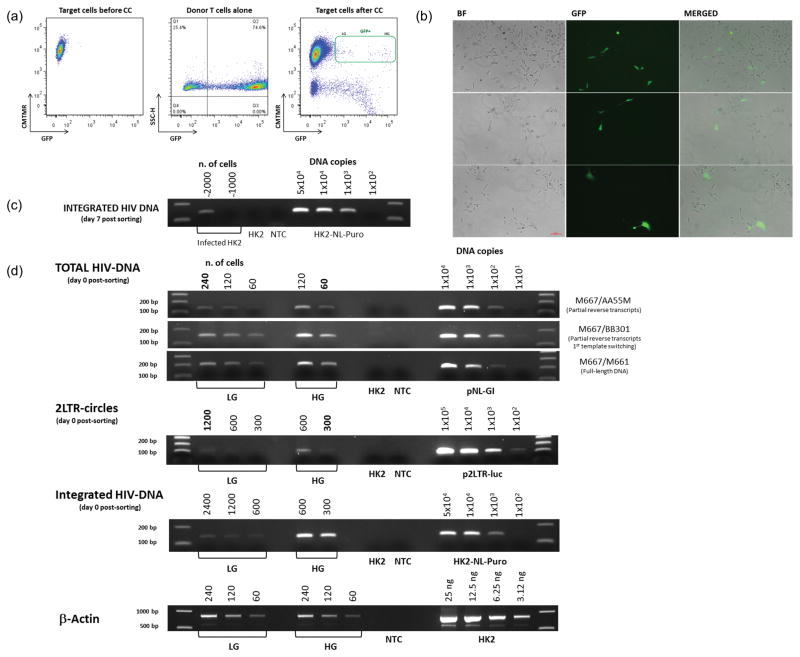
To determine the fate of internalized virus following cell-to-cell transfer, HK2 cells derived from overnight co-culture with infected T cells and double positive for GFP and CMTMR, were collected by flow sorting as shown in Figure 1a, re-plated and examined by fluorescence microscopy. Following co-cultivation, two distinct cell populations based on levels of GFP expression (High GFP VS Low GFP) were observed (Figure 1a). At day 4 post sorting only about 10%, of the sorted GFP positive HK2 cells remained green (Figure 1b). We hypothesized that the green cells in Figure 1b probably correspond to the high GFP (HG) population, while the negative ones correspond to the low GFP population (LG) and could either be cells that transiently express GFP from transferred RNA, un-integrated circular DNA, or cells in which the virus has become latent. To confirm HIV-1 integration in RTE cells, we performed an Alu-nested PCR [21]. HK2 cells (HK2/NL-Puro) stably transduced with a modified molecular clone of HIV-1 (NL-Puro) expressing the puromycin resistance gene, were used as a standard for evaluating integrated vector copies. As shown in Figure 1c, HIV-1 DNA stably integrates in the genome of renal epithelial cells since DNA extracted from cells at day 7 post sorting were positive by Alu-PCR. Quantification of integrated HIV-1 DNA copies in the sorted GFP positive HK2, by comparison with the standard curve, suggests that about 40 to 50% of the flow sorted cells contained integrated HIV-1 DNA. As shown in Figure 1b, only 10% of those cells showed high levels of GFP expression at day 4 post-sorting, suggesting either that integrated HIV-1 DNA is present also in the non-GFP expressing cells in a latent state, or that multiple copies of integrated viral DNA are present in the HG population. In another co-culture experiment the two GFP positive populations (LG and HG) were separated by flow sorting and the extracted DNA was analyzed for intermediates of reverse transcription, integrated and circular HIV-1 DNA using a semi-quantitative PCR approach as previously described [22,23]. As shown in Figure 1d, the total amount of HIV-1 DNA as well as the amount of integrated HIV-1 DNA and the number of 2-LTR circles, are much lower in the LG population compared to HG. In particular, when we compared the amount of total HIV-1 DNA and 2 LTR circles in the two populations, four times more HIV-1 DNA was found in the HG population than in the LG. The HG population contains a much higher number of integrated copies of DNA compared to the LG population.
