Summary
High-fidelity binding of transcription factors (TFs) to DNA target sites is fundamental for proper regulation of cellular processes, as well as for the maintenance of cell identity. Recognition of cognate binding motifs in the genome is attributed by and large to the DNA binding domains of TFs. As an additional mode of conferring binding specificity, noncoding RNAs (ncRNAs) have been proposed to assist associated TFs in finding their binding sites by interacting with either DNA or RNA in the vicinity of their target loci. However, a well-documented example of such a mechanism was lacking until we recently reported that a ncRNA made by Epstein-Barr virus uses an RNA-RNA interaction with nascent transcripts generated from the viral genome to facilitate the recruitment of an interacting TF, PAX5, to viral DNA. This proof-of-principle finding suggests that cellular ncRNAs may likewise function in guiding interacting TFs to chromatin target sites.
Keywords: noncoding RNA, transcription factor binding, trans-acting ncRNAs, transcription factor recruitment
Introduction
With the advent of deep sequencing technology, pervasive transcription of at least 80% of the genome has been uncovered [1]. Since protein-coding genes are limited to ∼2%, the vast majority of transcripts is considered to be non-protein coding. Several studies have mined the wealth of deep sequencing data and described a novel class of noncoding transcripts termed long noncoding RNAs (lncRNAs) [2-4], arbitrarily defined as transcripts longer than 200 nucleotides (nts). Even though the physiological relevance, as well as the mode of action, of only a tiny fraction of lncRNAs has been established [5], it is generally accepted that all transcripts are produced for a reason, as cells should not waste energy and resources to generate completely nonfunctional ncRNAs. An interesting feature of some lncRNAs is their association with transcription factors (TFs) [6-8], implicating them in transcription regulation. Three distinct modes of action can be envisaged whereby TF-interacting ncRNAs might affect transcription. They could act as: i) scaffolds (for one or more TFs) [9-11], ii) tethers that remain at the site of transcription and thus recruit an interacting TF [7], or iii) trans-acting guides for interacting TFs by base pairing with DNA or RNA in the vicinity of their target sites.
The ability to form complementary base pairs is a property of all nucleic acids and fundamental to the mode of action of all well-characterized classes of ncRNAs: ribosomal RNAs (rRNA), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and tiny RNAs (short-interfering/micro/PIWI-interacting RNAs). Notable instances of RNA-RNA interactions include the 16S rRNA sequence that base pairs with the Shine-Dalgarno sequence in bacterial mRNAs [12], tRNAs interacting with mRNA codons [13], snRNAs binding to exon-intron junctions of nascent pre-mRNAs during the process of splicing [14], snoRNAs base pairing with RNAs targeted for chemical modification [15], and si/mi/piRNAs base pairing with target mRNAs [16]. Thus, it is expected that lncRNAs as well may utilize complementary base pairing as an integral aspect of their function. However, cellular lncRNAs that interact with TFs have not yet been reported to act as guides via a base-pairing mechanism.
Examples of ncRNAs that recruit an effector protein, other than a TF, to a target site on the genome by base pairing with either DNA or RNA have recently been reported for smaller ncRNAs. One example is the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 (CRISPR associated gene 9) system, which has garnered great attention because of its powerful application in genome editing and transcriptional interference [17,18]. Here a ncRNA called CRISPR-RNA recruits its interacting effector protein, the endonuclease Cas9, to a target site by complementary base pairing with one strand of the DNA duplex. Short-interfering RNAs in fission yeast constitute another example of an effector protein-guiding ncRNA; they recruit the RNA-Induced Transcriptional Silencing (RITS) complex to heterochromatic regions by complementary base pairing with nascent transcripts from these sites [19]. This expanding number of ncRNAs that mediate targeting of effector proteins to chromatin via base pairing suggests that lncRNAs may likewise utilize base-pairing-mediated targeting of interacting TFs as well. We have recently described how a ncRNA from the Epstein-Barr virus (EBV) base pairs with nascent transcripts to facilitate recruitment of an interacting TF, PAX5, to binding sites on the EBV genome [20]. As effective mechanisms tend to be re-used, our results point to future identification of cellular lncRNAs that function in an analogous manner.
