Abstract
Activity-dependent changes in the strength of synaptic connections are fundamental to the formation and maintenance of memory. The mechanisms underlying persistent changes in synaptic strength in the hippocampus, specifically long-term potentiation and depression, depend on new protein synthesis. Such changes are thought to be orchestrated by engaging the signaling pathways that regulate mRNA translation in neurons. In this review, we discuss the key regulatory pathways that govern translational control in response to synaptic activity and the mRNA populations that are specifically targeted by these pathways. The critical contribution of regulatory control over new protein synthesis to proper cognitive function is underscored by human disorders associated with either silencing or mutation of genes encoding proteins that directly regulate translation. In light of these clinical implications, we also consider the therapeutic potential of targeting dysregulated translational control to treat cognitive disorders of synaptic dysfunction.
Keywords: eIF2α, mTOR, local protein synthesis, memory, autism, neurodegeneration
INTRODUCTION
Memory storage is thought to have a physical basis in long-lasting modifications of synaptic function in selective brain circuits. Pioneering studies from the Flexners revealed the requirement for protein synthesis as the first molecular distinction between labile short-term memory (STM), lasting from seconds to several minutes, and more stable long-term memory (LTM), which persists for many hours, days, years, or even a lifetime (reviewed in Dudai 2012, Kandel 2001, McGaugh 2000). Protein synthesis inhibitors selectively block LTM in various species, and these fundamental studies have been supported by more recent genetic manipulations (Costa-Mattioli et al. 2009, Richter & Klann 2009). Hence, the current dogma of the neurobiology of learning posits that the synthesis of specific proteins is what determines whether a synaptic or memory process remains transient or becomes persistently stored in the brain. These newly synthesized proteins are thought either to strengthen a preexisting synaptic connection, by inducing a structural remodeling or a functional change (e.g., insertion of receptors), or to form new synaptic connections. In this article, we focus on recent advances in our understanding of the role of protein synthesis in synaptic plasticity and cognitive dysfunction, emphasizing the different mechanisms by which protein synthesis regulates mnemonic processes, as well as potential pharmacological approaches to the treatment of cognitive disorders where translation is altered. Although translational control is crucial for many biological processes in the central nervous system, we limit our discussion to these specialized areas because we believe they are particularly important for future developments in the field.
TRANSLATIONAL CONTROL IN SYNAPTIC PLASTICITY
Synaptic plasticity, the activity-dependent modulation of the strength of synaptic connections, underlies changes in neuronal network dynamics and is therefore thought to be involved in the storage of LTM (Neves et al. 2008). An intriguing aspect of memory is that different types of learning are associated with either strengthening [long-term potentiation (LTP)] or weakening [long-term depression (LTD)] of synaptic efficacy (Malenka & Bear 2004). Repeated activity in a given neural pathway changes the efficacy of its synaptic connections. For instance, with high-frequency activation, an increase in efficacy can last for hours, days, or even weeks (hence LTP). The reverse is also true: Reduced activity lowers synaptic efficacy, resulting in LTD. Both processes are known to require new protein synthesis (Costa-Mattioli et al. 2009, Kandel 2001, Luscher & Huber 2010). Specifically, in ex vivo rodent hippocampal slices, the induction of late-phase LTP (L-LTP) resulting from 4 trains of 100-Hz high-frequency stimuli (4 × 100 Hz) requires de novo protein synthesis, whereas early-phase LTP (E-LTP) induced by 1 train of 100 Hz stimulus (1 × 100 Hz) is independent of protein synthesis (Kandel 2001). In the same preparation, LTD of CA1 synapses induced by activation of metabotropic glutamate receptors 1/5 (mGluR-LTD), but not N-methyl-D-aspartate receptor–dependent LTD (NMDAR-LTD), also requires new protein synthesis (Huber et al. 2000). We discuss below the translational control mechanisms that underlie both L-LTP and mGluR-LTD.