Figure 1. HIV-1 directed GFP expression and integration in renal epithelial cells.
(a) HK2 cells were co-cultivated overnight with CEM T cells infected with NL-GI. 48h post-coculture HK2 cells double positive for GFP and CMTMR were flow sorted and replated. Two distinct populations, low expressing GFP (LG) VS high expressing GFP (HG), were identified among the GFP positive HK2 cells. (b) Fluorescence microscopy analysis of flow sorted GFP positive HK2 at day 4 post-sorting. (c) DNA was extracted from cells cultivated for 7 days post sorting and analyzed for integrated HIV-1 DNA. (d) High GFP (HG) and the Low GFP (LG) expressing populations of infected HK2 cells were flow sorted separately. Genomic DNA was extracted from those cells and analyzed for the presence of total HIV-1 DNA, integrated HIV-1 DNA and 2-LTR circles. PCR reactions were performed as described in Table 1. The number of cells or DNA copy number for the standard curves is indicated on top of each lane. Negative controls include cells without HIV-1 and PCR reactions with no template (NTC). PCR products were identified on 2% agarose gels stained with ethidium bromide. PCR amplification of the reference gene β-Actin was used as loading control. Representative of four independent experiments.
HIV-1 gene expression in renal epithelial cells is sensitive to AZT and Ritonavir
HK2 cells were pre-treated with 100 μM of the reverse transcriptase inhibitor, AZT, before and during co-culture with infected T cells, as well as after T cell removal, to demonstrate that GFP expression by infected renal epithelial cells was the consequence of HIV-1 reverse transcription. Interestingly only the HG population was inhibited by the drug while GFP expression by the LG population was not affected (Figure 2a). We next examined whether the transfer of HIV-1 from infected T cell to renal epithelial cells required the formation of mature infectious virions. CEM T cells were pre-treated with 1μM of the protease inhibitor (PI) Ritonavir before HIV-1 infection and throughout the co-culture with HK2 cells. As shown in Figure 2b, there was no HG population when T cells were infected in the presence of Ritonavir suggesting that maturation of the virus particles was required. Interestingly, there was a LG population in the presence of ritonavir consistent with at least some of the expression being mediated by transfer of viral RNA.
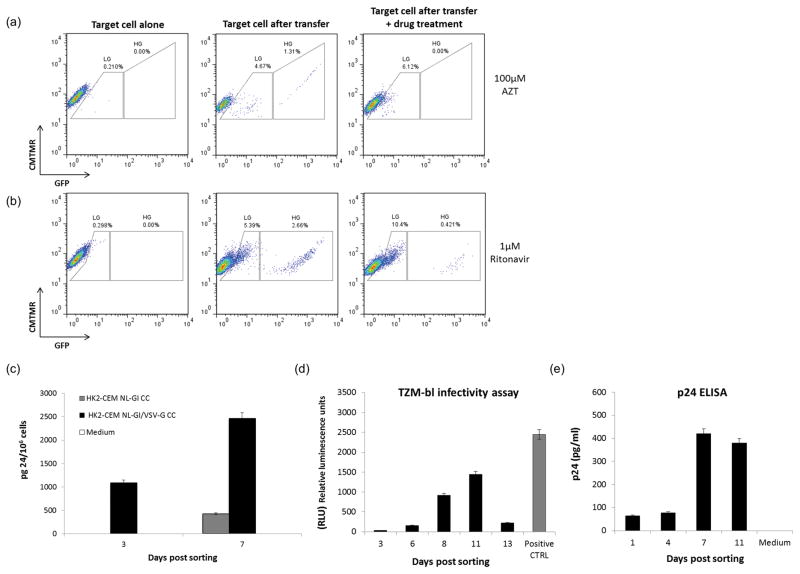
Figure 2. Antiretroviral drug inhibition and viral particles release from infected RTE.
