Abstract
Autism Spectrum Disorders (ASD) and Schizophrenia (SCZ) are cognitive disorders with complex genetic architectures but overlapping behavioral phenotypes, which suggests common pathway perturbations. Multiple lines of evidence implicate imbalances in excitatory and inhibitory activity (E/I imbalance) as a shared pathophysiological mechanism. Thus, understanding the molecular underpinnings of E/I imbalance may provide essential insight into the etiology of these disorders and may uncover novel targets for future drug discovery. Here, we review key genetic, physiological, neuropathological, functional, and pathway studies that suggest alterations to excitatory/inhibitory circuits are keys to ASD and SCZ pathogenesis.
Keywords: Autism, dendritic spine, E/I imbalance, GABAergic interneuron, glutamatergic, mTOR, NMDAR, schizophrenia
INTRODUCTION
Autism Spectrum Disorders (ASD) and Schizophrenia (SCZ) are two highly heritable spectrum disorders with complicated genetic etiologies but overlapping behavioral phenotypes [1]. ASDs are a group of neurodevelopmental disorders characterized by repetitive behaviors and problems with social interaction and communication while SCZ is a disease afflicting thought, perceptions of reality, affect, and cognition [2]. Genetic linkage and genome-wide association studies (GWAS) have discovered a plethora of disease-associated genes, but few have been widely reproducible because each individual gene likely holds small influence on the overall disease pathogenesis [3, 4] Moreover, given the conditions’ heterogeneous natures, many studies lack the required power to consistently reproduce these weak associations [5]. The behavioral symptoms of ASD and SCZ, however, are intriguingly similar; social interaction deficits, cognitive deficits, emotional processing problems, and executive functions dysfunction are all behavioral features commonly observed in both disorders [6, 7]. In addition, subsets of autism patients have auditory and visual hallucinations, similarly to acutely ill patients with SCZ [8]. Indeed, autism was initially believed to be an early manifestation of SCZ and was once referred to as “schizophrenia syndrome of childhood” [9, 10].
These observations suggest that while the genetics are diverse, the two diseases may convergence upon common pathophysiological mechanisms. One emerging hypothesis maintains that alterations in the ratio of excitatory to inhibitory cortical activity (E/I imbalance) could explain the diseases’ similar social and cognitive deficits. Such imbalances may arise from problems in initial neural circuit formation or maintenance, as many of the genes derived from linkage and association studies code for proteins involved with these processes [4, 11]. In addition, postmortem studies have discovered structural/functional changes in both glutamatergic excitatory and GABAergic inhibitory circuits in individuals with ASD and SCZ [12–14].
The end goal, however, is to provide biological evidence that risk variants impact the pathogenesis of the diseases through specific shared pathways, such as E/I imbalance, so that future therapies can be developed in a directed manner. For this purpose, we will briefly discuss the cellular physiology of E/I balance, review evidence of its dysregulation in ASD and SCZ, and categorize the implicated molecules into common signaling pathways involved with E/I homeostasis.
THE PHYSIOLOGY OF E/I BALANCE
Excitatory/Inhibitory Synapses and Dendrite Arborization
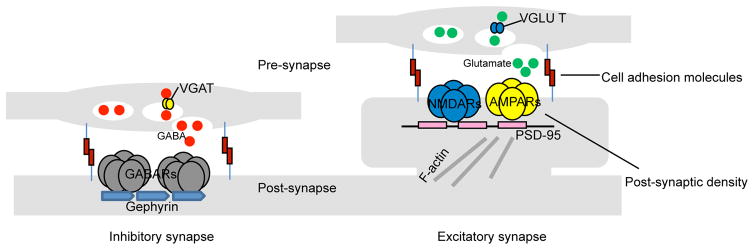
Excitatory and inhibitory synapses, although found on pyramidal and interneuron cells alike, have different architectures. While glutamatergic synapses are found almost exclusively on dendritic spines [2], GABAergic synapses are localized along the dendritic shaft, somata, and axon initial segments [15]. Excitatory synapses contain an electron-dense postsynaptic density (PSD) that directly opposes a pre-synaptic active zone while inhibitory synapses are missing this feature and are more symmetric. Likewise, the two also harbor different molecular constituents, which are often used as markers for identification purposes in immunofluorescent studies. Excitatory synapses maintain pre-synaptic vesicular glutamate transporters (VGLUTs), which pack the excitatory neurotransmitter glutamate into synaptic vesicles for release. The postsynaptic side harbors the postsynaptic density protein-95 kDa (PSD-95), a multimeric scaffold that anchor N-methyl-D-aspartate receptors (NMDARs) and 2-amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propanoic acid receptors (AMPARs) to the surface. Inhibitory synapses have pre-synaptic vesicular GABA transporters (VGATs), which pack the inhibitory neurotransmitter y-aminobutyric acid (GABA) into synaptic vesicles for release, while the adaptor protein gephyrin help cluster y-aminobutyric acid receptors (ionotropic GABAARs and metabotropic GABABRs) and glycine receptors (largely in spinal cord, brainstem, and retina [16]) onto the postsynaptic surface. Both synapse types are held together by trans-synaptic interactions between pre- and post-synaptic cell adhesion molecules [17] (Fig. 1).
Fig. 1.
Simplified architecture of excitatory and inhibitory synapses on pyramidal cells. Pre-synaptic terminals of excitatory synapses release glutamate neurotransmitters that are post-synaptically received by glutamate receptors (NMDARs and AMPARs) on dendritic spines - small membranous protrusion stemming from the dendrite. The tip of each spine contains an electrodense region called the PSD, which is comprised of cytoskeleton anchoring proteins, such as PSD-95, designed to scaffold glutamate receptors (NMDARs and AMPARs) to the spine’s surface and to dock hundreds of signaling molecules (not shown) to the underside. Inhibitory synapses are localized to the dendritic shaft. Pre-synaptic terminals release GABA to be received post-synaptically by GABARs anchored onto the post-synaptic surface by gephyrins (gephyrins also scaffold glycine receptors, which are activated by glycine neurotransmitters - not shown). Noticeably missing is the PSD. Both synapse types are structurally maintained by trans-synaptic adhesions through cell adhesion molecules.
The structure and arborization of dendrites has a profound impact on the processing of neuronal information because it determines the extent of a neuron’s synaptic field. The establishment of the dendritic arbor is highly plastic and involves dendrite extension, addition, elongation, retraction. Much of this early process is dependent on robust synapse formation and hence is particularly sensitive to experience and activity [18]. However, by adulthood, the arbor becomes stabilized and is no longer as dependent on synaptic activity. This mechanistic separation allows mature neurons the ability to fine-tune synaptic connections while retaining a stable dendrite field [19].
Pyramidal Neurons
Pyramidal cells comprise the majority of the neuronal population and are primarily responsible for long-range glutamatergic transmission in the mammalian forebrain. The dendrites of pyramidal neurons are usually regarded as input structures and receive excitatory synaptic contacts from other excitatory neurons on small protrusions called dendritic spines. Thousands of spines stud the pyramidal cell’s apical and basal dendritic branches, increasing the neuron’s receptive surface area and allowing for integration of thousands of excitatory signals to influence the output [20]. Due to this capability to control neuron activity, spines are morphologically regulated to meet the dynamic demands of the growing brain and thus intimately linked to cognitive function [21]. For instance, during development, spines actively participate in synaptogenesis and contribute to the formation of neuronal circuits [21]. Activity-dependent spine maintenance or elimination, on the other hand, is important for the remodeling of established neuronal circuits during post-natal and adolescence periods [22, 23]. These complicated dynamics are regulated by equally complex molecular pathways, involving cytoskeletal remodeling, trans-synaptic adhesion, receptor trafficking, protein translation, ubiquitination, and gene expression [24–26].
GABAergic Neurons
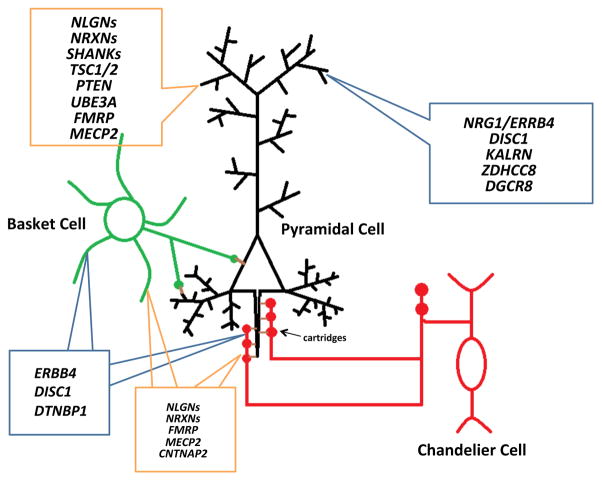
Cortical GABAergic neurons comprise only 10–15% of the entire neuron population but play crucial roles in regulating neuronal excitability due to their synaptic strengths, high firing rates, and abilities to synthesize the inhibitory neurotransmitter y-aminobutyric acid (GABA) by two isoforms of glutamic acid decarboxylase (GAD65 and GAD67) [27]. Unlike pyramidal neurons, which have long-projecting axons and homogenously characteristic spine-studded pyramidal-shaped arbors that can span several cortical layers, cortical interneurons communicate within spatially restricted regions and are relatively aspiny. Moreover, they are categorized into many subpopulations - classified by a combination of morphological, physiological and molecular properties - each of which possesses specialized functions for regulating pyramidal activity [27, 28]. Of these, interneurons possessing the calcium-binding protein parvalbumin greatly influence the generation and timing of a pyramidal cell’s action potentials due to the strategic location of their inhibitory synapses and their fast-spiking electrophysiological properties [29, 30]. For example, the chandelier subtypes electrophysiologically exhibit fast-spiking firing patterns [30] and have long linear axon terminals called cartridges that synapse onto the axon initial segment of pyramidal neurons [31]. Basket subtypes, on the other hand, have identical fast-spiking electrophysiological features [29], but their axons target the cell bodies and proximal dendrites of pyramidal neurons [32]. Thus, both parvalbumin-positive (PV+) subtypes are situated near the axon and have tight control over the excitatory cell’s firing rhythms. Moreover, their biophysical abilities to generate fast synchronized inhibition patterns onto and to receive rhythmic feedback inhibition from pyramidal cells provide the temporal synchrony necessary for generation of network oscillations [33] (Fig. 2). For the purposes of this review, we will focus largely on PV+ interneurons because they play an essential role in proper cognitive function and are noticeably afflicted in ASD and SCZ patients. For a more comprehensive review on interneuron classification and function, we would like to direct the reader to pertinent articles [27, 34].
Fig. 2.
Parvalbumin-containing interneurons regulate pyramidal cell firing. Both basket (green) and chandelier cells neurons (red) contain parvalbumin and have fast-spiking electrophysiological properties. Chandelier cells have long linear axon terminals called cartridges that synapse onto the axon initial segment of pyramidal neurons (black) while basket cell axons target the cell bodies and proximal dendrites of pyramidal neurons. Due to the strategic location of their synapses, both subtypes greatly influence the generation and timing of a pyramidal cell’s action potentials. The contents inside the blue and orange boxes respectively denote key SCZ and ASD-associated genes involved with dendritic spine or parvalbumin interneuron pathology, as discussed in this review.
Excitatory/Inhibitory Balance
Information transfer in the brain relies on a functional balance between excitatory and inhibitory networks. At the level of individual neurons, this balance involves the maintenance of appropriate ratios of excitatory versus inhibitory synaptic inputs [35]. Activation of an excitatory synapse by glutamate depolarizes the cell and increases the likelihood that the cell will fire an action potential while activation of an inhibitory synapse by GABA does the opposite. Thus, the E/I synaptic ratio is critical for keeping the cell’s overall firing patterns within a narrow range [36]. However, this ratio is highly asymmetric across development and between neuronal subtypes. For example, individual pyramidal cell dendrites in the cultured hippocampal cultures have E/I ratios of 2:1 at 14 days but 4:1 at 19 days [37] while hippocampal parvalbumin and calretinin-positive interneuron subtypes have 14:1 and 3:1 ratios respectively [38]. Surprisingly, the ratios themselves are tightly regulated with minimal variance even between branches on a single neuron. Yet how do neurons establish and maintain this long- term internal consistency despite such external variability? In a landmark paper, Turrigiano et al. [39] discovered chronic activity blockade with tetrodotoxin (TTX) and bicuculline increased and decreased the amplitude of miniature excitatory postsynaptic currents (mEPSC) respectively in cultured neurons, while drug washout restored firing rates to control levels. Thus, in response to outside influences forcibly altering activity, compensatory mechanisms are recruited to restore the initial circuit set point. These responses include modulation of excitatory and inhibitory postsynaptic strength, alterations in presynaptic neurotransmitter release probability, and adjustment of intrinsic membrane excitability [40], all of which affect and are influenced by neuronal architecture, including dendritic spines and arbors. Collectively, multiple levels of homeostatic control exist to ensure the neuron’s output is appropriately stable.
Since neural circuits are defined by inter-neuronal communications, output precision in individual cells becomes essential to network function. Cortical interneurons use rhythmic inhibition to create narrow windows for effective excitation, entraining excitatory pyramidal cells to fire certain oscillatory patterns [41–45]. Specific classes of interneurons contribute to the frequency of these particular patterns. For example, the synchronization of neuronal network activity in the human cortex and hippocampus at gamma frequencies (30–80 Hz) is essential for integrating information from different brain regions and is thus relevant for cognition, learning, and memory. Gamma oscillations emerge from the synchronized firing of interconnected excitatory glutamatergic and primarily inhibitory fast-spiking GABAergic PV+ interneurons (Fig. 2), and its power (i.e. amplitude) is modulated by the E/I balance at distinct synaptic sites in the circuit and the intrinsic excitable properties of the neurons [46]. Given the dynamic variability of early postnatal brain development and its activity-dependent shifts, such internal consistency - from single synapses to entire circuits - requires tight control and is essential for proper circuit formation and adaption to the onset and maturation of sensory input. Genetic perturbations to synaptic homeostatic mechanisms, if uncorrected, can serve as pathophysiological initiation points that lead to network destabilization, which in turn can impact behavior. To illustrate this point, Bateup et al. [47] showed loss of TSC1, a gene encoding a regulator of mTOR signaling (see mTOR and ASD) in hippocampal cultures resulted in a primary decrease of inhibitory synaptic transmission, followed closely by a futile secondary homeostatic response, and concluded with an overall increase in network hyperexcitability. Moreover, optogenetic manipulation of specific excitatory and inhibitory circuits directly caused changes in social and cognitive behavior in mice [48]. Thus, circuits tend to be malleable but vulnerable during postnatal development; notably, several neurodevelopmental disorders, such as ASD and SCZ, manifest during this plasticity period [49, 50].
NEUROPATHOLOGICAL FEATURES OF SCHIZOPHRENIA
One of the defining neuropathological features of SCZ is gray matter loss [51–53]. Postmortem studies have examined brain regions showing the highest indices of gray matter loss and have consistently found reductions in spine density in these parts. Intriguingly, these areas have also been associated with functions which are perturbed in SCZ. For example, the dorsolateral prefrontal cortex (DLPFC) is critical for working memory function and schizophrenic individuals show reduced activity of this region during working memory tasks. Spine loss in the DLPFC has been reproducibly reported, particularly in layer 3 neurons [54]. Moreover, there is also a profound reduction in spine density on pyramidal neurons in the primary auditory cortex, which could explain why schizophrenics suffer from auditory hallucinations [55]. Finally, reductions in hippocampal volume and reduced spine density on CA3 dendrites in SCZ [56, 57] could be the physiological reason why patients have problems with memory and spatial learning. Collectively, these studies reveal strong associations between brain region-specific loss of gray matter, reduced spine density and functional hypoactivity in SCZ.