Mechanisms of Translation
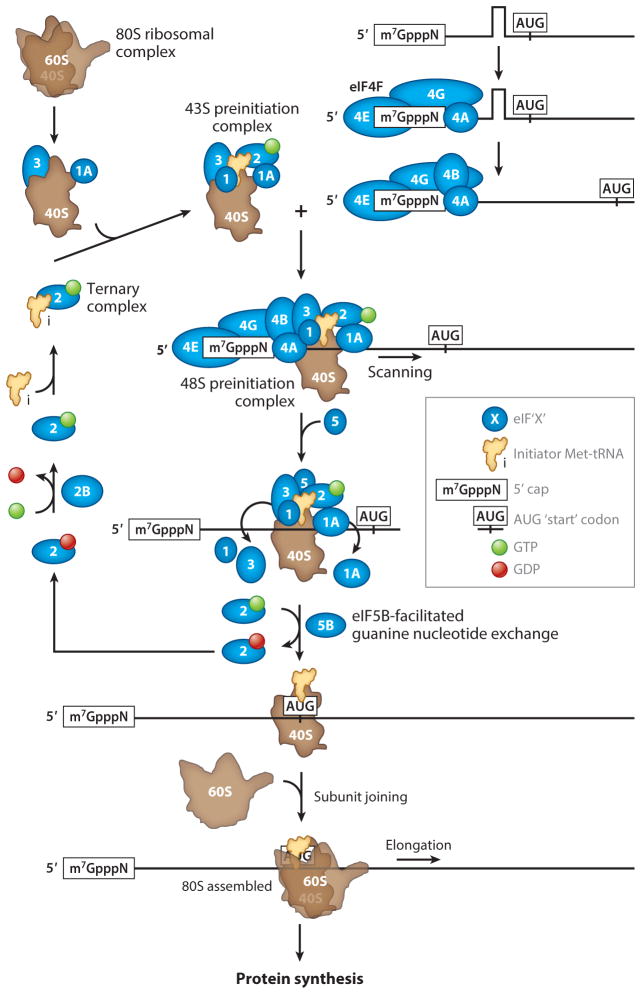
Protein synthesis occurs in three steps: initiation, elongation, and termination. Initiation, the rate-limiting step, is a major target for translational control (Sonenberg & Hinnebusch 2009). Translation initiation begins with the formation of the 43S preinitiation complex, which consists of the small 40S ribosome subunit and the ternary complex; this complex is formed by the initiator methionyl-tRNA (Met-tRNAiMet) and the GTP-bound form of eukaryotic initiation factor eIF2 (Figure 1). The 43S preinitiation complex binds to the 5′ end of the m7-G-capped messenger RNA (mRNA), a process that is promoted by eIF3, the poly(A)-binding protein (PABP) and eIF4F. eIF4F consists of the cap-binding protein eIF4E, eIF4A (the DEAD-box RNA helicase), and eIF4G—a scaffold protein that, through binding with eIF3, bridges the 40S ribosome to the 5′ end of the mRNA. eIF4G also serves as a scaffold to bind PABP, thus inducing the circularization of the mRNA (Jackson et al. 2010). After attachment, the 43S preinitiation complex scans along the mRNA in a 5′ to 3′ fashion until it encounters the initiation AUG codon in an optimum context [GCC(A/G)CCAUGG, with a purine at the −3 and a G at the +4 positions relative to the A of the AUG codon, which is designated +1] to form the 48S preinitiation complex. During scanning, the eIF4F complex catalyzes the unwinding of secondary structures in the 5′ untranslated region (UTR). The first step in initiation codon recognition is the base pairing between the AUG codon and the anticodon of Met-tRNAiMet in the peptidyl-tRNA (P) site of the 40S ribosome. Codon-anticodon recognition arrests the scanning ribosome and triggers the activity of the eIF2-specific GTPase activated protein (GAP) eIF5B in the ternary complex. eIF5B hydrolyzes the ternary complex GTP when it is bound to the 40S subunit, a process that reduces eIF2’s affinity for Met-tRNAiMet, thus leading eIF2-GDP to dissociate from the ribosome. The translation initiation factors eIF1, eIF1A, eIF3, and eIF2-GDP must dissociate from the complex for the 40S and 60S subunits to join, forming the 80S ribosomal complex, a process facilitated by eIF5B.
Figure 1.
Translation initiation in neurons. Translation intitiation, often the rate-limiting step in protein synthesis, involves multiple fundamental reactions. These include formation of the 43S preinitiation complex, ribosomal scanning along the mRNA, AUG initiation codon recognition, and subunit joining to form the 80S ribosomal complex. Following 80S formation, translation elongation factors are recruited to elongate the forming polypeptide chain. Once the stop codon is reached, the elongation factors are disengaged and release of the newly synthesized protein is orchestrated through the action of translation termination factors.
After initiation, translation elongation factors are recruited to elongate the polypeptide chain. The translation elongation factors eEF1A and -1B are required for the aminoacyl-tRNA to transfer onto the ribosome. eEF2 is a GTPase that mediates the translocation of the ribosome along the mRNA following peptide bond formation. Upon recognition of a stop codon, the polypeptide chain is released from the mRNA and ribosome through the coordination of termination factors (Jackson et al. 2012).