(a) HK2 cells were co-cultivated with infected CEM T cells in presence of 100 μM of AZT. (b) CEM T cells were pre-treated with 1 μM of Ritonavir before viral infection. Ritonavir was added again at the moment of the infection and throughout the co-culture with renal epithelial cells (c) GFP positive HK2 derived from a co-culture between T cells infected either with NL-GI (grey histogram) or with a VSV-G pseudotyped NL-GI (black histogram), were flow sorted and p24 measured in the cell supernatants. Supernatants were collected at day 3 and 7 post sorting (fresh medium was added to the cells after supernatant removal at day 3) and analyzed for p24 as described in Materials and Methods. The amount of p24 is expressed per 106 cells. (d) Supernatants from flow sorted GFP positive HK2 were collected at different time points and analyzed for their ability to infect TZM-bl cells as described in Material and Methods. e) p24 measurement in supernatants from flow sorted GFP positive HK2 over time. Representative of three independent experiments.
Infected renal epithelial cells produce new infectious particles
To determine if HIV-1 integration in renal epithelial cells results in new proviral synthesis, we collected supernatants from sorted GFP positive HK2 at different time points in order to analyze the presence of p24 by ELISA. Infected HK2 cells were derived from two different co-cultures, one using CEM T cells infected with NL-GI virus as donor cells and the other using CEM T cells infected with a VSV-G pseudotyped form of the same virus. Supernatants collected at day 3 post sorting were positive only with the VSV-G co-culture, but both became positive at day 7 (Figure 2c), consistent with new viral particle production over time. We repeated the same experiment collecting the supernatants/cell-culture medium at several time points to test their infectivity in the TZM-bl luciferase assay. The optimum rates of infection were obtained with the supernatants collected at day 8 and 11 post sorting consistent with the higher amount of viral p24 detected by ELISA (Figure 2d–e).
Bidirectional passage of HIV-1 between T cells and renal epithelial cells
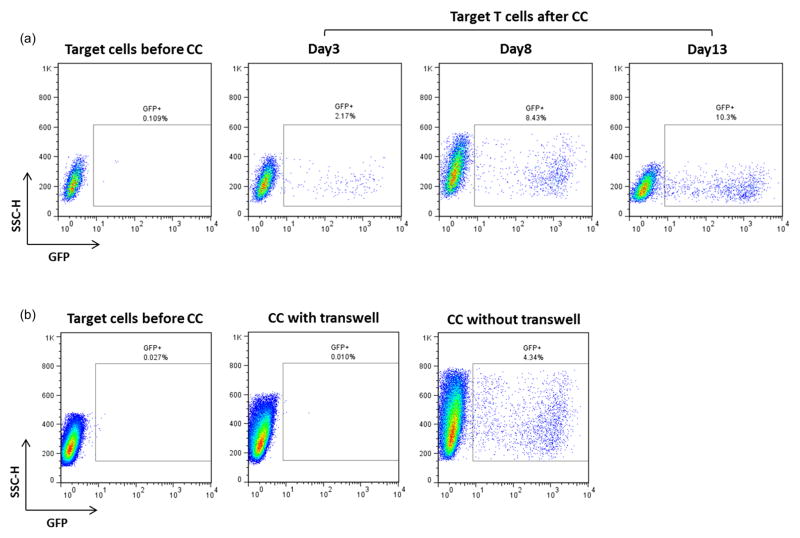
We then evaluated if infected renal epithelial cells can transfer the virus back to T cells. Infected HK2 cells were co-cultured overnight with uninfected CEM-SS T cells. CEM-SS T cells were chosen for this assay because they lack the cellular restriction factor APOBEC3G, and therefore are highly permissive to HIV-1 replication [25]. Target T cells were labeled with CMTMR to distinguish them from donor epithelial cells. At day 3 post co-culture, T cells were analyzed by flow cytometry and about 2% of them acquired HIV-1 from infected HK2. HIV-1 infection was amplified over time, as shown by the increasing number of GFP positive cells by day 8 and 13 post co-culture (Figure 3a). We also evaluated if T cell infection is cell-to-cell contact dependent, or if it is mainly due to the cell free virus contained in HK2 supernatant. To assess that, T cells were added on top of infected HK2 in fresh medium with and without a transwell membrane (0.4μm pore size) between the two cell populations, in order to block cell-cell contact. As shown in Figure 3b, HIV-1 transfer from infected HK2 cells to T cells required cell-to-cell contact, confirming the higher efficiency of cell-to cell viral transmission compared to cell free virus infection.