Dysfunction of DLPFC inhibitory PV+ basket cells is also thought to play a major role in SCZ’s working memory deficits. Coordinated firing of DLPFC pyramidal cells at the gamma frequency between a stimulus cue and behavioral response is essential for accurate working memory and relies largely on fast spiking PV+ GABAergic interneurons [58]. For example, PV+ neurons in the monkey DLPFC are active during the delay period of working memory tasks [59], and are responsible for spatial tuning of neuronal responses during this period [60]. In addition, injection of GABA antagonists into the DLPFC disrupts working memory [61]. Fittingly, gamma-frequency oscillations are abnormal in the prefrontal cortex of patients with SCZ who are performing working-memory tasks [45, 62, 63]. Interestingly, multiple postmortem studies have reproducibly found lower levels of parvalbumin mRNA and GAD67, the principal synthesizing enzyme for GABA, in DLPFC PV+ neurons, but found the absolute number of PV+ cells unchanged, suggesting a functional rather than a numerical deficit [64, 65]. Furthermore examination of PV+ chandelier axon terminals [66] revealed selective reduction of GAT1 - a GABA reuptake transporter - and upregulated levels of the GABAA receptor α2 subunit in the axon initial segments of pyramidal neurons, possibly as a compensation to the lowered GABA levels [67]. In conclusion, these findings suggest hyper-excitability and gamma wave disruption in particular brain regions can be etiological contributions of SCZ.
SCHIZOPHRENIA AT THE GLUTAMATERGIC CIRCUIT
As spiny synapses are the basic units of excitatory neural circuits and are intimately involved in neurotransmitter signal transduction, dendritic spine dysfunction may play an important etiological role in SCZ. Interestingly, the strongest results from genetic studies have been haplotypes, single nucleotide polymorphisms (SNPs), and de novo mutations associated with genes regulating neuronal excitability, synaptic structure, and plasticity [3, 5, 68]. Here, we will summarize recent findings on some of the most prominent ones and explore their roles in spine dynamics.
Neuregulin1/ErbB4
NRG1 and ERBB4 are two of the most reproducible SCZ candidate genes [69–76]. Neuregulins (NRGs) are trophic factors that can exist in membrane-bound or soluble forms [77]. ErbB4 is a post-synaptic tyrosine kinase receptor that predominantly binds to soluble NRG1 (type III), through NRG1’s epidermal growth factor-like domain [77, 78]. This interaction leads to stimulation of ErbB4’s tyrosine kinase activity and downstream changes leading to alterations in synaptic structure and function.
NRG1 and ErbB4 are expressed at the excitatory synapses [79–83] and regulate spine structure and function (although ErbB4’s role is more questionable, see next paragraph). Long-term NRG1 treatment increases pyramidal neuronal spine density and the preponderance of spines with mature phenotypes [84] and mice deficient in NRG1 (type III) show reductions in spine density in hippocampal neurons [85]. Similarly, ErbB4 over-expression increases spine density, area, and excitatory synaptic transmission while ErbB4 knockdown reduces spine density and size [86]. In addition, mice lacking ErbB4 in the CNS show reduced spine density in both the hippocampus and cortex and exhibit schizophrenia-related behavioral phenotypes [84].
Perplexingly, ErbB4 is found at much higher levels in interneuron excitatory synapses - especially belonging to the PV+ interneuron subtype - and a recent study used in vivo techniques to directly demonstrate ErbB4’s specificity in controlling cortical GABAergic circuit development [87]. Thus, it is possible that that ErbB4’s role in spines is indirect or an artifact of experimental conditions (see ErbB4 in Schizophrenia at the GABAergic Circuit for further discussion of this discrepancy).
Several SCZ SNPs have caused functional alterations in NRG1 processing, providing an additional level of validity to the NRG1/ErbB4-SCZ hypothesis. For instance, a SCZ-associated SNP in the transmembrane domain of NRG1 leads to disruptions in gamma-secretase dependent cleavage of the protein, which in turn reduces NRG1-mediated intracellular signaling and abolishes NRG1-regulated growth of pyramidal dendrites [88]. In addition, the second SCZ-associated polymorphism within the original HAPICE ‘at risk’ Icelandic haplotype [69] has been shown to transcriptionally regulate NRG1 III levels [89], thus providing a novel mechanism through which this haplotype may increase risk for SCZ.
DISC1
DISC1 was originally identified through a disruption in its open reading frame in a large Scottish pedigree [90] and later verified to be a legitimate SCZ risk gene through polymorphisms and frame shift mutations in other SCZ-afflicted lineages [91]. Further genetic studies have also associated DISC1 with bipolar, depression, and autism, making it an important psychiatric vulnerability gene [92–97].
DISC1 is a scaffold protein intimately involved with hippocampal development [98–100] and is found abundantly at the spines [101], where it facilitates the formation of protein complexes with its coiled-coil rich C-terminal domain. In SCZ patients carrying high risk DISC1 SNPs, DISC1-interacting protein expression levels, but not DISC1 mRNA levels, are drastically reduced [102], suggesting that DISC1 function might be affected in SCZ. Disruption of DISC1’s ability to scaffold proteins in spines would be expected to have deleterious consequences on spine morphogenesis.
Indeed, long-term DISC1 knockdown leads to reduced spine area in cortical neurons [103]. These effects have been hypothesized to be due to DISC1’s interaction with various regulators of spine morphogenesis. Recently, the Rac-GEF Kalirin-7 has been identified as a DISC1 interacting partner and has been shown to directly regulate DISC1’s effects on spine morphology through Rac1 [103]. Kalirin loss strongly correlates with spine loss in layer 3 prefrontal cortex neurons in postmortem schizophrenia samples [104]. Finally, a KALRN-knockout mouse shows severe reductions in spine density in the frontal cortex and schizophrenic-like behaviors during adolescent years [105]. This is interesting given the onset of SCZ symptoms in adolescence in humans, and points to a tight association between the onset of spine loss and the onset of behavioral impairments in these animals. Taken together, these evidence point to DISC1 as a facilitator in spine function and suggests a role of small GTPases in SCZ spine pathology.
22q11 microdeletion
The 22q11 microdeletion syndrome is the most common copy number variant (CNV) associated with SCZ and accounts for up to 1–2% of the cases [106]. Primary hippocampal neurons from mice engineered to carry hemizygous deletion of the 1.3-Mb orthologous chromosomal region (Df(16)A+/−) showed reduced spine density and sizes [107]. Interestingly, loss of either of two genes within this region (ZDHHC8 and DGCR8) was sufficient to impair spine and dendrite morphology [106, 107]. ZDHHC8 is a palmitoyl transferase which palmitoylates PSD-95; its loss results in reduced spine density and simpler dendrites, and its replacement into Df(16)A+/− neurons rescued spine and dendrite deficiency [107]. DGCR8 is involved in miRNA processing, and its loss results in smaller spines and simpler dendrites [106].
SCHIZOPHRENIA AT THE GABAergic CIRCUIT
Synchronization of cortical and hippocampal neural networks at gamma frequencies (30–80) Hz is important for proper cognition, memory, and learning [108], but are perturbed in schizophrenic patients [58, 109, 110]. Gamma oscillation amplitudes are reduced in patients with SCZ [111, 112], and multiple postmortem studies show schizophrenic adults have lower levels of GAD67 in DLPFC PV+ neurons [65, 113, 114], suggesting physiological impairment of these interneurons may disrupt working memory circuits in SCZ.
ErbB4
Through multiple mRNA studies, it has been established that ErbB4 is expressed predominantly in GABAergic cells in the neocortex and hippocampus [115–119]. Immunofluorescence studies using specific ErbB4 antibodies have pinpointed ErbB4 to be distributed in somatodendritic regions of several interneuronal subtypes in the neocortex [120, 121] and hippocampus [122, 123] of rodents and primates. More specifically, ErbB4 is expressed in nearly all PV+ interneurons in the neocortex [120] and over half in the hippocampus [123]. PV+ interneurons control E/I balance by either targeting the soma (basket cells) or the AIS (chandelier cells), as both locations have tremendous influence on the amplitude of the excitatory action potential.
Recently, it was demonstrated that perfusion of hippocampal slices with small doses of NRG1 led to increases in kainate-induced gamma oscillations. This effect was ErbB4-dependent, as it was blocked with an ErbB4 antagonist and was absent in ERBB4-knockout mice [123]. Similarly, mice harboring full or PV+ targeted ablation of ErbB4 exhibit reductions in kainate induced gamma oscillation power and exhibit severe SCZ-like behaviors [123–125].
A pressing question that arises from these results is if ErbB4 acts pre- or post-synaptically to regulate network activity. While ErbB4’s presence in the somatodendritic components of PV+ cells has been widely replicated, its role at the GABAergic pre-synaptic terminals has been disputed. Several electrophysiological studies show evidence of ErbB4’s presynaptic properties. For example, NRG1 treatment led to GABA release in basket cell terminals, purportedly by the activation of ErbB4 by NRG1 [124]. Moreover, PV+ chandelier cells harboring an ErbB4 conditional mutation were found to have decreases in miniature inhibitory postsynaptic potentials onto excitatory cells [124]. Oddly enough, other immunofluorescence studies using specific ErbB4 antibodies were unable to visually localize ErbB4 to the presynaptic terminal (possibly due to antibody or fixation issues) innervating either the soma or axon initial segment of pyramidal neurons in the hippocampus or frontal cortex of rodents and primates [120, 121].
Such discrepancy may be explained by another theory: ErbB4 is found post-synaptically on dendrites of PV+ interneurons and modulates network activity through the control of glutamatergic feedback. Indeed, targeted ablation of GluAs subunits at post-synaptic GABAergic sites produces intriguingly similar reductions in kainate-induced gamma oscillations [126]. Moreover, the C-terminus tail of ErbB4 directly interacts with PSD-95 [82, 83] and accumulates at synaptic puncta on inhibitory neurons [123, 127, 128]. Ultrastructural analysis in CA1 interneurons using immunoelectron microscopy revealed abundant ErbB4 expression at, and adjacent to, glutamatergic post-synaptic sites on interneurons [128].
These disparate observations were reconciled in vivo: an elegant set of experiments demonstrated that: 1) ErbB4 is expressed both in the axon terminals and post-synaptic excitatory synapses of PV+ chandelier and basket cells, but not in pyramidal cells. 2) ErbB4 cell-autonomously regulates the formation of excitatory synapses on these inhibitory subtypes and the formation of inhibitory synapses from PV+ subtypes onto neighboring pyramidal cells. 3) ErbB4 is not necessary for excitatory transmission between pyramidal neurons [87].
DISC1/Dysbindin
Although DISC1’s role on spine morphogenesis in excitatory neurons is well established, several lines of evidence have also suggested its requirement for proper cortical PV+ interneurons functioning. For instance, mutant mice that overexpress a dominant negative form of DISC1 have reduced PV+ immunoreactivity in the prefrontal cortex, suggesting DISC1 is necessary for proper functioning of GABAergic cortical interneurons [129]. Likewise, PV+ interneurons in transgenic mice expressing truncated or missense mutated DISC1 are reduced and mislocalized [130, 131].
Dysbindin is another SCZ susceptibility gene and is a possible binding partner to DISC1. Dysbindin is involved with intracellular trafficking and is downregulated in the prefrontal cortex and hippocampus of SCZ patients [132, 133]. DTNBP1-knockout mice have reductions in the excitability of cortical and striatal PV+ interneurons, which in turn lead to hyperexcitable pyramidal cells [134].
NEUROPATHOLOGICAL FEATURES OF ASD
Early brain overgrowth and neuronal hyperexcitability are two well-established phenotypes in ASD [135, 136]. About 25–30% of autistic children experience excessive increase in brain size between the first and second year of life when compared with healthy controls [137] while 30% struggle with epilepsy [138]. One possible biological mechanism connecting the two phenotypes is increased spine density, as recent evidence examining post-mortem ASD human brain tissue revealed an increase in spine density on apical dendrites of pyramidal neurons from cortical layer 2 in frontal, temporal and parietal lobes and layer 5 in the temporal lobe [14]. Furthermore, these trends are also observed in tissue from individuals with diseases co-morbid with autism. For instance, the fragile X brain is characterized by macrocephaly, elevated spine density and elongated, tortuous spine morphologies [139]. However, other studies have revealed contradicting findings: several case studies have found smaller neurons and simplified dendrite morphology in the limbic system, consistent with a curtailment of developmental maturity [140, 141]. While these discrepant results must be taken with caution due to lack of large samples (only around 120 postmortem ASD brains have been studied since 1980) and closely matched control groups [142], they also suggest a degree of heterogeneity in the autistic brain, which may be due to brain-region dependent changes caused by alterations in region-specific genetic factors [143]. Indeed, fMRI studies have consistently reported patterns of long-distance “under connectivity” with short-range “over connectivity” in brains of ASD adult patients [144].
Evidence for alterations in GABAergic circuits in ASD also come from postmortem studies showing significantly reduced GAD65/GAD67 levels in the parietal cortex and cerebellum [145, 146] and alterations in GABAA and GABAB receptors in postmortem brains of autistic subjects [147–149]. These alterations may be the result of widespread changes in GABA innervation and/or release, which may lower the threshold for developing seizures, given the high co-morbidity of ASD and epilepsy [150]. In support of this hypothesis, another study showed that ASD patient brains had lower numbers of PV+ interneurons in the prefrontal cortex [151]. As PV+ interneurons provide strong perisomatic innervations of pyramidal cells, a reduction in its absolute number could explain the aberrant GABAergic transmission and predisposition for epileptic symptoms in autism [152]. Together, these results suggest heterogenous changes in glutamatergic and GABAergic systems in the ASD brain can converge upon an overall increased ratio of excitation/inhibition, which can manifest in epileptic symptoms, macroscopic changes in brain volume, and behavioral alterations.
ASD AT THE GLUTAMATERGIC CIRCUIT
The high degree of heritability of ASD has inspired a great deal of research over potential mechanisms [153]. Multiple studies have identified rare mutations and CNVs in genes encoding synaptic proteins in autistic individuals, supporting the hypothesis that excitatory synapse dysfunction may be important in the etiology of ASD. Although each individual gene may not account for a large percentage of ASD cases, consistent with a polygenic mode, they do provide valuable insight into the molecular pathways important for the diseases’ pathogenesis [4]. For the same reason, functional investigations of individual genes may produce markedly different results in simplified experimental systems. However, in the context of a pathogenic brain, these molecules may actually work in conjunction to collectively disrupt common pathways (see section on Convergent Pathways of ASD and SCZ). Here, for the sake of simplicity, we will discuss the function of individual ASD risk genes in relation to spiny synapses, E/I imbalance, and behavioral phenotypes.
Neurexins/Neuroligins
Neuroligins (NL1-4) are a group of postsynaptic cell adhesion proteins [154] which interact with presynaptic Neurexins (Nrxns 1–3) to form trans-synaptic adhesions [155–158]. Knockout mice lacking all three α-Nrxns or NLs 1–3 show little change in total number of synapses but exhibit severe synaptic transmission phenotypes [159, 160], thereby implicating Nrxn-NLs in synaptic function, but not formation, in vivo. On the other hand, in vitro co-culture assays showed non-neuronal cells expressing NLs can trigger presynaptic formation on neighboring neurons and vice versa [161, 162]. These two disparate results were reconciled when Kwon et al. [163] showed that synapse formation in vivo was specifically dependent on transcellular differences in NL1 levels between neighboring neurons, suggesting that nascent neurons compete for limited presynaptic neurexin ligands, with cells expressing more NL1 forming synapses more readily. This theory explains why total genetic ablation of NLs in mice leads to an overall null effect while overexpression in cultured cells results in synapse formation [164]. While it is not yet known if other NL isoforms follow the same trend, it is likely Nrxn-NLs are important for both synaptic formation and function in vivo.
Both groups have been genetically linked with ASD: point mutants in NLGN3, NLGN4, or NRXN1 [165–169] truncations of NLGN4 or NRXN1 [170–172], chromosomal rearrangements of regions harboring NRXN1, NLGN1, or NLGN2 [173, 174], and a promoter mutation in NLGN4 [175] have been identified. More recently, a genome-wide copy number variation analysis also implicated NLGN1 among several candidate genes in ASD susceptibility [176].