Translational Control Mechanisms
By activating NMDA or TrkB receptors, synaptic activity leads to changes in general or gene-specific translation (Costa-Mattioli et al. 2009). The mechanisms regulating translation initiation fall into two categories: (a) those regulating the recruitment of the ribosome to the 5′ end of the mRNA through phosphorylation of translation initiation factors (such as eIF2α and 4E-BPs) and (b) those that control translation at the 3′ end (PABP and Paips) and impact the mRNA itself. In addition, translation can be regulated at the elongation level.
eIF2α-mediated translational control
Protein synthesis requires the recycling of inactive GDP-bound eIF2 to active GTP-bound eIF2. Phosphorylation of the alpha subunit of eIF2 (eIF2α) at Ser51 blocks the activity of eIF2B, the guanine nucleotide exchange factor (GEF) of eIF2 (Pavitt et al. 1998), thus reducing ternary complex formation and thereby the ability of the cell to synthesize new proteins. Paradoxically, it also results in the translational upregulation of a subset of mRNAs that contain upstream open reading frames (uORFs) in their 5′ UTRs. The molecular mechanism underlying this translational upregulation has been explained in great detail for the transcriptional activator GCN4 mRNA in yeast (Hinnebusch 2005) and the transcription factor ATF4 mRNA in mammalian cells (Lu et al. 2004, Vattem & Wek 2004).
The phosphorylation of eIF2α at Ser51 is tightly regulated by kinases and phosphatases (Figure 2a). Mammals have four eIF2α kinases: (a) heme-regulated kinase HRI (EIF2AK1), which is likely relevant only in erythroid cells; (b) the double-strand RNA-dependent kinase PKR (EIF2AK2) activated by viral double-stranded RNA and other stimuli (Garcia et al. 2007); (c) the PKR-like endoplasmic reticulum kinase (PERK, EIF2AK3), a transmembrane endoplasmic reticulum (ER) protein kinase enzyme that is activated by ER stress caused by misfolded proteins; and (d) the highly conserved eIF2α kinase GCN2 (EIF2AK4), which is activated by amino acid deprivation. Two phosphatase complexes are known to dephosphorylate eIF2α. The first complex, constituted by the catalytic subunit protein phosphatase 1 (PP1) and the regulatory subunit PPP1R15A/GADD34, is induced by phosphorylation of eIF2α (Ron & Harding 2007). The second complex, formed by PP1 and the regulatory protein PPP1R15B/CReP, is constitutively expressed.
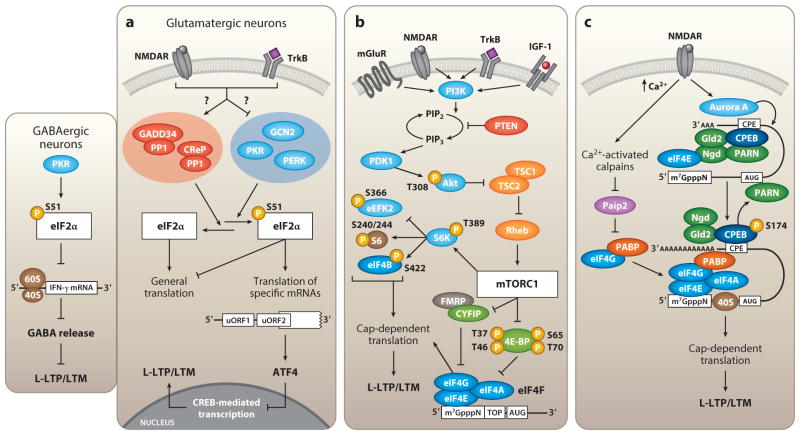
Figure 2.
The key signaling pathways that regulate activity-dependent translation initiation in neurons. (a) Synaptic activity triggers dephosphorylation of initiation factor eIF2α by either activation of eIF2α-specific phosphatase complexes or inhibition of the eIF2α kinases. Dephosphorylation of eIF2α is both sufficient and necessary to induce L-LTP and LTM formation. In glutamatergic neurons, phosphorylation of eIF2α promotes translation of ATF4, a CREB repressor. PKR-mediated phosphorylation of eIF2α in GABAergic neurons negatively regulates translation of the inflammatory cytokine IFN-γ, thus promoting GABA release and maintaining low network rhythmicity in the absence of significant stimulatory inputs. (b) Cap-dependent translation is mediated through mTORC1-dependent phosphorylation of its primary downstream effectors, the 4E-BPs and S6Ks. mTORC1 activity is driven by excitatory synaptic inputs, in addition to other cellular signals, that engage the PI3K/Akt signaling pathway. (c) Calcium influx following NMDAR activation triggers degradation of Paip2 by calcium-activated calpains, thus releasing PABP to bind eIF4G. The eIF4G-PABP complex then contributes to mRNA circularization by directly bridging the elongated 3′ poly(A) tail to the eIF4F complex at the 5′ cap. Cap-dependent translation is initiated upon subsequent recruitment of the 40S and 60S ribosomal complexes. Abbreviations: GADD34, growth arrest and DNA damage-inducible protein; CReP, constitutive reverter of eIF2α phosphorylation; CREB, cAMP response element-binding protein; PI3K, phosphatidylinositide 3-kinase; PDK1, phosphoinositide-dependent kinase-1.
