Abstract
The past two decades have seen an explosion in research on noncoding RNAs and their physiological and pathological functions. Several classes of small (20–30 nucleotides) and long (>200 nucleotides) noncoding RNAs have been firmly established as key regulators of gene expression in myriad processes ranging from embryonic development to innate immunity. In this review, we focus on our current understanding of the molecular mechanisms underlying the biogenesis and function of small interfering RNAs (siRNAs), microRNAs (miRNAs), and Piwi-interacting RNAs (piRNAs). In addition, we briefly review the relevance of small and long noncoding RNAs to human physiology and pathology and their potential to be exploited as therapeutic agents.
Introduction
For decades, the major cellular function of RNA was considered to be an intermediate molecule in the transfer of genetic information from DNA to proteins. While transcription is a pervasive process and the majority of the genome is transcribed (Clark et al., 2011), the scientific community was caught by a surprise when human genome sequencing revealed ~21,000 protein coding genes (Lander et al., 2001), a number that is similar to less complex species such as Caenorhabditis elegans. With the advent of robust HTS (High throughput sequencing) technologies and advances in bioinformatics, the ENCODE project has provided a detailed landscape of transcription in human cells (Frazer, 2012, Stamatoyannopoulos, 2012, Djebali et al., 2012). The human genome encodes tens of thousands of small and long RNA transcripts that do not code for proteins suggesting a notion that this non-coding “dark matter” of genome plays a role in physiological complexity of human and mammals. Biological functions of non-coding small RNAs started to emerge at a fast pace after the role of RNAs in gene silencing was discovered (Fire et al., 1998). In addition to small RNAs, recent genome-wide surveys have revealed that many regions encode long noncoding RNAs (lncRNAs) that may have important biological functions (Derrien et al., 2012). Here we discuss the molecular mechanisms of non-coding RNAs, their regulation of gene expression under physiological settings, and their involvement in the initiation and progression of disease. We will end with a brief summary of recent progress in exploiting ncRNAs as therapeutic agents.
Small noncoding RNAs
Small ncRNAs (~20–30 nucleotides [nt]) are central components of RNA interference (RNAi), an ancient gene regulation program that operates in a wide variety of organisms ranging from fission yeast to humans (Fire et al., 1998, Ambros, 2004, Cogoni and Macino, 1997, Hamilton and Baulcombe, 1999). Small ncRNAs can be grouped into three main categories based on their length, mode of biogenesis, and effector proteins: small interfering RNAs (siRNAs), microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs). By serving as guides for recognition of target RNAs, these molecules play a key role in modulating gene expression in a sequence-specific manner. In the following sections, we discuss the biogenesis and biological functions of these small regulatory RNAs.
Small interfering RNAs
Small interfering RNAs (siRNAs) are derived from long double-stranded precursor RNAs (dsRNAs). Endogenously formed dsRNAs are exported to the cytoplasm where they are cleaved into 20–25-nt duplexes by Dicer, a member of the dsRNA-specific ribonuclease (RNase) III family (Bernstein et al., 2001). Mammals and the nematode Caenorhabditis elegans each possess a single Dicer protein, whereas the Drosophila genome encodes two: Dicer-1 (Dcr-1) and Dicer-2 (Dcr-2) (Hutvagner et al., 2001, Grishok et al., 2001, Ketting et al., 2001, Knight and Bass, 2001, Lee et al., 2004b). In contrast to mammals and C. elegans, where the single Dicer is responsible for generating both siRNA and miRNAs, Drosophila Dcr-1 and Dcr-2 are dedicated to the production of miRNAs and siRNAs, respectively (Lee et al., 2004b). The siRNA duplexes formed by Dicer activity bear 5′ phosphates and 3′ hydroxyl groups on both strands, which are paired in a manner that leaves 2-nt overhangs at the 3′ ends (Figure 1). siRNAs are incorporated into multiprotein RNA-induced silencing complexes (RISCs) comprised of one of a family of Argonaute (AGO) proteins together with auxiliary proteins that extend or modify the function of the AGO protein (Hammond et al., 2001, Caudy et al., 2002, Ishizuka et al., 2002, Zhou et al., 2008). One strand of the siRNA duplex, the guide strand, is selectively retained in the siRISC and the second ‘passenger’ strand is discarded (Miyoshi et al., 2005, Rand et al., 2005, Matranga et al., 2005). AGO proteins contain a PAZ (Piwi, Argonaute, Zwille) domain and a Piwi domain. The PAZ domain accommodates the protruding 2-nt overhang of the siRNA duplex, and the PIWI domain, which is structurally similar to the catalytic domain of RNase H, carries endoribonuclease (or slicer) activity (Song et al., 2003, Ma et al., 2004, Parker et al., 2005, Song et al., 2004, Wang et al., 2009, Wang et al., 2008b). The AGO-bound siRNA guide strand directs the siRISC to target mRNAs through complementary base pairing, and the AGO PIWI domain endonuclease activity precisely cuts the target RNA at the phosphodiester linkage between the nucleotides base paired to the 10th and 11th residues of the siRNA guide strand. Among the four mammalian AGO proteins, only AGO2 possesses such slicer activity, whereas the Drosophila Ago1 and Ago2 proteins carry weak and robust slicer activities, respectively. AGO-mediated cleavage of target mRNAs generates products with 5′ monophosphates and 3′ hydroxyl termini, which are further degraded by exoribonucleases. This frees the siRISC, still containing the siRNA guide strand, to engage and cleave additional target mRNAs. This sequence of events is shown schematically in Figure 1. Chemical modification and mutational analyses of siRNAs have established the essential role of A-helical geometry in siRNA-mediated gene silencing (Chiu and Rana, 2002, Chiu and Rana, 2003, Chu and Rana, 2007, Rana, 2007). High-resolution crystal structures of Ago bound to a guide strand and its target RNA further highlighted the significance of the A-form helix in RISC catalysis (Wang et al., 2009). Since RNA can fold into complex secondary and tertiary structures, rates of RISC catalysis can be affected by the structure of target mRNA sequences because mRNA regions with strong secondary structures, such as hairpin and stem loops, are resistant to targeting by RISCs (Brown et al., 2005, Schubert et al., 2005, Overhoff et al., 2005).
Figure 1. siRNA-mediated gene regulation.
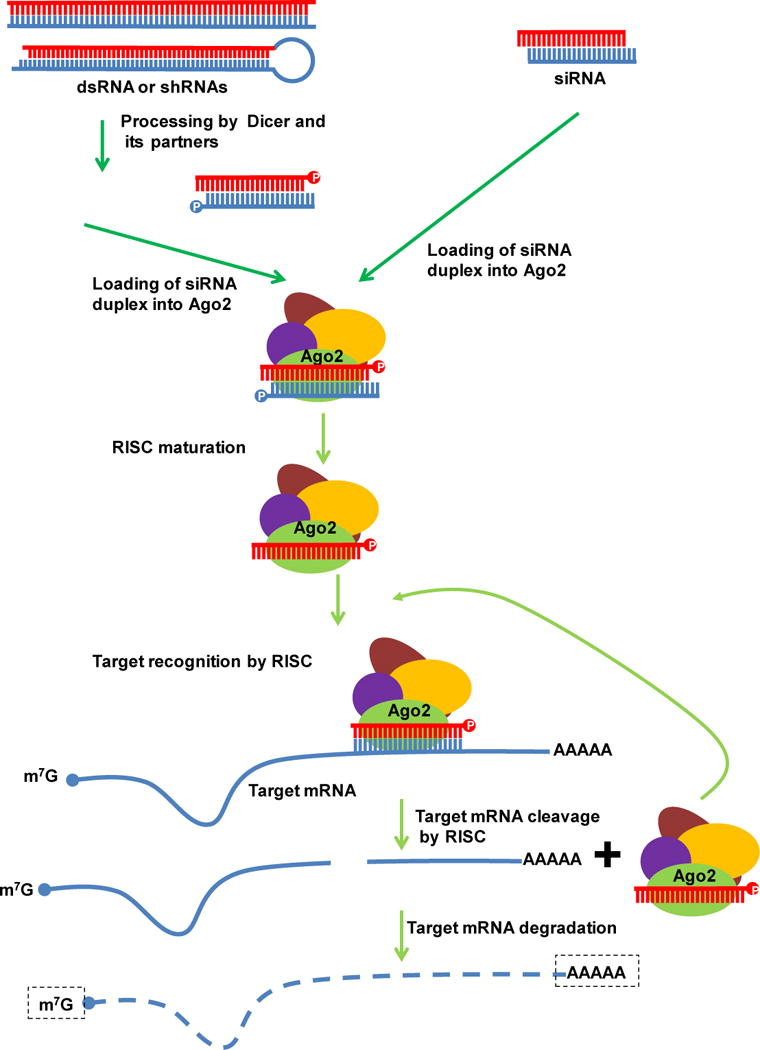
siRNAs can be introduced experimentally into the cell or processed by Dicer and partners from long dsRNAs or shRNAs. siRNAs interact with Ago2 and other proteins to form the RISC complex. During RISC assembly the siRNA guide strand is loaded onto Ago2 while the passenger strand is degraded. Once the active RISC is formed, the siRNA guide strand pairs with, cleaves, and degrades the target mRNA, and. the RISC complex is recycled.
Exo-siRNAs
Depending on the source of dsRNA precursor, siRNAs can be further divided into exogenous and endogenous siRNAs (exo- and endo-siRNAs, respectively). The siRNA pathway is best understood in flies, where exogenous dsRNAs are introduced physiologically as genomic replication intermediates of invading RNA viruses. In Drosophila, exo-siRNA duplexes are produced from the long dsRNAs through a series of cleavage events by Dcr-2 and its partner dsRNA binding protein (dsRBP), R2D2 (Liu et al., 2003, Miyoshi et al., 2010, Marques et al., 2010). The Dcr-2/R2D2 heterodimer gauges the thermodynamic stability of the 5′ ends of the siRNA duplex and binds asymmetrically (Tomari et al., 2004). The strand with lower 5′ thermodynamic stability becomes the guide strand. The Dcr-2/R2D2 complex then associates with Ago2 and the siRNA duplex is transferred, forming the pre-siRISC (Pham et al., 2004). This process requires ATP hydrolysis and is aided by the chaperone proteins Hsc70/Hsp90 (Iwasaki et al., 2010). Ago2 cleaves the siRNA passenger strand, which is then degraded by C3PO, a ribonuclease complex composed of Translin and Trax (Matranga et al., 2005). After removal of the passenger strand, the guide strand is 2′-O-methylated at the 3′ end by the S-adenosyl methionine-dependent methyl transferase HEN1 (Horwich et al., 2007, Pelisson et al., 2007, Saito et al., 2007). These steps convert the pre-RISC to a mature RISC, which then engages target mRNAs by complementary base-paring with the guide strand, leading to destabilization of target RNAs.
In contrast to flies, siRNA biogenesis and RISC assembly in mammals involves three proteins: DCR-1, Transactivation response element RNA-binding protein (TRBP), and AGO2, which form a complex even in the absence of the dsRNA trigger (Chendrimada et al., 2005, Gregory et al., 2005, Maniataki and Mourelatos, 2005, Lee et al., 2006). The complex cleaves long dsRNAs into duplex siRNAs, discards the passenger strand, and assembles into a functional RISC with the guide strand. Interestingly, Dcr-1–null mice can still assemble functional RISCs, suggesting that DCR-1 may be dispensable for RISC loading in mammals (Kanellopoulou et al., 2005, Murchison et al., 2005).
In flies, worms, and plants, the siRNA pathway is a key constituent of the antiviral defense mechanism. Infection of host cells by a wide array of RNA viruses initiates viral genome replication and the formation of long dsRNA intermediates that are specifically recognized and processed into virus-derived siRNAs (vsiRNAs) by Dicer proteins (Hamilton and Baulcombe, 1999, Wang et al., 2006, Li et al., 2002, Lu et al., 2005b). In addition to destroying key viral replication intermediates, this process also results in incorporation of vsiRNAs into AGO-containing siRISCs to further target viral genomic RNAs for degradation (Wang et al., 2006, van Rij et al., 2006). This two-pronged strategy to destroy viral RNA provides specific and effective defense against invading viral pathogens. Indeed, flies, worms, and plants that carry mutations in selected Dicer or Ago genes are hypersensitive to infection by RNA viruses and carry higher viral titers than do the wild-type organisms (Wang et al., 2006, van Rij et al., 2006). As a counterdefense mechanism, many viral genomes encode suppressors of RNAi that inhibit various steps of siRNA biogenesis and function (van Rij et al., 2006, Li et al., 2002, Nayak et al., 2010). vsiRNAs have also been identified in mammalian cells infected with RNA viruses (Schopman et al., 2012); however, it is not yet known whether such RNAs have gene silencing functions or whether the siRNA pathway is required for antiviral immunity in mammals.
Endo-siRNAs
siRNAs can also be derived from long dsRNAs transcribed from plant or animal genomes. Such endo-siRNAs were first detected in plants and C. elegans but were later found in flies and mammals, suggesting that they are common to most eukaryotes.