A viral ncRNA guides an interacting TF to its target site via RNA-RNA interaction with nascent transcripts
EBV is a human gamma-herpesvirus that infects B lymphocytes [21]. It is the causative agent of mononucleosis and is associated with several types of tumors, including lymphomas and carcinomas [22]. Two approximately 170 nt-long ncRNAs called EBV-encoded RNA 1 (EBER1) and EBER2 are expressed in infected cells at high levels (∼106 copies) [23,24] and localize exclusively to the nucleoplasm [25,26]. Their high copy number has made the EBERs an ideal diagnostic tool for EBV infection, as these viral RNAs can be readily detected by in situ hybridization in tissue samples [27]. Their abundance also suggests important functions in the life cycle of the virus, since cellular ncRNAs expressed at comparable levels are essential to the host cell, such as the small nuclear RNAs involved in the process of splicing. EBV strains with deletion in the EBER locus have been generated, but conflicting observations regarding their phenotype were reported. One study suggested that EBERs promote B-cell growth and transformation [28], whereas other studies did not find any phenotype for EBER deletions [29,30]. Thus, despite the abundance of the EBERs, their molecular modes of action have remained elusive until recently.
Given their nuclear localization, we probed EBERs for possible chromatin localization by Capture Hybridization Analysis of RNA Targets (CHART), a method comparable to chromatin immunoprecipitation but aimed at identification of RNA binding sites on the genome [31]. A prerequisite for CHART is the availability of an accessible region within the ncRNA of interest that can hybridize to an antisense oligonucleotide for selection. While EBER1 has no accessible sites and does not lend itself to CHART [32], EBER2 has two regions that are accessible for antisense oligonucleotide hybridization (Fig. 1A). To study whether EBER1 is also targeted to specific sites on either host or EBV genome, the conventional CHART protocol would require modification. One possibility would be to insert an aptamer into EBER1 to add an artificial selection surface [32]; since it would be impossible to know whether EBER1 function had been disrupted, ideally several such tags should be used.
Figure 1.
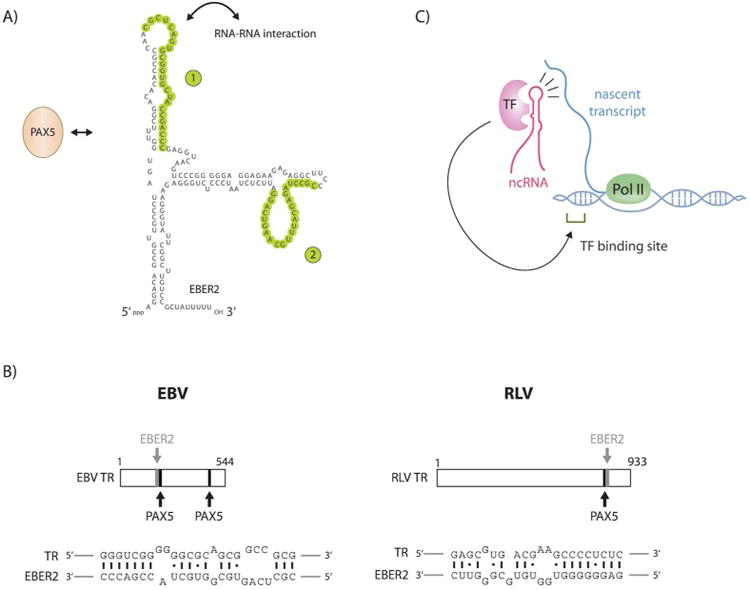
EBER2 facilitates binding of PAX5 to TRs via RNA-RNA interaction with nascent transcripts. A: The two accessible regions of EBER2 are marked in green, one of which engages in RNA-RNA interaction with nascent transcripts from the TR regions. PAX5 binding to EBER2 is indicated. B: A single TR unit of EBV and of RLV is shown; the EBER2-interacting region and PAX5 consensus sites are indicated (top). The predicted RNA-RNA interaction between EBER2 and the TR RNA is shown for EBV and RLV (bottom). C: Model for cellular ncRNAs in guiding interacting TFs to target sites by base pairing with nascent transcripts.
