Abstract
Introduction
Epithelial-mesenchymal transition (EMT) is a process whereby epithelial cells assume mesenchymal characteristics to facilitate cancer metastasis. However, EMT also contributes to the initiation and development of primary tumors. Prior studies that explored the hypothesis that EMT gene variants contribute to EOC risk have been based on small sample sizes and none have sought replication in an independent population.
Methods
We screened 1254 SNPs in 296 genes in a discovery phase using data from a genome-wide association study of EOC among women of European ancestry (1,947 cases and 2,009 controls) and identified 793 variants in 278 EMT-related genes that were nominally (p<0.05) associated with invasive EOC. These SNPs were then genotyped in a larger study of 14,525 invasive-cancer patients and 23,447 controls. A p-value <0.05 and a false discovery rate (FDR) <0.2 was considered statistically significant.
Results
In the larger dataset, GPC6/GPC5 rs17702471 was associated with the endometrioid subtype among Caucasians (OR=1.16, 95%CI=1.07–1.25, p=0.0003, FDR=0.19), while F8 rs7053448 (OR=1.69, 95%CI=1.27–2.24, p=0.0003, FDR=0.12), F8 rs7058826 (OR=1.69, 95%CI=1.27–2.24, p=0.0003, FDR=0.12), and CAPN13 rs1983383 (OR=0.79, 95%CI=0.69–0.90, p=0.0005, FDR=0.12) were associated with combined invasive EOC among Asians. In silico functional analyses revealed that GPC6/GPC5 rs17702471 coincided with DNA regulatory elements.
Conclusion
These results suggest that EMT gene variants do not appear to play a significant role in the susceptibility to EOC.
Keywords: ovarian cancer, epithelial-mesenchymal transition, single nucleotide polymorphisms
INTRODUCTION
Epithelial ovarian carcinoma (EOC) is the leading cause of gynecological cancer-related mortality in the western world. In the United States, 22,000 cases and 14,300 deaths are estimated in 2014 (American Cancer Society 2014). A family history of the disease is associated with a 2–3 fold increased risk of developing the disease (Auranen et al. 1996), implicating inherited genetic factors (Lichtenstein et al. 2000, Stratton et al. 1998). Germline mutations in highly penetrant genes such as BRCA1 and BRCA2 are estimated to account for about 40% of the excess familial risk (Narod and Foulkes 2004). Given little evidence for additional highly penetrant genes, the prevailing consensus is that the remaining familial risk reflects common susceptibility alleles that confer low risk. Indeed, genome-wide association studies (GWAS) and large-scale replication studies have identified some common susceptibility alleles that collectively account for another 4% of the excess familial risk (Pharoah et al. 2013). Thus, more variants remain to be identified.
Epithelial-mesenchymal transition (EMT) is a fundamental embryonic process whereby epithelial cells assume mesenchymal characteristics to facilitate migration through the extracellular matrix (ECM) into the interior of the embryo to contribute to the development of internal organs (Davidson, Trope and Reich 2012, Nieto 2013, Thiery et al. 2009). EMT is a complex process, accompanied by changes in the expression of multiple and diverse factors, including cell adhesion molecules, growth factors, metalloproteinases, transcription factors and epithelial markers (Thiery et al. 2009). A role for EMT in cancer has mostly been limited to cancer cell invasion and metastatic spread, where malignant cells lose their epithelial characteristics and assume mesenchymal properties that promote ECM invasion and metastasis (Thiery et al. 2009). However, several lines of evidence support the hypothesis that EMT contributes to the initiation and development of primary tumors: (i) the expression of inducers of EMT in non-invasive neoplastic lesions, (ii) drivers of EMT attenuate key tumor suppressive mechanisms in epithelial cells by affecting the functions of modulators of p53 and retinoblastoma dependent pathway, and (iii) the involvement of EMT in cell differentiation and tissue homeostasis (Nieto 2013, Puisieux, Brabletz and Caramel 2014). It is plausible that variants in EMT genes may alter gene expression and therefore contribute to inter-individual variation in the risk of ovarian cancer.
Previous studies of variants in EMT-related genes, such as vascular endothelial growth factor (VEGF) (Polterauer et al. 2007, Schultheis et al. 2008, Goode et al. 2010, Steffensen et al. 2010), matrix-metalloprotease genes (MMPs) (Schildkraut et al. 2009b), TP53 (Schildkraut et al. 2009a, Galic et al. 2007), E-cadherin (Li et al. 2008), nitric oxide synthase (NOS) (Hefler et al. 2002), heparanase (HPN) (Ralph et al. 2007) and PIK3CA (Quaye et al. 2009, Quaye et al. 2008) provide support for the hypothesis. However, the majority of the studies were based on small sample sizes with no replications. Furthermore, variation in association by histological subtype and race was not established. In this study, we selected 793 SNPs from 278 EMT-related genes based on nominal associations with EOC in a genome-wide association study among women of European ancestry and performed a replication study involving 14,525 case subjects with invasive disease and 23,447 controls from 43 sites in the Ovarian Cancer Association Consortium (OCAC), as part of the Collaborative Oncological Gene-environment Study (COGS). This large sample size of the replication study also provided an opportunity to explore associations by histological subtypes of EOC and by race.
MATERIALS AND METHODS
Discovery set
We initially selected SNPs based on screening of the results from the North American genome-wide association study (GWAS). Details of this study have been published elsewhere (Permuth-Wey et al. 2011). Briefly, five case-control studies contributed a total of 1,947 EOC cases and 2,009 controls (of European ancestry) to the North American GWAS. Genotyping for four of the studies was performed using the Illumina 610-quad Beadchip Array, while the fifth study was genotyped using the Illumna 317K and 370K arrays. Additional markers were imputed to account for non-overlapping markers in the two genotyping efforts and to improve genome coverage. A total of 2,508,744 (including the 1254 EMT-related SNPs) out of 2,543,887 SNPs (98.6%) passed quality control.