Stimuli known to induce a long-lasting change in synaptic strength, such as BDNF or forskolin application, repeated synaptic stimulation, or even behavioral training, all reduce eIF2α phosphorylation (Costa-Mattioli et al. 2009, Takei et al. 2001). In addition, L-LTP, contextual fear, and spatial and gustatory LTM are facilitated in eIF2α knockin heterozygous mice (where the single phosphorylatable Ser51 is replaced by alanine) or mice lacking the eIF2α kinase GCN2 or PKR, in which eIF2α phosphorylation is reduced in the hippocampus (Costa-Mattioli et al. 2005, 2007; Stern et al. 2013; Zhu et al. 2011). By contrast, Sal003, a small molecule inhibitor of eIF2α phosphatases, selectively promotes eIF2α phosphorylation and impairs L-LTP and LTM (Costa-Mattioli et al. 2007). Furthermore, Jiang et al. (2010) showed that L-LTP and LTM were blocked by an independent chemical genetic strategy that selectively activates the phosphorylation of eIF2α in CA1 in vivo. Together, these data demonstrate that eIF2α dephosphorylation is both sufficient and necessary for L-LTP and LTM storage. Consistent with these findings, treatment with integrated stress response inhibitor B (ISRIB), a new compound that blocks the translational effects mediated by eIF2α phosphorylation, or PKRi, a selective inhibitor of PKR, improves spatial and fear-associated LTM (Sidrauski et al. 2013, Zhu et al. 2011). Whether the activity-driven decrease in eIF2α phosphorylation occurs by promoting eIF2α phosphatase activity or by blocking eIF2α kinases remains unknown. Moreover, whether eIF2α phosphorylation regulates other forms of synaptic plasticity has yet to be determined.
How eIF2α phosphorylation controls long-lasting changes in synaptic function has just begun to be elucidated. eIF2α phosphorylation may have a differential role in glutamatergic versus GABAergic neurons. For instance, it is known that the elevation of eIF2α phosphorylation in glutamatergic neurons upregulates the translation of ATF4 mRNA (Costa-Mattioli et al. 2007, Jiang et al. 2010), which encodes a protein that acts as a repressor of cAMP response element-binding protein (CREB)-mediated L-LTP and LTM (Chen et al. 2003, Costa-Mattioli et al. 2005). By contrast, in GABAergic neurons PKR-mediated phosphorylation of eIF2α locally represses translation of interferon-γ (IFN-γ), resulting in enhanced GABAergic transmission and a consequent depression of neural network excitability (Figure 2a) (Zhu et al. 2011). Finally, mice lacking PERK in glutamatergic forebrain neurons display subtle memory deficits (reduced fear extinction) and some behavioral endophenotypes relevant to schizophrenia, as evidenced by behavioral perseveration and decreased prepulse inhibition (Trinh et al. 2012). The phenotype of PERK-deficient mice is remarkably different from that of other mouse models with reduced eIF2α phosphorylation. Thus, whether the schizophrenia-related behaviors in PERK-deficient mice are due to reduced eIF2α phosphorylation or the inability to cope with the ER stress caused by the accumulation of misfolded proteins remains to be determined.
mTORC1-mediated translational control
The mechanistic target of rapamycin complex 1 (mTORC1) regulates translation rates through phosphorylation of its main downstream effectors eIF4E–binding proteins (4E-BPs) and p70 S6 kinases (S6K1/2) (Hay & Sonenberg 2004, Ma & Blenis 2009). The defining component of mTORC1, Raptor, confers substrate specificity to mTORC1 and recruits its downstream targets. Specifically, the 4E-BPs and S6Ks interact with Raptor through a short amino acid sequence called the TOS (mTOR signaling) motif. Rapamycin binds to FKBP-12, and the FKBP-12-rapamycin complex directly binds to and inhibits mTORC1 (Laplante & Sabatini 2012, Wullschleger et al. 2006).