Figure 3. HIV-1 transfer from infected HK2 to uninfected CEM-SS T cells.
(a) Flow sorted GFP positive HK2 (day 6 post-sorting) were co-cultivated O.N. with uninfected CEM-SS T cells as described in Materials and Methods. At day 3 post co-culture CEM-SS T cells were analyzed for GFP expression by flow cytometry. Only the CMTMR-labeled population was gated for analysis. (b) Flow sorted GFP positive HK2 (day 4 post sorting) were co-cultivated O.N. with uninfected CEM-SS T cells in presence or absence of a transwell membrane (0.4μ pore size). At day 4 post co-culture, CEM-SS T cells were analyzed for GFP expression by flow cytometry. Representative of three independent experiments.
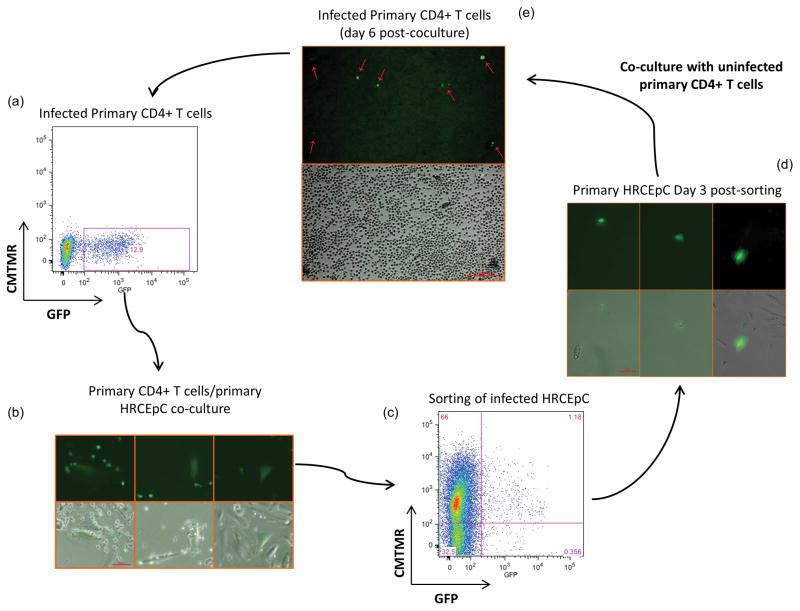
To confirm our observations in primary cells, infected primary CD4+ cells were co-cultivated with uninfected primary HRCEpCs. 48 hours post-co-culture, GFP+ positive HRCEpCs were flow sorted and re-plated. At day 3 post-sorting, uninfected PHA-activated CD4+ cells were added to infected HRCEpCs and co-cultured for 24 hours. Non-adherent CD4+ T cells were then collected, washed with PBS and cultured in presence of 25U/ml of IL-2. As shown in Figure 4, primary CD4+ T cells acquired the virus after co-cultivation with infected primary renal epithelial cells. GFP expression in the flow sorted HRCEpCs persisted over time (Sup. Fig. 1a) and Alu PCR confirmed integration in these cells at day 31 post-sorting (Sup. Fig. 1b).
Figure 4. Bidirectional passage of HIV-1 between primary CD4+ cells and primary renal epithelial cells.