Three classes of plant endo-siRNAs have been described that originate from different sources: cis-acting siRNAs (casiRNA), trans-acting siRNAs (tasiRNAs), and natural antisense-derived siRNAs (natsiRNAs) (Axtell, 2013). casiRNAs comprise the bulk of plant endo-siRNAs and are derived from transposons, repetitive elements, or tandem repeats such as 5S rRNA genes (Xie et al., 2004). These RNAs are predominantly 24-nt long, are methylated by HEN1, and are produced through the action of DCL3, RDR2, POL IV, and either AGO4 or AGO6. casiRNAs promote the formation of heterochromatin by directing histone modifications and DNA methylation at the loci from which they are derived. tasiRNAs are produced through a convergence of the miRNA and siRNA pathways (Vazquez et al., 2004, Peragine et al., 2004, Yoshikawa et al., 2005, Williams et al., 2005, Allen et al., 2005). In this process, miRNA-directed cleavage of certain transcripts recruits the RNA-dependent RNA polymerase RDR6, which copies the cleaved transcript into dsRNAs. The dsRNAs are then processed by DCL4 into tasiRNAs, which are capable of in trans repression of target transcripts distinct from the tasiRNA locus of origin. natsiRNAs are produced as part of the stress response in plants (Katiyar-Agarwal et al., 2006, Borsani et al., 2005). They are derived from a pair of cis-antisense transcripts in which one transcript is expressed constitutively and the other is expressed in response to environmental stress. In some cases, hybridization of pairs of transcripts derived from non-overlapping genes can give rise to dsRNA precursors. natsiRNA synthesis requires DCL1 and/or DCL2, RDR6, SGS3 (suppressor of gene silencing 3), and Pol IV (Katiyar-Agarwal et al., 2006, Borsani et al., 2005). natsiRNAs then direct the cleavage of one of the mRNAs of the pair and trigger the DCL1-dependent production of 21-nt secondary siRNAs.
Drosophila endo-siRNAs were identified by high-throughput sequencing of small RNAs from germline and somatic cells. They are derived primarily from transposon transcripts, long RNA transcripts with extensive dsRNA structures, and selected mRNAs (Ghildiyal et al., 2008, Czech et al., 2008, Okamura et al., 2008, Kawamura et al., 2008). Drosophila endo-siRNAs are 21-nt long, phased, and present in both sense and antisense orientations. Their production generally requires Dcr-2 and the dsRBP Loquacious (Loqs)-PD (Zhou et al., 2009, Hartig et al., 2009, Hartig and Forstemann, 2011). Drosophila endo-siRNAs are primarily loaded into Ago2-containing siRISCs and are modified by the methyltransferase DmHen1 at their 3′ ends (Horwich et al., 2007, Saito et al., 2007). The primary function of these endo-siRNAs is to control retrotransposon expression in the Drosophila soma and, in concert with piRNAs, the germline. In addition, a small group of cellular mRNAs are subject to regulation by endo-siRNAs (Czech et al., 2008).
The first mammalian endo-siRNA was identified against one of the retrotransposons, L1, a long interspersed nuclear element (LINEs) (Yang and Kazazian, 2006). Bidirectional transcription from the L1 loci is thought to yield dsRNAs that are processed into siRNAs by DCR-1. endo-siRNAs have also been identified in mouse oocytes (Tam et al., 2008, Watanabe et al., 2008). They originate from long dsRNAs derived from transposon transcripts or from hybridization between genic and pseudogenic transcripts. Similar to their Drosophila counterparts, the mouse endo-siRNAs are also 21-nt in length, depend on DCR-1 for biogenesis, and associate with AGO2.
microRNAs
The first miRNA to be discovered, lin-4, was identified in a screen for genes involved in post-embryonic development of C. elegans (Lee et al., 1993). The lin-4 locus produces a 22-nt RNA that is partially complementary to sequences in the 3′ UTR of its target mRNA, lin-14. Although lin-4 is specifically found in C. elegans, the second C elegans miRNA discovered, let-7, is conserved from flies to humans (Reinhart et al., 2000, Pasquinelli et al., 2000). Further studies using a combination of computational and genomic approaches have identified thousands of miRNAs in a variety of organisms, including humans, flies, worms, and plants (Aravin et al., 2003, Lau et al., 2001, Ghildiyal and Zamore, 2009). miRNAs are thought to be expressed in all cell types of higher organisms.
The miRNA genes are mainly clustered in the genome and are transcribed as polycistronic primary transcripts, although some animal miRNAs are produced from individual transcription units (Kim et al., 2009, Ambros, 2011, Li and Rana, 2012). In addition, miRNAs may be embedded in the exonic or intronic regions of host transcripts. miRNA genes are transcribed by RNA Pol II to generate primary miRNAs (pri-miRNA) that carry stem-loop structures containing the future mature miRNA. These transcripts resemble protein-coding transcripts in that they contain a 5′ 7-methylguanosine (m7G) cap and a 3′ poly(A) tail, and often contain introns (Cai et al., 2004, Lee et al., 2004a). Pri-miRNAs are cleaved into ~60–70-nt stem-loop precursor miRNAs (pre-miRNAs) in the nucleus by the microprocessor complex (Denli et al., 2004, Han et al., 2004, Han et al., 2006, Gregory et al., 2004), which consists of the RNase III endoribonuclease Drosha, its dsRBP DGCR8 (also known as Pasha in flies and worms), and auxiliary proteins (Lee et al., 2003, Denli et al., 2004). The Drosha–DGCR8 complex cleaves pri-miRNAs at the base of the stem structure to produce pre-miRNAs having a 5′ phosphate, a 3′ hydroxyl group, and a 2-nt overhang at the 3′ end (Han et al., 2006). The pre-miRNAs are then exported to the cytoplasm by the nuclear export factor Exportin-5 (EXPO-5) which partners with the GTP binding protein Ran to form a nuclear transport complex (Yi et al., 2003, Lund et al., 2004). In the cytoplasm, the pre-miRNAs are further processed by Dicer to generate 22–24-nt miRNA duplexes composed of the mature miRNA strand (containing the less thermostable 5′ end) and a partner strand (known as the miRNA star strand or miRNA*) with a 2-nt overhang at the 3′ terminus. Dicer is assisted by a number of dsRBPs to achieve a high degree of precision and efficiency in miRNA processing. For example, Dcr-1 interacts with the Loqs-PB dsRBP in flies (Zhou et al., 2009, Forstemann et al., 2005, Saito et al., 2005, Hartig et al., 2009) and with TRBP or PACT in mammals (Chendrimada et al., 2005, Lee et al., 2006). In addition to the canonical biogenesis pathway, a subset of miRNAs is derived from transfer RNAs, small nucleolar RNAs, and group II introns via non-canonical mechanisms (Yang and Lai, 2011).
Once the miRNA duplex is produced, only the mature strand is loaded into the RISC complex, whereas the miRNA* strand is typically degraded. However, in Drosophila, select miRNA* strands are loaded into Ago2 and repress the expression of reporter mRNAs containing perfectly complementary sequences (Czech et al., 2009, Okamura et al., 2009). Imperfectly base-paired miRNA duplexes are sorted so that the strand perfectly paired at nucleotide positions 9 and 10 associate with Ago2, whereas the strand that is mismatched in the central region predominantly associates with Ago1 (Czech et al., 2009, Okamura et al., 2009, Ghildiyal et al., 2010). The identity of the 5′ terminal nucleotide of the miRNA strands also affects the sorting process, with Ago1 favoring U and Ago2 C. In mammals, where miRNA sorting into Ago proteins is less well defined, the miRNA strands appear to be randomly partitioned among the four Ago proteins, all of which are equally competent to perform miRNA-mediated gene silencing (Li and Rana, 2012). Once the miRISC is formed, the miRNA imperfectly base pairs with the target mRNA, resulting in mismatch-induced bulges. Target recognition is primarily determined by the “seed sequence” (nucleotides 2–8) at the 5′ end of the miRNA guide strand (Doench and Sharp, 2004, Lewis et al., 2003) and the complementary sequences located predominantly in the 3′ UTR of the target mRNA. Accumulating evidence suggests that miRNA binding sites may also be present in the coding region and possibly the 5′ UTR of mRNAs. miRISC silencing of target mRNAs occurs by repressing protein translation, enhancing mRNA degradation, and/or sequestering target mRNAs to specific cellular compartments such as P bodies and stress granules (Chu and Rana, 2006, Chu and Rana, 2007, Fabian et al., 2010, Krol et al., 2010) (Figure 2). More than 50% of cellular mRNAs are thought to be regulated by miRNAs, emphasizing the far-reaching effects of miRNA-mediated gene regulation.
Figure 2. Biogenesis of miRNAs and their role in gene silencing.
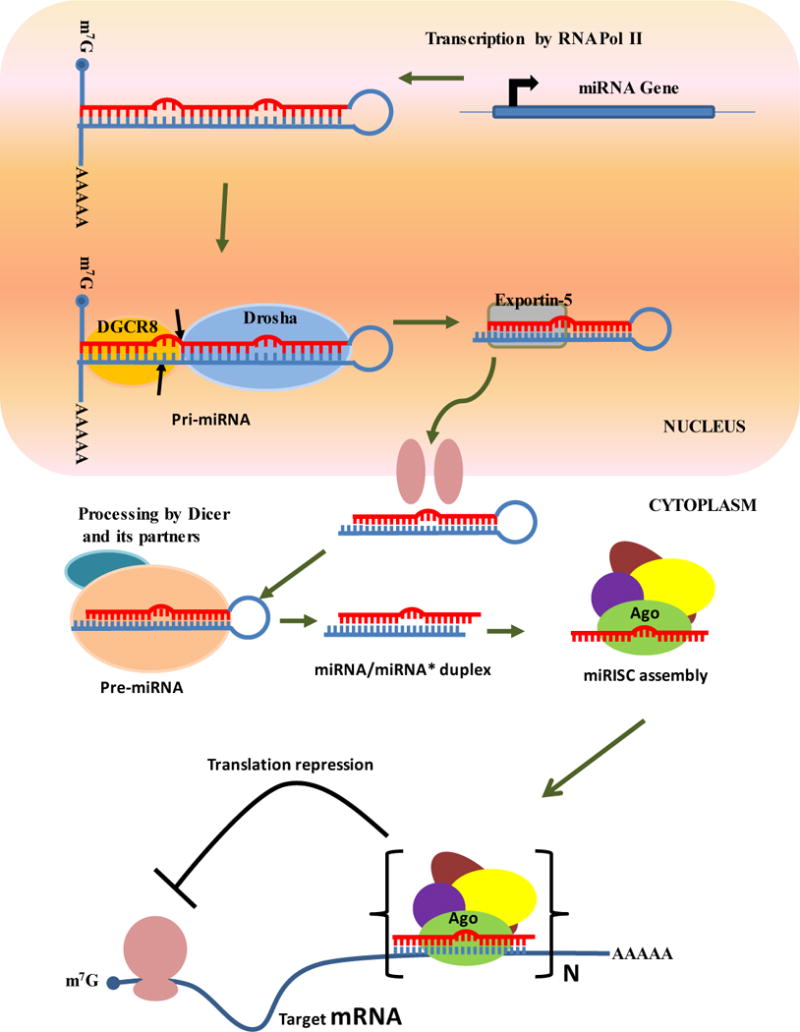
miRNAs are transcribed by RNA Pol II as long hairpin structures called primary miRNAs (pri-miRNAs), which are then processed into ~60–70-nt precursor miRNAs (pre-miRNAs) by the Drosha-DGCR8 complex. The pre-miRNAs are exported out of the nucleus by Exportin-5, and are processed into ~22-nt miRNA/miRNA* duplexes by Dicer and its partners. The 22-nt RNA duplexes associate with Ago and other proteins to form the miRISC complex. miRISCs bind at the 3′ UTR of the target mRNA, in which results translational repression of the target gene. The imperfect base pairing between the miRNA and the target mRNA leads to bulge structures that cannot be cleaved by Ago proteins.
Piwi-interacting RNAs
One of the most recently identified classes of small RNAs are piRNAs, which are the longest known (24–29 nt) small ncRNAs and are found mainly in the animal germline. piRNAs were initially identified in Drosophila by Aravin and coworkers during a study profiling small RNAs expressed at various developmental stages (Aravin et al., 2003). In addition to the 22-nt miRNAs, they identified a fraction of longer RNAs that lacked the complementary star strand sequences generally found in the miRNA/miRNA* precursor structure, suggesting they were a novel class of small ncRNA. The majority of piRNAs mapped to genomic repeats or transposons in Drosophila and hence were initially named repeat-associated small interfering RNAs (rasiRNAs). Later, these RNAs were shown to be involved in defending the germline genome against the deleterious effects of retrotransposons (Vagin et al., 2006). Soon after the discovery of rasiRNAs, several groups identified similar types of RNA in the gonads of many other organisms. Since all of these RNAs were found to associate with the Piwi subfamily of Ago proteins, they were collectively renamed as Piwi-interacting RNAs (piRNAs) (Brennecke et al., 2007, Aravin et al., 2007, Gunawardane et al., 2007, Houwing et al., 2007, Lau et al., 2006, Malone et al., 2009, Saito et al., 2006). piRNAs are produced from long single-stranded RNA precursors in a Dicer-independent manner and thus their biogenesis is quite distinct from that of siRNAs or miRNAs (Vagin et al., 2006, Houwing et al., 2007). To highlight the importance of piRNAs in safeguarding genomic integrity, mutations in genes encoding many components of the piRNA pathway affect fertility in animals (Pek et al., 2012b).
In Drosophila, the Piwi clade proteins associated with piRNAs are Piwi, Aub, and Ago3 (Brennecke et al., 2007, Gunawardane et al., 2007, Li et al., 2009, Malone et al., 2009). The mouse genome encodes three Piwi clade proteins, MIWI, MIWI2, and MILI (Kuramochi-Miyagawa et al., 2001), and piRNAs are classified as pre-pachytene or pachytene piRNAs based on the stage at which they are expressed and the specific Piwi proteins with which they associate (Aravin et al., 2006, Girard et al., 2006). MILI is broadly expressed in embryonic male germ cells at 12.5 dpc to postnatal haploid round spermatids, while MIWI and MIWI2 are expressed transiently during spermatogenesis. MIWI2 is expressed from 14.5 dpc to postnatal day 3 and MIWI is expressed during meiosis at pachytene spermatocytes and spermatid stage(Kuramochi-Miyagawa et al., 2004, Aravin et al., 2008, Aravin and Bourc’his, 2008). Pre-pachytene piRNAs are enriched in repeat-derived sequences and mainly associate with MIWI2 and MILI, whereas pachytene piRNAs are predominantly derived from intergenic unannotated sequences and associate with MIWI (Reuter et al., 2011, Aravin et al., 2006, Girard et al., 2006). The function of the pachytene piRNAs remains to be established.