The best-characterized process by which mTORC1 controls translation is by regulating eIF4F complex formation through phosphorylation of 4E-BPs (Figure 2b). 4E-BPs (4E-BP1, 4E-BP2, 4E-BP3) are small molecular weight proteins that compete with eIF4G for a common binding site on eIF4E. 4E-BP1, the best characterized 4E-BP, undergoes a hierarchical mTORC1-mediated phosphorylation first at Thr37 and Thr46, which serve as priming sites for the subsequent phosphorylation of Ser65 and Thr70. Phosphorylation of 4E-BP1 at these four sites prevents their binding to eIF4E, allowing it to assemble into the eIF4F complex and thereby stimulating translation rates. 4E-BP2 is the most abundant 4E-BP in the mammalian brain (Bidinosti et al. 2010, Tsukiyama-Kohara et al. 2001). Although compared with 4E-BP2, 4E-BP1 is expressed at lower levels in the brain, it plays an important role in the entrainment and synchrony of the master circadian clock (Cao et al. 2013).
An additional mechanism by which mTORC1 could regulate translation is through phosphorylation of S6Ks (two isoforms exist in vertebrates: S6K1 and S6K2). S6Ks regulate translation initiation, translation elongation, and ribosome biogenesis by phosphorylating eIF4B (a cofactor of eIF4A), eukaryotic elongation factor 2 kinase (eEF2K), and ribosomal protein S6, respectively (reviewed in Ma & Blenis 2009).
mTORC1 activity, as determined by the phosphorylation of its downstream targets S6Ks and 4E-BPs, is stimulated by both L-LTP and LTD-inducing stimuli (Cammalleri et al. 2003, Hou & Klann 2004, Tsokas et al. 2007). Inhibition of mTORC1 by rapamycin blocks L-LTP in rodent hippocampal slices (Cammalleri et al. 2003, Tang et al. 2002), as well as synaptic facilitation in Aplysia neuronal cultures (Casadio et al. 1999), highlighting the conserved role of mTORC1 in protein synthesis–dependent long-lasting synaptic potentiation. The contribution of mTORC1 to LTD remains controversial: Although experiments in hippocampal slices originally suggested that rapamycin-mediated blockage of mTORC1 impairs mGluR-LTD (Hou & Klann 2004), recent findings have challenged this view (Bhakar et al. 2012). Removal of upstream negative regulators of mTORC1, however, also blocks mGluR-LTD (Auerbach et al. 2011, Bateup et al. 2011).
Behavioral studies using rapamycin support the idea that mTORC1 is also required for LTM formation in mammals. Specifically, LTM, but not STM, is blocked by rapamycin treatment (Bekinschtein et al. 2007, Blundell et al. 2008). In a recent pharmacogenetic study, a low dose of rapamycin, subthreshold in wild-type mice, was effective in mTOR heterozygous mice. These findings demonstrate that direct inhibition of mTORC1 blocks L-LTP and LTM formation and rule out an off-target effect of rapamycin (Stoica et al. 2011).
mTORC1 is also crucial for the reconsolidation of memories associated with electric footshocks or addictive substances (Barak et al. 2013, Blundell et al. 2008, Stoica et al. 2011, Wang et al. 2010b), a process that depends on new protein synthesis (Barak et al. 2013, Milekic & Alberini 2002, Nader et al. 2000). Thus, mTORC1 inhibition or blockade of the translational program directed by mTORC1 holds particular therapeutic potential for conditions plagued by pathological memories, such as post-traumatic stress disorder (PTSD) and drug addiction.
By which mechanism (or mechanisms) does mTORC1 control L-LTP and LTM formation? Studies using mice lacking downstream targets of mTORC1 have begun to answer this question. In mice lacking 4E-BP2, which show enhanced eIF4F complex formation, an E-LTP-inducing protocol elicits L-LTP, but both L-LTP induced by 4 × 100 Hz and LTM formation are impaired (Banko et al. 2005). These mice exhibit various behavioral abnormalities (Banko et al. 2007), alterations in excitation/inhibition balance, and phenotypes associated with autism spectrum disorders (ASDs) (Gkogkas et al. 2013). Because 4E-BP2 also regulates structural plasticity during development (Ran et al. 2013) and undergoes a different posttranslational modification during adulthood (Bidinosti et al. 2010), an approach to decipher 4E-BP2’s function more clearly in the context of L-LTP and LTM may be conditional deletion of 4E-BP2 in the adult brain.