Multiple functional studies have implicated specific neurexin and neuroligins isoforms as key molecules for excitatory synaptic activity and normal behavior. Each of the three Nrxn genes encodes two principal types of neurexin proteins, α and β, each of which differs in length, domain composition, and number of splice variants. Therefore, the sheer number of isoforms, along with the lack of high-affinity antibodies and difficulty of studying presynaptic function, make Nrxns incredibly difficult to characterize [154]. Despite these challenges, culture studies have shown that particular splice variants influence the formation of specific synapses [158, 177–180]. In addition, knockout mice have been generated only for α-Nrxn isoforms, while no data are yet available for β-Nrxns; α-NRXN1-knockout mice have hippocampal E/I imbalance due to decreased spontaneous excitatory post-synaptic currents and decreased prepulse inhibitions, and also exhibited increases in grooming behaviors [181]. In vitro and in vivo studies show NL1, NL3, and NL4 localize to excitatory synapses [182–184]; here, Nrxn-NL trans-synaptic signaling recruits NMDARs and AMPARs to the synapse surface, possibly via intracellular mechanisms centered around the PSD-95 scaffold [185], although this has been disputed (see NMDAR Hypofunction and ASD section). Not surprisingly, over-expression of these NL isoforms increased excitatory synapse number and spine density in cultured neurons, while the NL3 Arg451Cys, NL4 D396X, and NL4 R87W variants - missense mutations found in affected in autistic members of several families - prevented this effect due to surface trafficking defects; conversely, knockdowns lead to excitatory synapse loss [163, 167, 186–188]. The mechanisms behind NL’s spinogenic effects are unclear, but may involve activity-dependent proteolytic cleavage [189, 190] and CaMKII phosphorylation [191]. Likewise, a NLGN1-knockout mouse exhibited deficits in spatial memory and increased repetitive behavior as a result of decreased long-term synaptic plasticity in area CA1 of the hippocampus and dorsal striatum [192]. Conversely, a transgenic mouse overexpressing NL1 also had disruptions in memory acquisition and LTP, but this time as a result of increased numbers of excitatory synapses [193]. NLGN3-knockout mice have reduced ultrasound vocalization, deficits in social novelty preference, and brain volume reduction while NLGN4-knockouts exhibit highly selective deficits in social interactions and communication [194, 195]. Unfortunately, investigation into the neuronal architecture has been lacking in these two models. However, Arg451Cys NL3 knock-in mice revealed a large increase in NMDAR and AMPAR-dependent excitatory transmission in the hippocampus, which were accompanied by decreases in spine size and increases in dendritic branching in the stratum radiatum of CA1 [196].
In summary, neurexins and neuroligins may contribute to ASD pathogenesis through its control of synaptic function; deregulation may result in neurodevelopmental imbalances of synaptic transmission and may progress to behavioral changes.
SHANK Family
The Shank family (Shank 1–3) is a group of scaffold proteins that organize extensive protein complexes at the postsynaptic density of excitatory synapses. All three members of the Shank family of have been linked with ASD susceptibility: a large number of de novo mutations in SHANK3 [197–199], in SHANK2 [200], and in SHANK1 [201] have been identified in individuals with ASD. This genetic association provides an immediate link between synaptic dysfunction and ASD pathophysiology and thus led to the creation of numerous knockout mice [202].
SHANK1-knockout mice have small dendritic spines, weakened synaptic transmission [203], and defects in social communication [204]. Mice with heterozygous or homozygous disruption of SHANK3 have self-injurious repetitive grooming, deficits in social interaction, alterations in learning and memory formation, and defects in synaptic transmission [205–207]. Finally, the SHANK2-knockout mice, like their SHANK3-knockout counterparts, show abnormalities in behavior tests, impairment in social activities, hyperactivity, and defects in synaptic transmission [208, 209].
The behavioral defects of SHANK3 mutant mice correlate with impaired basal synaptic transmission in CA3-CA1 connections, reduced GluR1 clusters and protein levels in the hippocampus, and major changes in striatal and corticostriatal synapses [205–207]. Interestingly, Shank3 interacts with the NLs through PSD95 and GKAP, suggesting the importance of this entire complex in ASD pathology [154]. Overall, these studies demonstrate that the SHANK genes cause alterations in excitatory synaptic morphology and signaling, as well as changes in behavior characteristics, suggesting they are good animal models for the study of synaptic pathology in ASD.
Syndromic Autisms
Studying syndromic autisms - monogenic disorders that are co-morbid with ASD - can shed light on ASD etiology because their phenotypes are entirely attributed to mutations on individual or particular sets of genes [210]. For example, individuals harboring mutations of tuberous sclerosis proteins 1 and 2 (TSC1, TSC2) or the phosphatase and tensin homolog (PTEN) gene are frequently diagnosed with autism [211, 212]. Fascinatingly, each of these proteins regulates synaptic structure [213–215]. PTEN deficiency results in hypertrophy of dendritic arbors and increased spine density [213, 216] while TSC1 or TSC2 loss causes enlarged spines [214]. Fragile X syndrome results from transcriptional silencing of the FMR1 gene, which encodes for the transcription regulator FMRP; loss of FMRP causes an upregulation in global dendritic translation rates that may contribute to the elevated spine density observed in the brains of affected individuals [217]. Similarly, MeCP2, a transcriptional regulator that is mutated in Rett syndrome, controls the function of excitatory synapses and spine morphology in an activity-dependent manner [218]. Lastly, maternal duplications of chromosome 15q11-q13, a region that encompasses the Angelman syndrome gene UBE3A, are associated with autism [219]. UBE3A encodes an E3 ubiquitin ligase whose loss reduces dendritic spine density and length in cerebellar and hippocampal pyramidal neurons [220, 221], suggesting a potential link between Angelman syndrome, autism and altered synaptic structure. These disorders, which have an autistic phenotype, can be useful in determining relevant molecular mechanisms in ASD pathogenesis. However, this information must be approached with caution; as these disorders are pathologically and genetically distinct from ‘pure’ autism, the relevance of specific molecular mechanisms and cellular alterations to pure autism may not be entirely accurate.
ASD AT THE GABAergic CIRCUIT
Converging evidence from genetic and neuropathological studies suggest pathological cellular alterations to GABAergic interneurons may play etiological roles in ASD. Below, we summarize animal model studies which mechanistically link ASD-associated genes to GABAergic interneuron function, E/I imbalance, and behavioral deficiencies.
Neurexins/Neuroligins
Autism is thought to arise from functional changes in neural circuitry and to be associated with an imbalance between excitatory and inhibitory synaptic transmission. Functional studies have linked neuroligin 2 (NL2) to proper inhibitory synaptic function [188, 222]. NL2 tightly regulates inhibitory synapse architecture and function by tethering pre-synaptic α-Nrxns or a subset of β-Nrxns with an insert in splice site 4 [158]. This interaction provides a physical link between pre- and post-synaptic membranes [158], regulates pre-synaptic neurotransmitter release, and post-synaptically recruits gephyrin which scaffolds GABAA receptors in precise locations [223]. Hence, due to its central role in synapse assembly, even slight alterations to NL2 can significantly alter E/I balance and result in particular behavioral deficits. More specifically, in vitro NL2 over-expression or knockdown specifically enhanced or decreased inhibitory circuit transmission respectively [188] and a transgenic mouse harboring an extra copy of the NLGN2 gene resulted in increased size of inhibitory synapses and a subsequent reduction in the E/I ratio. Moreover, these mice had stereotypied behavior and impaired social interactions [224]. Interestingly, a small population of NL2 is also endogenously present in excitatory synapses and NLGN2-overexpressing mice also exhibit small but significant changes in excitatory synapse morphology [224], suggesting NL2 may also have some overlapping and redundant functions at the excitatory synapse.
Similarly, while NL3 is predominantly localized to excitatory synapses [184], it is also found to a lesser extent in GABAergic synapses [183]. A knock-in mouse harboring an autism-associated mutation in a conserved amino acid residue (R451C) of NLGN3 was found have increased inhibitory transmission but no changes in excitatory transmission in the somatosensory cortex [225]. These mice, similar to the NLGN2-overexpression mice, also exhibited impaired social interactions. Thus, the R451C mutation may lead to a gain-of-function shift of NL3 activity to the inhibitory synapse, but additional studies unexpectedly revealed a large increase in excitatory transmission in the hippocampus [196]. Therefore, NL3 R451C mutation causes region-specific alterations in either excitatory or inhibitory neural circuits that accompany the ASD-relevant mouse behaviors.
While previous studies pinpoint NL4 to the excitatory synapse, the conclusions were based on over-expression data [167, 184, 186]. More recently, Hoon et al. [226] showed NL4 was endogenously localized to inhibitory glycinergic synapses in the retina and other areas of the CNS, associates with gephyrin but not PSD-95 in vivo, and is important for glycinergic-dependent inhibitory transmission in the retina. This conflicting data, along with the fact that NLGN4-knockout mice are accurate models of certain monogenic heritable ASDs [195], mandates a more extensive investigation of the physiological properties of this protein.
In conclusion, these studies provide evidence that both reduced and enhanced expression of ASD genes such as NLs can intimately affect the balance between excitatory and inhibitory transmission, and hence provides strong evidence that a change in the E/I balance contributes to the pathogenesis of ASDs.
MeCP2
Rett Syndrome is a neurodevelopmental disorder that presents at around 6–18 months with autistic-like deficiencies in speech, social and motor skills - particularly repetitive movements - which results from the functional loss of one copy of the MECP2 gene [227, 228]. MeCP2 is a transcriptional regulator which is highly expressed in GABAergic neurons in the brain [229]; interestingly, epilepsy is co-morbid in 80% of cases of Rett Syndrome, suggestive of an underlying inhibitory transmission problem [230].
MECP2-null mice exhibit most of the neurological phenotypes of Rett syndrome [231, 232], but region-specific deletion of MECP2 can also recapitulate many of these behaviors. For example, mice with specific deletion of MECP2 from forebrain GABAergic-interneurons exhibit repetitive behaviors, increased sociability, cognitive deficits, impaired motor coordination and neuronal hyperexcitability [233]. Moreover, GABAergic abnormalities in the thalamus [234] and medulla [235] occur in MECP2-null mice prior symptom onset.
Given the plethora of MeCP2 binding targets throughout the genome [236], it is particularly difficult to tease apart how MeCP2 specifically impairs inhibitory function [227]. However, it is best to begin with MeCP2-targeted genes that are particularly important in GABAergic neurons. For instance, brain-derived neurotrophic factor (BDNF), which regulates development and maturation of inhibitory circuits in the brain, is a MeCP2 target [237]. Moreover MeCP2 controls the expression of GAD65 and GAD67 in GABAergic neurons in a cell-autonomous manner [233]. Loss of MeCP2 leads to inhibitory neurons containing lower than normal levels of GABA, which may explain the abnormal cellular hyperexcitability observed in these mice [233]. Taken together, these results suggest that MeCP2 function may be more crucial in some inhibitory circuits than others and that behavioral symptoms may be the downstream result of region-specific MeCP2-associated defects in GABAergic neurons.
FMRP
Fragile X Syndrome (FXS) is a monogenetic neurodevelopmental disorder caused by an abnormal expansion of a CGG trinucleotide repeat within the promoter region of the gene FMR1, which prevents expression of the encoded protein FMRP [238]. Around 20% of those with FXS meet formal criteria for an ASD [239–242] and 25% exhibit epileptic symptoms [243], thus implicating FMR1 both as an autism susceptibility gene and a regulator of cortical inhibitory function. Indeed, FMRP is broadly expressed in GABAergic neuron populations [244] indicating that it is involved interneuron function.
This theory is backed up by evidence from the FMR1-knockout mice, which exhibit autistic-like behaviors [245]. Furthermore, knockout mice have significant reductions and drastic lamina redistributions of PV+ interneurons in the neocortex, but not hippocampus, suggesting these effects may involve region-specific gamma oscillation deficiencies [246]. Likewise, other studies reported the reduction of several GABA subunits’ mRNA in the cortex but not in the cerebellum in FMR1-null mice [247, 248] and various other reports discovered region-specific increases or decreases of GAD 65/67 levels [244, 248–251]. Finally, FXS specifically exhibit profound network hyperexcitability in the amygdala - an area of the brain responsible for emotional processing - due to the loss of GAD65 and GAD67 expression and reduced GABAergic synapses [244]. In conclusion, FXS mice are good animal models analyze how FMRP links region-specific GABA dysregulation to autistic symptoms; however, caution must be taken as that FXS is not a “pure” autism and thus cannot entirely be relied upon to understand the latter’s complex pathophysiology.
CNTNAP2
Genetic studies over the past decade have reproducibly associated CNTNAP2 variants with ASD [252–256]. Contactin-associated protein-like 2 (CNTNAP2) is a neurexin cell-adhesion molecule that is highly expressed throughout the central nervous system, with highest expression in the frontal and anterior lobes, striatum and dorsal thalamus [256], which recapitulates the corticostriato-thalamic circuitry known to control higher order functions such as speech and language.
Recent evidence suggests CNTNAP2 regulates neural networks through E/I balance. CNTNAP2 knockdown led to decreases in dendrite arborization and impaired synaptic transmission in mouse cortical neurons [257]. CNTNAP2-knockout mice have reduced numbers of PV+ GABAergic interneurons, as well as neuronal synchrony defects, epileptic seizures, and altered social behavior [258]. Thus, CNTNAP2 loss may lead to impairments in connectivity and reductions in the interneuron population, particularly the PV+ subtype, resulting in a hyper-excitable network that promotes downstream epileptic symptoms and behavioral abnormalities. However, it is unclear if the interneuron phenotype is a result of migratory defects, as CNTNAP2 is expressed at the ganglionic eminences during development [258] and interacts to TAG-1 [259], a protein involved in interneuron migration [260]. Thus further work needs to be done to dissect the relationships of these phenotypes to one another as well as to further tease apart the cellular and neurobiological effects of CNTNAP2 loss on neural networks.
CONVERGENT PATHWAYS OF ASD AND SCZ
SCZ and ASD have been characterized as polygenic disorders, meaning multiple susceptibility genes may work together - possibly along different points of common pathways - to trigger disease onset [3]. Indeed, genetic studies have consistently identified reoccurring CNVs mutual overlapping both disorders. Many of these candidates - such as DISC1, NRXN1, NLGN3-4, SHANK3, and CNTNAP2 - encode for synaptic proteins (NRXN1, NLGN3-4, and SHANK3, see ASD at the Glutamatergic Circuit) or molecules involved with neuronal development (DISC1 [261], CNTNAP2 [258, 262]) and thus support the existence of shared biologic pathways between these two neurodevelopmental disorders. In this section of the review, we will describe the role of two major signaling pathways (through their regulations of neuronal architecture and synaptic function) on E/I homeostasis, explore the functional relationship of risk molecules to these pathways, and discuss how disease-associated disruption of such molecules can disrupt these pathways and contribute to E/I imbalance.
mTOR SIGNALING
The mammalian target of rapamycin (mTOR) is a serine/threonine kinase that responds to environmental signals by controlling cell growth and proliferation [263, 264], synaptogenesis [213], and dendrite [265, 266] and axon growth [213, 267] in neurons via regulation of protein translation and hence plays an important role in brain development. mTOR is controlled by the tuberous sclerosis complex (TSC1/TSC2), a heteromer which binds and constitutively inhibits the GTPase Ras-homolog enriched in brain (Rheb), a direct activator of mTOR. Phosphorylation of TSC1/TSC2 dissociates with and thereby activates Rheb. Upstream kinases - many of which are downstream effectors of extracellular signals - thereby exert control on mTOR through TSC1/TSC2, making the latter a focal point for integration of cellular energy status, nutritional availability, and growth factor signaling. For instance, in the highly conserved PI3K-Akt-mTOR signaling pathway, binding of various extracellular factors (mitogens, trophic factors, and glutamate) to respective receptors lead to the activation of phosphatidylinositol 3-kinase (PI3K) at the plasma membrane. Activated PI3K promotes plasma membrane translocation and subsequent activation of Protein Kinase B (Akt) via production of phosphatidylinositol (3,4,5)- triphosphate (PIP3); meanwhile, PTEN opposes PI3K by depleting PIP3. Active Akt phosphorylates TSC1/TSC2 to disinhibit Rheb, leading to protein translation through mTOR’s (as a member of mTORC1) phosphorylation of S6 kinase (S6K) and eukaryotic initiation factor (eIF-4B) [268]. For a more comprehensive picture of mTOR’s function and other pertinent molecular players and pathways, we direct the reader to a more comprehensive review [269].