A number of studies have identified C. elegans 21U RNAs to be the functional equivalents of fly and vertebrate piRNAs (Ruby et al., 2006, Batista et al., 2008, Das et al., 2008, Wang and Reinke, 2008). Although shorter than typical piRNAs, 21U RNAs bear many of the same characteristics: they associate with the Piwi protein PRG-1, start with a 5′ uridine, have modified 3′ ends, and are required for the maintenance of germline integrity and fertility (Ruby et al., 2006, Batista et al., 2008, Das et al., 2008, Wang and Reinke, 2008).
Biogenesis of piRNAs
The biogenesis of piRNAs is best understood in Drosophila, where they are produced from active transposons and piRNA clusters, the specific genomic regions that are populated with fragmented transposon sequences incapable of mobilization (reviewed in (Malone and Hannon, 2009)). The majority of transposons in the piRNA cluster are bidirectionally transcribed (reviewed in (Pek et al., 2012b). piRNA biogenesis occurs by two pathways; primary processing occurs in both somatic and germline cells and secondary processing, which occurs via a feed-forward loop called the ping-pong amplification cycle, occurs only in germline cells (reviewed in (Malone and Hannon, 2009). The primary processing pathway takes place predominantly in follicle cells, a thin layer of somatic cells surrounding the oocytes in the Drosophila ovary. The piRNAs are derived from long single-stranded antisense precursor RNAs that are loaded into Piwi, the only Piwi clade protein expressed in Drosophila follicle cells. Further processing is performed by Trimmer, a 3′ to 5′ progressive exoribonuclease (Kawaoka et al., 2011). piRNAs derived from the flamenco locus are generated via this pathway. Upon biogenesis of primary piRNAs and the formation of Piwi-containing piRISCs, primary piRNAs control the expression of sense transposon-derived transcripts, probably through the slicer activity of Piwi. Piwi-bound primary piRNAs can also silence the expression of transposons by forming heterochromatic structures at the transposon loci (Le Thomas et al., 2013). Primary piRNAs also serve to initiate the ping-pong amplification cycle described below. Most of our current understanding of the primary processing pathway comes from studies with somatic follicle cells, and the primary processing pathway in germ cells remains elusive (Figure 3). Genome-wide RNAi screens both in whole flies and in cultured ovarian follicle cells have recently identified a collection of proteins involved in primary piRNA biogenesis and function (Muerdter et al., 2013, Handler et al., 2013).
Figure 3. piRNA biogenesis pathway.
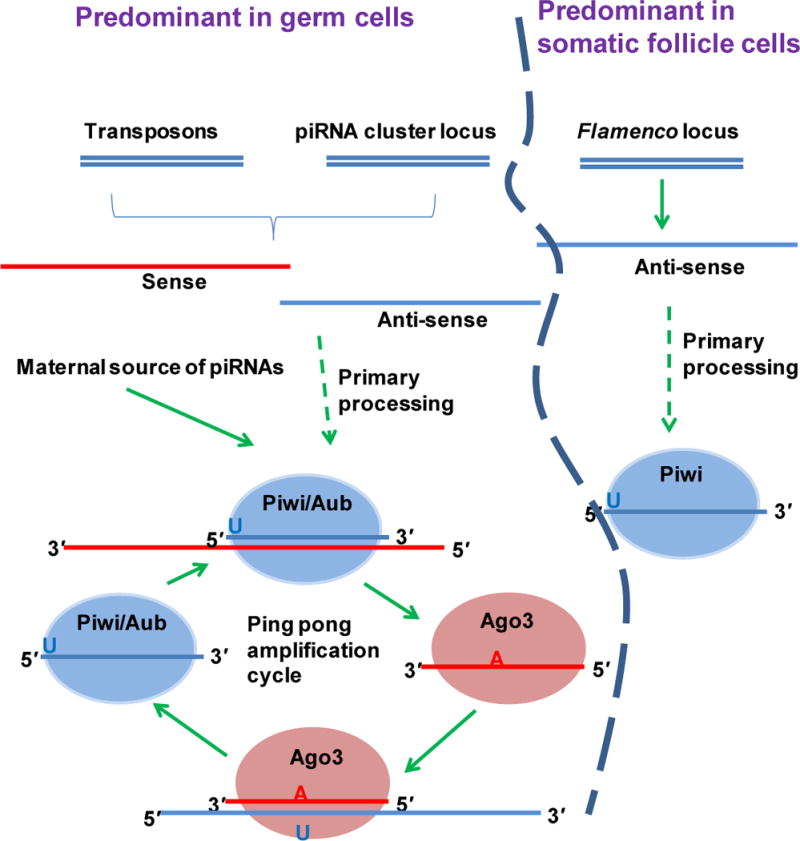
An unknown primary processing event generates antisense piRNAs from the piRNA loci and/or from active transposons. Antisense piRNAs associate with Aub or Piwi, which cleave and process the target mRNA to generate sense piRNAs. Sense piRNAs associate with Ago3, which then binds the antisense transcripts. This form of repetitive binding and cleavage creates an amplification loop that maintains a constant level of sense and antisense piRNAs. The flamenco-derived piRNAs, on the other hand, are generated by an unknown primary processing pathway that does not involve an amplification cycle. Flamenco piRNAs appear to be restricted to somatic follicle cells and are associated with Piwi.
The ping-pong amplification pathway operates exclusively in the germline. Two groups (Brennecke et al., 2007, Gunawardane et al., 2007) concurrently proposed the model based on the following observations: 1) Piwi and Aub bind antisense piRNAs and Ago3 binds sense piRNAs. 2) Piwi- and Aub-bound piRNAs carry a U at the 5′ end, whereas Ago3-bound piRNAs carry A at the 10th nucleotide from the 5′ end. 3) The first 10 nt of Aub- and Ago3-associated piRNAs are complementary to each other. 4) All three Piwi family proteins retain their slicer activity allowing them to cleave an RNA substrate opposite to position ten of the bound piRNA. Aub-bound antisense piRNAs initiate the ping-pong amplification cycle by triggering the cleavage of the sense transposon transcript to produce sense piRNAs. The sense piRNAs are in turn loaded into Ago3, engage antisense transposon transcripts via complementary base pairing, and slice the antisense transposon transcript via the slicer activity of Ago3. Ago3-mediated cleavage defines the 5′ end of antisense piRNAs, whereas 3′ end maturation is mediated by a yet-to-be-identified ribonuclease(s) (Brennecke et al., 2007, Gunawardane et al., 2007, Malone et al., 2009, Li et al., 2009). Upon biogenesis, Ago3-bound sense piRNAs in turn engage with antisense transposon transcripts, thereby sustaining the feed-forward amplification loop. The piRNAs derived from the 42AB cluster are generated in an Aub- and Ago3-dependent manner via the ping-pong cycle (Malone et al., 2009, Li et al., 2009). Interestingly, piRNAs can be transmitted vertically through maternal inheritance, thereby providing a consistent defense against retrotransposons (Brennecke et al., 2008).
Protein components of the piRNA pathway
Although PIWI proteins form the core of the piRNA biogenesis pathway, additional proteins, including Tudor domain-containing proteins, RNA helicases, and nucleases, also play a key role in the piRNA pathway. Indeed, loss of function mutations in these genes often results in severe defects in gametogenesis and piRNA production. Many of these proteins interact with each other both genetically and physically (reviewed in (Pek et al., 2012a, Pek et al., 2012b). In particular, several piRNA pathway components localize to nuage/chromatoid bodies, unique germline-specific structures found at the perinuclear region of germ cells. Several studies have suggested that the nuage is the probable site of ping-pong amplification (reviewed in (Pek et al., 2012a, Pek et al., 2012b).
In Drosophila, nuage components display genetic hierarchy with respect to their localization to the nuage (Patil and Kai, 2010, Lim and Kai, 2007, Anand and Kai, 2012). Similarly, localization of the Tudor domain proteins Tdrd1, Tdrd6, and Tdrd7 to the nuage depends on the mouse Vas homolog, mvh (Hosokawa et al., 2007). The same set of proteins was also found to colocalize with either one or all of the Piwi family proteins Miwi/Mili/Miwi2, and Mvh (Vagin et al., 2009, Shoji et al., 2009, Hosokawa et al., 2007). In mili mutants, localization of Tdrd1 and Tdrd9 to nuage/chromatoid bodies is compromised, while in miwi mutants the localization of Tdrd1, Tdrd6, Tdrd7, Tdrd9, and Mvh remains normal (Vagin et al., 2009, Shoji et al., 2009). Tdrd7 colocalizes with Vas in zebrafish (Strasser et al., 2008). Biochemical studies showed that many of these proteins physically interact with each other (Pek et al., 2012a, Pek et al., 2012b). Taken together, these reports suggest that such genetic hierarchy underlying the assembly of protein (sub)complexes is necessary for piRNA biogenesis and function and the phenomenon might be conserved across species. Biogenesis and functions of piRNAs have been recently reviewed (Guzzardo et al., 2013, Ishizu et al., 2012).
Long noncoding RNAs
The first reported example of a long noncoding RNA (lncRNA) was the H19 transcript, which lacked large open reading frames and was not translated into protein (Pachnis et al., 1988, Brannan et al., 1990). Later work revealed the existence of thousands of lncRNAs in the human genome. This unexpected abundance was initially thought to be transcriptional noise arising from the low fidelity of RNA polymerase (Struhl, 2007). However, the precise expression of lncRNAs at particular developmental stages and specific subcellular localization patterns argue otherwise. lncRNAs are typically >200-nt in length and bear several features of typical protein-coding transcripts, such as a 5′ cap, poly(A) tail, and introns (Carninci et al., 2005). Although some lncRNAs are located within intergenic sequences, the majority is transcribed as complex, interlaced networks of overlapping sense and antisense transcripts that often include protein-coding genes (Kapranov et al., 2007). Genomic sequences within these transcriptional foci are often shared within a number of different coding and noncoding transcripts in the sense and antisense directions giving rise to a complex hierarchy of overlapping isoforms (Derrien et al., 2012, Harrow et al., 2012).
lncRNAs have been implicated in the regulation of diverse biological processes including transcription, splicing, translation, chromosome gene dosage compensation, imprinting, epigenetic regulation, cell cycle control, cytoplasmic and nuclear trafficking, and cell differentiation (Lee and Bartolomei, 2013). For example, Air plays a key role in imprinting of the Igf2r locus, and Xist is critical for X chromosome inactivation. HOTAIR and HOTTIP affect the expression of HOXD and HOXA respectively. lncRNA-RoR affects reprogramming efficiency, whereas NRON influences the activity of the transcription factor NFAT. Tug1 is involved in retina development through regulation of the cell cycle (reviewed in (Guttman and Rinn, 2012)). In the following section we discuss in detail a few examples that highlight the various functions of lncRNAs.
lncRNAs in transcriptional and post-transcriptional regulation
lncRNAs regulate transcription by targeting transcriptional activators or repressors (Goodrich and Kugel, 2006), complexing with components of the transcriptional machinery, acting as cofactors for transcription factors, or by regulating the association and activity of transcription factor coregulators. One example is regulation of the cell cycle protein Cyclin D1 (CCND1) (Wang et al., 2008b). In human cell lines, DNA damage induces the expression of lncRNAs associated with the CCND1 gene promoter, which function cooperatively to modulate the activity of the RNA-binding protein TLS. In turn, TLS inhibits the histone acetyltransferase activities of CREB-binding protein and p300 to repress CCND1 transcription. In another example, the lncRNA Evf2 functions as a co-activator for the homeobox transcription factor Dlx2, which plays a key role in forebrain development and neurogenesis (Feng et al., 2006, Panganiban and Rubenstein, 2002). lncRNAs have also been shown to regulate RNA Pol II activity. In humans, a lncRNA transcribed from an upstream promoter of the dihydrofolate reductase gene (DHFR) forms a stable RNA–DNA triplex in the major promoter of DHFR to prevent binding of the transcriptional cofactor TFIID (Martianov et al., 2007). The lncRNA lincRNA-p21 is transcribed as a 3.1 kb transcript with two exons and is located ~15 kb upstream of the cell cycle regulator gene p21/Cdkn1a, a canonical target of p53. lincRNA-p21 functions as an inhibitor of the p53-dependent transcriptional response by repressing genes that interfere with apoptosis (Huarte et al., 2010). DNA damage caused by doxorubicin induces lincRNA-p21 transcription in a p53-dependent manner in mouse embryonic fibroblasts and several tumor-derived cell lines (lung tumor, sarcoma, and lymphoma). RNA pull-down experiments indicated that lincRNA-p21 interacts through its 5′ terminal region with heterogeneous nuclear ribonucleoprotein K (hnRNP-K). hnRNP-K binds to the promoters of genes that are corepressed by lincRNA-p21 and p53 (Huarte et al., 2010), suggesting that lincRNA-p21 acts as a transcriptional repressor of the p53 response genes by recruiting hnRNP-K to the promoters of target genes (Figure 4).
Figure 4. Transcriptional control mediated by lncRNAs Gas-5 and lincRNA-p21.