S6Ks are likely the main downstream effectors of mTORC1 activity necessary for long-term facilitation (LTF) in Aplysia. Expression of dominant-negative S6K blocks LTF in Aplysia sensory neurons, whereas expression of dominant-negative 4E-BP does not (Weatherill et al. 2010). If S6Ks are likewise the major effectors of mTORC1 in the mammalian brain, one would expect that in mice lacking S6Ks, L-LTP and LTM (but not E-LTP and STM) should be blocked. However, mice lacking S6K1 or S6K2 display relatively mild memory deficits, and L-LTP is surprisingly normal (Antion et al. 2008). Although compensation by the remaining S6K could be taking place in the single knockout mice, S6K1/2 double knockout mice have yet to be characterized. Alternatively, studies of conditional S6K1/2 double knockout mice or mutant mice in which all five phosphorylatable serine residues in ribosomal protein S6 are replaced by alanine (Ruvinsky et al. 2005) will determine whether the S6Ks and its major target S6 are major players in mTORC1-mediated plasticity and memory processes in the mammalian brain. That said, recent advances using transcriptome-scale ribosome profiling convincingly show that in nonneuronal cells 4E-BPs are the main effectors by which mTORC1 regulates translation (Hsieh et al. 2012, Thoreen et al. 2012). Specifically, 4E-BPs control translation of mRNAs containing 5′ terminal oligopyrimidine (TOP)- and TOP-like motifs. However, in adult neurons, unlike developing neurons, the major posttranslational modification of 4E-BP2 seems to be deamidation (the conversion of asparagine to aspartic acid), not phosphorylation (Bidinosti et al. 2010). Moreover, in the brain 4E-BPs seem to control translation of GluA1, GluA2, neuroligins, and Vip mRNAs (Cao et al. 2013, Gkogkas et al. 2013, Ran et al. 2013).
These findings raise several interesting conceptual issues. First, is 4E-BP1 or 4E-BP2 the major effector of mTORC1 in the adult brain? Second, do GluA1, GluA2, neuroligins, and Vip mRNAs contain TOP-like motifs? If not, how are they regulated by 4E-BPs? Third, how is 4E-BP2 regulated during L-LTP and LTM formation? Fourth, does mTORC1 control 4E-BP2 deamidation? Finally, given that disruption of eIF4F complex formation inhibits L-LTP and LTM (Hoeffer et al. 2011, 2013), one wonders how mTORC1 regulates 4E-BP2-mediated eIF4F complex formation and translation in the adult brain.
Poly(A)-mediated translational control
The cytoplasmic polyadenylation element-binding protein (CPEB) regulates translation of specific mRNAs containing cytoplasmic polyadenylation elements (CPE) in their 3′ UTR (Richter & Klann 2009). Vertebrates have four CPEB paralogs: CPEB1–CPEB4.
CPEB1 localizes at dendrites of mammalian neurons where it regulates translation together with its associated partners: poly(A) polymerase Gld2, deadenylase PARN, and the translation inhibitory factor neuroguidin (Ngd) (Richter & Klann 2009). These proteins comprise a dendritic polyadenylation apparatus (Udagawa et al. 2012) that is subject to regulation by neuronal activity (Figure 2c). For instance, NMDAR activation stimulates the phosphorylation of CPEB, which is thought to remove PARN from the RNA complex. Consequently, the poly(A) tail is elongated by Gld2, thus allowing binding of the poly(A)-binding protein (PABP). PABP promotes the recruitment of the eIF4F complex to the 5′ end of the mRNA and promotes translation of CamKIIα and NR2A mRNAs (Huang et al. 2002, Udagawa et al. 2012, Wu et al. 1998). CPEB-1 knockout mice are surprisingly defective in protein synthesis–independent E-LTP and display reduced extinction of hippocampus-dependent spatial and fear memory (Alarcon et al. 2004, Berger-Sweeney et al. 2006). Modulation of CPEB partners also alters LTP: Knockdown of Gld2 impairs LTP at dentate gyrus synapses in vivo; in contrast, this LTP is facilitated in vivo by depletion of the inhibitor Ngd (Udagawa et al. 2012). CPEB2–4, which unlike CPEB1 do not bind to CPE and regulate polyadenylation, bind to another consensus RNA sequence (Huang et al. 2006). These proteins are also expressed in neurons, but their role in mammals remains to be determined.
Significant bodies of work demonstrate that the Drosophila and Aplysia CPEB homologs, Orb2 and ApCPEB, respectively, also regulate long-term synaptic plasticity and LTM. Orb2 and ApCPEB are thought to act as prion-like proteins that form multimers essential for LTM, but not STM (Kruttner et al. 2012, Majumdar et al. 2012), and the maintenance phase, but not the initiation phase, of LTF (Miniaci et al. 2008; Si et al. 2003a, b, 2010), respectively.