Throughout its development, a neuron is faced with changing environmental conditions and must appropriately adapt its energy expenditure for optimal function. For instance, the critical period in early postnatal brain development represents a particularly harsh period of metabolic demand, as neurons undergo neurogenesis, migration, synapse formation, and the establishment of synaptic fields. Individual neurons therefore must perform frenzied anabolic processes to keep up with these rapid changes and mTOR activity becomes essential during this time by controlling key components of homeostasis such as synaptogenesis, spinogenesis, and dendrite arborization [270]. However, sustained periods of high demand can also become periods of enhanced vulnerability if lesions occur in these metabolic pathways. Thus if protein production cannot be in tune with critical developmentally driven shifts, neuronal networks may be pathogenically imbalanced.
mTOR and SCZ
Recent investigations have linked SCZ pathology to the PI3K-Akt-mTOR signaling cascade [268, 271]. Moreover, genetic linkage and association studies have identified AKT as a candidate susceptibility gene related for SCZ [272, 273] and the level of Akt-1 protein and its kinase activity decreased significantly both in white blood cells and in postmortem brain tissue of schizophrenic patients [274]. Repeated treatments of MK-801, a non-competitive NMDAR antagonist known for its psychotomimetic effects, led to modulation of Akt, mTOR, and its downstream targets and induced schizophrenia-like behavioral changes in neonatal rats [275, 276].
Recent studies show DISC1 knockdown of adult-born neurons in the dentate gyrus led to acceleration of dendrite development, network hyperexcitability, and an increase in phosphorylated Akt. These effects were rescued when either NKCC1 - a neuronal co-transporter responsible for depolarizing GABA signaling via chloride influx - or GABAAR γ2 subunit was simultaneously knocked down with DISC1, but enhanced when DISC1 knockdown neurons were treated with GABA agonists [277]. Moreover, it was discovered DISC1 interacts with and suppressed KIAA1212, an Akt signaling enhancer, and rapamycin treatment of DISC1 knockdown neurons prevented enhanced Akt phosphorylation and hypertrophic dendritic growth [261]. Together, these results suggest a specific requirement of depolarizing GABA signaling in DISC1-dependent regulation of Akt/mTOR signaling and dendritic growth during adult neurogenesis. Finally, mice with dentate gyrus-specific DISC1 knockdown exhibited schizophrenic-like symptoms including cognitive deficits and anxiety, but rapamycin injection was able to rescue these defects in a time-sensitive manner [278]. Pathological GABA signaling, therefore, may be indirectly responsible for the hallmark alterations in inhibitory circuits and behavior in SCZ via the DISC1-Akt-mTOR signaling pathway.
NRG-1 induces the phosphorylation of the ErbB4 receptors, which then become docking sites for upstream mTOR molecules such as PI3K [279]. Lymphoblast lines from schizophrenic patients have upregulated expression of the ErbB4 receptor isoform CYT-1, which contains a PI3K-binding site and triggers activation of the PI3K pathway [280, 281]. Further examination of these cell lines show enhanced transcripts for p110δ (a catalytic subunit of PI3K) and dampened PI3K-catalyzed second messenger PIP3 levels [281], suggesting enhanced signaling from the ErbB4 isoform CYT-1 lowers PI3K activity. As Akt is a prominent target of PI3K [282], this study also found targeted inhibition of p110δ increased Akt Thr308 phosphorylation and reversed PPI deficits in a neurodevelopmental rat model of SCZ [281], adding evidence that enhanced NRG1-ErbB4 signaling may diminish PI3K-Akt signaling in SCZ. In corroboration, elevated NRG1-ErbB4 activity has been observed in SCZ postmortem brain tissue [283] while analysis from peripheral blood leucocytes from SCZ patients confirmed a 1.3 fold decrease in Akt levels [274]. Since NRG1-ErbB4 and PI3K-Akt are involved in neural circuit development and synaptic plasticity [87, 284], this discovery may have joined two previously disparate pathways.
Finally, over-expression of dysbindin-1 in cortical neurons has a neuroprotective effect against apoptosis via PI3K/Akt pathway activation while knockdown has the opposite effect [285]. Likewise, dysbindin expression in the brain of schizophrenic patients is reduced [132, 133]. These findings suggest that dysbindin-1 deficits may contribute to the pathophysiology of SCZ by affecting neuronal viability of E/I circuits via regulation the PI3K/Akt signaling cascade.
mTOR and ASD
As mTOR regulates a variety of neuronal functions, including cell proliferation, growth, and synaptogenesis, disrupted network homeostasis via mTOR perturbation has been proposed as a pathophysiology mechanism contributing to autism spectrum disorders due to the disease’s synaptopathology, E/I imbalance, and the observations of macrocephaly and epilepsy in 10–30% of the patients with ASD [286].
Individuals with dominant mutations in TSC1/TSC2 or PTEN - genes encoding repressors of mTOR - are often diagnosed with syndromic forms of ASD and are also co-morbid for epilepsy, suggesting a relationship between mTOR hyperactivity and alterations of network circuits [270]. As such, conditional knockout (as germline deletions lead to early mortality) mouse models consistently produced seizure phenotypes. For example, mice with PTEN ablation from mature CA3 neurons, dentate granule cells, and layer III-V cortical neurons exhibit spontaneous seizures [213]. Similarly, removal of TSC1 throughout the cortex, subcortical gray matter, and hippocampal CA3 and hilar neurons produced mice with spontaneous seizures by day 5 and death within 3–5 weeks [287]. Investigations into the pathophysiological mechanisms of these phenotypes revealed mTOR-dependent alterations in synaptic transmission. Hippocampal TSC1-knockout neurons exhibited mTOR hyperactivity, decreased inhibitory transmission (yet no changes in excitatory transmission), and network hyperexcitability [47] but normal spine density and size [288]. Loss of PTEN hyperactivated mTOR, but surprisingly also enhanced both excitatory and inhibitory neurotransmission in the hippocampus [289] and increased spine density [213]. Thus, despite being functionally similar in some aspects, PTEN and TSC1 may have distinct mechanisms for E/I regulation [289].
It was recently reported loss of FMRP resulted in enhanced mTOR signaling and subsequent increased formation of the eukaryotic initiation factor complex 4F (eIF4F) in a mouse model of FXS [290]. These findings suggest that in addition to its RNA-binding activity, FMRP also plays a role in the regulation of translation initiation and subsequent protein synthesis. Similarly, a MECP2-overexpression mouse also has elevated mTOR signaling [291] while a MECP2-knockout model displays the opposite effects [292]. Indeed, eIF4E - a cap-binding protein that is a member of the eIF4F complex - was confirmed to be hyperactivated via mTOR phosphorylation in fragile-X syndrome (FXS) patients diagnosed with autism [293]. Moreover, transgenic mice harboring knockouts of eukaryotic translation initiation factor 4E-binding protein 2 (4E-BP2) - an eIF4E repressor - or eIF4E overexpression exhibit autistic-like behaviors, E/I imbalance, and increased translation of neuroligins [294, 295]. Thus, while neuroligins mutations have been associated with ASD, these studies provide a framework for this process - namely, the disruption of a synaptic homeostasis process. Taken together, several lines of evidence suggest prominent ASD risk genes converge upon the mTOR signaling cascade to disrupt E/I balance via abnormal synaptic function owing to aberrations in synaptic protein translation.
NMDAR SIGNALING
N-methyl-D-aspartate receptors (NMDARs) are ionotropic glutamate receptors essential for mediating ion flux and hence are clearly positioned to control E/I balance at the synaptic level. Structurally tetrameric, they contain two NR1 obligatory subunits and either two NR2 (A, B, C or D) or two NR3 (A or B) subunits, with the latter two being interchangeable. As a result, NMDARs exist as multiple subtypes - with each having uniquely physiological and pharmacological properties - and are spatially and developmentally regulated and differentially distributed across neuronal subtypes [296]. This complex diversity, coupled with a subtype-dependent permeability to calcium influx, make NMDARs the predominant molecular devices for controlling complex experience-dependent synaptic remodeling and long-lasting synaptic changes, which are crucial for associative learning, working memory, behavioral flexibility or attention [297]. Moreover, NMDAR-associated proteins have key roles in the receptor’s trafficking, stability, subunit composition and function through post-translational modifications such as phosphorylation, ubiquitination, and palmitoylation. Dysregulation of these processes can impact channel function and expression and alter synaptic transmission [298, 299]. Thus, NMDAR dysfunction could be a convergence point in ASD and SCZ pathophysiology and may be responsible for disease progression in both disorders.
NMDAR Hypofunction and SCZ
A prominent hypothesis for SCZ pathophysiology posits that NMDARs on PV+ neurons are hypofunctional, thus resulting in lowered GABAergic signaling, overstimulation of excitatory neurons, and a consequent loss of gamma wave oscillations [58]. This theory is based on observations that NMDAR antagonist (e.g. PCP, MK-801, ketamine) administration in normal subjects produce metabolic, neurochemical, and cognitive deficits almost identical to SCZ patients [300], on functional studies showing NMDAR antagonist treatment specifically decreases GAD67 and parvalbumin in PV+ interneurons [301–303], on post-mortem studies reporting abnormalities in NMDAR density and subunit composition in SCZ patients [304, 305], and on a PV-specific GRIN1-knockout mouse model displaying pyramidal cell hyperexcitability, neural synchrony reductions, and schizophrenic-like behaviors [306]. Supporting this assertion, several SCZ-specific polymorphisms in GRIN1 have been discovered [307, 308] and a recent large study from the Consortium on the Genetics of Schizophrenia verified this connection and also discovered several genes encoding for NR1 interacting proteins associated with the disease [309]. Moreover, other studies have found NMDAR subunits tend to be hotspots for de novo SCZ-associated mutation: one identified two de novo mutations in patients with sporadic SCZ in NR2A (GRIN2A) [310] while others show an over-representation of de novo mutations in glutamatergic postsynaptic proteins associated with NMDAR complexes in SCZ patients [68, 311], leading to the notion that NMDAR signaling could be a point of convergence for various SCZ-associated pathways.
NRG1 and ErbB4 accumulate at synapses in the adult brain, suggesting its signaling is involved with synaptic maintenance or function (see Schizophrenia at the Glutamatergic Circuit and Schizophrenia at the GABAergic Circuit). Indeed, NRG1-ErbB4 activity has been shown to down-regulate LTP via modifications in NMDAR surface trafficking: bath perfusion of mouse prefrontal cortex (PFC) brain slices with NRG1 decreased NMDAR-mediated EPSCs via internalization of the NR1 subunit, with this effect being blocked by ErbB4 inhibitors [312]. ErbB4 is enriched in the PSD and its association with PSD-95 results in enhancement of NRG1-dependent signaling [83]. Similarly, chronic MK-801 treatment in rats enhanced ErbB4’s association with PSD-95 and increased ErbB4’s phosphorylation levels [313]. NRG1’s regulation of NMDAR activity, therefore, may involve ErbB4-controlled postsynaptic mechanisms. Interestingly, the NR2 subunit has a large C-terminus with many putative phosphorylation sites, which affect synaptic targeting and channel physiology [298]. Recent data showed that NRG1-ErbB4 signaling blocks Src kinase-specific phosphorylation of this subunit, leading to suppression of Src-mediated enhancement of NMDAR activity in the mouse prefrontal cortex and hippocampus [314]. Thus, aberrant increases in NRG1-ErbB4 signaling could contribute to E/I imbalance by attenuating NMDAR function via different postsynaptic pathways (e.g. NR1 internalization or hypophosphorylation of NR2 subunits). Supporting this assertion, another study showed schizophrenic patients have up-regulated NRG1-ErbB4 signaling and more pronounced NMDAR attenuation in the PFC as compared with control subjects [283].
DISC1 may regulate NMDAR function through various pathways. It interacts directly with PDE4B, a regulator of cAMP and thus can indirectly influence PKA activity [315]. NMDAR phosphorylation by PKA, in turn, can affect the receptor’s release from the endoplasmic reticulum onto the surface [316]. DISC1 also stabilizes serine racemase (SR), the enzyme that generates D-serine, an endogenous NMDAR co-agonist. Mutant DISC1 cannot bind to SR, leading to the latter’s degradation and resulting in D-serine deficiency and hypofunctional NMDAR neurotransmission [317]. NR1 knockdown mice also exhibit decreased DISC1 levels within synapses [318].
Maintenance of appropriate levels of synaptic NMDARs via a balance between receptor insertion and endocytosis is also necessary for the receptor’s proper function. Specialized endocytic zones involving clathrin-coated pits have been described for glutamatergic synapses and serve to internalize NMDARs [319, 320]; altered dysbindin expression can modify NMDAR surface expression through this mechanism [321]. Moreover, palmitoylation can either promote NMDAR synaptic stabilization or Golgi apparatus sequestration [322]. Interestingly, altered protein palmitoylation was found in a mouse model of 22q11.2 deletion, a high risk factor of developing SCZ [107], but the connection between NMDAR palmitoylation and SCZ has thus far been relatively underexplored.
NMDAR Hypofunction and ASD
A similar NMDAR hypofunction theory has been proposed for ASD [323]. Likewise, genes encoding NR2A, NR2B, and NR2C [255, 324, 325] have also been associated with ASD and differential splicing of NR1 has been reported in an ASD post-mortem study [326]. Pathway analysis of autism associated genes have also revealed reduced NMDAR-mediated neurotransmission [324] and several studies found SCZ and autism patients share common de novo genetic mutations, some of which are involved with NMDAR trafficking [68]. Knockout mice and molecular studies of several ASD risk genes discussed in this review - SHANK3, NLGN1, FMR1, and MECP2 - revealed disrupted NMDAR activity, altered synaptic transmission and autistic-like symptoms.
RNAi knockdown of Shank3 in culture leads to reduction of NMDAR-mediated synaptic current as well as loss of NR1 surface expression. This effect was blocked by an actin stabilizer but mimicked with an actin destabilizer and a Rac1 or PAK inhibitor [327], thereby defining Shank3’s role for NMDAR surface trafficking via the actin cytoskeleton mechanisms. In a similar study, neurons derived from IPS cells generated from Phelan-McDermid syndrome (PMDS) - a syndrome with autistic-like features caused by heterozygous deletions of chromosome 22q13.3, which includes SHANK3 - patients had reduced Shank3 expression and major defects in excitatory transmission due partially to decreased NMDA receptors and reduced excitatory synapses, both of which were rescued with exogenous Shank3 expression [328]. Recapitulating the in vitro data, SHANK3-knockdown mouse exhibited a decrease in NMDA/AMPA ratio in area CA1 of the hippocampus, leading to reduced CA1 LTP, and deficits in hippocampus-dependent spatial learning and memory [329]. Similarly, SHANK2 mutant mice have autistic-like behaviors and decreased functional NMDARs, but both deficits are rescued with NMDAR partial agonist D-cycloserine or through enhancement of NMDAR activity via a positive allosteric modulator of mGluR5 [209].
NLGN1-knockout mice have deficits in spatial learning and memory as well as repetitive behavior, which corresponded with impaired hippocampal LTP and decreased NMDA/AMPA ratio [192]. Moreover, the repetitive behavior was ameliorated with NMDA co-partial agonist D-cycloserine, suggesting NL1 is required for NMDAR-mediated currents, normal LTP function, and specific behavioral tasks. Mechanistic studies show NL1 is selectively localized to excitatory synapses [182] and over-expression leads to clustering of synaptic NMDARs in excitatory synapses, suggesting NL1 recruits and retains NMDAR to postsynaptic sites [330]. This was verified using immunoelectron microscopy images and with a MK-801 wash-out paradigm [330]. NL1 may interact with NMDARs through PSD-95 via its C-terminal PDZ domain [331], and the association between NL1, PSD-95, and other PSD-95-associated proteins such as Shank3 - a scaffold protein which acts to traffic NMDAR to synapses - may be necessary for the precise incorporation of NMDAR into synapses. This theory, however, has recently been in contention - how can NL1-PSD95 specifically recruit NMDARs to excitatory synapses [188] when all NL isoforms have C-terminal PSD-95 binding sites? A recent study showed NL1-PSD95 interactions to be neither necessary nor sufficient for NMDARs recruitment to postsynaptic sites; rather, the extracellular NL1 cholinesterase domain, which differs between isoforms [154], interacts specifically with the NR1 subunit [330].