Collaborative Oncological Gene-environment Study
SNPs that were nominally (P<0.05) associated with ovarian cancer in the discovery set were genotyped in the Collaborative Oncological Gene-environment Study (COGS) that included 43 individual studies from OCAC. Details of this study have been published elsewhere (Pharoah et al. 2013). Briefly, 34 case-control studies contributed 14,525 EOC cases and 23,447 controls of white-European ancestry, 150 cases and 200 controls of African ancestry, and 714 cases and 1574 controls of Asian ancestry. Ancestry was assigned using Local Ancestry in adMixed Populations (Sankararaman, 2008) based on genotype frequency for intercontinental ancestry. Participants with >90% white-European ancestry were defined as European and those with >80% African or Asian ancestry were defined as African or Asian, respectively. Population substructure within each ancestry group was determined by principal components analyses using a set of 37,000 uncorrelated markers. The first five principal components were employed for analyses of Europeans and Asians and the first one for Africans. The COGS dataset was used to perform SNP-specific analyses for each of the four main histological subtypes (serous, endometroid, clear cell and mucinous), and each race.
Independent Replication set
The COGS dataset included 62% (1,207 cases and 1,246 controls) of the samples from women of white-European ancestry that were genotyped in the discovery set. Therefore we excluded all these samples from the COGS dataset to create an independent replication set. This independent replication set was used to replicate SNPs identified for all invasive EOC among women of European ancestry in the discovery dataset.
SNP selection, genotyping and quality control
EMT-related genes were identified through review of the published literature (www.pubmed.gov) and pathways described in the Cancer Genome Anatomy Project. Genotype data from HapMap, Perlegen and the NIEHS resequencing projects were then used to select tagSNPs (r2 > 0.8), non-synonymous coding SNPs and putative functional SNPs from unrelated Caucasian samples. A total of 15,816 SNPs in 296 EMT genes had results available in the discovery dataset. The final selection of EMT-related SNPs for genotyping in the large COGS dataset was informed by data from the discovery set and design scores from Illumina as previously described (6). SNPs were ranked based on P-value in the discovery set and the 793 SNPs associated with risk of combined invasive EOC or serous ovarian cancer at a nominal P < 0.05 were selected (Supplementary Table 1). All 793 SNPs were successfully designed for genotyping and included as part of the “candidate SNPs” on the custom Illumina Infinium iSelect array that had a total of 211,155 SNPs designed for COGS (Pharoah et al. 2013).
Genotyping was performed using an Illumina Infinium iSelect BeadChip at McGill University-Génome Québec Innovation Centre and the Mayo Clinic Medical Genome Facility following established quality control criteria (Pharoah et al. 2013). The concordance for 1,251 duplicate samples was 99.6%. Out of the 211,155 SNPs included on the array, 94.5% passed quality control (QC). All 793 SNPs in the present study were genotyped successfully and passed QC.
In silico analysis using publicly available datasets
We used publicly available data to test for evidence of functional effects of the SNPs using Haploreg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php), ENCODE (http://genome.ucsc.edu/ENCODE/) and ovary super-enhancer datasets (Ward & Kellis 2012, Hnisz et al. 2013). These datasets were used to identify epigenetic marks coinciding with risk associated SNPs.
Functional Analyses
An in vitro model of early-stage ovarian cancer has been previously described (Lawrenson et al. 2011). Briefly, Illumina HT12 gene expression microarrays were used to profile the transcriptome of 3D models of normal ovarian cells immortalized with TERT and overexpressing cMYC and a mutant KRAS or BRAF allele.
Statistical methods
Participants’ demographic and clinical characteristics by cancer status were compared using t-tests for continuous variables and chi-square tests for categorical variables. Statistical analysis methods for the discovery dataset have been described previously (Permuth-Wey et al. 2011). Analyses of the independent replication set and the COGS dataset were performed as follows. Associations between each SNP and case-control status were estimated as log-linear per allele odds ratios (ORs) with associated 95% confidence intervals (95% CIs) using unconditional logistic regression that treated the number of alleles of interest as ordinal variables. The ORs specific for each histological subtype was estimated by comparing cases of that subtype to all available controls as reference.
All models included variables for study site and for the first five (for Europeans and Asians) or one (for Africans) ancestry-specific principal components as covariates. False discovery rates (FDR) were calculated to control for multiple testing. Statistical tests were two-sided with an alpha level < 0.05 and an FDR < 0.2 considered statistically significant. All statistical analyses were implemented with SAS/Genetics version 9.2 (SAS Institute, NC).
RESULTS
A detailed description of the OCAC studies and study participants in the discovery (Permuth-Wey et al. 2011) and COGS (Pharoah et al. 2013) datasets are provided elsewhere. In the large COGS dataset, the majority of women (93.5%, n=37,972) were of European ancestry; only 0.9% (n=350) and 5.6% (n=2288) were of African or Asian ancestry, respectively (Table 1). As expected, the proportion of serous histological subtype was higher than the proportions for other subtypes for all race groups.
Table 1.