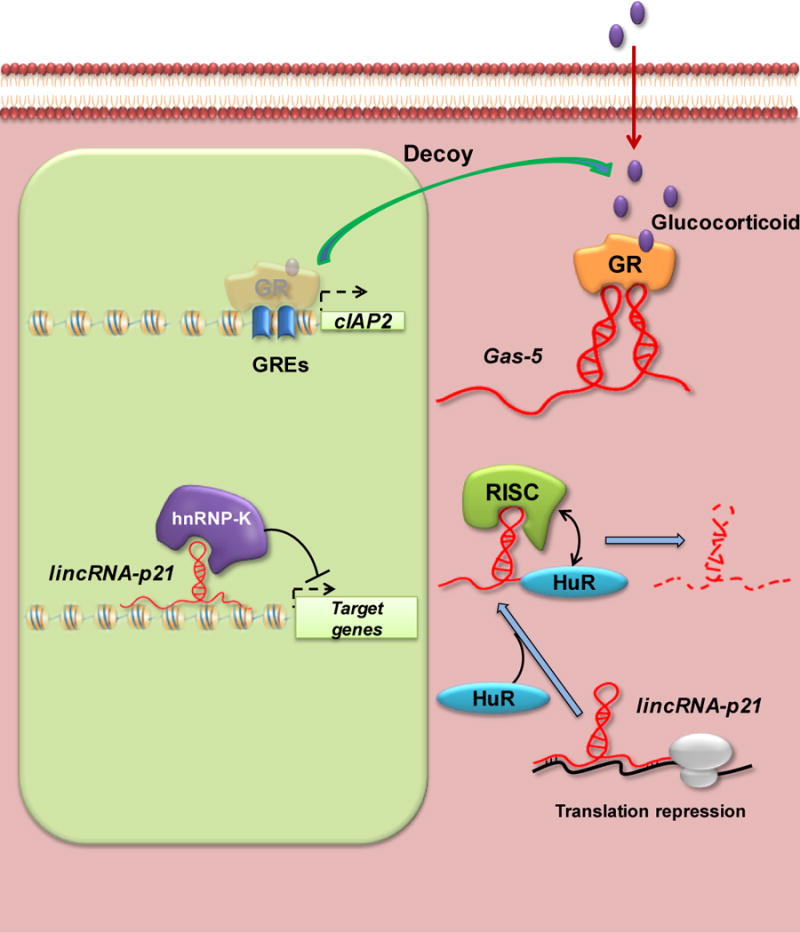
The lncRNA Gas-5 functions as a molecular decoy to prevent binding of the nuclear transcription factor GR to response elements in the cIAP2 promoter region. LincRNA-p21 acts as a repressor at both the transcriptional and translational levels.
lncRNAs have also been reported to regulate various aspects of post-transcriptional RNA processing, including pre-mRNA splicing, cytoplasmic transport, translation, and degradation, which often involve base pairing between lncRNAs and the target mRNAs. One example is the lncRNA MALAT-1, which interacts with and influences the phosphorylation state of serine/arginine rich (SR) splicing proteins (Ji et al., 2003, Bernard et al., 2010, Tripathi et al., 2010) (Figure 5).
Figure 5. Post-transcriptional and translational regulation mediated by the lncRNAs MALAT-1 and BACE1-AS, respectively.
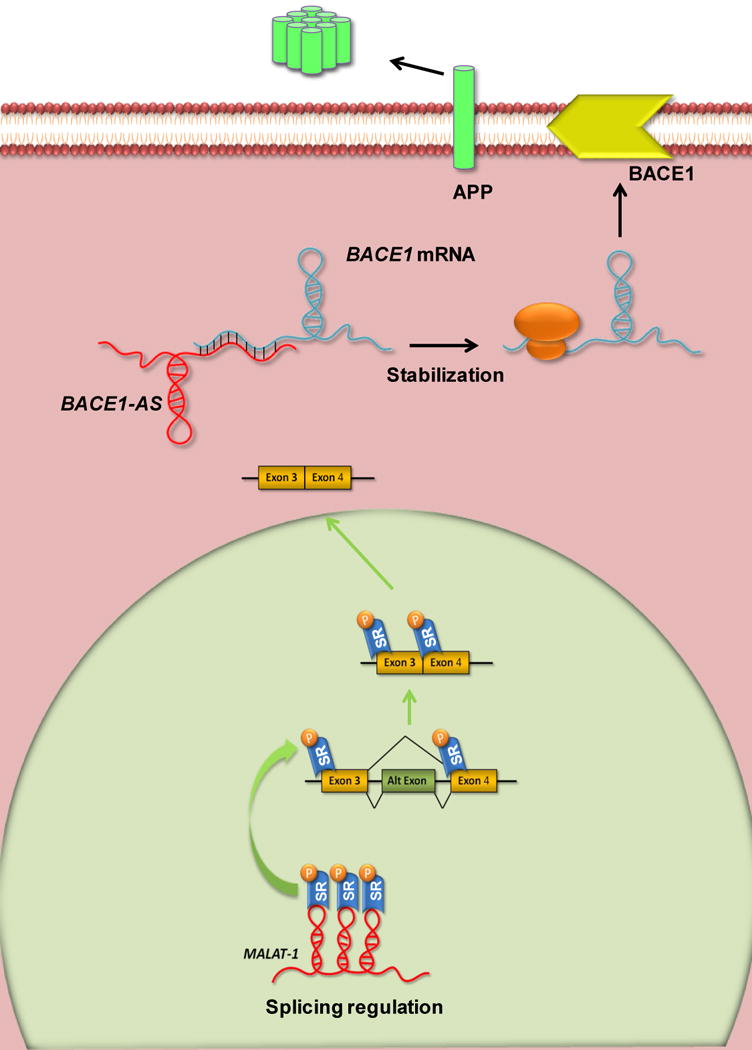
MALAT-1 is retained in the nucleus and modulates alternative splicing by forming an RNP complex with serine/arginine (SR) splicing factors and their recruitment to the transcription site. BACE1 and BACE1-AS form RNA duplexes that stabilize the mRNA. Altering expression of BACE1-AS decreases or increases the stability of BACE1 mRNA.
Translational regulation by lncRNAs
Several studies have demonstrated the involvement of lncRNAs in control of mRNA translation. The lncRNAs BC1 and BC200 are transcribed in the central nervous system of mice and humans, respectively, by RNA Pol III. BC1 controls the efficiency of dopamine D2 receptor-mediated transmission in dendrites in the striatum, and BC1-deficient mice display reduced exploration behavior and increased anxiety (Centonze et al., 2007, Lewejohann et al., 2004). BC1 shows some sequence complementarity with various neuron-specific mRNAs, suggesting a possible role for this lncRNA in targeted translational repression (Wang et al., 2005).
BACE1-AS (β-site amyloid precursor protein (APP)-cleaving enzyme) is an antisense lncRNA transcribed from the opposite strand of BACE1, an aspartyl protease that cleaves APP at the β-site to produce amyloid β-peptide (Aβ). Aβ accumulates in the brains of patients with Alzheimer’s disease, highlighting the importance of regulation of BACE1 expression for normal brain function (Faghihi et al., 2008) (Figure 5).
lncRNAs in epigenetic regulation
Many lncRNAs have been implicated in epigenetic regulation of gene expression. In flies, lncRNAs induce the expression of the homeotic gene Ubx by recruiting the trithorax protein Ash1 and directing its chromatin modifying functions to Hox regulatory elements (Sanchez-Elsner et al., 2006). Certain mammalian Hox genes are also regulated by lncRNAs (Mazo et al., 2007, Rinn et al., 2007). The expression of these lncRNAs is spatially and temporally regulated throughout human development and defines chromatin domains of differential histone methylation and accessibility by RNA polymerase (Rinn et al., 2007). The 40 kb lncRNA HOTAIR originating from the HOXC locus represses transcription across the HOXD locus in trans by associating with the Polycomb repressive complex 2 (PRC2) and directing its recruitment to the target loci, where it influences heterochromatin formation (Tsai et al., 2010) (Figure 6).
Figure 6. Epigenetic regulation mediated by the lncRNAs HOTAIR and ANRIL.
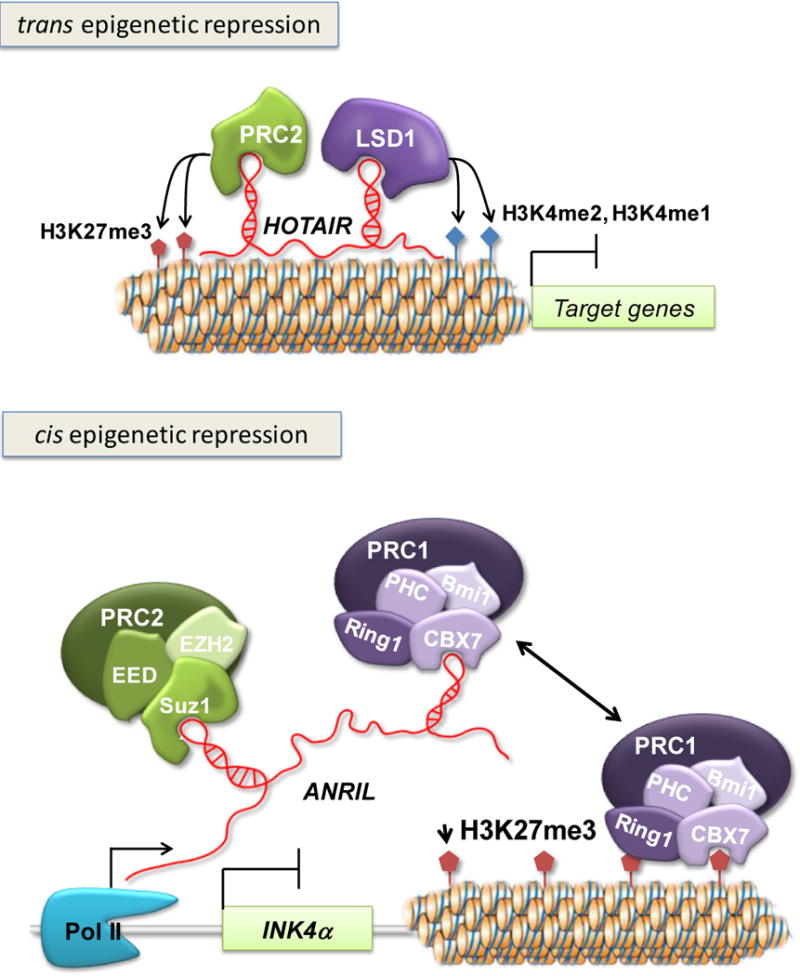
HOTAIR, one of the best-characterized lncRNAs, is a 2.2 kb antisense transcript residing in the HOXC locus. HOTAIR mediates epigenetic silencing by interacting with the PRC2 and LSD1-CoREST complexes via its 5′ and 3′ domains, respectively. The EZH2 subunit of PRC2 has histone methyltransferase activity and trimethylates histone 3 at lysine 27, whereas LSD1 demethylates H3K4me2 and H3K4me1. The lncRNA ANRIL is located within the INK4b/ARF/INK4a locus that encodes three tumor suppressor genes: INK4b encodes p15/CDKN2B (cyclin-dependent kinase inhibitor 2B), ARF encodes p14/ARF (alternative reading frame), and INK4a encodes p16/CDKN2A (cyclin-dependent kinase 2A). ANRIL regulates transcriptional silencing of INK4a by recruiting the PcG protein Chromobox 7 (CBX7) to the INK4b/ARF/INK4a locus. CBX7 is a component of PRC1, which binds to the H3K27me3 repressive mark and is required for maintenance of epigenetic gene silencing.
The lncRNA ANRIL transcriptionally represses the tumor suppressor gene INK4α in cis through a PRC1 member, CBX7 (Pc/chromobox7). INK4α maps to a locus that encompasses two additional tumor suppressor genes linked to various types of cancers, INK4β and ARF. Both INK4β and INK4α are involved in cell cycle regulation while ARF in apoptotic pathway and cell cycle arrest by promoting MDM2 degradation. The ANRIL transcript is antisense to INK4β and correlates with INK4α epigenetic regulation (Yap et al., 2010). Increased levels of ANRIL and CBX7 and decreased levels of INK4α have been reported in prostate cancer tissues (Yap et al., 2010) (Figure 6).
lncRNA in imprinting
Imprinting is the phenomenon by which genes are expressed from only one of the maternally or paternally inherited alleles and the second allele is inactivated. Many imprinted genes are clustered on the chromosome and are often associated with lncRNAs. A possible role for these lncRNAs in imprinting was suggested by the observation that lncRNA expression correlates with repression of the imprinted genes (Pauler et al., 2007). Two examples are the lncRNAs Kcnqot1 and Air (Braidotti et al., 2004). Most of the genes at the Kcnq1 locus are inherited maternally, the exception being the antisense lncRNA Kcnqot1 (Mitsuya et al., 1999). Genetic studies in mice expressing a truncated Kcnqot1 transgene show that it is crucial for imprinting of genes on the paternal chromosome (Mancini-Dinardo et al., 2006). Kcnqot1 is thought to direct the trimethylation of lysine 9 and 27 of histone 3 (H3K9me3 and H3K27me3) at an imprinting center that overlaps with the Kcnqot1 promoter and resides within a Kcnq1 sense exon (Umlauf et al., 2004). The PRC2 components Eed and Ezh2 are recruited to the Kcnq1 locus on the paternal chromosome, possibly by Kcnqot1, where they may mediate gene silencing through repressive histone methylation (Umlauf et al., 2004). A differentially methylated imprinting center also overlaps with the promoter of the long antisense ncRNA Air, which is responsible for silencing of neighboring genes at the Igf2r locus on the paternal chromosome (Sleutels et al., 2002, Zwart et al., 2001). The Igf2r locus displays an allele-specific histone methylation pattern, suggesting that Air also mediates silencing via chromatin modification.
lncRNAs in cell cycle regulation and apoptosis
Several lncRNAs contribute to regulation of the cell cycle and apoptosis. One example, described above, is lincRNA-p21, which regulates the cell cycle through its interaction with hnRNP-K and p53 (Mourtada-Maarabouni et al., 2009, Huarte et al., 2010) (Figure 4). Another example is the lncRNA Gas5 (growth arrest specific 5), which regulates the activity of glucocorticoids in response to nutrient starvation and sensitizes cells to apoptosis (Smith and Steitz, 1998). Gas5 binds to glucocorticoid receptors (GR) and prevents them from associating with glucocorticoid response elements in GR target genes such as cIAP2 (cellular inhibitor of apoptosis 2), which inhibits caspase activity and suppresses apoptosis. Thus, Gas5 promotes apoptosis by blocking transcription of cIAP2 (Webster et al., 2002) (Figure 4).