Employing CHART, EBER2 was found to localize to the so-called terminal repeats (TRs) of the latent EBV genome [20]. TRs are tandem direct repeat units of approximately 550 base pairs that flank both ends of the linear genome found in virions and are the site of circularization to form the viral episome after its entry into the host cell [33]. Interestingly, the EBER2 binding sites at the TRs coincide with previously identified binding sites for the B-cell TF PAX5 (Fig. 1B). PAX5 and EBF1 are the two major B-cell specificity-determining TFs [34,35]. Thus, EBV evolved to select PAX5, one of the master regulators of the cell type it primarily infects, as an interacting partner for one of its own gene products. The chromatin co-localization of EBER2 and PAX5 suggested a collaborative action, and indeed these factors interact with each other, probably indirectly through an as-yet unidentified bridging factor. Functionally, knockdown of either factor has two outcomes: i) viral genes located nearest their binding sites become de-regulated during latency; and ii) when the viral lytic cycle is induced, EBV genome replication is compromised [20].
Most unexpected is the recruitment mechanism of the EBER2-PAX5 complex to its binding sites in the TRs of the EBV genome. Since EBER2 depletion abrogates PAX5 binding to the TRs, but PAX5 depletion has no effect on EBER2 localization to the TRs, EBER2 appears to be the key recruiting component of the complex. Indeed, EBER2 base pairs with nascent transcripts generated from the TR regions, thus potentially guiding the interacting PAX5 TF to its consensus sequence sites within the TRs and facilitating its binding. Consistent with recruitment being mediated by RNA-RNA interactions, one of the accessible regions within EBER2 (marked as site 1 in Fig. 1A) was predicted in silico and subsequently shown in vitro to interact with nascent transcripts from the TR region (Fig. 1C). Another piece of evidence suggesting an RNA-RNA based mechanism, rather than EBER2 interaction with DNA, is our observation that RNase H treatment prior to CHART did not abrogate EBER2 localization to the TRs. The precise function of the second accessible region in EBER2 (site 2 in Fig. 1A), which we targeted by antisense oligonucleotides during the CHART procedure, remains to be determined. Possibly the second site represents an additional interaction interface, for either another RNA or a protein. EBER2 binding to chromatin appears to be restricted to the TR regions of the EBV genome, as no apparent binding sites on host chromatin were observed by CHART.
Evolutionary conservation of a molecular mechanism across related viral species provides important evidence of functionality. The rhesus lymphocryptovirus (RLV), a gamma-herpesvirus related to EBV [36,37], harbors in its genome an EBER2 homolog that can be modeled to adopt an equivalent secondary structure to EBV EBER2. Even though the primary sequences of the EBER2 homologs are only 65% conserved and the TR sequences between the two species are not conserved except for a high GC-content, prediction of RNA-RNA interactions between the RLV-EBER2 and the RLV TR RNA sequence revealed a potential duplex. It exhibits remarkably divergent base pairing as compared to EBV (Fig. 1B, bottom), but nonetheless displays two striking common features: i) the region in RLV-EBER2 predicted to form base pairs is in the same relative location within the ncRNA (top stem-loop region/site 1 in Fig. 1A); ii) the predicted interaction site on the TR RNA is adjacent to the PAX5 consensus sequence site on the TR DNA sequence (Fig. 1B) [20]. Whether other gamma-herpesviruses that contain an EBER2 homolog employ such RNA-RNA interaction remains to be studied once their complete genome sequences become available.