Demographic and clinical characteristics of study participants in the COGS replication study
| European descent | Asian descent | African descent | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Controls (n=23,447) N (%) |
Invasive Cases (n=14,525) N (%) |
p- value2 |
Controls (n=1,574) N (%) |
Invasive Cases (n=714) N (%) |
p- value2 |
Controls (n=200) N (%) |
Invasive Cases (n=150) N (%) |
p- value2 |
| Age (years) | |||||||||
| Mean ± SD | 55.6± 11.9 | 58.1± 11.3 | <.0001 | 53.5± 11.2 | 54.2± 12.1 | 0.15 | 53.3± 11.6 | 56.6± 11.4 | 0.009 |
| <40 | 2027 (8.7) | 748 (5.2) | <.0001 | 101 (6.4) | 69 (9.7) | <0.0001 | 23 (11.5) | 9 (6.0) | 0.021 |
| 40–49 | 4771 (20.6) | 2544 (17.6) | 565 (35.9) | 187 (26.2) | 56 (28.0) | 25 (16.7) | |||
| 50–59 | 7403 (31.9) | 4537 (31.3) | 459 (29.2) | 228 (32.0) | 60 (30.0) | 54 (36.0) | |||
| 60–69 | 6098 (26.3) | 4324 (29.8) | 346 (22.0) | 147 (20.6) | 41 (20.5) | 42 (28.0) | |||
| ≥70 | 2892 (12.5) | 2343 (16.2) | 103 (6.5) | 82 (11.5) | 20 (10.0) | 20 (13.3) | |||
| Family history of ovarian cancer1 | |||||||||
| No | 15425 (92.0) | 8634 (82.4) | <.0001 | 382 (61.0) | 370 (75.5) | <.0001 | 152 (91.6) | 97 (89.8) | 0.67 |
| Yes | 1351 (8.0) | 1849 (17.6) | 244 (39.0) | 120 (24.5) | 14 (8.4) | 11 (10.2) | |||
| Histological subtypes | |||||||||
| Serous | N/A | 8368 (57.6) | N/A | 249 (34.9) | N/A | 89 (59.3) | |||
| Endometroid | 2067 (14.2) | 112 (15.7) | 16 (10.7) | ||||||
| Clear Cell | 1024 (7.1) | 103 (14.4) | 6 (4.0) | ||||||
| Mucinous | 944 (6.5) | 60 (8.4) | 11 (7.3) | ||||||
| Others3 | 2122 (14.6) | 190 (26.6) | 28 (18.7) | ||||||
for the first degree relatives
t-test for a continuous variable and chi-square test for a categorical variable
include mixed cell, other specified epithelial, undifferentiated, unknown (but known to be epithelial)
In the first step, we screened 15,816 SNPs in 296 EMT-related candidate genes in the GWAS data set, which consisted of women of European ancestry only. A total of 793 SNPs in 278 genes were nominally (p<0.05) associated with the risk of all invasive EOC. Seven of the 793 SNPs showed a nominal consistent (in direction) association with risk of invasive EOC among women of European ancestry in an independent replication dataset (Table 2). However, none of the associations for these SNPs or additional SNPs in the corresponding genes remained statistically significant after controlling for multiple testing (Table 2).
Table 2.
EMT SNPs significantly associated with invasive ovarian cancer risk among Caucasians
| Discovery dataset | Replication dataset | COGs dataseta | FDR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Nearest gene | Location | Chr | Minor allele |
MAF | P | OR(95%CI) | MAF | P | OR(95%CI) | MAF | P | OR(95%CI) | |
| rs3771044 | NRP2 | intron | 2 | G | 0.13 | 0.022 | 1.17 (1.02–1.34) | 0.11 | 0.014 | 1.06 (1.01–1.12) | 0.12 | 0.002 | 1.08 (1.03–1.13) | 0.5 |
| rs10460829 | HEG1/MUC13 | Intergenic | 3 | A | 0.21 | 0.015 | 1.15 (1.03–1.29) | 0.21 | 0.023 | 1.05 (1.01–1.09) | 0.21 | 0.005 | 1.06 (1.02–1.10) | 0.55 |
| rs2100143 | GHSR/TNFSF10 | Intergenic | 3 | G | 0.48 | 0.028 | 0.9 (0.82–0.99) | 0.49 | 0.044 | 0.97 (0.94–1.00) | 0.48 | 0.013 | 0.96 (0.93–0.99) | 0.55 |
| rs10954593 | SEMA3C/LOC100128317 | Intergenic | 7 | G | 0.19 | 0.033 | 1.14 (1.01–1.27) | 0.19 | 0.034 | 1.05 (1.00–1.09) | 0.19 | 0.011 | 1.05 (1.01–1.09) | 0.55 |
| rs172310 | SHH | flanking_5 | 7 | A | 0.31 | 0.039 | 1.11 (1.01–1.23) | 0.30 | 0.01 | 1.05 (1.01–1.08) | 0.30 | 0.002 | 1.05 (1.02–1.09) | 0.55 |
| rs8030039 | SEMA4B | intron | 15 | A | 0.35 | 0.019 | 1.12 (1.02–1.23) | 0.35 | 0.033 | 1.04 (1.00–1.07) | 0.35 | 0.021 | 1.04 (1.01–1.07) | 0.55 |
| rs10794486 | IGF1R | intron | 15 | G | 0.3 | 0.043 | 1.11 (1.00–1.22) | 0.30 | 0.004 | 1.05 (1.02–1.09) | 0.30 | 0.002 | 1.06 (1.02–1.09) | 0.5 |
Abbreviations: Chr=chromosome; MAF=minor allele frequency; OR=odds ratio; CI=confidence interval.
Includes the replication dataset and 62% of the discovery dataset
Previous studies have revealed that genetic associations with EOC can differ, sometimes in the opposite direction, by histology. Since this effect can mask associations, we performed analyses for the four main histological subtypes, separately in the COGs dataset only because the discovery set did not have information on all the histological subtypes (Supplementary Table 1). This analysis revealed a statistically significant association, after controlling for multiple testing, at glypican 6/glypican 5 (GPC6/GPC5) rs17702471 (OR=1.16, 95%CI=1.07–1.25, p=0.0003, FDR=0.19) for the endometroid subtype among Caucasians (Table 3).
Table 3.