Disease relevance of ncRNAs
Given the array of essential biological processes influenced by long and short ncRNAs, it is not surprising that dysregulation of ncRNA biogenesis and function is associated with a number of pathological conditions in humans.
miRNAs in disease
miRNAs are thought to regulate the expression of ~50% of cellular mRNAs, and each miRNA can potentially regulate hundreds of target genes. Thus, dysregulation of the expression of specific miRNAs or of genes encoding canonical miRNA factors has a profound effect on cell homeostasis.
Studies in Drosophila S2 cells have shown that FMR1, the fly ortholog of human fragile X mental retardation protein (FMR1P in human) binds to the RNAi components Ago2 and VIG in the RISC complex (Caudy et al., 2002). Fragile X syndrome (FXS) is one of the most common inherited mental retardations. The disease is associated with expansion of a CGG trinucleotide repeat in the 5′ UTR of the FMR1 gene, leading to hypermethylation and loss of FMR1 expression (Jin et al., 2004a, Jin et al., 2004b). Studies using human HeLa cells revealed that Ago2 associates not only with FMR1P but also with the fragile X-related proteins FXR1P and FXR2P. FMR1P, FXR1P, and FXR2P form complex(es) with miRNA-sized nucleotides in cells from control subjects but not from FXS patients (Jin et al., 2004a, Jin et al., 2004b). The proteins are thought to act as acceptor molecules for Dicer-processed miRNAs and facilitate miRISC formation and engagement of target mRNAs (Plante et al., 2006). In addition, DGCR8, the regulatory subunit of the miRNA microprocessor, has been implicated in DiGeorge syndrome, a congenital disease characterized by facial dysmorphology, cardiac defects, and thymic aplasia in humans (Sullivan, 2004a, Sullivan, 2004b).
Several miRNA pathway components are reported to be differentially expressed in multiple neoplasias compared with normal tissues, and may serve as potential biomarkers for those diseases. For example, reduced DCR-1 and Drosha expression are associated with poor post-operative prognosis in non-small cell lung cancer (NSCLC) patients (Karube et al., 2005). In addition, the miRNA pathway components Ago2, XPO-5, Ago1, Mov10, and TNRC6B are upregulated in metastatic prostate cancer (Chiosea et al., 2006).
A high-throughput miRNA profiling study revealed differential expression of ~21 miRNAs in tumor samples (Lu et al., 2005a, Volinia et al., 2006). Some miRNAs, such as miR-21, miR-17-5p, and miR-191 are overexpressed in tumors of embryonic origin (Volinia et al., 2006). In many tumors, individual miRNAs that reside in the same cluster are not concordantly expressed, suggesting they may be differentially regulated at the post-transcriptional level. For instance, of the six miRNAs contained in the miR-17-92 cluster, two are upregulated (miR-19 and miR-92-1) and two are downregulated (miR-17 and miR-20) in chronic lymphocytic leukemia (Calin et al., 2004).
Several studies have focused on profiling miRNA expression in non-neoplastic diseases. For example, 16 miRNAs were found to be differentially expressed in the prefrontal cortex of patients with schizophrenia, with 15 being downregulated compared to normal control samples (Perkins et al., 2007). Two of these miRNAs (miR-9 and miR-128a) were found to be upregulated in the brains of Alzheimer’s disease patients (Lukiw, 2007). A microarray-based approach to profile miRNA expression under stress conditions found that several miRNAs are upregulated in response to low oxygen levels (Kulshreshtha et al., 2007a, Kulshreshtha et al., 2007b). In addition, several studies have been carried out to identify the role of individual miRNAs in diseases using cell lines and mouse models (reviewed in (Calin and Croce, 2006, Esquela-Kerscher and Slack, 2006, Soifer et al., 2007).
lncRNAs in disease
Recent studies have started to uncover roles for several lncRNAs in development and cell biology. Although a few studies have shown the association of lncRNAs with specific diseases, we have little understanding of their role in disease etiology. The expression of lncRNAs has been shown to change in diseases such as various types of cancers, neurodegenerative diseases, and coronary disease (Wapinski and Chang, 2011). Here, we discuss selected examples.
lncRNAs in cancer
A number of lncRNAs have been reported to show aberrant expression in cancer. The lncRNA MALAT-1 is abundantly expressed in NSCLC and is present at three-fold higher levels in metastasizing than non-metastasizing tumors. Moreover, MALAT-1 expression correlates with poor prognosis in patients with stage I disease and thus serves as a prognostic marker for survival (Ji et al., 2003). The mouse homolog of MALAT1 is highly expressed in hepatocellular carcinoma (Lin et al., 2007). The lncRNA PCGEM1 is overexpressed in prostate tumors and correlates with increased proliferation and colony formation, suggesting its probable involvement in cell growth regulation (Fu et al., 2006). Elevated levels of the lncRNA ANRIL have also been reported in prostate cancer tissues (Pasmant et al., 2011, Yap et al., 2010). A large germline deletion of the INK4/ARF locus and lncRNA ANRIL has been associated with hereditary cutaneous malignant melanoma and neural system tumors syndrome (Pasmant et al., 2011). A recent study of leukemia and colorectal cancer identified both germline and somatic mutations in lncRNA genes (Wojcik et al., 2010). The lncRNAs HIS-1 and BIC have been implicated in oncogenesis and growth control but their function in normal cells is unknown (Eis et al., 2005, Li et al., 1997). The lncRNA HOTAIR shows higher expression in breast cancer and is correlated with poor prognosis and metastasis (Burd et al., 2010). Genome-wide profiling has revealed many transcribed noncoding ultraconserved regions with distinct profiles in various human cancer states (Calin et al., 2007). Colorectal carcinoma, chronic lymphocytic leukemia, and hepatocellular carcinoma show aberrant expression profiles for ultraconserved ncRNAs compared to normal cells. Further analysis of one ultraconserved ncRNA suggested it behaved like an oncogene by mitigating apoptosis and subsequently expanding the number of malignant cells in colorectal cancers (Calin et al., 2007). Many of the transcribed ultraconserved sites that exhibit distinct signatures in cancer are found at fragile sites and genomic regions associated with cancer. It seems likely that the aberrant expression of these ultraconserved ncRNAs within malignant processes results from important functions they fulfill in normal human development. The lncRNA GAS5 transcript is expressed at reduced levels in breast cancer compared to normal breast epithelia (Huarte et al., 2010), and chromosomal translocations encompassing the GAS5 locus are associated with melanoma, B cell lymphoma, and prostate and breast cancer (Mourtada-Maarabouni et al., 2009). Although lincRNA-p21 has not been directly associated with cancer, its association with p53 and putative role in regulation of the cell cycle suggest it may be involved in cancer initiation.
Small RNA-based therapeutics
Although the clinical utility of RNAi is yet to be established, there are a number of ongoing clinical trials that could indicate the potential for success (reviewed in (Davidson and McCray, 2011)), many RNAi-based treatments for metabolic diseases, cancer, liver fibrosis, and viral infection are in progress (Hu and Xie, 2009). Preclinical and clinical trials to lower plasma LDLs are currently in progress with siRNAs targeting the expression of apolipoprotein B (APOB) and proprotein convertase subtilisin/kexin type 9 (PCSK9). The liver was one of the first organs to be tested for effectiveness of RNAi in vivo (Xia et al., 2002, McCaffrey et al., 2002), and was also one of the first organs targeted for cancer. Effective RNA-based therapies require identification of target-specific siRNA sequences and development of efficient vehicles to deliver siRNA to the appropriate cells in vivo. RNA delivery is by far the major roadblock in developing RNA-based drugs.
A number of approaches have been developed and tested for in vivo delivery of RNA drugs. For example, in vivo delivery with SNALPs (stable nucleic acid lipid particles), in which siRNA is complexed to carriers or embedded in liposomal particles effectively silences target mRNAs. SNALPs were used to target PLK1, a cell cycle protein required for phosphorylation of many cell cycle proteins. Hepatic tumor-bearing mice treated with such SNALPs showed significant improvement in survival (Judge et al., 2009). Other strategies employed synthesis of cholesterol-modified lipids (Ghosh et al., 2010), interfering nanoparticles (iNOPs) (Baigude and Rana, 2012, Su et al., 2011, Baigude and Rana, 2009, Baigude et al., 2007, Baigude et al., 2013), and functionalized nanotubes (McCarroll et al., 2010) as new siRNA delivery agents. iNOPs can be quickly assembled by mixing siRNA with functionalized poly-L-lysine dendrimers without the need for complex liposomal formulation procedures or harsh covalent reaction to label RNA with lipids or other delivery agents. At a clinically feasible dose of 1 mg kg−1, apolipoprotein B (apoB) siRNA–iNOP complexes achieved ~40–45% reduction of liver apoB mRNA and plasma apoB protein levels within 48 h of administration to mice, without apparent toxicity (Baigude et al., 2013).
Synthetic siRNAs or expressed shRNAs can be used to inhibit viral infections, including influenza A, respiratory syncytial virus, severe acute respiratory syndrome coronavirus (SARS), and parainfluenza virus (Bitko et al., 2005, Tompkins et al., 2004, Ge et al., 2004a, Ge et al., 2004b, Li et al., 2005). RNAi-based treatment strategies have also been exploited to target HIV. A clinical trial has been performed with lentiviral vectors expressing shRNA targeting an exon shared by HIV tat and rev genes combined with HIV specific RNA-based inhibitors. Hematopoietic progenitor cells were transduced ex vivo and then reinfused into patients. The phase I trial showed that the cells were successfully engrafted within 11 days and there were no treatment-related toxicities (DiGiusto et al., 2010). Some neurological diseases may also benefit from RNAi treatments. The mutant genes that have gained undefined properties that harm cells in the nervous system cause a number of inherited neurodegenerative diseases, leading to neurodegeneration and clinical phenotypes. Some examples of these diseases include Huntington disease and subset of Alzheimer disease, Parkinson disease, and amyotrophic lateral sclerosis. Thus, RNAi can be used to lower the concentration of mutant proteins. A chemically stabilized siRNA against human Cu,Zn-superoxide dismutase (SOD1) in a mouse model for amyotrophic lateral sclerosis have been reported to knocked down the mutant SOD1 expression, slowed the disease progression, and extended the survival (Wang et al., 2008a). Work in Huntington’s disease animal models showed that partial knockdown of both huntingtin alleles by targeting SNPs with RNAi was tolerated and provided clinical benefit (Boudreau et al., 2009). Recently, chemically modified single stranded siRNAs have been employed to potently and allele-selectively inhibit mutant huntingtin expression (Yu et al., 2012, Lima et al., 2012, Davidson and Monteys, 2012). It is quite feasible that in coming years, we will witness RNA-based therapies in a number of disease indications.
miRNAs as therapeutic targets
miRNA expression profiles are likely to become important diagnostic and prognostic tools. Identification of these misregulated miRNAs in cellular transformation and malignant states has tremendous implications in cancer therapy. Inhibition of oncogenic miRNAs known to regulate multiple targets might switch off several cancer-promoting signals. While some approaches have used siRNA to target misregulated miRNAs, others have used miRNA sponges that sequester miRNAs and inhibit their ability to bind and suppress their targets (Brown and Naldini, 2009). A number of strategies utilizing miRNA-based anticancer therapies are being developed including combinations with current anti-cancer therapies (Garzon et al., 2010). These approaches may improve the efficacy or rates to cure various cancers. Antagomirs or anti-miRs, chemically modified RNA oligonucleotides antisense to miRNAs, are being developed to treat a variety of disease states (Pedersen et al., 2007, Krutzfeldt et al., 2005). Similar to siRNAs, these anti-miRs can also be assembled with nanoparticles to inhibit miR functions in vivo (Baigude and Rana, 2012, Su et al., 2011). Anti-miRs, has sparked enthusiasm for miRNAs as novel therapeutic targets. Indeed, in a primate model of HCV infection, oligonucleotides aimed at sequestering miR-122 inhibited virus replication (Lanford et al., 2010). An anti-miR-122 drug, miravirsen, is being evaluated in Phase IIa trial clinical Santaris Pharma in a number of countries. In addition, anti-miRs targeting a number of miRNAs are being developed and tested in cardiovascular indications (reviewed in (van Rooij and Olson, 2012)). Given these anti-miRs exhibit favorable pharmacokinetic and pharmacodynamic properties, they can provide new therapeutic modalities to treat disease where no efficacious small molecule drugs are available.
Acknowledgments
We are grateful to members of the Rana laboratory for helpful discussions. This work was supported in part by grants from the National Institutes of Health to T.M.R.
Footnotes
Declaration of interest
The authors declare no conflicts of interest.