MECP2-knockout (RTT) mice have abnormal NMDAR incorporation at the synapse. Using whole-brain fractionation techniques, one study found MECP2-null mice to have decreases in overall NR2A/NR2B ratio and NR1 levels at the synapses compared to wildtype [332], which is consistent with attenuated synaptic maturation [333, 334] and fewer overall functional NMDA receptors respectively, supporting the possible involvement of a malfunctioning NMDAR system in Rett syndrome pathophysiology. However, because the technique involved a whole-brain analysis, this does not provide detailed information to which specific brain region these alterations are taking place. Other studies of RTT mice confirm there are regional differences within neural circuits in regards to connectivity [335], which is also consistent with previous postmortem binding studies showing significant alterations in total NMDA receptor complexes only in particular regions of the Rett syndrome brain [336, 337].
Like many developmental disorders, FXS is associated with alterations in synaptic plasticity that may impair learning and memory processes in the brain. FMRP normally binds to NMDAR subunits NR1, NR2A, and NR2B and components of the postsynaptic density, such as PSD-95 and CaMKIIα [338, 339]. Expectedly, FMR1-knockout mice had diminished LTP and lowered NMDA/AMPA ratio in the dentate gyrus (DG) and exhibited defunct context discrimination, a behavior reliant on intact NMDAR function in the DG [340]. DG-specific deficits are also accompanied by a significant reduction in NR1, NR2A, and NR2B subunit levels while treatment with NMDAR co-agonists (glycine or D-serine) independently rescued synaptic impairments in this region [341]. Thus, FMRP differentially affects separate regions of the hippocampus, as the CA1 region of was not impaired in these mice [341].
CONCLUSION
Disorders such as ASD and SCZ have complex genetic etiologies and behavioral phenotypes. However, despite their heterogeneous nature, a significant overlap in symptoms exists, raising the idea that these historically distinct disorders might share overlapping pathogenic mechanisms. The pathways that control E/I balance provide a framework for understanding how different genetic perturbations from two distinct disorders can interact in a convergent way to disrupt excitatory and inhibitory neuron function, neuronal circuit organization, and behavior. Yet how do disruptions of shared mechanisms result in such heterogeneous outcomes? Rather than global impairments, disease-related changes might be subtly specific - affecting only a subset of synapses in a selective group of neurons, for instance. As a result, differential manifestations of alterations in shared cellular substrates might underlie the phenotypic variability, which are then classified as distinct cognitive diseases. Such findings highlight the importance of looking beyond individual genes and into comprehensive mechanisms of molecular convergence. Elucidating the shared functions of newly identified disease-associated genes is a key step to translating genetic findings into clinical applications.
Footnotes
CONFLICT OF INTEREST
The authors certify that they have NO affiliations with or involvement in any organization or entity with any financial interest in the subject matter or materials discussed in this manuscript.
References
- 1.de Lacy N, King BH. Revisiting the relationship between autism and schizophrenia: toward an integrated neurobiology. Annu Rev Clin Psychol. 2013;9:555–87. doi: 10.1146/annurev-clinpsy-050212-185627. [DOI] [PubMed] [Google Scholar]
- 2.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–93. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10(1):40–68. doi: 10.1038/sj.mp.4001558. image 5. [DOI] [PubMed] [Google Scholar]
- 4.Toro R, Konyukh M, Delorme R, et al. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010;26(8):363–72. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Owen MJ, Williams NM, O’Donovan MC. The molecular genetics of schizophrenia: new findings promise new insights. Mol Psychiatry. 2004;9(1):14–27. doi: 10.1038/sj.mp.4001444. [DOI] [PubMed] [Google Scholar]
- 6.Rapin I, Tuchman RF. Autism: definition, neurobiology, screening, diagnosis. Pediatr Clin North Am. 2008;55(5):1129–46. viii. doi: 10.1016/j.pcl.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Cheung C, Yu K, Fung G, et al. Autistic disorders and schizophrenia: related or remote? An anatomical likelihood estimation. PLoS One. 2010;5(8):e12233. doi: 10.1371/journal.pone.0012233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toal F, Bloemen OJ, Deeley Q, et al. Psychosis and autism: magnetic resonance imaging study of brain anatomy. Br J Psychiatry. 2009;194(5):418–25. doi: 10.1192/bjp.bp.107.049007. [DOI] [PubMed] [Google Scholar]
- 9.Kolvin I. Studies in the childhood psychoses. I. Diagnostic criteria and classification. Br J Psychiatry. 1971;118(545):381–4. doi: 10.1192/bjp.118.545.381. [DOI] [PubMed] [Google Scholar]
- 10.Kolvin I, Ounsted C, Humphrey M, McNay A. Studies in the childhood psychoses. II. The phenomenology of childhood psychoses. Br J Psychiatry. 1971;118(545):385–95. doi: 10.1192/bjp.118.545.385. [DOI] [PubMed] [Google Scholar]
- 11.Penzes P, Buonanno A, Passafaro M, Sala C, Sweet RA. Developmental vulnerability of synapses and circuits associated with neuropsychiatric disorders. J Neurochem. 2013;126(2):165–82. doi: 10.1111/jnc.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyaya B, Cristo GD. GABAergic circuit dysfunctions in neurodevelopmental disorders. Front Psychiatry. 2012;3:51. doi: 10.3389/fpsyt.2012.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- 15.Fritschy JM, Brunig I. Formation and plasticity of GABAergic synapses: physiological mechanisms and pathophysiological implications. Pharmacol Ther. 2003;98(3):299–323. doi: 10.1016/s0163-7258(03)00037-8. [DOI] [PubMed] [Google Scholar]
- 16.Lynch JW. Molecular structure and function of the glycine receptor chloride channel. Physiol Rev. 2004;84(4):1051–95. doi: 10.1152/physrev.00042.2003. [DOI] [PubMed] [Google Scholar]
- 17.Gatto CL, Broadie K. Genetic controls balancing excitatory and inhibitory synaptogenesis in neurodevelopmental disorder models. Front Synaptic Neurosci. 2010;2:4. doi: 10.3389/fnsyn.2010.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu GY, Zou DJ, Rajan I, Cline H. Dendritic dynamics in vivo change during neuronal maturation. J Neurosci. 1999;19(11):4472–83. doi: 10.1523/JNEUROSCI.19-11-04472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koleske AJ. Molecular mechanisms of dendrite stability. Nat Rev Neurosci. 2013;14(8):536–50. doi: 10.1038/nrn3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9(3):206–21. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- 21.Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10(9):647–58. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 22.Zuo Y, Lin A, Chang P, Gan WB. Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron. 2005;46(2):181–9. doi: 10.1016/j.neuron.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annu Rev Neurosci. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- 24.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16(1):95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Penzes P, Jones KA. Dendritic spine dynamics--a key role for kalirin-7. Trends Neurosci. 2008;31(8):419–27. doi: 10.1016/j.tins.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penzes P, Cahill ME, Jones KA, Srivastava DP. Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol. 2008;18(9):405–13. doi: 10.1016/j.tcb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5(10):793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 28.McBain CJ, Fisahn A. Interneurons unbound. Nat Rev Neurosci. 2001;2(1):11–23. doi: 10.1038/35049047. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Burgos G, Krimer LS, Povysheva NV, Barrionuevo G, Lewis DA. Functional properties of fast spiking interneurons and their synaptic connections with pyramidal cells in primate dorsolateral prefrontal cortex. J Neurophysiol. 2005;93(2):942–53. doi: 10.1152/jn.00787.2004. [DOI] [PubMed] [Google Scholar]
- 30.Kawaguchi Y. Physiological subgroups of nonpyramidal cells with specific morphological characteristics in layer II/III of rat frontal cortex. J Neurosci. 1995;15(4):2638–55. doi: 10.1523/JNEUROSCI.15-04-02638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis DA, Lund JS. Heterogeneity of chandelier neurons in monkey neocortex: corticotropin-releasing factor- and parvalbumin-immunoreactive populations. J Comp Neurol. 1990;293(4):599–615. doi: 10.1002/cne.902930406. [DOI] [PubMed] [Google Scholar]
- 32.Freund TF, Katona I. Perisomatic inhibition. Neuron. 2007;56(1):33–42. doi: 10.1016/j.neuron.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Williams S, Boksa P. Gamma oscillations and schizophrenia. J Psychiatry Neurosci. 2010;35(2):75–7. doi: 10.1503/jpn.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeFelipe J, Lopez-Cruz PL, Benavides-Piccione R, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14(3):202–16. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Megias M, Emri Z, Freund TF, Gulyas AI. Total number and distribution of inhibitory and excitatory synapses on hippocampal CA1 pyramidal cells. Neuroscience. 2001;102(3):527–40. doi: 10.1016/s0306-4522(00)00496-6. [DOI] [PubMed] [Google Scholar]
- 36.Eichler SA, Meier JC. E-I balance and human diseases - from molecules to networking. Front Mol Neurosci. 2008;1:2. doi: 10.3389/neuro.02.002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu G. Local structural balance and functional interaction of excitatory and inhibitory synapses in hippocampal dendrites. Nat Neurosci. 2004;7(4):373–9. doi: 10.1038/nn1206. [DOI] [PubMed] [Google Scholar]
- 38.Gulyas AI, Megias M, Emri Z, Freund TF. Total number and ratio of excitatory and inhibitory synapses converging onto single interneurons of different types in the CA1 area of the rat hippocampus. J Neurosci. 1999;19(22):10082–97. doi: 10.1523/JNEUROSCI.19-22-10082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391(6670):892–6. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- 40.Turrigiano G. Too many cooks? Intrinsic and synaptic homeostatic mechanisms in cortical circuit refinement. Annu Rev Neurosci. 2011;34:89–103. doi: 10.1146/annurev-neuro-060909-153238. [DOI] [PubMed] [Google Scholar]
- 41.Beierlein M, Gibson JR, Connors BW. A network of electrically coupled interneurons drives synchronized inhibition in neocortex. Nat Neurosci. 2000;3(9):904–10. doi: 10.1038/78809. [DOI] [PubMed] [Google Scholar]
- 42.Moran LV, Hong LE. High vs low frequency neural oscillations in schizophrenia. Schizophr Bull. 2011;37(4):659–63. doi: 10.1093/schbul/sbr056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szabadics J, Lorincz A, Tamas G. Beta and gamma frequency synchronization by dendritic gabaergic synapses and gap junctions in a network of cortical interneurons. J Neurosci. 2001;21(15):5824–31. doi: 10.1523/JNEUROSCI.21-15-05824.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamas G, Buhl EH, Lorincz A, Somogyi P. Proximally targeted GABAergic synapses and gap junctions synchronize cortical interneurons. Nat Neurosci. 2000;3(4):366–71. doi: 10.1038/73936. [DOI] [PubMed] [Google Scholar]
- 45.Uhlhaas PJ, Singer W. Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci. 2010;11(2):100–13. doi: 10.1038/nrn2774. [DOI] [PubMed] [Google Scholar]
- 46.Bartos M, Vida I, Jonas P. Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat Rev Neurosci. 2007;8(1):45–56. doi: 10.1038/nrn2044. [DOI] [PubMed] [Google Scholar]
- 47.Bateup HS, Johnson CA, Denefrio CL, Saulnier JL, Kornacker K, Sabatini BL. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78(3):510–22. doi: 10.1016/j.neuron.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yizhar O, Fenno LE, Prigge M, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477(7363):171–8. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302(5646):826–30. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 50.Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull. 2009;35(3):528–48. doi: 10.1093/schbul/sbn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selemon LD, Goldman-Rakic PS. The reduced neuropil hypothesis: a circuit based model of schizophrenia. Biological psychiatry. 1999;45(1):17–25. doi: 10.1016/s0006-3223(98)00281-9. [DOI] [PubMed] [Google Scholar]
- 52.Thompson PM, Vidal C, Giedd JN, et al. Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early-onset schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(20):11650–5. doi: 10.1073/pnas.201243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vita A, De Peri L, Deste G, Sacchetti E. Progressive loss of cortical gray matter in schizophrenia: a meta-analysis and meta-regression of longitudinal MRI studies. Transl Psychiatry. 2012;2:e190. doi: 10.1038/tp.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57(1):65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 55.Sweet RA, Henteleff RA, Zhang W, Sampson AR, Lewis DA. Reduced dendritic spine density in auditory cortex of subjects with schizophrenia. Neuropsychopharmacology. 2009;34(2):374–89. doi: 10.1038/npp.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steen RG, Mull C, McClure R, Hamer RM, Lieberman JA. Brain volume in first-episode schizophrenia: systematic review and meta-analysis of magnetic resonance imaging studies. Br J Psychiatry. 2006;188:510–8. doi: 10.1192/bjp.188.6.510. [DOI] [PubMed] [Google Scholar]
- 57.Kolomeets NS, Orlovskaya DD, Rachmanova VI, Uranova NA. Ultrastructural alterations in hippocampal mossy fiber synapses in schizophrenia: a postmortem morphometric study. Synapse. 2005;57(1):47–55. doi: 10.1002/syn.20153. [DOI] [PubMed] [Google Scholar]
- 58.Gonzalez-Burgos G, Lewis DA. GABA neurons and the mechanisms of network oscillations: implications for understanding cortical dysfunction in schizophrenia. Schizophr Bull. 2008;34(5):944–61. doi: 10.1093/schbul/sbn070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson FA, O’Scalaidhe SP, Goldman-Rakic PS. Functional synergism between putative gamma-aminobutyrate-containing neurons and pyramidal neurons in prefrontal cortex. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(9):4009–13. doi: 10.1073/pnas.91.9.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao SG, Williams GV, Goldman-Rakic PS. Destruction and creation of spatial tuning by disinhibition: GABA(A) blockade of prefrontal cortical neurons engaged by working memory. J Neurosci. 2000;20(1):485–94. doi: 10.1523/JNEUROSCI.20-01-00485.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sawaguchi T, Matsumura M, Kubota K. Delayed response deficits produced by local injection of bicuculline into the dorsolateral prefrontal cortex in Japanese macaque monkeys. Exp Brain Res. 1989;75(3):457–69. doi: 10.1007/BF00249897. [DOI] [PubMed] [Google Scholar]
- 62.Haenschel C, Bittner RA, Waltz J, et al. Cortical oscillatory activity is critical for working memory as revealed by deficits in early-onset schizophrenia. J Neurosci. 2009;29(30):9481–9. doi: 10.1523/JNEUROSCI.1428-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barr MS, Farzan F, Tran LC, Chen R, Fitzgerald PB, Daskalakis ZJ. Evidence for excessive frontal evoked gamma oscillatory activity in schizophrenia during working memory. Schizophr Res. 2010;121(1–3):146–52. doi: 10.1016/j.schres.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 64.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6(4):312–24. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 65.Hashimoto T, Volk DW, Eggan SM, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23(15):6315–26. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woo TU, Whitehead RE, Melchitzky DS, Lewis DA. A subclass of prefrontal gamma-aminobutyric acid axon terminals are selectively altered in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(9):5341–6. doi: 10.1073/pnas.95.9.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volk DW, Pierri JN, Fritschy JM, Auh S, Sampson AR, Lewis DA. Reciprocal alterations in pre- and postsynaptic inhibitory markers at chandelier cell inputs to pyramidal neurons in schizophrenia. Cereb Cortex. 2002;12(10):1063–70. doi: 10.1093/cercor/12.10.1063. [DOI] [PubMed] [Google Scholar]
- 68.Fromer M, Pocklington AJ, Kavanagh DH, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–84. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71(4):877–92. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hall J, Whalley HC, Job DE, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9(12):1477–8. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- 71.Munafo MR, Thiselton DL, Clark TG, Flint J. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11(6):539–46. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- 72.Munafo MR, Attwood AS, Flint J. Neuregulin 1 genotype and schizophrenia. Schizophr Bull. 2008;34(1):9–12. doi: 10.1093/schbul/sbm129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silberberg G, Darvasi A, Pinkas-Kramarski R, Navon R. The involvement of ErbB4 with schizophrenia: association and expression studies. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(2):142–8. doi: 10.1002/ajmg.b.30275. [DOI] [PubMed] [Google Scholar]
- 74.Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16(2):129–41. doi: 10.1093/hmg/ddl449. [DOI] [PubMed] [Google Scholar]