Association between GPC6/GPC5 rs17702471 and histological subtypes of epithelial ovarian cancer among Caucasians
| Histological subtype | P | OR(95%CI) | FDR |
|---|---|---|---|
| Endometroid | 0.00026 | 1.16 (1.07–1.25) | 0.19 |
| Clear Cell | 0.013 | 1.15 (1.03–1.28) | 0.80 |
| Mucinous | 0.13 | 1.09 (0.98–1.22) | 0.94 |
| Serous | 0.21 | 1.03 (0.98–1.08) | 0.77 |
| All invasive | 0.019 | 1.05 (1.01–1.09) | 0.55 |
Analyses for Asians and African Americans separately in the COGs dataset because the discovery set did not have information on these two races. We observed associations at F8 rs7053448 (OR=1.69, 95%CI=1.27–2.24, p=0.0003, FDR=0.12), F8 rs7058826 (OR=1.69, 95%CI=1.27–2.24, p=0.0003, FDR=0.12), and CAPN13 rs1983383 (OR=0.79, 95%CI=0.69–0.90, p=0.0005, FDR=0.12) for combined invasive EOC among Asians (Table 4). However, F8 rs7053448 and rs7058826 were highly correlated (r2=0.7). No statistical significant associations were observed for women of African ancestry.
Table 4.
Associations between F8 rs77053448 and rs7058826 and CAPN13 rs1983383 and all invasive epithelial ovarian cancer by race
| SNP | Nearest gene |
Chr | Race | Minor allele |
MAF | P | OR(95%CI) | FDR |
|---|---|---|---|---|---|---|---|---|
| rs7053448 | F8 | 23 | Asian | G | 0.06 | 0.00034 | 1.69 (1.27–2.24) | 0.12 |
| African American | G | 0.13 | 0.91 | 0.98 (0.63–1.52) | 0.97 | |||
| Caucasian | G | 0.18 | 0.45 | 0.98 (0.95–1.03) | 0.79 | |||
| rs7058826 | F8 | 23 | Asian | A | 0.06 | 0.00034 | 1.69 (1.27–2.24) | 0.12 |
| African American | A | 0.10 | 0.47 | 0.83 (0.51–1.36) | 0.94 | |||
| Caucasian | A | 0.15 | 0.74 | 0.99 (0.95–1.04) | 0.84 | |||
| rs1983383 | CAPN13 | 2 | Asian | A | 0.45 | 0.00049 | 0.79 (0.69–0.90) | 0.12 |
| African American | A | 0.49 | 0.11 | 0.79 (0.59–1.05) | 0.93 | |||
| Caucasian | A | 0.34 | 0.67 | 0.99 (0.96–1.03) | 0.83 |
In silico functional analysis of variants and candidate genes
We used publicly available data, including Haploreg, ENCODE and ovary super-enhancer datasets, to test for evidence of functionality of the four SNPs (GPC6/GPC5 rs17702471, F8 rs7053448, F8 rs7058826 and CAPN13 rs1983383) that were significant after controlling for multiple testing. GPC6/GPC5 rs17702471 coincided with functional elements in the datasets (Table 5).
Table 5.
Identification of regulatory DNA elements coinciding with risk variants
| rsID | Chr | Position | Nearest Gene |
Location | ENCODE | ROADMAP | DNAse |
† Super Enhancer |
Motifs | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Promoter | Enhancer | Promoter | Enhancer | ||||||||
| rs3771044 | 2 | 206577411 | NRP2 | Intronic | BATF; ERα;Maf | ||||||
| rs10460829 | 3 | 124665905 | MUC13 | Intergenic | Gmeb1; p300 | ||||||
| rs2100143 | 3 | 172202876 | TNFSF10 | Intergenic | ERα;HNF4;LRH1; MIF1;RXR::LXR; RXRA; SF1 | ||||||
| rs10513702 | 3 | 172186953 | GHSR | Intergenic | |||||||
| rs172310 | 7 | 155615627 | SHH | Intergenic | RXRA;SP1;TR4 | ||||||
| rs10954593 | 7 | 81036747 | AC008163.4 | Intergenic | GR;HMG-IY; Hoxa3;Myc; Nkx6-1;Pax-4;SIX5;XBP-1 | ||||||
| rs288761 | 7 | 155633481 | SHH | Intergenic | |||||||
| rs1233556 | 7 | 155600417 | SHH | Intronic | Ets; NF-kappaB | ||||||
| rs17702471 | 13 | 93877117 | GPC6 | Intergenic | RXRA | ||||||
| rs10794486 | 15 | 99301535 | IGF1R | Intronic | Esx1; GR;OTX;Pax7 | ||||||
| rs8030039 | 15 | 90767489 | SEMA4B | Intronic | Irf;SR | ||||||
| rs7053448 | X | 154193211 | F8 | Intronic | Foxa;Foxf2; Foxj2;Foxk1; Foxl1;Pou3f1; Pou5f1;Sox15;Sox19;Sox2; TATA;Zfp105 | ||||||
| rs7058826 | X | 154194989 | F8 | Intronic | DMRT1; ERα;Esr2;Foxa;RREB-1 | ||||||
Using data from Haploreg, ENCODE and and † Hnisz et al (2013).
Identification of regulatory DNA elements coinciding with risk variants, using data from Haploreg, ENCODE and Hnisz et al (2013). TF, transcription factor.