References
- ALLEN E, XIE Z, GUSTAFSON AM, CARRINGTON JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–21. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- AMBROS V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- AMBROS V. MicroRNAs and developmental timing. Current opinion in genetics & development. 2011;21:511–7. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANAND A, KAI T. The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J. 2012;31:870–82. doi: 10.1038/emboj.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARAVIN A, GAIDATZIS D, PFEFFER S, LAGOS-QUINTANA M, LANDGRAF P, IOVINO N, MORRIS P, BROWNSTEIN MJ, KURAMOCHI-MIYAGAWA S, NAKANO T, CHIEN M, RUSSO JJ, JU J, SHERIDAN R, SANDER C, ZAVOLAN M, TUSCHL T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- ARAVIN AA, BOURC’HIS D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–5. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARAVIN AA, LAGOS-QUINTANA M, YALCIN A, ZAVOLAN M, MARKS D, SNYDER B, GAASTERLAND T, MEYER J, TUSCHL T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–50. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- ARAVIN AA, SACHIDANANDAM R, BOURC’HIS D, SCHAEFER C, PEZIC D, TOTH KF, BESTOR T, HANNON GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–99. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARAVIN AA, SACHIDANANDAM R, GIRARD A, FEJES-TOTH K, HANNON GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–7. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- AXTELL MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013;64:137–59. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- BAIGUDE H, MCCARROLL J, YANG CS, SWAIN PM, RANA TM. Design and creation of new nanomaterials for therapeutic RNAi. ACS chemical biology. 2007;2:237–41. doi: 10.1021/cb7000582. [DOI] [PubMed] [Google Scholar]
- BAIGUDE H, RANA TM. Delivery of therapeutic RNAi by nanovehicles. Chembiochem: a European journal of chemical biology. 2009;10:2449–54. doi: 10.1002/cbic.200900252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAIGUDE H, RANA TM. Interfering nanoparticles for silencing microRNAs. Methods in enzymology. 2012;509:339–53. doi: 10.1016/B978-0-12-391858-1.00017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAIGUDE H, SU J, MCCARROLL J, RANA TM. In Vivo Delivery of RNAi by Reducible Interfering Nanoparticles (iNOPs) ACS Medicinal Chemistry Letters. 2013;4 doi: 10.1021/ml4001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BATISTA PJ, RUBY JG, CLAYCOMB JM, CHIANG R, FAHLGREN N, KASSCHAU KD, CHAVES DA, GU W, VASALE JJ, DUAN S, CONTE D, JR, LUO S, SCHROTH GP, CARRINGTON JC, BARTEL DP, MELLO CC. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERNARD D, PRASANTH KV, TRIPATHI V, COLASSE S, NAKAMURA T, XUAN Z, ZHANG MQ, SEDEL F, JOURDREN L, COULPIER F, TRILLER A, SPECTOR DL, BESSIS A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–93. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERNSTEIN E, CAUDY AA, HAMMOND SM, HANNON GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- BITKO V, MUSIYENKO A, SHULYAYEVA O, BARIK S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–5. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- BORSANI O, ZHU J, VERSLUES PE, SUNKAR R, ZHU JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–91. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOUDREAU RL, MCBRIDE JL, MARTINS I, SHEN S, XING Y, CARTER BJ, DAVIDSON BL. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol Ther. 2009;17:1053–63. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAIDOTTI G, BAUBEC T, PAULER F, SEIDL C, SMRZKA O, STRICKER S, YOTOVA I, BARLOW DP. The Air noncoding RNA: an imprinted cis-silencing transcript. Cold Spring Harb Symp Quant Biol. 2004;69:55–66. doi: 10.1101/sqb.2004.69.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRANNAN CI, DEES EC, INGRAM RS, TILGHMAN SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNECKE J, ARAVIN AA, STARK A, DUS M, KELLIS M, SACHIDANANDAM R, HANNON GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- BRENNECKE J, MALONE CD, ARAVIN AA, SACHIDANANDAM R, STARK A, HANNON GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–92. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROWN BD, NALDINI L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–85. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- BROWN KM, CHU CY, RANA TM. Target accessibility dictates the potency of human RISC. Nat Struct Mol Biol. 2005;12:469–70. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- BURD CE, JECK WR, LIU Y, SANOFF HK, WANG Z, SHARPLESS NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAI X, HAGEDORN CH, CULLEN BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALIN GA, CROCE CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- CALIN GA, LIU CG, FERRACIN M, HYSLOP T, SPIZZO R, SEVIGNANI C, FABBRI M, CIMMINO A, LEE EJ, WOJCIK SE, SHIMIZU M, TILI E, ROSSI S, TACCIOLI C, PICHIORRI F, LIU X, ZUPO S, HERLEA V, GRAMANTIERI L, LANZA G, ALDER H, RASSENTI L, VOLINIA S, SCHMITTGEN TD, KIPPS TJ, NEGRINI M, CROCE CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–29. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- CALIN GA, LIU CG, SEVIGNANI C, FERRACIN M, FELLI N, DUMITRU CD, SHIMIZU M, CIMMINO A, ZUPO S, DONO M, DELL’AQUILA ML, ALDER H, RASSENTI L, KIPPS TJ, BULLRICH F, NEGRINI M, CROCE CM. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–60. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARNINCI P, KASUKAWA T, KATAYAMA S, GOUGH J, FRITH MC, MAEDA N, OYAMA R, RAVASI T, LENHARD B, WELLS C, KODZIUS R, SHIMOKAWA K, BAJIC VB, BRENNER SE, BATALOV S, FORREST AR, ZAVOLAN M, DAVIS MJ, WILMING LG, AIDINIS V, ALLEN JE, AMBESI-IMPIOMBATO A, APWEILER R, ATURALIYA RN, BAILEY TL, BANSAL M, BAXTER L, BEISEL KW, BERSANO T, BONO H, CHALK AM, CHIU KP, CHOUDHARY V, CHRISTOFFELS A, CLUTTERBUCK DR, CROWE ML, DALLA E, DALRYMPLE BP, DE BONO B, DELLA GATTA G, DI BERNARDO D, DOWN T, ENGSTROM P, FAGIOLINI M, FAULKNER G, FLETCHER CF, FUKUSHIMA T, FURUNO M, FUTAKI S, GARIBOLDI M, GEORGII-HEMMING P, GINGERAS TR, GOJOBORI T, GREEN RE, GUSTINCICH S, HARBERS M, HAYASHI Y, HENSCH TK, HIROKAWA N, HILL D, HUMINIECKI L, IACONO M, IKEO K, IWAMA A, ISHIKAWA T, JAKT M, KANAPIN A, KATOH M, KAWASAWA Y, KELSO J, KITAMURA H, KITANO H, KOLLIAS G, KRISHNAN SP, KRUGER A, KUMMERFELD SK, KUROCHKIN IV, LAREAU LF, LAZAREVIC D, LIPOVICH L, LIU J, LIUNI S, MCWILLIAM S, MADAN BABU M, MADERA M, MARCHIONNI L, MATSUDA H, MATSUZAWA S, MIKI H, MIGNONE F, MIYAKE S, MORRIS K, MOTTAGUI-TABAR S, MULDER N, NAKANO N, NAKAUCHI H, NG P, NILSSON R, NISHIGUCHI S, NISHIKAWA S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- CAUDY AA, MYERS M, HANNON GJ, HAMMOND SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–6. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CENTONZE D, ROSSI S, NAPOLI I, MERCALDO V, LACOUX C, FERRARI F, CIOTTI MT, DE CHIARA V, PROSPERETTI C, MACCARRONE M, FEZZA F, CALABRESI P, BERNARDI G, BAGNI C. The brain cytoplasmic RNA BC1 regulates dopamine D2 receptor-mediated transmission in the striatum. J Neurosci. 2007;27:8885–92. doi: 10.1523/JNEUROSCI.0548-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHENDRIMADA TP, GREGORY RI, KUMARASWAMY E, NORMAN J, COOCH N, NISHIKURA K, SHIEKHATTAR R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIOSEA S, JELEZCOVA E, CHANDRAN U, ACQUAFONDATA M, MCHALE T, SOBOL RW, DHIR R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIU YL, RANA TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Molecular cell. 2002;10:549–61. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- CHIU YL, RANA TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–48. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHU CY, RANA TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHU CY, RANA TM. Small RNAs: regulators and guardians of the genome. J Cell Physiol. 2007;213:412–9. doi: 10.1002/jcp.21230. [DOI] [PubMed] [Google Scholar]
- CLARK MB, AMARAL PP, SCHLESINGER FJ, DINGER ME, TAFT RJ, RINN JL, PONTING CP, STADLER PF, MORRIS KV, MORILLON A, ROZOWSKY JS, GERSTEIN MB, WAHLESTEDT C, HAYASHIZAKI Y, CARNINCI P, GINGERAS TR, MATTICK JS. The reality of pervasive transcription. PLoS biology. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COGONI C, MACINO G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci U S A. 1997;94:10233–8. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CZECH B, MALONE CD, ZHOU R, STARK A, SCHLINGEHEYDE C, DUS M, PERRIMON N, KELLIS M, WOHLSCHLEGEL JA, SACHIDANANDAM R, HANNON GJ, BRENNECKE J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CZECH B, ZHOU R, ERLICH Y, BRENNECKE J, BINARI R, VILLALTA C, GORDON A, PERRIMON N, HANNON GJ. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36:445–56. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAS PP, BAGIJN MP, GOLDSTEIN LD, WOOLFORD JR, LEHRBACH NJ, SAPETSCHNIG A, BUHECHA HR, GILCHRIST MJ, HOWE KL, STARK R, MATTHEWS N, BEREZIKOV E, KETTING RF, TAVARE S, MISKA EA. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIDSON BL, MCCRAY PB., JR Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–40. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIDSON BL, MONTEYS AM. Singles engage the RNA interference pathway. Cell. 2012;150:873–5. doi: 10.1016/j.cell.2012.08.008. [DOI] [PubMed] [Google Scholar]
- DENLI AM, TOPS BB, PLASTERK RH, KETTING RF, HANNON GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- DERRIEN T, JOHNSON R, BUSSOTTI G, TANZER A, DJEBALI S, TILGNER H, GUERNEC G, MARTIN D, MERKEL A, KNOWLES DG, LAGARDE J, VEERAVALLI L, RUAN X, RUAN Y, LASSMANN T, CARNINCI P, BROWN JB, LIPOVICH L, GONZALEZ JM, THOMAS M, DAVIS CA, SHIEKHATTAR R, GINGERAS TR, HUBBARD TJ, NOTREDAME C, HARROW J, GUIGO R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIGIUSTO DL, KRISHNAN A, LI L, LI H, LI S, RAO A, MI S, YAM P, STINSON S, KALOS M, ALVARNAS J, LACEY SF, YEE JK, LI M, COUTURE L, HSU D, FORMAN SJ, ROSSI JJ, ZAIA JA. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DJEBALI S, DAVIS CA, MERKEL A, DOBIN A, LASSMANN T, MORTAZAVI A, TANZER A, LAGARDE J, LIN W, SCHLESINGER F, XUE C, MARINOV GK, KHATUN J, WILLIAMS BA, ZALESKI C, ROZOWSKY J, RODER M, KOKOCINSKI F, ABDELHAMID RF, ALIOTO T, ANTOSHECHKIN I, BAER MT, BAR NS, BATUT P, BELL K, BELL I, CHAKRABORTTY S, CHEN X, CHRAST J, CURADO J, DERRIEN T, DRENKOW J, DUMAIS E, DUMAIS J, DUTTAGUPTA R, FALCONNET E, FASTUCA M, FEJES-TOTH K, FERREIRA P, FOISSAC S, FULLWOOD MJ, GAO H, GONZALEZ D, GORDON A, GUNAWARDENA H, HOWALD C, JHA S, JOHNSON R, KAPRANOV P, KING B, KINGSWOOD C, LUO OJ, PARK E, PERSAUD K, PREALL JB, RIBECA P, RISK B, ROBYR D, SAMMETH M, SCHAFFER L, SEE LH, SHAHAB A, SKANCKE J, SUZUKI AM, TAKAHASHI H, TILGNER H, TROUT D, WALTERS N, WANG H, WROBEL J, YU Y, RUAN X, HAYASHIZAKI Y, HARROW J, GERSTEIN M, HUBBARD T, REYMOND A, ANTONARAKIS SE, HANNON G, GIDDINGS MC, RUAN Y, WOLD B, CARNINCI P, GUIGO R, GINGERAS TR. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOENCH JG, SHARP PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EIS PS, TAM W, SUN L, CHADBURN A, LI Z, GOMEZ MF, LUND E, DAHLBERG JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESQUELA-KERSCHER A, SLACK FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- FABIAN MR, SONENBERG N, FILIPOWICZ W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- FAGHIHI MA, MODARRESI F, KHALIL AM, WOOD DE, SAHAGAN BG, MORGAN TE, FINCH CE, ST LAURENT G, 3RD, KENNY PJ, WAHLESTEDT C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FENG J, BI C, CLARK BS, MADY R, SHAH P, KOHTZ JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–84. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIRE A, XU S, MONTGOMERY MK, KOSTAS SA, DRIVER SE, MELLO CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- FORSTEMANN K, TOMARI Y, DU T, VAGIN VV, DENLI AM, BRATU DP, KLATTENHOFF C, THEURKAUF WE, ZAMORE PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRAZER KA. Decoding the human genome. Genome research. 2012;22:1599–601. doi: 10.1101/gr.146175.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FU X, RAVINDRANATH L, TRAN N, PETROVICS G, SRIVASTAVA S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA and cell biology. 2006;25:135–41. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- GARZON R, MARCUCCI G, CROCE CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature reviews. Drug discovery. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GE Q, EISEN HN, CHEN J. Use of siRNAs to prevent and treat influenza virus infection. Virus Res. 2004a;102:37–42. doi: 10.1016/j.virusres.2004.01.013. [DOI] [PubMed] [Google Scholar]