(a) Primary CD4+ T cell were infected overnight with NL-GI and analyzed for GFP expression by flow cytometry at day 3 post-infection; (b) Infected CD4+ T cells from (a) were co-cultured overnight with uninfected primary HRCEpC. Transfer of virus from CD4+T cells to HRCEpC cells was analyzed by fluorescence microscope. (c) GFP and CMTMR double positive HRCEpC from (b) were flow sorted and re-plated (d) GFP expression by cultured HRCEpC from (c) was evaluated at day 3 post- sorting; (e) Uninfected CD4+ T cells were co-cultivated overnight with HIV-1 infected HRCEpC from (d) and analyzed for GFP expression at day 6 post-sorting. (Representative of three experiments performed with PBMC from three different healthy donors).
Prolonged HIV gene expression and cell-to-cell transfer require proviral DNA integration
To test whether proviral DNA integration in renal epithelial cells was required for prolonged gene expression and virus transfer to T cells, HK2 cells were pretreated with 1 μM of the integrase inhibitor Raltegravir before infection with a VSV-G pseudotyped HIV. By day 7 post-infection the raltegravir treated cells completely lost GFP expression while in absence of the drug GFP expression persisted over time (Sup. Fig. 2 a–b). At 2 weeks post-infection, uninfected CEM-SS T cells were added to both infected RTE cultures (with or without Raltegravir). No viral transfer was observed in the co-culture conducted in presence of the integrase inhibitor (Sup Figure 2 c–d). Furthermore, p24 antigen was detected only in the supernatants from the RTE culture conducted in absence of Raltegravir at day 12 post-infection (data not shown). Taken together these data suggest that HIV DNA integration is required for prolonged gene expression as well as for the establishment of a productive infection in RTE.
DISCUSSION
Previous studies have shown that HIV-1 is efficiently transferred to renal epithelial cells after cell contact with infected T cells [11,12]. This study aimed to further characterize the fate of HIV-1 after its internalization into renal epithelial cells. Using the HK2 cells that have been employed in several studies as an in vitro model system for HIVAN pathology [25,26,27], we showed that renal epithelial cells can be productively infected by HIV-1 following co-culture with infected T cells. We identified two distinct populations among the cells that acquired the virus, one expressing GFP at lower levels (LG) than the other (HG). Interestingly, when the co-culture between infected T cells and renal epithelial cells was conducted in presence of a reverse transcriptase inhibitor, only the HG population was affected by the presence of the drug, while the LG population remained mostly insensitive. We found similar results when a protease inhibitor was used to inhibit viral particles maturation in infected T cells. Therefore, infection resulting in high GFP expression requires a mature virus particle and goes through reverse transcription and integration. The analysis of these two GFP-positive populations provides further insights into our previously observed results, showing relative AZT and PI insensitivity of gene expression in renal tubule epithelial cells following cell to cell transfer. Only the less abundant HG population is inhibited, while the LG is not sensitive to the antiretroviral treatment of renal tubule cells and to PI treatment of donor T cells [11]. Since the content of HIV-1 DNA in the LG population is much lower than in the HG one, we hypothesize that the initial GFP expression in the LG population is due to the transfer of viral proteins and/or RNA subsequently lost after days in culture. Since our prior work demonstrated that protein synthesis is required for detection of viral proteins in the RTEs [11], most likely the bulk of this population represents expression from viral RNAs that are subsequently degraded. Given the relatively high integrated copy number in the HG, these results suggest that during cell-to-cell infection a variable number of genomes can be transmitted to the target cells. These findings in renal tubule cells resemble studies in cell-to-cell infection of T cells where infection by multiple variants of HIV was observed during transmission through virological synapse [28]. More interestingly we showed, using both primary and immortalized cells, that when infected renal epithelial cells are co-cultivated with uninfected T cells, the latter became infected and the infection in those cells was propagated over time, delineating a cyclic mechanism for intrarenal virus propagation. The pattern of renal tubule infection in HIVAN biopsies, where all cells of a single tubule appear to be infected, is consistent with cell to cell transfer of the virus to neighboring epithelial cells [2,4,29]. Further studies are required to determine the mechanism(s) involved in intra-tubule spread of the virus. Interestingly, a recent study performed on HIV-1 positive patients who underwent kidney transplantation showed that renal epithelial cells became infected in the transplanted kidney, including podocytes (32%) and/or tubular cells (68%), in 13 out of 19 patients despite non detectable plasma HIV-1 RNA with a similar distribution as previously described in native kidneys. This not only emphasizes the importance of the kidney as a viral reservoir, but suggests that direct T cell to renal epithelial cell infection could be an important route of infection of the transplant [30].