Such a guide function, bringing an interacting TF to its DNA target site, has not previously been described for trans-acting ncRNAs. As viruses often adopt host mechanisms and pathways to enhance their own replication, our results suggest that cellular ncRNAs comparable to EBER2 may exist. These would engage in RNA-RNA interactions with nascent transcripts to guide other TFs to their target sites on host chromatin, possibly employing the same protein components that participate in EBER2's mode of action (Fig. 1C). Further analysis of the composition of the EBER2-PAX5 complex will enable the identification of these cellular guide ncRNAs.
Do ncRNAs enhance transcription factor binding to target sites?
Since PAX5 contains a paired domain that recognizes a cognate DNA motif present in the TRs [38], why should a ncRNA, EBER2, be required to facilitate PAX5 binding to these sites on the viral genome? Genome-wide location analysis of Pax5 in murine B cells has identified, based on the ∼8000 genomic binding sites, a consensus sequence comprising a 15-bp degenerate motif with sequence variations at each position [39]. This degenerate motif occurs more frequently in the genome than the ∼8000 reported Pax5 binding sites. Thus, bona fide PAX5 binding to the EBV TR regions, each of which contains two perfect consensus sequence sites (Fig. 1B), may require EBER2 to enhance the binding specificity or increase the affinity of PAX5 once bound.
The concept of co-factor mediated enhancement of binding specificity is well established for TFs, as exemplified by the Hox gene-encoded homeodomain-containing TFs that play a role in determining cell identity along the anterior-posterior axis [40]. Each Hox TF regulates a different set of target genes, yet their DNA binding homeodomain, encoded by the homeobox, is highly conserved, resulting in Hox TFs recognizing nearly identical DNA sequences in vitro [41]. Target gene specificity of Hox TFs in vivo is achieved through the protein co-factor Extradenticle (Exd) [42,43]. Interaction with Exd alters DNA recognition and ensures proper recruitment of Hox TFs. A similar scenario can be envisaged for EBER2 and PAX5 binding at the TRs. EBER2's interaction with nascent transcripts from the TR regions might be necessary to bring PAX5 to the vicinity of its consensus sequence sites within the TRs, whereupon it binds to these sites with high affinity. The role of EBER2 could thus be compared to the role of a tugboat that maneuvers PAX5 to its appropriate landing pad, ignoring the many low-affinity sites in the genome. Here, EBV chose to utilize a ncRNA, EBER2, rather than a viral protein as a specificity-enhancing co-factor. The advantages of employing an RNA are twofold: i) given the non-immunogenic properties of RNAs, this approach enables EBV-infected cells to slip under the radar of the host immune defense system; ii) less genetic information needs to be stored in the viral genome, which is a concern for viruses where space constraints are dictated by the size of the viral capsid. Encoding a DNA binding protein consumes significantly more genetic information, as a typical DNA binding domain that recognizes 5-10 bp of DNA comprises ∼60 amino acids, requiring 180 bp of DNA. In contrast, a ncRNA requires only the same number of complementary nucleotides as the DNA/RNA region it recognizes.
PAX5 binding sites within the EBV TR regions differs from canonical PAX5 sites found at cellular promoters, as the TR regions are located within an intron of a viral latent gene (LMP2) [44]. While promoters are binding hotspots for multiple TFs and auxiliary factors (e.g. components of the general transcription machinery that synergize for efficient transcription initiation and elongation), Arvey et al. reported that the EBV TR regions are bound solely by PAX5 and devoid of binding sites for any other host transcription regulatory factor studied by the ENCODE project [45]. Additional binding specificity of PAX5 for promoter regions may be conferred by chromatin regulatory factors, some of which have been shown to interact with PAX5, such as the components of the BAF chromatin remodeling complex and histone 3 lysine 4 methyltransferase complexes [46]. At the TR regions, EBER2, via base pairing with nascent transcript, could substitute for a protein co-factor that boosts PAX5 binding specificity by bringing it to the vicinity of a PAX5 consensus sequence or stabilizing the interaction with DNA once formed.