Poly(A)-mediated translational control in neurons can also be regulated by controlling PABP function. The inhibitory PABP-interacting protein 2 (Paip2, of which there are two isoforms Paip2a and Paip2b) controls translation by a dual mechanism: (a) decreasing the affinity of PABP for the polyadenylated RNA (Khaleghpour et al. 2001) and (b) competing with eIF4G for PABP binding (Karim et al. 2006). Although investigators have not observed functional or mechanistic differences between Paip2A and Paip2B in vitro or in vivo, evidence shows that their tissue distribution in mice differs at both the mRNA and protein levels (Berlanga et al. 2006). In neurons, Paip2a is rapidly degraded by calpains following neuronal stimulation or contextual fear conditioning, and mice lacking Paip2 show a facilitated LTP and LTM after weak behavioral training (Khoutorsky et al. 2013). Paip2a seems to control translation of CamKIIα mRNA, an important regulator of LTP and LTM (Silva et al. 1992a, b). Because mRNAs are polyadenylated to different extents in response to synaptic activity (Huang et al. 2002), it will be important to identify the mRNAs regulated by PABP and to determine whether the length of the poly(A) tail makes the mRNA more susceptible or resistant to the PABP–Paip2 ratio.
Translational control during elongation
Translation elongation has also been implicated in synaptic plasticity. One of the best-characterized pathways in translational control at the elongation level is that of eEF2K-eEF2 (Dever & Green 2012). eEF2 is phosphorylated by its kinase eEF2K, a Ca2+-activated protein, in response to neuronal activity. Specifically, activation of α-amino-3-hydroxy-5-methyl-4-isoxazoleproprionic acid receptors (AMPAR), NMDAR, and mGluR1/5 all increase eEF2 phosphorylation (Taha et al. 2013).
Phosphorylation of eEF2 is thought to block general translation by impairing its binding to the ribosome. In cultured hippocampal neurons, eEF2 phosphorylation is maintained at relatively low levels by intrinsic network activity, whereas miniature synaptic excitation by spontaneous glutamate release promotes eEF2 phosphorylation locally at dendrites (Sutton et al. 2007). This observation, together with emerging evidence for compartmentalized regulation of the eEF2K-eEF2 pathway in neurons (Weatherill et al. 2011), points to an intriguing model for synapse-specific modification of translation. In addition, p-eEF2 promotes translation of certain mRNAs by yet unknown mechanisms. In the case of mGluR-LTD, translation of Map1b (microtubule associated protein 1B) and Arc (activity-regulated cytoskeleton-associated protein) mRNAs, two molecules essential to this process, is selectively upregulated when eEF2 is phosphorylated. RNAi-knockdown of eEF2K in primary neurons (Davidkova & Carroll 2007) and genetic knockout of eEF2K (Park et al. 2008) block the dihydroxyphenylglycine (DHPG)-induced increases in MAP1B and Arc synthesis, respectively, both resulting in an impaired LTD. However, the mechanism by which a particular mRNA can be specifically upregulated by elongating its amino acid chain remains to be determined.
Studies have also documented eEF2’s involvement in LTM formation. Phosphorylation of eEF2 in gustatory cortex is elevated within 30 min of novel taste learning during the conditioned taste aversion task (Belelovsky et al. 2005). In eEF2K knockin mice, where eEF2 phosphorylation levels are reduced, associative but not incidental taste learning is impaired (Gildish et al. 2012).
Local translational control at the synapse
Almost a decade after the discovery of LTP (Bliss & Lømo 1973), the identification of synapse-associated polyribosome complexes (Steward & Levy 1982) led to the revolutionary theory that synapses could act as quasi-independent plastic units through local protein synthesis. Colocalization of specific mRNAs and translational machinery at postsynaptic sites suggests that local mRNA translation could play an important role in synaptic processes (Steward 1997, Steward & Schuman 2001, Sutton & Schuman 2006). In a landmark study, Kang & Schuman (1996) observed protein synthesis–dependent LTP in CA1 dendrites that had been surgically severed from their cell bodies. These data provided the first evidence for a functional role of local protein synthesis in synaptic plasticity. Subsequent studies by the Schuman lab and others have not only visualized local translation at dendrites (Sutton & Schuman 2006), but also shown that activity-dependent mRNA translation in dendrites also plays a critical role in other forms of long-lasting synaptic plasticity, such as mGluR-LTD (Huber et al. 2000) and homeostatic plasticity (Sutton et al. 2004, 2006). In the latter, mTORC1 promotes local translation of BDNF in response to AMPAR blockade. BDNF is then released as a retrograde signal to drive the homeostatic upregulation of presynaptic function (Jakawich et al. 2010). This mechanism is conserved between mice and flies (Henry et al. 2012, Penney et al. 2012).