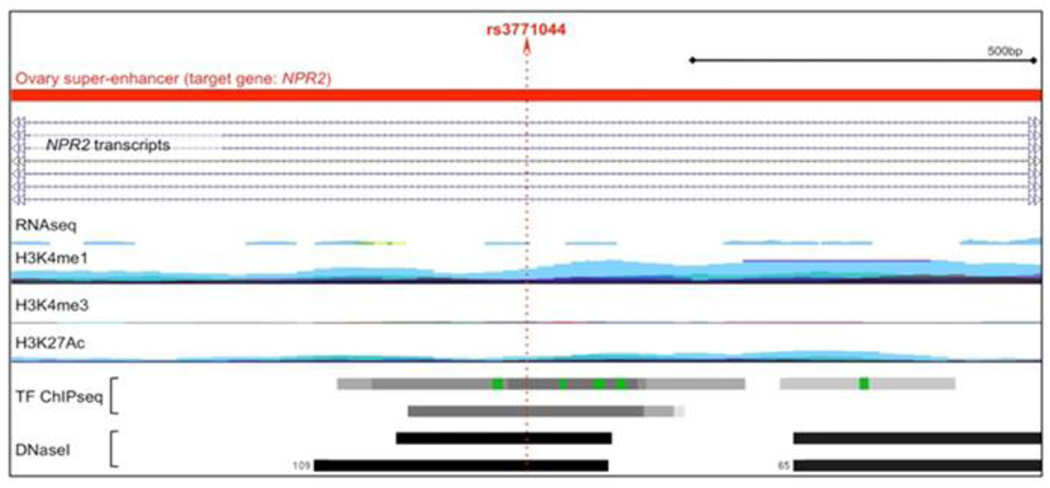
Although none of the seven SNPs (NRP2 rs3771044, HEG1/MUC13 rs10460829, GHSP/TNFSF10 rs2100143, SEMA3C/LOC100128317 rs10954593, SHH rs172310, SEMA4B rs8030039 and IGF1R rs10794486) that showed consistent association between the discovery and independent replication datasets remained statistically significant after multiple testing, we evaluated their functionality because of their consistent associations. Functional analyses indicated a strong evidence of regulatory activity at the NRP2 rs3771044 polymorphism, which coincided with DNase I hypersensitivity peaks and transcription factor binding sites in 62 different cell lines (Hnisz et al, 2013), plus the footprint of a super-enhancer (Figure 1), active in ovarian tissue and known to target NRP2. NRP2 (neurophilin 2) is a transmembrane glycoprotein that interacts with TGF-β1 signaling to promote cancer progression (Grandclement et al. 2011).
Figure 1. Annotation of regulatory elements at rs3771044.
This variant lies within a super-enhancer detected in ovarian tissues (data from Hsinz et al). Using data from ENCODE Regulation tracks, we identified evidence of cell type specific H3K4me1 marks. This SNP also coincides with ChIPseq marks for multiple transactions factors (TFs) including EP300, NFKB and RUNX3. This variant lies with a DNasel hypersensitive region detected in 109 samples.
DISCUSSION
The current study sought to test the hypothesis that genetic variants in the EMT process influence risk of EOC. Seven variants showed nominal consistent associations with EOC in the discovery and replication sets among women of European ancestry, but none of them reached the threshold for statistical significance after controlling for multiple testing in the combined COGS dataset. However, four SNPs showed significant associations, after controlling for multiple testing, in the endometrioid subtype among Caucasians (GPC6/GPC5 rs17702471), and for combined invasive EOC among Asians (F8 rs7053448, F8 rs7058826 and CAPN13 rs1983383). Rs17702471, an intergenic SNP located between GPC5 and GPC6 was associated with the endometroid subtype among women of European-ancestry. Glypicans (GPC) are a family of heparan sulphate proteoglycans (HSPGs) that are attached to the plasma membrane and regulate cell proliferation and division (De Cat and David 2001, Filmus and Selleck 2001) and have been previously shown to be involved in the development and metastasis of various types of human cancer ((Yang et al. 2013, Zhang et al. 2011). Variants in GPC5/GPC6 have been implicated in lung (Li et al. 2010, Liu et al. 2014), but not ovarian cancer. Rs17702471 coincides with potential regulatory elements and further studies evaluating its role in the endometroid histological subtype of ovarian cancer are warranted.
F8 rs7053448, F8 rs7058826 and CAPN13 rs1983383 were associated with all invasive EOC in Asians. Coagulation factor VIII (F8) participates in the intrinsic pathway of blood coagulation. A direct role for F8 has not been established for cancer. Calpain (CAPN) 13 is part of a family of cytosolic calcium-activated proteases involved in apoptosis, cell division and modulation of integrin-cytoskeletal interaction. Although the associations for these SNPs met our threshold for statistical significance, the analyses were exploratory and also based on a small sample size. Thus future studies are warranted to confirm these initial findings.
Although statistical evidence did not support SNPs that showed consistent nominal associations in the discovery and the independent replication datasets, functional data for NRP2 rs3771044 may suggest a role in ovarian cancer. This SNP coincides with DNase I hypersensitivity peaks and transcription factor binding sites and a super-enhancer active in ovarian tissues (Hnisz et al. 2013). Analysis of the expression of NRP2 using TCGA data revealed a significantly higher expression in high-grade serous tumors compared to normal control tissues (data not shown). Neurophilins are transmembrane glycoproteins that are involved in several signaling pathways leading to cytoskeletal organization, angiogenesis and cancer progression (Prahst et al. 2008, Soker et al. 2002, Sulpice et al. 2008). A role for NRP2 in ovarian cancer has not been reported previously, thus warranting further evaluation of its role in ovarian cancer risk.
Our study is the largest one to date to evaluate EMT-related gene variants in ovarian cancer. The study also benefits from other strengths including discovery and replication phases, control for population substructure within each ethnicity, biological plausibility of selected genes, evaluation of a comprehensive list of SNPs in EMT-related genes, evaluation of histological subtypes and race (albeit exploratory) and the tight quality control on the genotype data. A limitation of our study is that the analysis in the other ethnic/racial groups was based on small numbers and the associations may not be reliable. Future studies with larger numbers of African and Asians are therefore warranted.