Why does the EBV life cycle need a ncRNA?
Depletion of either EBER2 or PAX5 results in transcriptional upregulation of EBV genes located nearby the TRs, indicating a suppressive role for EBER2-PAX5. The effect of EBER2, or PAX5, depletion on transcription of these EBV genes becomes apparent only after three days of knockdown, even though EBER2 levels are markedly reduced (<20% its original level) after several hours. This finding suggests that the EBER2-PAX5 complex interacts stably with the TR regions and only prolonged depletion disturbs its regulatory role. We thus speculate that EBER2-PAX5 is involved in properly organizing the viral chromatin, especially the direct tandem repeats of the TR regions, which are present at up to 20 copies per episome[47]. As EBV episomes are also present in multiple copies (up to 50) in an infected cell [48], improper genome organization could result in deleterious genome instability; the repeat regions could potentially recombine with those from other episomes [49].
Support for the idea that the EBER2-PAX5 complex orchestrates the formation of heterochromatin around the TR regions comes from observations on the murine major satellite repeats. Here, TFs also of the PAX family, Pax3 and Pax9, bind and recruit a histone methyltransferase, Suv39h, which deposits repressive histone marks (trimethylation of histone H3 on lysine 9), resulting in heterochromatinization [50]. Interestingly, ncRNAs generated from the major satellite repeats of mouse DNA have been proposed to be essential for this process as well. These nascent transcripts might function similarly to those corresponding to the EBV-TR intron in recruiting an RNA-TF complex. Thus, the parallels with our findings for the EBV TR regions are striking: both situations involve repeated DNA sequences that contain TF binding sites, nascent transcripts from these repeat regions, and participation of the PAX family of TFs. The transcriptional upregulation we observe upon loss of EBER2 or PAX5 binding to the TRs suggests that the EBER2-PAX5 complex might likewise recruit writers of repressive histone marks to induce heterochromatinization, an idea that is yet to be tested. Even though only repeat regions have been implicated in ncRNA-mediated recruitment of TFs so far, non-repeat genomic binding sites, as well as TFs other than those of the PAX family, could also employ an RNA-guide mechanism.
Conclusions and outlook
NcRNAs utilize base pairing as a fundamental aspect of their mode of action. We hypothesize that cellular (l)ncRNAs recruit and/or enhance TF binding to target sites by base pairing with nascent transcripts generated in the vicinity of their binding sites. Many reported binding motifs of TFs contain degenerate sequences, implying the existence of an additional layer of regulation to find their proper binding sites in the genome. Protein co-factors have been shown to modulate TF binding specificity; ncRNAs could act similarly and substitute for protein co-factors. By base pairing with complementary sequences within nascent transcripts generated nearby target sites, ncRNAs are ideally suited to increase the specificity of TF binding and to provide an expanded landing platform for the ncRNA-TF complex. Given the precedent of the viral EBER2 ncRNA facilitating the binding of its interacting TF PAX5, cellular ncRNAs may employ similar recruitment mechanisms. EBER2 and PAX5 interact indirectly through as-yet unidentified bridging factor(s). The same set of protein factors might be involved in the mode of action of cellular guide ncRNAs. Future studies will reveal how prevalent such ncRNA-mediated recruitment of TFs is in host cells.
Acknowledgments
We thank Dr. Johanna Withers for comments and Angela Miccinello for editorial assistance. JAS is an investigator of the Howard Hughes Medical Institute and supported by grant CA16038 from the NCI.