- 75.Nicodemus KK, Law AJ, Radulescu E, et al. Biological validation of increased schizophrenia risk with NRG1, ERBB4, and AKT1 epistasis via functional neuroimaging in healthy controls. Arch Gen Psychiatry. 2010;67(10):991–1001. doi: 10.1001/archgenpsychiatry.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan W, Dean M, Law AJ. Molecular cloning and characterization of the human ErbB4 gene: identification of novel splice isoforms in the developing and adult brain. PLoS One. 2010;5(9):e12924. doi: 10.1371/journal.pone.0012924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9(6):437–52. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buonanno A. The neuregulin signaling pathway and schizophrenia: from genes to synapses and neural circuits. Brain Res Bull. 2010;83(3–4):122–31. doi: 10.1016/j.brainresbull.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chaudhury AR, Gerecke KM, Wyss JM, Morgan DG, Gordon MN, Carroll SL. Neuregulin-1 and erbB4 immunoreactivity is associated with neuritic plaques in Alzheimer disease brain and in a transgenic model of Alzheimer disease. J Neuropathol Exp Neurol. 2003;62(1):42–54. doi: 10.1093/jnen/62.1.42. [DOI] [PubMed] [Google Scholar]
- 80.Law AJ, Shannon Weickert C, Hyde TM, Kleinman JE, Harrison PJ. Neuregulin-1 (NRG-1) mRNA and protein in the adult human brain. Neuroscience. 2004;127(1):125–36. doi: 10.1016/j.neuroscience.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 81.Ozaki M, Tohyama K, Kishida H, Buonanno A, Yano R, Hashikawa T. Roles of neuregulin in synaptogenesis between mossy fibers and cerebellar granule cells. J Neurosci Res. 2000;59(5):612–23. doi: 10.1002/(SICI)1097-4547(20000301)59:5<612::AID-JNR4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 82.Garcia RA, Vasudevan K, Buonanno A. The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(7):3596–601. doi: 10.1073/pnas.070042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang YZ, Won S, Ali DW, et al. Regulation of neuregulin signaling by PSD-95 interacting with ErbB4 at CNS synapses. Neuron. 2000;26(2):443–55. doi: 10.1016/s0896-6273(00)81176-9. [DOI] [PubMed] [Google Scholar]
- 84.Barros CS, Calabrese B, Chamero P, et al. Impaired maturation of dendritic spines without disorganization of cortical cell layers in mice lacking NRG1/ErbB signaling in the central nervous system. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(11):4507–12. doi: 10.1073/pnas.0900355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen YJ, Johnson MA, Lieberman MD, et al. Type III neuregulin-1 is required for normal sensorimotor gating, memory-related behaviors, and corticostriatal circuit components. J Neurosci. 2008;28(27):6872–83. doi: 10.1523/JNEUROSCI.1815-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li B, Woo RS, Mei L, Malinow R. The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron. 2007;54(4):583–97. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fazzari P, Paternain AV, Valiente M, et al. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464(7293):1376–80. doi: 10.1038/nature08928. [DOI] [PubMed] [Google Scholar]
- 88.Chen Y, Hancock ML, Role LW, Talmage DA. Intramembranous valine linked to schizophrenia is required for neuregulin 1 regulation of the morphological development of cortical neurons. J Neurosci. 2010;30(27):9199–208. doi: 10.1523/JNEUROSCI.0605-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weickert CS, Tiwari Y, Schofield PR, Mowry BJ, Fullerton JM. Schizophrenia-associated HapICE haplotype is associated with increased NRG1 type III expression and high nucleotide diversity. Transl Psychiatry. 2012;2:e104. doi: 10.1038/tp.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Millar JK, Wilson-Annan JC, Anderson S, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9(9):1415–23. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 91.Schumacher J, Laje G, Abou Jamra R, et al. The DISC locus and schizophrenia: evidence from an association study in a central European sample and from a meta-analysis across different European populations. Hum Mol Genet. 2009;18(14):2719–27. doi: 10.1093/hmg/ddp204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hodgkinson CA, Goldman D, Jaeger J, et al. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am J Hum Genet. 2004;75(5):862–72. doi: 10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hennah W, Thomson P, McQuillin A, et al. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14(9):865–73. doi: 10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- 94.Hashimoto R, Numakawa T, Ohnishi T, et al. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum Mol Genet. 2006;15(20):3024–33. doi: 10.1093/hmg/ddl244. [DOI] [PubMed] [Google Scholar]
- 95.Harris SE, Hennah W, Thomson PA, et al. Variation in DISC1 is associated with anxiety, depression and emotional stability in elderly women. Mol Psychiatry. 2010;15(3):232–4. doi: 10.1038/mp.2009.88. [DOI] [PubMed] [Google Scholar]
- 96.Williams JM, Beck TF, Pearson DM, Proud MB, Cheung SW, Scott DA. A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am J Med Genet A. 2009;149A(8):1758–62. doi: 10.1002/ajmg.a.32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kilpinen H, Ylisaukko-Oja T, Hennah W, et al. Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 2008;13(2):187–96. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- 98.Austin CP, Ky B, Ma L, Morris JA, Shughrue PJ. Expression of Disrupted-In-Schizophrenia-1, a schizophrenia-associated gene, is prominent in the mouse hippocampus throughout brain development. Neuroscience. 2004;124(1):3–10. doi: 10.1016/j.neuroscience.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 99.Enomoto A, Asai N, Namba T, et al. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63(6):774–87. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 100.Meyer KD, Morris JA. Disc1 regulates granule cell migration in the developing hippocampus. Hum Mol Genet. 2009;18(17):3286–97. doi: 10.1093/hmg/ddp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kirkpatrick B, Xu L, Cascella N, Ozeki Y, Sawa A, Roberts RC. DISC1 immunoreactivity at the light and ultrastructural level in the human neocortex. J Comp Neurol. 2006;497(3):436–50. doi: 10.1002/cne.21007. [DOI] [PubMed] [Google Scholar]
- 102.Lipska BK, Peters T, Hyde TM, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum Mol Genet. 2006;15(8):1245–58. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 103.Hayashi-Takagi A, Takaki M, Graziane N, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13(3):327–32. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2006;11(6):557–66. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- 105.Cahill ME, Xie Z, Day M, et al. Kalirin regulates cortical spine morphogenesis and disease-related behavioral phenotypes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(31):13058–63. doi: 10.1073/pnas.0904636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stark KL, Xu B, Bagchi A, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40(6):751–60. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 107.Mukai J, Dhilla A, Drew LJ, et al. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci. 2008;11(11):1302–10. doi: 10.1038/nn.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Engel AK, Singer W. Temporal binding and the neural correlates of sensory awareness. Trends Cogn Sci. 2001;5(1):16–25. doi: 10.1016/s1364-6613(00)01568-0. [DOI] [PubMed] [Google Scholar]
- 109.Herrmann CS, Demiralp T. Human EEG gamma oscillations in neuropsychiatric disorders. Clin Neurophysiol. 2005;116(12):2719–33. doi: 10.1016/j.clinph.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 110.Spencer KM. The functional consequences of cortical circuit abnormalities on gamma oscillations in schizophrenia: insights from computational modeling. Front Hum Neurosci. 2009;3:33. doi: 10.3389/neuro.09.033.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kwon JS, O’Donnell BF, Wallenstein GV, et al. Gamma frequency-range abnormalities to auditory stimulation in schizophrenia. Arch Gen Psychiatry. 1999;56(11):1001–5. doi: 10.1001/archpsyc.56.11.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wilson TW, Hernandez OO, Asherin RM, Teale PD, Reite ML, Rojas DC. Cortical gamma generators suggest abnormal auditory circuitry in early-onset psychosis. Cereb Cortex. 2008;18(2):371–8. doi: 10.1093/cercor/bhm062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57(3):237–45. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- 114.Akbarian S, Kim JJ, Potkin SG, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52(4):258–66. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- 115.Lai C, Lemke G. An extended family of protein-tyrosine kinase genes differentially expressed in the vertebrate nervous system. Neuron. 1991;6(5):691–704. doi: 10.1016/0896-6273(91)90167-x. [DOI] [PubMed] [Google Scholar]
- 116.Steiner H, Blum M, Kitai ST, Fedi P. Differential expression of ErbB3 and ErbB4 neuregulin receptors in dopamine neurons and forebrain areas of the adult rat. Exp Neurol. 1999;159(2):494–503. doi: 10.1006/exnr.1999.7163. [DOI] [PubMed] [Google Scholar]
- 117.Gerecke KM, Wyss JM, Karavanova I, Buonanno A, Carroll SL. ErbB transmembrane tyrosine kinase receptors are differentially expressed throughout the adult rat central nervous system. J Comp Neurol. 2001;433(1):86–100. doi: 10.1002/cne.1127. [DOI] [PubMed] [Google Scholar]
- 118.Fox IJ, Kornblum HI. Developmental profile of ErbB receptors in murine central nervous system: implications for functional interactions. J Neurosci Res. 2005;79(5):584–97. doi: 10.1002/jnr.20381. [DOI] [PubMed] [Google Scholar]
- 119.Thompson M, Lauderdale S, Webster MJ, et al. Widespread expression of ErbB2, ErbB3 and ErbB4 in non-human primate brain. Brain Res. 2007;1139:95–109. doi: 10.1016/j.brainres.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 120.Neddens J, Buonanno A. Expression of the neuregulin receptor ErbB4 in the brain of the rhesus monkey (Macaca mulatta) PLoS One. 2011;6(11):e27337. doi: 10.1371/journal.pone.0027337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neddens J, Fish KN, Tricoire L, et al. Conserved interneuron-specific ErbB4 expression in frontal cortex of rodents, monkeys, and humans: implications for schizophrenia. Biological psychiatry. 2011;70(7):636–45. doi: 10.1016/j.biopsych.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yau HJ, Wang HF, Lai C, Liu FC. Neural development of the neuregulin receptor ErbB4 in the cerebral cortex and the hippocampus: preferential expression by interneurons tangentially migrating from the ganglionic eminences. Cereb Cortex. 2003;13(3):252–64. doi: 10.1093/cercor/13.3.252. [DOI] [PubMed] [Google Scholar]
- 123.Fisahn A, Neddens J, Yan L, Buonanno A. Neuregulin-1 modulates hippocampal gamma oscillations: implications for schizophrenia. Cereb Cortex. 2009;19(3):612–8. doi: 10.1093/cercor/bhn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wen L, Lu YS, Zhu XH, et al. Neuregulin 1 regulates pyramidal neuron activity via ErbB4 in parvalbumin-positive interneurons. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(3):1211–6. doi: 10.1073/pnas.0910302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shamir A, Kwon OB, Karavanova I, et al. The importance of the NRG-1/ErbB4 pathway for synaptic plasticity and behaviors associated with psychiatric disorders. J Neurosci. 2012;32(9):2988–97. doi: 10.1523/JNEUROSCI.1899-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fuchs EC, Zivkovic AR, Cunningham MO, et al. Recruitment of parvalbumin-positive interneurons determines hippocampal function and associated behavior. Neuron. 2007;53(4):591–604. doi: 10.1016/j.neuron.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 127.Longart M, Chatani-Hinze M, Gonzalez CM, Vullhorst D, Buonanno A. Regulation of ErbB-4 endocytosis by neuregulin in GABAergic hippocampal interneurons. Brain Res Bull. 2007;73(4–6):210–9. doi: 10.1016/j.brainresbull.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vullhorst D, Neddens J, Karavanova I, et al. Selective expression of ErbB4 in interneurons, but not pyramidal cells, of the rodent hippocampus. J Neurosci. 2009;29(39):12255–64. doi: 10.1523/JNEUROSCI.2454-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hikida T, Jaaro-Peled H, Seshadri S, et al. Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(36):14501–6. doi: 10.1073/pnas.0704774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee FH, Zai CC, Cordes SP, Roder JC, Wong AH. Abnormal interneuron development in disrupted-in-schizophrenia-1 L100P mutant mice. Mol Brain. 2013;6:20. doi: 10.1186/1756-6606-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shen S, Lang B, Nakamoto C, et al. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated Disc1. J Neurosci. 2008;28(43):10893–904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weickert CS, Straub RE, McClintock BW, et al. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry. 2004;61(6):544–55. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- 133.Talbot K, Eidem WL, Tinsley CL, et al. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest. 2004;113(9):1353–63. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ji Y, Yang F, Papaleo F, et al. Role of dysbindin in dopamine receptor trafficking and cortical GABA function. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(46):19593–8. doi: 10.1073/pnas.0904289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Parellada M, Penzol MJ, Pina L, et al. The neurobiology of autism spectrum disorders. Eur Psychiatry. 2014;29(1):11–9. doi: 10.1016/j.eurpsy.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 136.Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes, brain, and behavior. 2003;2(5):255–67. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Courchesne E, Redcay E, Kennedy DP. The autistic brain: birth through adulthood. Curr Opin Neurol. 2004;17(4):489–96. doi: 10.1097/01.wco.0000137542.14610.b4. [DOI] [PubMed] [Google Scholar]
- 138.Tuchman R, Rapin I. Epilepsy in autism. Lancet Neurol. 2002;1(6):352–8. doi: 10.1016/s1474-4422(02)00160-6. [DOI] [PubMed] [Google Scholar]
- 139.Irwin SA, Patel B, Idupulapati M, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98(2):161–7. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 140.Guerin P, Lyon G, Barthelemy C, et al. Neuropathological study of a case of autistic syndrome with severe mental retardation. Dev Med Child Neurol. 1996;38(3):203–11. doi: 10.1111/j.1469-8749.1996.tb15082.x. [DOI] [PubMed] [Google Scholar]
- 141.Raymond GV, Bauman ML, Kemper TL. Hippocampus in autism: a Golgi analysis. Acta Neuropathol. 1996;91(1):117–9. doi: 10.1007/s004010050401. [DOI] [PubMed] [Google Scholar]
- 142.Gadad BS, Hewitson L, Young KA, German DC. Neuropathology and animal models of autism: genetic and environmental factors. Autism Res Treat. 2013;2013:731935. doi: 10.1155/2013/731935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Geschwind DH. Autism: many genes, common pathways? Cell. 2008;135(3):391–5. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wass S. Distortions and disconnections: disrupted brain connectivity in autism. Brain Cogn. 2011;75(1):18–28. doi: 10.1016/j.bandc.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 145.Fatemi SH, Halt AR, Stary JM, Kanodia R, Schulz SC, Realmuto GR. Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biological psychiatry. 2002;52(8):805–10. doi: 10.1016/s0006-3223(02)01430-0. [DOI] [PubMed] [Google Scholar]
- 146.Yip J, Soghomonian JJ, Blatt GJ. Decreased GAD67 mRNA levels in cerebellar Purkinje cells in autism: pathophysiological implications. Acta Neuropathol. 2007;113(5):559–68. doi: 10.1007/s00401-006-0176-3. [DOI] [PubMed] [Google Scholar]
- 147.Collins AL, Ma D, Whitehead PL, et al. Investigation of autism and GABA receptor subunit genes in multiple ethnic groups. Neurogenetics. 2006;7(3):167–74. doi: 10.1007/s10048-006-0045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fatemi SH, Reutiman TJ, Folsom TD, Thuras PD. GABA(A) receptor downregulation in brains of subjects with autism. J Autism Dev Disord. 2009;39(2):223–30. doi: 10.1007/s10803-008-0646-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oblak AL, Gibbs TT, Blatt GJ. Decreased GABA(B) receptors in the cingulate cortex and fusiform gyrus in autism. J Neurochem. 2010;114(5):1414–23. doi: 10.1111/j.1471-4159.2010.06858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brooks-Kayal A. Epilepsy and autism spectrum disorders: are there common developmental mechanisms? Brain Dev. 2010;32(9):731–8. doi: 10.1016/j.braindev.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 151.Zikopoulos B, Barbas H. Altered neural connectivity in excitatory and inhibitory cortical circuits in autism. Front Hum Neurosci. 2013;7:609. doi: 10.3389/fnhum.2013.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.DeFelipe J. Chandelier cells and epilepsy. Brain. 1999;122(Pt 10):1807–22. doi: 10.1093/brain/122.10.1807. [DOI] [PubMed] [Google Scholar]
- 153.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9(5):341–55. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455(7215):903–11. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ichtchenko K, Hata Y, Nguyen T, et al. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81(3):435–43. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- 156.Ichtchenko K, Nguyen T, Sudhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271(5):2676–82. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- 157.Comoletti D, Flynn RE, Boucard AA, et al. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry. 2006;45(42):12816–27. doi: 10.1021/bi0614131. [DOI] [PubMed] [Google Scholar]