In summary, we showed for the first time that T cell mediated infection of renal epithelial cells results in productive infection. In an envelope independent mechanism, HIV-1 is transferred to renal epithelial cells, followed by reverse transcription and integration resulting in stable and persistent infection of renal epithelial cells. As a consequence of integration, renal epithelial cells produce new infectious virions and transfer newly produced viral particles to permissive CD4+ T cells, in a contact dependent manner. In this scenario, the presence of HIV-1 and the expression of viral genes in epithelial cells may favor the recruitment of additional lymphocytes through induction of chemokine and cytokine expression, potentiating a positive feedback cycle, where viral transfer enhances recruitment of inflammatory cells [31]. These findings further support a role for cells intrinsic to the kidney as a viral reservoir that can fuel local and/or systemic viral rebound. Reservoirs include infected cells that can either harbor transcriptionally silent virus and/or support persistent low level virus replication, often as a consequence of sub-optimal drug concentrations [32]. Further studies are required to investigate the nature of HIV persistence in RTE cells.
Supplementary Material
(a) GFP expression from primary renal epithelial cells that had acquired the virus following co-culture with HIV+ primary CD4+ cells was assessed at day 30 days post-sorting. (b) DNA was extracted from primary HRCEpC cultivated for 31 days post-sorting and analyzed for integrated HIV-1 DNA.
(a–b) Time course GFP expression from HK2 infected in absence (a) or presence (b) of 1μM of the integrase inhibitor Raltegravir. (c) GFP expression from CEM-SS T cells co-cultivated with HIV-infected HK2 cells (with or without Raltegravir) at 2 weeks post-infection. (d) Flow cytometry analysis of CEM-SS from panel (c) at day 6 post co-culture.
Acknowledgments
The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc [33,34].
The authors thank Josh Eudailey for technical assistance with the fluorescence microscope and John Whitesides for assistance with the flow cytometry sorting.
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases [P01DK056492]
Footnotes
Author contributions: M.B. designed and performed the majority of the experiments and wrote the initial draft of the manuscript. B.B. assisted with the microscopic analysis and the initial draft of the manuscript. P.C. and B.K.C. developed the initial study protocol, provided reagents and assisted with the design and analysis of the experiments. A.C. and D.R.M. helped with the design and analysis of the experiments focused on viral quantitation. M.E.K. oversaw the planning and direction of the project including analysis and interpretation of the data and editing of the manuscript. All authors have provided input to the analyses, have participated in the development of the manuscript and have seen and approved the final version.