- GE Q, FILIP L, BAI A, NGUYEN T, EISEN HN, CHEN J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc Natl Acad Sci U S A. 2004b;101:8676–81. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHILDIYAL M, SEITZ H, HORWICH MD, LI C, DU T, LEE S, XU J, KITTLER EL, ZAPP ML, WENG Z, ZAMORE PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–81. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHILDIYAL M, XU J, SEITZ H, WENG Z, ZAMORE PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHILDIYAL M, ZAMORE PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHOSH A, MUKHERJEE K, JIANG X, ZHOU Y, MCCARROLL J, QU J, SWAIN PM, BAIGUDE H, RANA TM. Design and assembly of new nonviral RNAi delivery agents by microwave-assisted quaternization (MAQ) of tertiary amines. Bioconjugate chemistry. 2010;21:1581–7. doi: 10.1021/bc900482r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIRARD A, SACHIDANANDAM R, HANNON GJ, CARMELL MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- GOODRICH JA, KUGEL JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7:612–6. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- GREGORY RI, CHENDRIMADA TP, COOCH N, SHIEKHATTAR R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- GREGORY RI, YAN KP, AMUTHAN G, CHENDRIMADA T, DORATOTAJ B, COOCH N, SHIEKHATTAR R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- GRISHOK A, PASQUINELLI AE, CONTE D, LI N, PARRISH S, HA I, BAILLIE DL, FIRE A, RUVKUN G, MELLO CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- GUNAWARDANE LS, SAITO K, NISHIDA KM, MIYOSHI K, KAWAMURA Y, NAGAMI T, SIOMI H, SIOMI MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- GUTTMAN M, RINN JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUZZARDO PM, MUERDTER F, HANNON GJ. The piRNA pathway in flies: highlights and future directions. Current opinion in genetics & development. 2013;23:44–52. doi: 10.1016/j.gde.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMILTON AJ, BAULCOMBE DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–2. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- HAMMOND SM, BOETTCHER S, CAUDY AA, KOBAYASHI R, HANNON GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–50. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- HAN J, LEE Y, YEOM KH, KIM YK, JIN H, KIM VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN J, LEE Y, YEOM KH, NAM JW, HEO I, RHEE JK, SOHN SY, CHO Y, ZHANG BT, KIM VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- HANDLER D, MEIXNER K, PIZKA M, LAUSS K, SCHMIED C, GRUBER FS, BRENNECKE J. The Genetic Makeup of the Drosophila piRNA Pathway. Mol Cell. 2013;50:762–77. doi: 10.1016/j.molcel.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARROW J, FRANKISH A, GONZALEZ JM, TAPANARI E, DIEKHANS M, KOKOCINSKI F, AKEN BL, BARRELL D, ZADISSA A, SEARLE S, BARNES I, BIGNELL A, BOYCHENKO V, HUNT T, KAY M, MUKHERJEE G, RAJAN J, DESPACIO-REYES G, SAUNDERS G, STEWARD C, HARTE R, LIN M, HOWALD C, TANZER A, DERRIEN T, CHRAST J, WALTERS N, BALASUBRAMANIAN S, PEI B, TRESS M, RODRIGUEZ JM, EZKURDIA I, VAN BAREN J, BRENT M, HAUSSLER D, KELLIS M, VALENCIA A, REYMOND A, GERSTEIN M, GUIGO R, HUBBARD TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARTIG JV, ESSLINGER S, BOTTCHER R, SAITO K, FORSTEMANN K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. Embo J. 2009;28:2932–44. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARTIG JV, FORSTEMANN K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011;39:3836–51. doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORWICH MD, LI C, MATRANGA C, VAGIN V, FARLEY G, WANG P, ZAMORE PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–72. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- HOSOKAWA M, SHOJI M, KITAMURA K, TANAKA T, NOCE T, CHUMA S, NAKATSUJI N. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev Biol. 2007;301:38–52. doi: 10.1016/j.ydbio.2006.10.046. [DOI] [PubMed] [Google Scholar]
- HOUWING S, KAMMINGA LM, BEREZIKOV E, CRONEMBOLD D, GIRARD A, VAN DEN ELST H, FILIPPOV DV, BLASER H, RAZ E, MOENS CB, PLASTERK RH, HANNON GJ, DRAPER BW, KETTING RF. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- HU PF, XIE WF. Targeted RNA interference for hepatic fibrosis. Expert Opin Biol Ther. 2009;9:1305–12. doi: 10.1517/14712590903213677. [DOI] [PubMed] [Google Scholar]
- HUARTE M, GUTTMAN M, FELDSER D, GARBER M, KOZIOL MJ, KENZELMANN-BROZ D, KHALIL AM, ZUK O, AMIT I, RABANI M, ATTARDI LD, REGEV A, LANDER ES, JACKS T, RINN JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUTVAGNER G, MCLACHLAN J, PASQUINELLI AE, BALINT E, TUSCHL T, ZAMORE PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- ISHIZU H, SIOMI H, SIOMI MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes & development. 2012;26:2361–73. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISHIZUKA A, SIOMI MC, SIOMI H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IWASAKI S, KOBAYASHI M, YODA M, SAKAGUCHI Y, KATSUMA S, SUZUKI T, TOMARI Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39:292–9. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- JI P, DIEDERICHS S, WANG W, BOING S, METZGER R, SCHNEIDER PM, TIDOW N, BRANDT B, BUERGER H, BULK E, THOMAS M, BERDEL WE, SERVE H, MULLER-TIDOW C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- JIN P, ALISCH RS, WARREN ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004a;6:1048–53. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- JIN P, ZARNESCU DC, CEMAN S, NAKAMOTO M, MOWREY J, JONGENS TA, NELSON DL, MOSES K, WARREN ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004b;7:113–7. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- JUDGE AD, ROBBINS M, TAVAKOLI I, LEVI J, HU L, FRONDA A, AMBEGIA E, MCCLINTOCK K, MACLACHLAN I. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Invest. 2009;119:661–73. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANELLOPOULOU C, MULJO SA, KUNG AL, GANESAN S, DRAPKIN R, JENUWEIN T, LIVINGSTON DM, RAJEWSKY K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAPRANOV P, CHENG J, DIKE S, NIX DA, DUTTAGUPTA R, WILLINGHAM AT, STADLER PF, HERTEL J, HACKERMULLER J, HOFACKER IL, BELL I, CHEUNG E, DRENKOW J, DUMAIS E, PATEL S, HELT G, GANESH M, GHOSH S, PICCOLBONI A, SEMENTCHENKO V, TAMMANA H, GINGERAS TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- KARUBE Y, TANAKA H, OSADA H, TOMIDA S, TATEMATSU Y, YANAGISAWA K, YATABE Y, TAKAMIZAWA J, MIYOSHI S, MITSUDOMI T, TAKAHASHI T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATIYAR-AGARWAL S, MORGAN R, DAHLBECK D, BORSANI O, VILLEGAS A, JR, ZHU JK, STASKAWICZ BJ, JIN H. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci U S A. 2006;103:18002–7. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWAMURA Y, SAITO K, KIN T, ONO Y, ASAI K, SUNOHARA T, OKADA TN, SIOMI MC, SIOMI H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–7. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- KAWAOKA S, IZUMI N, KATSUMA S, TOMARI Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43:1015–22. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- KETTING RF, FISCHER SE, BERNSTEIN E, SIJEN T, HANNON GJ, PLASTERK RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM VN, HAN J, SIOMI MC. Biogenesis of small RNAs in animals. Nature reviews. Molecular cell biology. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- KNIGHT SW, BASS BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–71. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KROL J, LOEDIGE I, FILIPOWICZ W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- KRUTZFELDT J, RAJEWSKY N, BRAICH R, RAJEEV KG, TUSCHL T, MANOHARAN M, STOFFEL M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- KULSHRESHTHA R, FERRACIN M, NEGRINI M, CALIN GA, DAVULURI RV, IVAN M. Regulation of microRNA expression: the hypoxic component. Cell Cycle. 2007a;6:1426–31. [PubMed] [Google Scholar]
- KULSHRESHTHA R, FERRACIN M, WOJCIK SE, GARZON R, ALDER H, AGOSTO-PEREZ FJ, DAVULURI R, LIU CG, CROCE CM, NEGRINI M, CALIN GA, IVAN M. A microRNA signature of hypoxia. Mol Cell Biol. 2007b;27:1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KURAMOCHI-MIYAGAWA S, KIMURA T, IJIRI TW, ISOBE T, ASADA N, FUJITA Y, IKAWA M, IWAI N, OKABE M, DENG W, LIN H, MATSUDA Y, NAKANO T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–49. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- KURAMOCHI-MIYAGAWA S, KIMURA T, YOMOGIDA K, KUROIWA A, TADOKORO Y, FUJITA Y, SATO M, MATSUDA Y, NAKANO T. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–33. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- LANDER ES, LINTON LM, BIRREN B, NUSBAUM C, ZODY MC, BALDWIN J, DEVON K, DEWAR K, DOYLE M, FITZHUGH W, FUNKE R, GAGE D, HARRIS K, HEAFORD A, HOWLAND J, KANN L, LEHOCZKY J, LEVINE R, MCEWAN P, MCKERNAN K, MELDRIM J, MESIROV JP, MIRANDA C, MORRIS W, NAYLOR J, RAYMOND C, ROSETTI M, SANTOS R, SHERIDAN A, SOUGNEZ C, STANGE-THOMANN N, STOJANOVIC N, SUBRAMANIAN A, WYMAN D, ROGERS J, SULSTON J, AINSCOUGH R, BECK S, BENTLEY D, BURTON J, CLEE C, CARTER N, COULSON A, DEADMAN R, DELOUKAS P, DUNHAM A, DUNHAM I, DURBIN R, FRENCH L, GRAFHAM D, GREGORY S, HUBBARD T, HUMPHRAY S, HUNT A, JONES M, LLOYD C, MCMURRAY A, MATTHEWS L, MERCER S, MILNE S, MULLIKIN JC, MUNGALL A, PLUMB R, ROSS M, SHOWNKEEN R, SIMS S, WATERSTON RH, WILSON RK, HILLIER LW, MCPHERSON JD, MARRA MA, MARDIS ER, FULTON LA, CHINWALLA AT, PEPIN KH, GISH WR, CHISSOE SL, WENDL MC, DELEHAUNTY KD, MINER TL, DELEHAUNTY A, KRAMER JB, COOK LL, FULTON RS, JOHNSON DL, MINX PJ, CLIFTON SW, HAWKINS T, BRANSCOMB E, PREDKI P, RICHARDSON P, WENNING S, SLEZAK T, DOGGETT N, CHENG JF, OLSEN A, LUCAS S, ELKIN C, UBERBACHER E, FRAZIER M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- LANFORD RE, HILDEBRANDT-ERIKSEN ES, PETRI A, PERSSON R, LINDOW M, MUNK ME, KAUPPINEN S, ORUM H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAU NC, LIM LP, WEINSTEIN EG, BARTEL DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- LAU NC, SETO AG, KIM J, KURAMOCHI-MIYAGAWA S, NAKANO T, BARTEL DP, KINGSTON RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–7. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- LE THOMAS A, ROGERS AK, WEBSTER A, MARINOV GK, LIAO SE, PERKINS EM, HUR JK, ARAVIN AA, TOTH KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–9. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE JT, BARTOLOMEI MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–23. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- LEE RC, FEINBAUM RL, AMBROS V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- LEE Y, AHN C, HAN J, CHOI H, KIM J, YIM J, LEE J, PROVOST P, RADMARK O, KIM S, KIM VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- LEE Y, HUR I, PARK SY, KIM YK, SUH MR, KIM VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–32. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE Y, KIM M, HAN J, YEOM KH, LEE S, BAEK SH, KIM VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004a;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE YS, NAKAHARA K, PHAM JW, KIM K, HE Z, SONTHEIMER EJ, CARTHEW RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004b;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- LEWEJOHANN L, SKRYABIN BV, SACHSER N, PREHN C, HEIDUSCHKA P, THANOS S, JORDAN U, DELL’OMO G, VYSSOTSKI AL, PLESKACHEVA MG, LIPP HP, TIEDGE H, BROSIUS J, PRIOR H. Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav Brain Res. 2004;154:273–89. doi: 10.1016/j.bbr.2004.02.015. [DOI] [PubMed] [Google Scholar]
- LEWIS BP, SHIH IH, JONES-RHOADES MW, BARTEL DP, BURGE CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- LI BJ, TANG Q, CHENG D, QIN C, XIE FY, WEI Q, XU J, LIU Y, ZHENG BJ, WOODLE MC, ZHONG N, LU PY. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2005;11:944–51. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI C, VAGIN VV, LEE S, XU J, MA S, XI H, SEITZ H, HORWICH MD, SYRZYCKA M, HONDA BM, KITTLER EL, ZAPP ML, KLATTENHOFF C, SCHULZ N, THEURKAUF WE, WENG Z, ZAMORE PD. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–21. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI H, LI WX, DING SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–21. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- LI J, WITTE DP, VAN DYKE T, ASKEW DS. Expression of the putative proto-oncogene His-1 in normal and neoplastic tissues. Am J Pathol. 1997;150:1297–305. [PMC free article] [PubMed] [Google Scholar]
- LI Z, RANA TM. Molecular mechanisms of RNA-triggered gene silencing machineries. Accounts of chemical research. 2012;45:1122–31. doi: 10.1021/ar200253u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIM AK, KAI T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–9. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIMA WF, PRAKASH TP, MURRAY HM, KINBERGER GA, LI W, CHAPPELL AE, LI CS, MURRAY SF, GAUS H, SETH PP, SWAYZE EE, CROOKE ST. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–94. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- LIN R, MAEDA S, LIU C, KARIN M, EDGINGTON TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–8. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- LIU Q, RAND TA, KALIDAS S, DU F, KIM HE, SMITH DP, WANG X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–5. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- LU J, GETZ G, MISKA EA, ALVAREZ-SAAVEDRA E, LAMB J, PECK D, SWEET-CORDERO A, EBERT BL, MAK RH, FERRANDO AA, DOWNING JR, JACKS T, HORVITZ HR, GOLUB TR. MicroRNA expression profiles classify human cancers. Nature. 2005a;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- LU R, MADURO M, LI F, LI HW, BROITMAN-MADURO G, LI WX, DING SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005b;436:1040–3. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUKIW WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- LUND E, GUTTINGER S, CALADO A, DAHLBERG JE, KUTAY U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- MA JB, YE K, PATEL DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–22. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MALONE CD, BRENNECKE J, DUS M, STARK A, MCCOMBIE WR, SACHIDANANDAM R, HANNON GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–35. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MALONE CD, HANNON GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANCINI-DINARDO D, STEELE SJ, LEVORSE JM, INGRAM RS, TILGHMAN SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–82. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANIATAKI E, MOURELATOS Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–90. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARQUES JT, KIM K, WU PH, ALLEYNE TM, JAFARI N, CARTHEW RW. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat Struct Mol Biol. 2010;17:24–30. doi: 10.1038/nsmb.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIANOV I, RAMADASS A, SERRA BARROS A, CHOW N, AKOULITCHEV A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- MATRANGA C, TOMARI Y, SHIN C, BARTEL DP, ZAMORE PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- MAZO A, HODGSON JW, PETRUK S, SEDKOV Y, BROCK HW. Transcriptional interference: an unexpected layer of complexity in gene regulation. J Cell Sci. 2007;120:2755–61. doi: 10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- MCCAFFREY AP, MEUSE L, PHAM TT, CONKLIN DS, HANNON GJ, KAY MA. RNA interference in adult mice. Nature. 2002;418:38–9. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- MCCARROLL J, BAIGUDE H, YANG CS, RANA TM. Nanotubes functionalized with lipids and natural amino acid dendrimers: a new strategy to create nanomaterials for delivering systemic RNAi. Bioconjugate chemistry. 2010;21:56–63. doi: 10.1021/bc900296z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MITSUYA K, MEGURO M, LEE MP, KATOH M, SCHULZ TC, KUGOH H, YOSHIDA MA, NIIKAWA N, FEINBERG AP, OSHIMURA M. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet. 1999;8:1209–17. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- MIYOSHI K, MIYOSHI T, HARTIG JV, SIOMI H, SIOMI MC. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. 2010;16:506–15. doi: 10.1261/rna.1952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIYOSHI K, TSUKUMO H, NAGAMI T, SIOMI H, SIOMI MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–48. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOURTADA-MAARABOUNI M, PICKARD MR, HEDGE VL, FARZANEH F, WILLIAMS GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- MUERDTER F, GUZZARDO PM, GILLIS J, LUO Y, YU Y, CHEN C, FEKETE R, HANNON GJ. A Genome-wide RNAi Screen Draws a Genetic Framework for Transposon Control and Primary piRNA Biogenesis in Drosophila. Mol Cell. 2013;50:736–48. doi: 10.1016/j.molcel.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURCHISON EP, PARTRIDGE JF, TAM OH, CHELOUFI S, HANNON GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAYAK A, BERRY B, TASSETTO M, KUNITOMI M, ACEVEDO A, DENG C, KRUTCHINSKY A, GROSS J, ANTONIEWSKI C, ANDINO R. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat Struct Mol Biol. 2010;17:547–54. doi: 10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKAMURA K, BALLA S, MARTIN R, LIU N, LAI EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998c. [DOI] [PubMed] [Google Scholar]