The results suggest that EMT gene variants do not appear to play a significant role in the susceptibility to EOC.
Supplementary Material
Acknowledgments
Individual acknowledgements by study:
We thank all the individuals who took part in this study and all the researchers, clinicians and technical and administrative staff who have made possible the many studies contributing to this work. In particular, we thank: D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward, P. Webb and D. Whiteman (AUS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); the staff of the genotyping unit, S LaBoissiere and F Robidoux (Genome Quebec); U. Eilber (GER); L. Gacucova (HMO); P. Schurmann, F. Kramer, W. Zheng, T. W. Park, Simon, K. Beer- Grondke and D. Schmidt (HJO); S. Windebank, C. Hilker and J. Vollenweider (MAY); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WYL (NHS); L. Paddock, M. King, L. Rodriguez-Rodriguez, A. Samoila, and Y. Bensman (NJO); M. Sherman, A. Hutchinson,N. Szeszenia--- Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao and M. Stagner (POL); C. Luccarini,P. Harrington the SEARCH team and ECRIC (SEA); R. Royer, S. Zhang (TOR); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan and J. Ford (UKO); Carole Pye (UKR); A. Amin Al Olama, K. Michilaidou, K. Kuchenbaker (COGS).
Main funding:
The scientific development and funding for this project were funded by the following: NIH R01 CA-1491429 (Phelan PI); the US National Cancer Institute (R01-CA076016); the COGS project is funded through a European Commission's Seventh Framework Programme grant (agreement number 223175 HEALTH F2 2009-223175); the Genetic Associations and Mechanisms in Oncology (GAME-ON): a NCI Cancer Post-GWAS Initiative (U19-CA148112); the Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07).
Investigator-specific funding:
E.K.A was supported by a Cancer prevention fellowship from the National Cancer Institute (R25T CA147832) during the conduct of this study. K.L. is supported by a K99/R00 grant from the National Cancer Institute (Grant number 1K99CA184415-01). C.-T. is supported by the National Health and Medical Research Council; B.K. holds an ACS Early Detection Professorship (SIOP---06---258---01---COUN); L.E.K. is supported by a Canadian Institute of Health Research New Investigator Award (MSH---87734). AWL is supported by NIEHS T32 training grant (T32ES013678).
Funding of included studies:
Funding of the constituent studies was provided by the California Cancer Research Program (00---01389V---20170, N01---CN25403, 2II0200); the Canadian Institutes of Health Research (MOP---86727); Cancer Australia; Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124); the Danish Cancer Society (94---222---52); the ELAN Program of the University of Erlangen---Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Helse Vest; the Norwegian Cancer Society; the Norwegian Research Council; the Ovarian Cancer Research Fund; Nationaal Kankerplan of Belgium; Grant---in---Aid for the Third Term Comprehensive 10---Year Strategy For Cancer Control from the Ministry of Health Labour and Welfare of Japan; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07---CA095666, K07---CA143047,K22---CA138563, N01---CN55424, N01---PC67001, N01---PC067010, N01---PC035137, P01---CA017054, P01---CA087696, P30---CA072720, P50---CA105009, P50-CA136393, R01---CA014089, R01---CA016056, R01---CA017054, R01---CA049449, R01---CA050385, R01---CA054419, R01--- CA058598, R01---CA058860, R01---CA061107, R01---CA061132, R01-CA063678, R01-CA063682, R01---CA067262, R01---CA071766, R01---CA074850, R01---CA080742, R01---CA080978, R01---CA083918, R01---CA087538, R01---CA092044, R01---095023, R01---CA122443, R01---CA112523, R01---CA114343, R01---CA126841, R01---CA136924, R03---CA113148, R03---CA115195, U01---CA069417, U01---CA071966 and Intramural research funds); the US Army Medical Research and Material Command (DAMD17---01---1---0729, DAMD17---02---1---0666, DAMD17---02---1---0669, W81XWH---07---0449, W81XWH---10---1---02802); the US Public Health Service (PSA---042205); The National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01GB 9401); the State of Baden---Wurttemberg through Medical Faculty of the University of Ulm (P.685); the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation (LVS---39420); the Oak Foundation; the OHSU Foundation; the Mermaid I project; the Rudolf---Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital “Womens Health Theme” and the Royal Marsden Hospital; WorkSafeBC 14.
Footnotes
The co-authors have no conflicts of interest to declare.