Footnotes
The authors have declared no conflict of interest.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guttman M, Amit I, Garber M, French C, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer MK, Niknafs YS, Malik R, Singhal U, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabili MN, Trapnell C, Goff L, Koziol M, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Khalil AM, Guttman M, Huarte M, Garber M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Sun BK, Erwin JA, Song JJ, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–5. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 10.Tsai MC, Manor O, Wan Y, Mosammaparast N, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–22. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 12.Steitz JA, Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975;72:4734–8. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthaei JH, Nirenberg MW. Characteristics and stabilization of DNAase-sensitive protein synthesis in E. coli extracts. Proc Natl Acad Sci U S A. 1961;47:1580–8. doi: 10.1073/pnas.47.10.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 16.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert LA, Larson MH, Morsut L, Liu Z, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 19.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160:607–18. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 22.Delecluse HJ, Feederle R, O'Sullivan B, Taniere P. Epstein Barr virus-associated tumours: an update for the attention of the working pathologist. J Clin Pathol. 2007;60:1358–64. doi: 10.1136/jcp.2006.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1981;78:805–9. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tycowski KT, Guo YE, Lee N, Moss WN, et al. Viral noncoding RNAs: more surprises. Genes Dev. 2015;29:567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fok V, Friend K, Steitz JA. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J Cell Biol. 2006;173:319–25. doi: 10.1083/jcb.200601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howe JG, Steitz JA. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc Natl Acad Sci U S A. 1986;83:9006–10. doi: 10.1073/pnas.83.23.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulley ML, Tang W. Laboratory assays for Epstein-Barr virus-related disease. J Mol Diagn. 2008;10:279–92. doi: 10.2353/jmoldx.2008.080023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yajima M, Kanda T, Takada K. Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J Virol. 2005;79:4298–307. doi: 10.1128/JVI.79.7.4298-4307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregorovic G, Bosshard R, Karstegl CE, White RE, et al. Cellular gene expression that correlates with EBER expression in Epstein-Barr Virus-infected lymphoblastoid cell lines. J Virol. 2011;85:3535–45. doi: 10.1128/JVI.02086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci U S A. 1991;88:1546–50. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon MD, Wang CI, Kharchenko PV, West JA, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee N, Pimienta G, Steitz JA. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA. 2012;18:2073–82. doi: 10.1261/rna.034900.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindahl T, Adams A, Bjursell G, Bornkamm GW, et al. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976;102:511–30. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- 34.Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol. 2011;111:179–206. doi: 10.1016/B978-0-12-385991-4.00005-2. [DOI] [PubMed] [Google Scholar]
- 35.Nechanitzky R, Akbas D, Scherer S, Gyory I, et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14:867–75. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 36.Lacoste V, Lavergne A, de Thoisy B, Pouliquen JF, et al. Genetic diversity and molecular evolution of human and non-human primate Gammaherpesvirinae. Infect Genet Evol. 2010;10:1–13. doi: 10.1016/j.meegid.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Rivailler P, Jiang H, Cho YG, Quink C, et al. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J Virol. 2002;76:421–6. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Busslinger M, Urbanek P. The role of BSAP (Pax-5) in B-cell development. Curr Opin Genet Dev. 1995;5:595–601. doi: 10.1016/0959-437x(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 39.Revilla IDR, Bilic I, Vilagos B, Tagoh H, et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31:3130–46. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 41.Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan SK, Jaffe L, Capovilla M, Botas J, et al. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–15. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 43.Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, et al. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–74. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 44.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–9. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 45.Arvey A, Tempera I, Tsai K, Chen HS, et al. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12:233–45. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McManus S, Ebert A, Salvagiotto G, Medvedovic J, et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30:2388–404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown NA, Liu CR, Wang YF, Garcia CR. B-cell lymphoproliferation and lymphomagenesis are associated with clonotypic intracellular terminal regions of the Epstein-Barr virus. J Virol. 1988;62:962–9. doi: 10.1128/jvi.62.3.962-969.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugden B, Phelps M, Domoradzki J. Epstein-Barr virus DNA is amplified in transformed lymphocytes. J Virol. 1979;31:590–5. doi: 10.1128/jvi.31.3.590-595.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bzymek M, Lovett ST. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A. 2001;98:8319–25. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bulut-Karslioglu A, Perrera V, Scaranaro M, de la Rosa-Velazquez IA, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol. 2012;19:1023–30. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]