- OKAMURA K, LIU N, LAI EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–44. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OVERHOFF M, ALKEN M, FAR RK, LEMAITRE M, LEBLEU B, SCZAKIEL G, ROBBINS I. Local RNA target structure influences siRNA efficacy: a systematic global analysis. J Mol Biol. 2005;348:871–81. doi: 10.1016/j.jmb.2005.03.012. [DOI] [PubMed] [Google Scholar]
- PACHNIS V, BRANNAN CI, TILGHMAN SM. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 1988;7:673–81. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PANGANIBAN G, RUBENSTEIN JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–86. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- PARKER JS, ROE SM, BARFORD D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–6. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASMANT E, SABBAGH A, VIDAUD M, BIECHE I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–8. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- PASQUINELLI AE, REINHART BJ, SLACK F, MARTINDALE MQ, KURODA MI, MALLER B, HAYWARD DC, BALL EE, DEGNAN B, MULLER P, SPRING J, SRINIVASAN A, FISHMAN M, FINNERTY J, CORBO J, LEVINE M, LEAHY P, DAVIDSON E, RUVKUN G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- PATIL VS, KAI T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol. 2010;20:724–30. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- PAULER FM, KOERNER MV, BARLOW DP. Silencing by imprinted noncoding RNAs: is transcription the answer? Trends Genet. 2007;23:284–92. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEDERSEN IM, CHENG G, WIELAND S, VOLINIA S, CROCE CM, CHISARI FV, DAVID M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–22. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEK JW, ANAND A, KAI T. Tudor domain proteins in development. Development. 2012a;139:2255–66. doi: 10.1242/dev.073304. [DOI] [PubMed] [Google Scholar]
- PEK JW, PATIL VS, KAI T. piRNA pathway and the potential processing site, the nuage, in the Drosophila germline. Dev Growth Differ. 2012b;54:66–77. doi: 10.1111/j.1440-169x.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- PELISSON A, SAROT E, PAYEN-GROSCHENE G, BUCHETON A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–60. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PERAGINE A, YOSHIKAWA M, WU G, ALBRECHT HL, POETHIG RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–79. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PERKINS DO, JEFFRIES CD, JARSKOG LF, THOMSON JM, WOODS K, NEWMAN MA, PARKER JS, JIN J, HAMMOND SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHAM JW, PELLINO JL, LEE YS, CARTHEW RW, SONTHEIMER EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- PLANTE I, DAVIDOVIC L, OUELLET DL, GOBEIL LA, TREMBLAY S, KHANDJIAN EW, PROVOST P. Dicer-derived microRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RANA TM. Illuminating the silence: understanding the structure and function of small RNAs. Nature reviews. Molecular cell biology. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- RAND TA, PETERSEN S, DU F, WANG X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–9. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- REINHART BJ, SLACK FJ, BASSON M, PASQUINELLI AE, BETTINGER JC, ROUGVIE AE, HORVITZ HR, RUVKUN G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- REUTER M, BERNINGER P, CHUMA S, SHAH H, HOSOKAWA M, FUNAYA C, ANTONY C, SACHIDANANDAM R, PILLAI RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–7. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- RINN JL, KERTESZ M, WANG JK, SQUAZZO SL, XU X, BRUGMANN SA, GOODNOUGH LH, HELMS JA, FARNHAM PJ, SEGAL E, CHANG HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUBY JG, JAN C, PLAYER C, AXTELL MJ, LEE W, NUSBAUM C, GE H, BARTEL DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- SAITO K, ISHIZUKA A, SIOMI H, SIOMI MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO K, NISHIDA KM, MORI T, KAWAMURA Y, MIYOSHI K, NAGAMI T, SIOMI H, SIOMI MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–22. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO K, SAKAGUCHI Y, SUZUKI T, SIOMI H, SIOMI MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–8. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANCHEZ-ELSNER T, GOU D, KREMMER E, SAUER F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–23. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- SCHOPMAN NC, WILLEMSEN M, LIU YP, BRADLEY T, VAN KAMPEN A, BAAS F, BERKHOUT B, HAASNOOT J. Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res. 2012;40:414–27. doi: 10.1093/nar/gkr719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHUBERT S, GRUNWELLER A, ERDMANN VA, KURRECK J. Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J Mol Biol. 2005;348:883–93. doi: 10.1016/j.jmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- SHOJI M, TANAKA T, HOSOKAWA M, REUTER M, STARK A, KATO Y, KONDOH G, OKAWA K, CHUJO T, SUZUKI T, HATA K, MARTIN SL, NOCE T, KURAMOCHI-MIYAGAWA S, NAKANO T, SASAKI H, PILLAI RS, NAKATSUJI N, CHUMA S. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–87. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- SLEUTELS F, ZWART R, BARLOW DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- SMITH CM, STEITZ JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOIFER HS, ROSSI JJ, SAETROM P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15:2070–9. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- SONG JJ, LIU J, TOLIA NH, SCHNEIDERMAN J, SMITH SK, MARTIENSSEN RA, HANNON GJ, JOSHUA-TOR L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–32. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- SONG JJ, SMITH SK, HANNON GJ, JOSHUA-TOR L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–7. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- STAMATOYANNOPOULOS JA. What does our genome encode? Genome research. 2012;22:1602–11. doi: 10.1101/gr.146506.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRASSER MJ, MACKENZIE NC, DUMSTREI K, NAKKRASAE LI, STEBLER J, RAZ E. Control over the morphology and segregation of Zebrafish germ cell granules during embryonic development. BMC Dev Biol. 2008;8:58. doi: 10.1186/1471-213X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRUHL K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–5. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- SU J, BAIGUDE H, MCCARROLL J, RANA TM. Silencing microRNA by interfering nanoparticles in mice. Nucleic acids research. 2011;39:e38. doi: 10.1093/nar/gkq1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SULLIVAN KE. The clinical, immunological, and molecular spectrum of chromosome 22q11.2 deletion syndrome and DiGeorge syndrome. Curr Opin Allergy Clin Immunol. 2004a;4:505–12. doi: 10.1097/00130832-200412000-00006. [DOI] [PubMed] [Google Scholar]
- SULLIVAN KE. Live viral vaccines in patients with DiGeorge syndrome. Clin Immunol. 2004b;113:3. doi: 10.1016/j.clim.2004.04.004. [DOI] [PubMed] [Google Scholar]
- TAM OH, ARAVIN AA, STEIN P, GIRARD A, MURCHISON EP, CHELOUFI S, HODGES E, ANGER M, SACHIDANANDAM R, SCHULTZ RM, HANNON GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOMARI Y, MATRANGA C, HALEY B, MARTINEZ N, ZAMORE PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–80. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- TOMPKINS SM, LO CY, TUMPEY TM, EPSTEIN SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci U S A. 2004;101:8682–6. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRIPATHI V, ELLIS JD, SHEN Z, SONG DY, PAN Q, WATT AT, FREIER SM, BENNETT CF, SHARMA A, BUBULYA PA, BLENCOWE BJ, PRASANTH SG, PRASANTH KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAI MC, MANOR O, WAN Y, MOSAMMAPARAST N, WANG JK, LAN F, SHI Y, SEGAL E, CHANG HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UMLAUF D, GOTO Y, CAO R, CERQUEIRA F, WAGSCHAL A, ZHANG Y, FEIL R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- VAGIN VV, SIGOVA A, LI C, SEITZ H, GVOZDEV V, ZAMORE PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- VAGIN VV, WOHLSCHLEGEL J, QU J, JONSSON Z, HUANG X, CHUMA S, GIRARD A, SACHIDANANDAM R, HANNON GJ, ARAVIN AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–62. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN RIJ RP, SALEH MC, BERRY B, FOO C, HOUK A, ANTONIEWSKI C, ANDINO R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–95. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN ROOIJ E, OLSON EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nature reviews. Drug discovery. 2012;11:860–72. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAZQUEZ F, VAUCHERET H, RAJAGOPALAN R, LEPERS C, GASCIOLLI V, MALLORY AC, HILBERT JL, BARTEL DP, CRETE P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- VOLINIA S, CALIN GA, LIU CG, AMBS S, CIMMINO A, PETROCCA F, VISONE R, IORIO M, ROLDO C, FERRACIN M, PRUEITT RL, YANAIHARA N, LANZA G, SCARPA A, VECCHIONE A, NEGRINI M, HARRIS CC, CROCE CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG G, REINKE V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–7. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG H, GHOSH A, BAIGUDE H, YANG CS, QIU L, XIA X, ZHOU H, RANA TM, XU Z. Therapeutic gene silencing delivered by a chemically modified small interfering RNA against mutant SOD1 slows amyotrophic lateral sclerosis progression. The Journal of biological chemistry. 2008a;283:15845–52. doi: 10.1074/jbc.M800834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG H, IACOANGELI A, LIN D, WILLIAMS K, DENMAN RB, HELLEN CU, TIEDGE H. Dendritic BC1 RNA in translational control mechanisms. J Cell Biol. 2005;171:811–21. doi: 10.1083/jcb.200506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG X, ARAI S, SONG X, REICHART D, DU K, PASCUAL G, TEMPST P, ROSENFELD MG, GLASS CK, KUROKAWA R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008b;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG XH, ALIYARI R, LI WX, LI HW, KIM K, CARTHEW R, ATKINSON P, DING SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–4. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Y, JURANEK S, LI H, SHENG G, WARDLE GS, TUSCHL T, PATEL DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–61. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAPINSKI O, CHANG HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- WATANABE T, TOTOKI Y, TOYODA A, KANEDA M, KURAMOCHI-MIYAGAWA S, OBATA Y, CHIBA H, KOHARA Y, KONO T, NAKANO T, SURANI MA, SAKAKI Y, SASAKI H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- WEBSTER JC, HUBER RM, HANSON RL, COLLIER PM, HAWS TF, MILLS JK, BURN TC, ALLEGRETTO EA. Dexamethasone and tumor necrosis factor-alpha act together to induce the cellular inhibitor of apoptosis-2 gene and prevent apoptosis in a variety of cell types. Endocrinology. 2002;143:3866–74. doi: 10.1210/en.2002-220188. [DOI] [PubMed] [Google Scholar]
- WILLIAMS L, CARLES CC, OSMONT KS, FLETCHER JC. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci U S A. 2005;102:9703–8. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOJCIK SE, ROSSI S, SHIMIZU M, NICOLOSO MS, CIMMINO A, ALDER H, HERLEA V, RASSENTI LZ, RAI KR, KIPPS TJ, KEATING MJ, CROCE CM, CALIN GA. Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis. 2010;31:208–15. doi: 10.1093/carcin/bgp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIA H, MAO Q, PAULSON HL, DAVIDSON BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–10. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- XIE Z, JOHANSEN LK, GUSTAFSON AM, KASSCHAU KD, LELLIS AD, ZILBERMAN D, JACOBSEN SE, CARRINGTON JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG JS, LAI EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG N, KAZAZIAN HH., JR L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- YAP KL, LI S, MUNOZ-CABELLO AM, RAGUZ S, ZENG L, MUJTABA S, GIL J, WALSH MJ, ZHOU MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YI R, QIN Y, MACARA IG, CULLEN BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOSHIKAWA M, PERAGINE A, PARK MY, POETHIG RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–75. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YU D, PENDERGRAFF H, LIU J, KORDASIEWICZ HB, CLEVELAND DW, SWAYZE EE, LIMA WF, CROOKE ST, PRAKASH TP, COREY DR. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU R, CZECH B, BRENNECKE J, SACHIDANANDAM R, WOHLSCHLEGEL JA, PERRIMON N, HANNON GJ. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–95. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU R, HOTTA I, DENLI AM, HONG P, PERRIMON N, HANNON GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–9. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZWART R, SLEUTELS F, WUTZ A, SCHINKEL AH, BARLOW DP. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 2001;15:2361–6. doi: 10.1101/gad.206201. [DOI] [PMC free article] [PubMed] [Google Scholar]