- 158.Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51(2):171–8. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 159.Missler M, Zhang W, Rohlmann A, et al. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423(6943):939–48. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- 160.Varoqueaux F, Aramuni G, Rawson RL, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51(6):741–54. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 161.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101(6):657–69. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 162.Nam CI, Chen L. Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(17):6137–42. doi: 10.1073/pnas.0502038102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kwon HB, Kozorovitskiy Y, Oh WJ, et al. Neuroligin-1-dependent competition regulates cortical synaptogenesis and synapse number. Nat Neurosci. 2012;15(12):1667–74. doi: 10.1038/nn.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Barrow SL, McAllister AK. Neuroligins help dendrites keep up with the Joneses. Nat Neurosci. 2012;15(12):1609–11. doi: 10.1038/nn.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Comoletti D, De Jaco A, Jennings LL, et al. The Arg451Cys-neuroligin-3 mutation associated with autism reveals a defect in protein processing. J Neurosci. 2004;24(20):4889–93. doi: 10.1523/JNEUROSCI.0468-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Etherton MR, Tabuchi K, Sharma M, Ko J, Sudhof TC. An autism-associated point mutation in the neuroligin cytoplasmic tail selectively impairs AMPA receptor-mediated synaptic transmission in hippocampus. EMBO J. 2011;30(14):2908–19. doi: 10.1038/emboj.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang C, Milunsky JM, Newton S, et al. A neuroligin-4 missense mutation associated with autism impairs neuroligin-4 folding and endoplasmic reticulum export. J Neurosci. 2009;29(35):10843–54. doi: 10.1523/JNEUROSCI.1248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Feng J, Schroer R, Yan J, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409(1):10–3. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 169.Jamain S, Quach H, Betancur C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34(1):27–9. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lawson-Yuen A, Saldivar JS, Sommer S, Picker J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur J Hum Genet. 2008;16(5):614–8. doi: 10.1038/sj.ejhg.5202006. [DOI] [PubMed] [Google Scholar]
- 171.Zahir FR, Baross A, Delaney AD, et al. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2008;45(4):239–43. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- 172.Gauthier J, Siddiqui TJ, Huashan P, et al. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet. 2011;130(4):563–73. doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Konstantareas MM, Homatidis S. Chromosomal abnormalities in a series of children with autistic disorder. J Autism Dev Disord. 1999;29(4):275–85. doi: 10.1023/a:1022155201662. [DOI] [PubMed] [Google Scholar]
- 174.Kim HG, Kishikawa S, Higgins AW, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82(1):199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Daoud H, Bonnet-Brilhault F, Vedrine S, et al. Autism and nonsyndromic mental retardation associated with a de novo mutation in the NLGN4X gene promoter causing an increased expression level. Biological psychiatry. 2009;66(10):906–10. doi: 10.1016/j.biopsych.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 176.Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459(7246):569–73. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48(2):229–36. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 178.Graf ER, Kang Y, Hauner AM, Craig AM. Structure function and splice site analysis of the synaptogenic activity of the neurexin-1 beta LNS domain. J Neurosci. 2006;26(16):4256–65. doi: 10.1523/JNEUROSCI.1253-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dean C, Scholl FG, Choih J, et al. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6(7):708–16. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Koehnke J, Katsamba PS, Ahlsen G, et al. Splice form dependence of beta-neurexin/neuroligin binding interactions. Neuron. 2010;67(1):61–74. doi: 10.1016/j.neuron.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(42):17998–8003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Song JY, Ichtchenko K, Sudhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(3):1100–5. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Budreck EC, Scheiffele P. Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. Eur J Neurosci. 2007;26(7):1738–48. doi: 10.1111/j.1460-9568.2007.05842.x. [DOI] [PubMed] [Google Scholar]
- 184.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119(7):1013–26. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci. 2007;8(3):206–20. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chih B, Afridi SK, Clark L, Scheiffele P. Disorder-associated mutations lead to functional inactivation of neuroligins. Hum Mol Genet. 2004;13(14):1471–7. doi: 10.1093/hmg/ddh158. [DOI] [PubMed] [Google Scholar]
- 187.Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307(5713):1324–8. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- 188.Chubykin AA, Atasoy D, Etherton MR, et al. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54(6):919–31. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Suzuki K, Hayashi Y, Nakahara S, et al. Activity-dependent proteolytic cleavage of neuroligin-1. Neuron. 2012;76(2):410–22. doi: 10.1016/j.neuron.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 190.Peixoto RT, Kunz PA, Kwon H, et al. Transsynaptic signaling by activity-dependent cleavage of neuroligin-1. Neuron. 2012;76(2):396–409. doi: 10.1016/j.neuron.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bemben MA, Shipman SL, Hirai T, et al. CaMKII phosphorylation of neuroligin-1 regulates excitatory synapses. Nat Neurosci. 2014;17(1):56–64. doi: 10.1038/nn.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Blundell J, Blaiss CA, Etherton MR, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30(6):2115–29. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dahlhaus R, Hines RM, Eadie BD, et al. Overexpression of the cell adhesion protein neuroligin-1 induces learning deficits and impairs synaptic plasticity by altering the ratio of excitation to inhibition in the hippocampus. Hippocampus. 2010;20(2):305–22. doi: 10.1002/hipo.20630. [DOI] [PubMed] [Google Scholar]
- 194.Radyushkin K, Hammerschmidt K, Boretius S, et al. Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes, brain, and behavior. 2009;8(4):416–25. doi: 10.1111/j.1601-183X.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- 195.Jamain S, Radyushkin K, Hammerschmidt K, et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1710–5. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Etherton M, Foldy C, Sharma M, et al. Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(33):13764–9. doi: 10.1073/pnas.1111093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39(1):25–7. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Moessner R, Marshall CR, Sutcliffe JS, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81(6):1289–97. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gauthier J, Spiegelman D, Piton A, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):421–4. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 200.Berkel S, Marshall CR, Weiss B, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42(6):489–91. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- 201.Sato D, Lionel AC, Leblond CS, et al. SHANK1 Deletions in Males with Autism Spectrum Disorder. Am J Hum Genet. 2012;90(5):879–87. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78(1):8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hung AY, Futai K, Sala C, et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28(7):1697–708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wohr M, Roullet FI, Hung AY, Sheng M, Crawley JN. Communication impairments in mice lacking Shank1: reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS One. 2011;6(6):e20631. doi: 10.1371/journal.pone.0020631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bozdagi O, Sakurai T, Papapetrou D, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1(1):15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Peca J, Feliciano C, Ting JT, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472(7344):437–42. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wang X, McCoy PA, Rodriguiz RM, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20(15):3093–108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Schmeisser MJ, Ey E, Wegener S, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486(7402):256–60. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 209.Won H, Lee HR, Gee HY, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486(7402):261–5. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- 210.Singh SK, Eroglu C. Neuroligins provide molecular links between syndromic and nonsyndromic autism. Sci Signal. 2013;6(283):re4. doi: 10.1126/scisignal.2004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Prather P, de Vries PJ. Behavioral and cognitive aspects of tuberous sclerosis complex. J Child Neurol. 2004;19(9):666–74. doi: 10.1177/08830738040190090601. [DOI] [PubMed] [Google Scholar]
- 212.Butler MG, Dasouki MJ, Zhou XP, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42(4):318–21. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kwon CH, Luikart BW, Powell CM, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50(3):377–88. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005;8(12):1727–34. doi: 10.1038/nn1566. [DOI] [PubMed] [Google Scholar]
- 215.Fraser MM, Bayazitov IT, Zakharenko SS, Baker SJ. Phosphatase and tensin homolog, deleted on chromosome 10 deficiency in brain causes defects in synaptic structure, transmission and plasticity, and myelination abnormalities. Neuroscience. 2008;151(2):476–88. doi: 10.1016/j.neuroscience.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Pun RY, Rolle IJ, Lasarge CL, et al. Excessive activation of mTOR in postnatally generated granule cells is sufficient to cause epilepsy. Neuron. 2012;75(6):1022–34. doi: 10.1016/j.neuron.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6(5):376–87. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 218.Zhou Z, Hong EJ, Cohen S, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52(2):255–69. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cook EH, Jr, Lindgren V, Leventhal BL, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60(4):928–34. [PMC free article] [PubMed] [Google Scholar]
- 220.Dindot SV, Antalffy BA, Bhattacharjee MB, Beaud, et al. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17(1):111–8. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- 221.Greer PL, Hanayama R, Bloodgood BL, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140(5):704–16. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol. 2004;83(9):449–56. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
- 223.Huang ZJ, Scheiffele P. GABA and neuroligin signaling: linking synaptic activity and adhesion in inhibitory synapse development. Curr Opin Neurobiol. 2008;18(1):77–83. doi: 10.1016/j.conb.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hines RM, Wu L, Hines DJ, et al. Synaptic imbalance, stereotypies, and impaired social interactions in mice with altered neuroligin 2 expression. J Neurosci. 2008;28(24):6055–67. doi: 10.1523/JNEUROSCI.0032-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tabuchi K, Blundell J, Etherton MR, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318(5847):71–6. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hoon M, Soykan T, Falkenburger B, et al. Neuroligin-4 is localized to glycinergic postsynapses and regulates inhibition in the retina. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(7):3053–8. doi: 10.1073/pnas.1006946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56(3):422–37. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 228.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol. 2011;27:631–52. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 229.Akbarian S, Chen RZ, Gribnau J, et al. Expression pattern of the Rett syndrome gene MeCP2 in primate prefrontal cortex. Neurobiol Dis. 2001;8(5):784–91. doi: 10.1006/nbdi.2001.0420. [DOI] [PubMed] [Google Scholar]
- 230.Jian L, Nagarajan L, de Klerk N, et al. Predictors of seizure onset in Rett syndrome. J Pediatr. 2006;149(4):542–7. doi: 10.1016/j.jpeds.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 231.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27(3):322–6. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 232.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27(3):327–31. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 233.Chao HT, Chen H, Samaco RC, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468(7321):263–9. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zhang ZW, Zak JD, Liu H. MeCP2 is required for normal development of GABAergic circuits in the thalamus. J Neurophysiol. 2010;103(5):2470–81. doi: 10.1152/jn.00601.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Medrihan L, Tantalaki E, Aramuni G, et al. Early defects of GABAergic synapses in the brain stem of a MeCP2 mouse model of Rett syndrome. J Neurophysiol. 2008;99(1):112–21. doi: 10.1152/jn.00826.2007. [DOI] [PubMed] [Google Scholar]
- 236.Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–9. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Martinowich K, Hattori D, Wu H, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–3. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 238.Wisniewski KE, Segan SM, Miezejeski CM, Sersen EA, Rudelli RD. The Fra(X) syndrome: neurological, electrophysiological, and neuropathological abnormalities. Am J Med Genet. 1991;38(2–3):476–80. doi: 10.1002/ajmg.1320380267. [DOI] [PubMed] [Google Scholar]
- 239.Clifford S, Dissanayake C, Bui QM, Huggins R, Taylor AK, Loesch DZ. Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J Autism Dev Disord. 2007;37(4):738–47. doi: 10.1007/s10803-006-0205-z. [DOI] [PubMed] [Google Scholar]
- 240.Harris SW, Hessl D, Goodlin-Jones B, et al. Autism profiles of males with fragile X syndrome. Am J Ment Retard. 2008;113(6):427–38. doi: 10.1352/2008.113:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hatton DD, Sideris J, Skinner M, et al. Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am J Med Genet A. 2006;140A(17):1804–13. doi: 10.1002/ajmg.a.31286. [DOI] [PubMed] [Google Scholar]
- 242.Wang LW, Berry-Kravis E, Hagerman RJ. Fragile X: leading the way for targeted treatments in autism. Neurotherapeutics. 2010;7(3):264–74. doi: 10.1016/j.nurt.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Coghlan S, Horder J, Inkster B, Mendez MA, Murphy DG, Nutt DJ. GABA system dysfunction in autism and related disorders: from synapse to symptoms. Neurosci Biobehav Rev. 2012;36(9):2044–55. doi: 10.1016/j.neubiorev.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Olmos-Serrano JL, Paluszkiewicz SM, Martin BS, Kaufmann WE, Corbin JG, Huntsman MM. Defective GABAergic neurotransmission and pharmacological rescue of neuronal hyperexcitability in the amygdala in a mouse model of fragile X syndrome. J Neurosci. 2010;30(29):9929–38. doi: 10.1523/JNEUROSCI.1714-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Spencer CM, Alekseyenko O, Serysheva E, Yuva-Paylor LA, Paylor R. Altered anxiety-related and social behaviors in the Fmr1 knockout mouse model of fragile X syndrome. Genes, brain, and behavior. 2005;4(7):420–30. doi: 10.1111/j.1601-183X.2005.00123.x. [DOI] [PubMed] [Google Scholar]
- 246.Selby L, Zhang C, Sun QQ. Major defects in neocortical GABAergic inhibitory circuits in mice lacking the fragile X mental retardation protein. Neurosci Lett. 2007;412(3):227–32. doi: 10.1016/j.neulet.2006.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.D’Hulst C, De Geest N, Reeve SP, et al. Decreased expression of the GABAA receptor in fragile X syndrome. Brain Res. 2006;1121(1):238–45. doi: 10.1016/j.brainres.2006.08.115. [DOI] [PubMed] [Google Scholar]
- 248.D’Hulst C, Heulens I, Brouwer JR, et al. Expression of the GABAergic system in animal models for fragile X syndrome and fragile X associated tremor/ataxia syndrome (FXTAS) Brain Res. 2009;1253:176–83. doi: 10.1016/j.brainres.2008.11.075. [DOI] [PubMed] [Google Scholar]
- 249.Adusei DC, Pacey LK, Chen D, Hampson DR. Early developmental alterations in GABAergic protein expression in fragile X knockout mice. Neuropharmacology. 2010;59(3):167–71. doi: 10.1016/j.neuropharm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 250.El Idrissi A, Ding XH, Scalia J, Trenkner E, Brown WT, Dobkin C. Decreased GABA(A) receptor expression in the seizure-prone fragile X mouse. Neurosci Lett. 2005;377(3):141–6. doi: 10.1016/j.neulet.2004.11.087. [DOI] [PubMed] [Google Scholar]
- 251.Paluszkiewicz SM, Martin BS, Huntsman MM. Fragile X syndrome: the GABAergic system and circuit dysfunction. Dev Neurosci. 2011;33(5):349–64. doi: 10.1159/000329420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bakkaloglu B, O’Roak BJ, Louvi A, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82(1):165–73. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Rossi E, Verri AP, Patricelli MG, et al. A 12Mb deletion at 7q33-q35 associated with autism spectrum disorders and primary amenorrhea. Eur J Med Genet. 2008;51(6):631–8. doi: 10.1016/j.ejmg.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 254.Poot M, Beyer V, Schwaab I, et al. Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics. 2010;11(1):81–9. doi: 10.1007/s10048-009-0205-1. [DOI] [PubMed] [Google Scholar]