References
- 1.Wyatt CM, Rosenstiel PE, Klotman PE. HIV-associated nephropathy. Contrib Nephrol. 2008;159:151–61. doi: 10.1159/000125831. [DOI] [PubMed] [Google Scholar]
- 2.Bruggeman LA, Ross MD, Tanji N, Cara A, Dikman S, Gordon RE, et al. Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol. 2000 Nov;11(11):2079–87. doi: 10.1681/ASN.V11112079. [DOI] [PubMed] [Google Scholar]
- 3.Tanji N, Ross MD, Tanji K, Bruggeman LA, Markowitz GS, Klotman PE, et al. Detection and localization of HIV-1 DNA in renal tissues by in situ polymerase chain reaction. Histol Histopathol. 2006;21:393–401. doi: 10.14670/HH-21.393. [DOI] [PubMed] [Google Scholar]
- 4.Marras D, Bruggeman LA, Gao F, Tanji N, Mansukhani MM, Cara A, et al. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med. 2002;8:522–526. doi: 10.1038/nm0502-522. [DOI] [PubMed] [Google Scholar]
- 5.Chen P, Hübner W, Spinelli MA, Chen BK. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007 Nov;81(22):12582–95. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hübner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FY, et al. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009 Mar 27;323(5922):1743–7. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale BM, Alvarez RA, Chen BK. Mechanisms of enhanced HIV spread through T-cell virological synapses. Immunol Rev. 2013 Jan;251(1):113–24. doi: 10.1111/imr.12022. [DOI] [PubMed] [Google Scholar]
- 8.Conaldi PG, Biancone L, Bottelli A, Wade-Evans A, Racusen LC, Boccellino M, et al. HIV-1 kills renal tubular epithelial cells in vitro by triggering an apoptotic pathway involving caspase activation and Fas upregulation. J Clin Invest. 1998 Dec 15;102(12):2041–9. doi: 10.1172/JCI3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ray PE, Liu XH, Henry D, Dye L, 3rd, Xu L, Orenstein JM, et al. Infection of human primary renal epithelial cells with HIV-1 from children with HIV-associated nephropathy. Kidney Int. 1998;53:1217–1229. doi: 10.1046/j.1523-1755.1998.00900.x. [DOI] [PubMed] [Google Scholar]
- 10.Eitner F, Cui Y, Hudkins KL, Stokes MB, Segerer S, Mack M, et al. Chemokine receptor CCR5 and CXCR4 expression in HIV-associated kidney disease. J Am Soc Nephrol. 2000 May;11(5):856–67. doi: 10.1681/ASN.V115856. [DOI] [PubMed] [Google Scholar]
- 11.Chen P, Chen BK, Mosoian A, Hays T, Ross MJ, Klotman PE, et al. Virological synapses allow HIV-1 uptake and gene expression in renal tubular epithelial cells. J Am Soc Nephrol. 2011 Mar;22(3):496–507. doi: 10.1681/ASN.2010040379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatsukari I, Singh P, Hitosugi N, Messmer D, Valderrama E, Teichberg S, et al. DEC-205-mediated internalization of HIV-1 results in the establishment of silent infection in renal tubular cells. J Am Soc Nephrol. 2007 Mar;18(3):780–7. doi: 10.1681/ASN.2006121307. [DOI] [PubMed] [Google Scholar]
- 13.Saphire AC, Bobardt MD, Zhang Z, David G, Gallay PA. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J Virol. 2001 Oct;75(19):9187–200. doi: 10.1128/JVI.75.19.9187-9200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray PE, Liu XH, Henry D, Dye L, 3rd, Xu L, Orenstein JM, et al. Infection of human primary renal epithelial cells with HIV-1 from children with HIV-associated nephropathy. Kidney Int. 1998 May;53(5):1217–29. doi: 10.1046/j.1523-1755.1998.00900.x. [DOI] [PubMed] [Google Scholar]
- 15.Pearce-Pratt R, Phillips DM. Sulfated polysaccharides inhibit lymphocyte-to-epithelial transmission of human immunodeficiency virus-1. Biol Reprod. 1996 Jan;54(1):173–82. doi: 10.1095/biolreprod54.1.173. [DOI] [PubMed] [Google Scholar]