References
- American Cancer Society. Cancer Facts and Figures 2014. Atlanta: American Cancer Society; 2014. [Google Scholar]
- Auranen A, Pukkala E, Makinen J, Sankila R, Grenman S, Salmi T. Cancer incidence in the first-degree relatives of ovarian cancer patients. Br J Cancer. 1996;74:280–284. doi: 10.1038/bjc.1996.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Trope CG, Reich R. Epithelial-mesenchymal transition in ovarian carcinoma. Front Oncol. 2012;2:33. doi: 10.3389/fonc.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cat B, David G. Developmental roles of the glypicans. Semin Cell Dev Biol. 2001;12:117–125. doi: 10.1006/scdb.2000.0240. [DOI] [PubMed] [Google Scholar]
- Filmus J, Selleck SB. Glypicans: proteoglycans with a surprise. J Clin Invest. 2001;108:497–501. doi: 10.1172/JCI13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galic V, Willner J, Wollan M, Garg R, Garcia R, Goff BA, Gray HJ, Swisher EM. Common polymorphisms in TP53 and MDM2 and the relationship to TP53 mutations and clinical outcomes in women with ovarian and peritoneal carcinomas. Genes, chromosomes & cancer. 2007;46:239–247. doi: 10.1002/gcc.20407. [DOI] [PubMed] [Google Scholar]
- Goode EL, Maurer MJ, Sellers TA, Phelan CM, Kalli KR, Fridley BL, Vierkant RA, Armasu SM, White KL, Keeney GL, Cliby WA, Rider DN, Kelemen LE, Jones MB, Peethambaram PP, Lancaster JM, Olson JE, Schildkraut JM, Cunningham JM, Hartmann LC. Inherited determinants of ovarian cancer survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:995–1007. doi: 10.1158/1078-0432.CCR-09-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandclement C, Pallandre JR, Valmary Degano S, Viel E, Bouard A, Balland J, Remy-Martin JP, Simon B, Rouleau A, Boireau W, Klagsbrun M, Ferrand C, Borg C. Neuropilin-2 expression promotes TGF-beta1-mediated epithelial to mesenchymal transition in colorectal cancer cells. PLoS One. 2011;6:e20444. doi: 10.1371/journal.pone.0020444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefler LA, Ludwig E, Lampe D, Zeillinger R, Leodolter S, Gitsch G, Koelbl H, Tempfer CB. Polymorphisms of the endothelial nitric oxide synthase gene in ovarian cancer. Gynecologic oncology. 2002;86:134–137. doi: 10.1006/gyno.2002.6749. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenson K, Sproul D, Grun B, Notaridou M, Benjamin E, Jacobs IJ, Dafou D, Sims AH, Gayther SA. Modelling genetic and clinical heterogeneity in epithelial ovarian cancers. Carcinogenesis. 2011;32:1540–1549. doi: 10.1093/carcin/bgr140. [DOI] [PubMed] [Google Scholar]
- Li Y, Liang J, Kang S, Dong Z, Wang N, Xing H, Zhou R, Li X, Zhao X. E-cadherin gene polymorphisms and haplotype associated with the occurrence of epithelial ovarian cancer in Chinese. Gynecologic oncology. 2008;108:409–414. doi: 10.1016/j.ygyno.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Li Y, Sheu CC, Ye Y, de Andrade M, Wang L, Chang SC, Aubry MC, Aakre JA, Allen MS, Chen F, Cunningham JM, Deschamps C, Jiang R, Lin J, Marks RS, Pankratz VS, Su L, Li Y, Sun Z, Tang H, Vasmatzis G, Harris CC, Spitz MR, Jen J, Wang R, Zhang ZF, Christiani DC, Wu X, Yang P. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321–330. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhong R, Zou L, Fu J, Zhu B, Chen W, Ye X, Gao Y, Yang Y, Christiani DC, Chen S, Miao X. Variants in the 5'-upstream region of GPC5 confer risk of lung cancer in never smokers. Cancer Epidemiol. 2014;38:66–72. doi: 10.1016/j.canep.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- Permuth-Wey J, Kim D, Tsai YY, Lin HY, Chen YA, Barnholtz-Sloan J, Birrer MJ, Bloom G, Chanock SJ, Chen Z, Cramer DW, Cunningham JM, Dagne G, Ebbert-Syfrett J, Fenstermacher D, Fridley BL, Garcia-Closas M, Gayther SA, Ge W, Gentry-Maharaj A, Gonzalez-Bosquet J, Goode EL, Iversen E, Jim H, Kong W, McLaughlin J, Menon U, Monteiro AN, Narod SA, Pharoah PD, Phelan CM, Qu X, Ramus SJ, Risch H, Schildkraut JM, Song H, Stockwell H, Sutphen R, Terry KL, Tyrer J, Vierkant RA, Wentzensen N, Lancaster JM, Cheng JQ, Sellers TA. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer research. 2011;71:3896–3903. doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, Weber R, Karevan R, Larson MC, Song H, Tessier DC, Bacot F, Vincent D, Cunningham JM, Dennis J, Dicks E, Aben KK, Anton-Culver H, Antonenkova N, Armasu SM, Baglietto L, Bandera EV, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brenton JD, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Campbell I, Carney ME, Carvalho RS, Chang-Claude J, Chen YA, Chen Z, Chow WH, Cicek MS, Coetzee G, Cook LS, Cramer DW, Cybulski C, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dork T, du Bois A, Durst M, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher D, Flanagan J, Gao YT, Garcia-Closas M, Gentry-Maharaj A, Giles G, Gjyshi A, Gore M, Gronwald J, Guo Q, Halle MK, Harter P, Hein A, Heitz F, Hillemanns P, Hoatlin M, Hogdall E, Hogdall CK, Hosono S, Jakubowska A, Jensen A, Kalli KR, Karlan BY, Kelemen LE, Kiemeney LA, Kjaer SK, Konecny GE, Krakstad C, Kupryjanczyk J, Lambrechts D, Lambrechts S, Le ND, Lee N, Lee J, Leminen A, Lim BK, Lissowska J, Lubinski J, Lundvall L, Lurie G, Massuger LF, Matsuo K, McGuire V, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45:362–370. 370e1–370e2. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polterauer S, Grimm C, Mustea A, Concin N, Tanner B, Thiel F, Heinze G, Reinthaller A, Zeillinger R, Hefler LA. Vascular endothelial growth factor gene polymorphisms in ovarian cancer. Gynecologic oncology. 