- 255.O’Roak BJ, Deriziotis P, Lee C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43(6):585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Alarcon M, Abrahams BS, Stone JL, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–9. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Anderson GR, Galfin T, Xu W, Aoto J, Malenka RC, Sudhof TC. Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(44):18120–5. doi: 10.1073/pnas.1216398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Penagarikano O, Abrahams BS, Herman EI, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147(1):235–46. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Traka M, Goutebroze L, Denisenko N, et al. Association of TAG-1 with Caspr2 is essential for the molecular organization of juxtaparanodal regions of myelinated fibers. J Cell Biol. 2003;162(6):1161–72. doi: 10.1083/jcb.200305078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Denaxa M, Chan CH, Schachner M, Parnavelas JG, Karagogeos D. The adhesion molecule TAG-1 mediates the migration of cortical interneurons from the ganglionic eminence along the corticofugal fiber system. Development. 2001;128(22):4635–44. doi: 10.1242/dev.128.22.4635. [DOI] [PubMed] [Google Scholar]
- 261.Kim JY, Duan X, Liu CY, et al. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63(6):761–73. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Strauss KA, Puffenberger EG, Huentelman MJ, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354(13):1370–7. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 263.Murakami M, Ichisaka T, Maeda M, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24(15):6710–8. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 265.Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25(49):11300–12. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci. 2005;25(49):11288–99. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Choi YJ, Di Nardo A, Kramvis I, et al. Tuberous sclerosis complex proteins control axon formation. Genes Dev. 2008;22(18):2485–95. doi: 10.1101/gad.1685008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Gururajan A, van den Buuse M. Is the mTOR-signalling cascade disrupted in Schizophrenia? J Neurochem. 2014;129(3):377–87. doi: 10.1111/jnc.12622. [DOI] [PubMed] [Google Scholar]
- 269.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–45. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 270.Lasarge CL, Danzer SC. Mechanisms regulating neuronal excitability and seizure development following mTOR pathway hyperactivation. Front Mol Neurosci. 2014;7:18. doi: 10.3389/fnmol.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kalkman HO. The role of the phosphatidylinositide 3-kinase-protein kinase B pathway in schizophrenia. Pharmacol Ther. 2006;110(1):117–34. doi: 10.1016/j.pharmthera.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 272.Mathur A, Law MH, Megson IL, Shaw DJ, Wei J. Genetic association of the AKT1 gene with schizophrenia in a British population. Psychiatr Genet. 2010;20(3):118–22. doi: 10.1097/YPG.0b013e32833a2234. [DOI] [PubMed] [Google Scholar]
- 273.Karege F, Perroud N, Schurhoff F, et al. Association of AKT1 gene variants and protein expression in both schizophrenia and bipolar disorder. Genes, brain, and behavior. 2010;9(5):503–11. doi: 10.1111/j.1601-183X.2010.00578.x. [DOI] [PubMed] [Google Scholar]
- 274.Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36(2):131–7. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 275.Yoon SC, Seo MS, Kim SH, et al. The effect of MK-801 on mTOR/p70S6K and translation-related proteins in rat frontal cortex. Neurosci Lett. 2008;434(1):23–8. doi: 10.1016/j.neulet.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 276.Kim SH, Park HG, Kim HS, Ahn YM, Kim YS. Effects of neonatal MK-801 treatment on p70S6K-S6/eIF4B signal pathways and protein translation in the frontal cortex of the developing rat brain. Int J Neuropsychopharmacol. 2010;13(9):1233–46. doi: 10.1017/S1461145709991192. [DOI] [PubMed] [Google Scholar]
- 277.Kim JY, Liu CY, Zhang F, et al. Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell. 2012;148(5):1051–64. doi: 10.1016/j.cell.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Zhou M, Li W, Huang S, et al. mTOR Inhibition ameliorates cognitive and affective deficits caused by Disc1 knockdown in adult-born dentate granule neurons. Neuron. 2013;77(4):647–54. doi: 10.1016/j.neuron.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Fuller SJ, Sivarajah K, Sugden PH. ErbB receptors, their ligands, and the consequences of their activation and inhibition in the myocardium. J Mol Cell Cardiol. 2008;44(5):831–54. doi: 10.1016/j.yjmcc.2008.02.278. [DOI] [PubMed] [Google Scholar]
- 280.Junttila TT, Sundvall M, Maatta JA, Elenius K. Erbb4 and its isoforms: selective regulation of growth factor responses by naturally occurring receptor variants. Trends Cardiovasc Med. 2000;10(7):304–10. doi: 10.1016/s1050-1738(01)00065-2. [DOI] [PubMed] [Google Scholar]
- 281.Law AJ, Wang Y, Sei Y, et al. Neuregulin 1-ErbB4-PI3K signaling in schizophrenia and phosphoinositide 3-kinase-p110delta inhibition as a potential therapeutic strategy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(30):12165–70. doi: 10.1073/pnas.1206118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Keri S, Seres I, Kelemen O, Benedek G. Neuregulin 1-stimulated phosphorylation of AKT in psychotic disorders and its relationship with neurocognitive functions. Neurochem Int. 2009;55(7):606–9. doi: 10.1016/j.neuint.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 283.Hahn CG, Wang HY, Cho DS, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12(7):824–8. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- 284.Cuesto G, Enriquez-Barreto L, Carames C, et al. Phosphoinositide-3-kinase activation controls synaptogenesis and spinogenesis in hippocampal neurons. J Neurosci. 2011;31(8):2721–33. doi: 10.1523/JNEUROSCI.4477-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Numakawa T, Yagasaki Y, Ishimoto T, et al. Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum Mol Genet. 2004;13(21):2699–708. doi: 10.1093/hmg/ddh280. [DOI] [PubMed] [Google Scholar]
- 286.Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19(2):231–4. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 287.Meikle L, Talos DM, Onda H, et al. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J Neurosci. 2007;27(21):5546–58. doi: 10.1523/JNEUROSCI.5540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J Neurosci. 2011;31(24):8862–9. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Weston MC, Chen H, Swann JW. Loss of mTOR repressors Tsc1 or Pten has divergent effects on excitatory and inhibitory synaptic transmission in single hippocampal neuron cultures. Front Mol Neurosci. 2014;7:1. doi: 10.3389/fnmol.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Sharma A, Hoeffer CA, Takayasu Y, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30(2):694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Jiang M, Ash RT, Baker SA, et al. Dendritic arborization and spine dynamics are abnormal in the mouse model of MECP2 duplication syndrome. J Neurosci. 2013;33(50):19518–33. doi: 10.1523/JNEUROSCI.1745-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Ricciardi S, Boggio EM, Grosso S, et al. Reduced AKT/mTOR signaling and protein synthesis dysregulation in a Rett syndrome animal model. Hum Mol Genet. 2011;20(6):1182–96. doi: 10.1093/hmg/ddq563. [DOI] [PubMed] [Google Scholar]
- 293.Hoeffer CA, Sanchez E, Hagerman RJ, et al. Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes, brain, and behavior. 2012;11(3):332–41. doi: 10.1111/j.1601-183X.2012.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Gkogkas CG, Khoutorsky A, Ran I, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493(7432):371–7. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Santini E, Huynh TN, MacAskill AF, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493(7432):411–5. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14(6):383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 297.Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3(3):175–90. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- 298.Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8(6):413–26. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- 299.Gladding CM, Raymond LA. Mechanisms underlying NMDA receptor synaptic/extrasynaptic distribution and function. Mol Cell Neurosci. 2011;48(4):308–20. doi: 10.1016/j.mcn.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 300.Morris BJ, Cochran SM, Pratt JA. PCP: from pharmacology to modelling schizophrenia. Curr Opin Pharmacol. 2005;5(1):101–6. doi: 10.1016/j.coph.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 301.Cochran SM, Fujimura M, Morris BJ, Pratt JA. Acute and delayed effects of phencyclidine upon mRNA levels of markers of glutamatergic and GABAergic neurotransmitter function in the rat brain. Synapse. 2002;46(3):206–14. doi: 10.1002/syn.10126. [DOI] [PubMed] [Google Scholar]
- 302.Kinney JW, Davis CN, Tabarean I, Conti B, Bartfai T, Behrens MM. A specific role for NR2A-containing NMDA receptors in the maintenance of parvalbumin and GAD67 immunoreactivity in cultured interneurons. J Neurosci. 2006;26(5):1604–15. doi: 10.1523/JNEUROSCI.4722-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Behrens MM, Ali SS, Dao DN, et al. Ketamine-induced loss of phenotype of fast-spiking interneurons is mediated by NADPH-oxidase. Science. 2007;318(5856):1645–7. doi: 10.1126/science.1148045. [DOI] [PubMed] [Google Scholar]
- 304.Akbarian S, Sucher NJ, Bradley D, et al. Selective alterations in gene expression for NMDA receptor subunits in prefrontal cortex of schizophrenics. J Neurosci. 1996;16(1):19–30. doi: 10.1523/JNEUROSCI.16-01-00019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kristiansen LV, Beneyto M, Haroutunian V, Meador-Woodruff JH. Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia. Mol Psychiatry. 2006;11(8):737–47. 05. doi: 10.1038/sj.mp.4001844. [DOI] [PubMed] [Google Scholar]
- 306.Belforte JE, Zsiros V, Sklar ER, et al. Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nat Neurosci. 2010;13(1):76–83. doi: 10.1038/nn.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Begni S, Moraschi S, Bignotti S, et al. Association between the G1001C polymorphism in the GRIN1 gene promoter region and schizophrenia. Biological psychiatry. 2003;53(7):617–9. doi: 10.1016/s0006-3223(02)01783-3. [DOI] [PubMed] [Google Scholar]
- 308.Zhao X, Li H, Shi Y, et al. Significant association between the genetic variations in the 5′ end of the N-methyl-D-aspartate receptor subunit gene GRIN1 and schizophrenia. Biological psychiatry. 2006;59(8):747–53. doi: 10.1016/j.biopsych.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 309.Greenwood TA, Lazzeroni LC, Murray SS, et al. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2011;168(9):930–46. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Tarabeux J, Kebir O, Gauthier J, et al. Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry. 2011;1:e55. doi: 10.1038/tp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Kirov G, Pocklington AJ, Holmans P, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17(2):142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Gu Z, Jiang Q, Fu AK, Ip NY, Yan Z. Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J Neurosci. 2005;25(20):4974–84. doi: 10.1523/JNEUROSCI.1086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Li JT, Feng Y, Su YA, Wang XD, Si TM. Enhanced interaction among ErbB4, PSD-95 and NMDAR by chronic MK-801 treatment is associated with behavioral abnormalities. Pharmacol Biochem Behav. 2013;108:44–53. doi: 10.1016/j.pbb.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 314.Pitcher GM, Kalia LV, Ng D, et al. Schizophrenia susceptibility pathway neuregulin 1-ErbB4 suppresses Src upregulation of NMDA receptors. Nat Med. 2011;17(4):470–8. doi: 10.1038/nm.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Millar JK, Pickard BS, Mackie S, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310(5751):1187–91. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 316.Scott DB, Blanpied TA, Ehlers MD. Coordinated PKA and PKC phosphorylation suppresses RXR-mediated ER retention and regulates the surface delivery of NMDA receptors. Neuropharmacology. 2003;45(6):755–67. doi: 10.1016/s0028-3908(03)00250-8. [DOI] [PubMed] [Google Scholar]
- 317.Ma TM, Abazyan S, Abazyan B, et al. Pathogenic disruption of DISC1-serine racemase binding elicits schizophrenia-like behavior via D-serine depletion. Mol Psychiatry. 2013;18(5):557–67. doi: 10.1038/mp.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Ramsey AJ, Milenkovic M, Oliveira AF, et al. Impaired NMDA receptor transmission alters striatal synapses and DISC1 protein in an age-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(14):5795–800. doi: 10.1073/pnas.1012621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Blanpied TA, Scott DB, Ehlers MD. Dynamics and regulation of clathrin coats at specialized endocytic zones of dendrites and spines. Neuron. 2002;36(3):435–49. doi: 10.1016/s0896-6273(02)00979-0. [DOI] [PubMed] [Google Scholar]
- 320.Nong Y, Huang YQ, Salter MW. NMDA receptors are movin’ in. Curr Opin Neurobiol. 2004;14(3):353–61. doi: 10.1016/j.conb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 321.Jeans A, Malins R, Padamsey Z, Reinhart M, Emptage N. Increased expression of dysbindin-1A leads to a selective deficit in NMDA receptor signaling in the hippocampus. Neuropharmacology. 2011;61(8):1345–53. doi: 10.1016/j.neuropharm.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 322.Hayashi T, Thomas GM, Huganir RL. Dual palmitoylation of NR2 subunits regulates NMDA receptor trafficking. Neuron. 2009;64(2):213–26. doi: 10.1016/j.neuron.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Brock J, Brown CC, Boucher J, Rippon G. The temporal binding deficit hypothesis of autism. Dev Psychopathol. 2002;14(2):209–24. doi: 10.1017/s0954579402002018. [DOI] [PubMed] [Google Scholar]
- 324.Gai X, Xie HM, Perin JC, et al. Rare structural variation of synapse and neurotransmission genes in autism. Mol Psychiatry. 2012;17(4):402–11. doi: 10.1038/mp.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Barnby G, Abbott A, Sykes N, et al. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. Am J Hum Genet. 2005;76(6):950–66. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–4. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Duffney LJ, Wei J, Cheng J, et al. Shank3 deficiency induces NMDA receptor hypofunction via an actin-dependent mechanism. J Neurosci. 2013;33(40):15767–78. doi: 10.1523/JNEUROSCI.1175-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Shcheglovitov A, Shcheglovitova O, Yazawa M, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503(7475):267–71. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kouser M, Speed HE, Dewey CM, et al. Loss of predominant Shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. J Neurosci. 2013;33(47):18448–68. doi: 10.1523/JNEUROSCI.3017-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Budreck EC, Kwon OB, Jung JH, et al. Neuroligin-1 controls synaptic abundance of NMDA-type glutamate receptors through extracellular coupling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(2):725–30. doi: 10.1073/pnas.1214718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Irie M, Hata Y, Takeuchi M, et al. Binding of neuroligins to PSD-95. Science. 1997;277(5331):1511–5. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 332.Maliszewska-Cyna E, Bawa D, Eubanks JH. Diminished prevalence but preserved synaptic distribution of N-methyl-D-aspartate receptor subunits in the methyl CpG binding protein 2(MeCP2)-null mouse brain. Neuroscience. 2010;168(3):624–32. doi: 10.1016/j.neuroscience.2010.03.065. [DOI] [PubMed] [Google Scholar]
- 333.Townsend M, Yoshii A, Mishina M, Constantine-Paton M. Developmental loss of miniature N-methyl-D-aspartate receptor currents in NR2A knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(3):1340–5. doi: 10.1073/pnas.0335786100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Fu Z, Logan SM, Vicini S. Deletion of the NR2A subunit prevents developmental changes of NMDA-mEPSCs in cultured mouse cerebellar granule neurones. J Physiol. 2005;563(Pt 3):867–81. doi: 10.1113/jphysiol.2004.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Shepherd GM, Katz DM. Synaptic microcircuit dysfunction in genetic models of neurodevelopmental disorders: focus on Mecp2 and Met. Curr Opin Neurobiol. 2011;21(6):827–33. doi: 10.1016/j.conb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Blue ME, Naidu S, Johnston MV. Development of amino acid receptors in frontal cortex from girls with Rett syndrome. Ann Neurol. 1999;45(4):541–5. doi: 10.1002/1531-8249(199904)45:4<541::aid-ana21>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 337.Blue ME, Naidu S, Johnston MV. Altered development of glutamate and GABA receptors in the basal ganglia of girls with Rett syndrome. Exp Neurol. 1999;156(2):345–52. doi: 10.1006/exnr.1999.7030. [DOI] [PubMed] [Google Scholar]
- 338.Zalfa F, Eleuteri B, Dickson KS, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10(5):578–87. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Schutt J, Falley K, Richter D, Kreienkamp HJ, Kindler S. Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. J Biol Chem. 2009;284(38):25479–87. doi: 10.1074/jbc.M109.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Eadie BD, Cushman J, Kannangara TS, Fanselow MS, Christie BR. NMDA receptor hypofunction in the dentate gyrus and impaired context discrimination in adult Fmr1 knockout mice. Hippocampus. 2012;22(2):241–54. doi: 10.1002/hipo.20890. [DOI] [PubMed] [Google Scholar]
- 341.Bostrom CA, Majaess NM, Morch K, White E, Eadie BD, Christie BR. Rescue of NMDAR-dependent synaptic plasticity in Fmr1 knock-out mice. Cereb Cortex. 2015;25(1):271–9. doi: 10.1093/cercor/bht237. [DOI] [PubMed] [Google Scholar]