- 16.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, et al. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 17.Mochizuki H, Schwartz JP, Tanaka K, Brady RO, Reiser J. High-Titer Human Immunodeficiency Virus Type 1-Based Vector Systems for Gene Delivery into Nondividing Cells. J Virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990 Apr 20;61(2):213–22. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 19.Pierson TC, Zhou Y, Kieffer TL, Ruff CT, Buck C, Siliciano RF. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol. 2002 Sep;76(17):8518–31. doi: 10.1128/JVI.76.17.8518-8531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cara A, Cereseto A, Lori F, Reitz MS., Jr HIV-1 protein expression from synthetic circles of DNA mimicking the extrachromosomal forms of viral DNA. J Biol Chem. 1996 Mar 8;271(10):5393–7. doi: 10.1074/jbc.271.10.5393. [DOI] [PubMed] [Google Scholar]
- 21.Brussel A, Delelis O, Sonigo P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol Biol. 2005;304:139–54. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- 22.Cara A, Vargas J, Jr, Keller M, Jones S, Mosoian A, Gurtman A, et al. Circular viral DNA and anomalous junction sequence in PBMC of HIV-infected individuals with no detectable plasma HIV RNA. Virology. 2002 Jan 5;292(1):1–5. doi: 10.1006/viro.2001.1243. [DOI] [PubMed] [Google Scholar]
- 23.Lori F, Malykh A, Cara A, Sun D, Weinstein JN, Lisziewicz J, et al. Hydroxyurea as an inhibitor of human immunodeficiency virus-type 1 replication. Science. 1994 Nov 4;266(5186):801–5. doi: 10.1126/science.7973634. [DOI] [PubMed] [Google Scholar]
- 24.Wiegand HL1, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004 Jun 16;23(12):2451–8. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenstiel PE, Gruosso T, Letourneau AM, Chan JJ, Leblanc A, Husain M, et al. HIV-1 Vpr inhibits cytokinesis in human proximal tubule cells. Kidney Int. 2008;74:1049–1058. doi: 10.1038/ki.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder A, Alsauskas Z, Gong P, Rosenstiel PE, Klotman ME, Klotman PE, et al. FAT10: a novel mediator of Vpr-induced apoptosis in human immunodeficiency virus-associated nephropathy. J Virol. 2009;83:11983–11988. doi: 10.1128/JVI.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenstiel PE, Chan J, Snyder A, Planelles V, D’Agati VD, Klotman PE, et al. HIV-1 Vpr activates the DNA damage response in renal tubule epithelial cells. AIDS. 2009;23:2054–2056. doi: 10.1097/QAD.0b013e32833088a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Del Portillo A, Tripodi J, Najfeld V, Wodarz D, Levy DN, Chen BK. Multiploid inheritance of HIV-1 during cell-to-cell infection. J Virol. 2011 Jul;85(14):7169–76. doi: 10.1128/JVI.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross MJ, Bruggeman LA, Wilson PD, Klotman PE. Microcyst formation and HIV-1 gene expression occur in multiple nephron segments in HIV-associated nephropathy. J Am Soc Nephrol. 2001 Dec;12(12):2645–51. doi: 10.1681/ASN.V12122645. [DOI] [PubMed] [Google Scholar]
- 30.Canaud G, Dejucq-Rainsford N, Avettand-Fenoël V, Viard JP, Anglicheau D, Bienaimé F, et al. The Kidney as a Reservoir for HIV-1 after Renal Transplantation. J Am Soc Nephrol. 2014 Feb;25(2):407–19. doi: 10.1681/ASN.2013050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross MJ, Fan C, Ross MD, Chu TH, Shi Y, Kaufman L, et al. HIV-1 infection initiates an inflammatory cascade in human renal tubular epithelial cells. J Acquir Immune Defic Syndr. 2006 May;42(1):1–11. doi: 10.1097/01.qai.0000218353.60099.4f. [DOI] [PubMed] [Google Scholar]
- 32.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A. 2014 Feb 11;111(6):2307–12. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infection by macrophage tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) GFP expression from primary renal epithelial cells that had acquired the virus following co-culture with HIV+ primary CD4+ cells was assessed at day 30 days post-sorting. (b) DNA was extracted from primary HRCEpC cultivated for 31 days post-sorting and analyzed for integrated HIV-1 DNA.
(a–b) Time course GFP expression from HK2 infected in absence (a) or presence (b) of 1μM of the integrase inhibitor Raltegravir. (c) GFP expression from CEM-SS T cells co-cultivated with HIV-infected HK2 cells (with or without Raltegravir) at 2 weeks post-infection. (d) Flow cytometry analysis of CEM-SS from panel (c) at day 6 post co-culture.