2007;105:385–389. doi: 10.1016/j.ygyno.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Prahst C, Heroult M, Lanahan AA, Uziel N, Kessler O, Shraga-Heled N, Simons M, Neufeld G, Augustin HG. Neuropilin-1-VEGFR-2 complexing requires the PDZ-binding domain of neuropilin-1. J Biol Chem. 2008;283:25110–25114. doi: 10.1074/jbc.C800137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- Quaye L, Gayther SA, Ramus SJ, Di Cioccio RA, McGuire V, Hogdall E, Hogdall C, Blaakr J, Easton DF, Ponder BA, Jacobs I, Kjaer SK, Whittemore AS, Pearce CL, Pharoah PD, Song H. The effects of common genetic variants in oncogenes on ovarian cancer survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:5833–5839. doi: 10.1158/1078-0432.CCR-08-0819. [DOI] [PubMed] [Google Scholar]
- Quaye L, Song H, Ramus SJ, Gentry-Maharaj A, Hogdall E, DiCioccio RA, McGuire V, Wu AH, Van Den Berg DJ, Pike MC, Wozniak E, Doherty JA, Rossing MA, Ness RB, Moysich KB, Hogdall C, Blaakaer J, Easton DF, Ponder BA, Jacobs IJ, Menon U, Whittemore AS, Kruger-Kjaer S, Pearce CL, Pharoah PD, Gayther SA. Tagging single-nucleotide polymorphisms in candidate oncogenes and susceptibility to ovarian cancer. British journal of cancer. 2009;100:993–1001. doi: 10.1038/sj.bjc.6604947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph S, Brenchley PE, Summers A, Rosa DD, Swindell R, Jayson GC. Heparanase gene haplotype (CGC) is associated with stage of disease in patients with ovarian carcinoma. Cancer science. 2007;98:844–849. doi: 10.1111/j.1349-7006.2007.00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. A. Am J Hum Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildkraut JM, Goode EL, Clyde MA, Iversen ES, Moorman PG, Berchuck A, Marks JR, Lissowska J, Brinton L, Peplonska B, Cunningham JM, Vierkant RA, Rider DN, Chenevix-Trench G, Webb PM, Beesley J, Chen X, Phelan C, Sutphen R, Sellers TA, Pearce L, Wu AH, Van Den Berg D, Conti D, Elund CK, Anderson R, Goodman MT, Lurie G, Carney ME, Thompson PJ, Gayther SA, Ramus SJ, Jacobs I, Kruger Kjaer S, Hogdall E, Blaakaer J, Hogdall C, Easton DF, Song H, Pharoah PD, Whittemore AS, McGuire V, Quaye L, Anton-Culver H, Ziogas A, Terry KL, Cramer DW, Hankinson SE, Tworoger SS, Calingaert B, Chanock S, Sherman M, Garcia-Closas M. Single nucleotide polymorphisms in the TP53 region and susceptibility to invasive epithelial ovarian cancer. Cancer research. 2009a;69:2349–2357. doi: 10.1158/0008-5472.CAN-08-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildkraut JM, Goode EL, Clyde MA, Iversen ES, Moorman PG, Berchuck A, Marks JR, Lissowska J, Brinton L, Peplonska B, Cunningham JM, Vierkant RA, Rider DN, Chenevix-Trench G, Webb PM, Beesley J, Chen X, Phelan C, Sutphen R, Sellers TA, Pearce L, Wu AH, Van Den Berg D, Conti D, Elund CK, Anderson R, Goodman MT, Lurie G, Carney ME, Thompson PJ, Gayther SA, Ramus SJ, Jacobs I, Kruger Kjaer S, Hogdall E, Blaakaer J, Hogdall C, Easton DF, Song H, Pharoah PD, Whittemore AS, McGuire V, Quaye L, Anton-Culver H, Ziogas A, Terry KL, Cramer DW, Hankinson SE, Tworoger SS, Calingaert B, Chanock S, Sherman M, Garcia-Closas M G. Australian Ovarian Cancer Study. Single nucleotide polymorphisms in the TP53 region and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2009b;69:2349–2357. doi: 10.1158/0008-5472.CAN-08-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis AM, Lurje G, Rhodes KE, Zhang W, Yang D, Garcia AA, Morgan R, Gandara D, Scudder S, Oza A, Hirte H, Fleming G, Roman L, Lenz HJ. Polymorphisms and clinical outcome in recurrent ovarian cancer treated with cyclophosphamide and bevacizumab. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:7554–7563. doi: 10.1158/1078-0432.CCR-08-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soker S, Miao HQ, Nomi M, Takashima S, Klagsbrun M. VEGF165 mediates formation of complexes containing VEGFR-2 and neuropilin-1 that enhance VEGF165-receptor binding. J Cell Biochem. 2002;85:357–368. doi: 10.1002/jcb.10140. [DOI] [PubMed] [Google Scholar]
- Steffensen KD, Waldstrom M, Brandslund I, Jakobsen A. The relationship of VEGF polymorphisms with serum VEGF levels and progression-free survival in patients with epithelial ovarian cancer. Gynecologic oncology. 2010;117:109–116. doi: 10.1016/j.ygyno.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Stratton JF, Pharoah P, Smith SK, Easton D, Ponder BA. A systematic review and meta-analysis of family history and risk of ovarian cancer. Br J Obstet Gynaecol. 1998;105:493–499. doi: 10.1111/j.1471-0528.1998.tb10148.x. [DOI] [PubMed] [Google Scholar]
- Sulpice E, Plouet J, Berge M, Allanic D, Tobelem G, Merkulova-Rainon T. Neuropilin-1 and neuropilin-2 act as coreceptors, potentiating proangiogenic activity. Blood. 2008;111:2036–2045. doi: 10.1182/blood-2007-04-084269. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhang Z, Qiu M, Hu J, Fan X, Wang J, Xu L, Yin R. Glypican-5 is a novel metastasis suppressor gene in non-small cell lung cancer. Cancer Lett. 2013;341:265–273. doi: 10.1016/j.canlet.2013.08.020. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang S, Zhang D, Zhang Z, Xu Y, Liu S. A lung cancer gene GPC5 could also be crucial in breast cancer. Mol Genet Metab. 2011;103:104–105. doi: 10.1016/j.ymgme.2011.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.