Protein synthesis in dendrites involves trafficking of translationally repressed mRNAs in RNA granules (see Martin & Ephrussi 2009, Swanger & Bassell 2011). The selective derepression of mRNA translation at activated synapses is thought to be fundamental to long-lasting synaptic plasticity (Wang et al. 2010a). Proteins synthesized locally in dendrites could be captured by synapses that have been “tagged” by previous activity, in what is defined as the synaptic tagging hypothesis (Frey & Morris 1997, Martin et al. 1997, Redondo & Morris 2011). Although synaptic tagging occurs at single spines, as recently shown by two-photon imaging in hippocampal slice preparations (Govindarajan et al. 2011), and in behaving animals (Shires et al. 2012), the identities of the locally synthesized proteins and the “tag(s)” remain largely unknown. That said, a limited number of proteins and several dendritically localized candidate mRNAs have been identified. Studies employing in situ hybridization (Eberwine et al. 2002, Lein et al. 2007, Poon et al. 2006) and, more recently, deep RNA sequencing have revealed thousands of dendritically localized mRNAs, including many that encode key regulators of synaptic plasticity (Cajigas et al. 2012).
Dendritic mRNAs translated in response to synaptic stimulation include CaMKIIα, the postsynaptic scaffolding molecule PSD-95, and the GluR1 subunit of AMPA receptors. The importance of local translation of these particular mRNAs is underscored by their collective dysregulation in fragile X syndrome (FXS) (Muddashetty et al. 2007). In addition, mRNAs encoding components of the translational machinery, such as elongation factor 1 alpha, are also dendritically localized and translated in response to stimuli that induce long-term changes in synaptic strength (Huang et al. 2005, Tsokas et al. 2005). Given the number of synaptic mRNAs and local translational machinery, and the spatial limitations of dendritic spines, it will be interesting to determine the number of mRNAs that can be cotranslated at spines and whether this varies between synapses.
Application of novel technologies including high-resolution ribosome profiling (Ingolia et al. 2009) and translating ribosome affinity purification (TRAP) (Doyle et al. 2008) in combination with next-generation sequencing hold great promise for the identification of mRNAs whose translation is specifically upregulated in response to synaptic activation. In fact, simple comparative analysis of data reported in two recent studies employing these technologies reveals that 93 of the 100 mRNAs most sensitive to translational repression by the ATP-competitive inhibitor of mTORC1 Torin 1 (Thoreen et al. 2012) were identified among the neuropil transcriptome (Cajigas et al. 2012). Thus, translational regulation of dendritically localized TOP and TOP-like mRNAs by the mTOR pathway may play an underappreciated role in compartmentalized new protein synthesis and synaptic plasticity.
DYSREGULATION OF TRANSLATIONAL CONTROL IN DISEASE
Dysregulation of the signaling pathways that engage protein synthesis have been linked to the pathophysiology of several neurodegenerative diseases and neurodevelopmental disorders (Bagni et al. 2012, Darnell & Klann 2013, Swanger & Bassell 2011). Indeed, disease progression in Alzheimer’s disease (AD) and other neurodegenerative disorders has recently been linked to dysregulation of eIF2α-mediated translational control, whereas several components of the mTORC1 signaling pathway are implicated in the etiology of syndromic ASDs. Finally, loss-of-function mutations in the RNA binding protein and putative translational repressor FMRP cause FXS.
Translational Dysregulation in Neurodegenerative Diseases
Suppression of polysomal mRNA translation in the brains of AD patients was first documented more than 20 years ago (Langstrom et al. 1989). A prevailing hypothesis postulates that the deficits in translational control in AD could be due to aberrant eIF2α phosphorylation. Consistent with this hypothesis, increased phosphorylation of eIF2α has been observed in postmortem analysis of AD patients’ brains (Chang et al. 2002a, b; Page et al. 2006) and also in numerous experimental preparations, including cultured neurons treated with synthetic Aβ peptides (Chang et al. 2002a) and brain sections of AD mouse models (Ma et al. 2013, O’Connor et al. 2008, Page et al. 2006). The eIF2α kinases PKR and PERK are activated under similar conditions (Chang et al. 2002a, b; O’Connor et al. 2008; Onuki et al. 2004; Page et al. 2006). As general translation is depressed, translation of ATF4 and β-secretase (BACE1) mRNAs, both of which contain uORFs in the 5′ UTR, appear to be elevated by p-eIF2α in AD brains (O’Connor et al. 2008). Because BACE1 promotes amyloidogenesis by initiating cleavage of amyloid precursor protein (APP) to form Aβ, investigators believe that dysregulated translation in AD, via elevated p-eIF2α, establishes a feedforward loop that progressively diminishes the overall structure and function of the affected neuronal population.
Significant efforts have been directed toward restoring physiological translational control in AD in hopes of hindering disease progression. In a recent study, deletion of PERK or GCN2 in APP/PS1 mice (APPswe, PSEN1dE9) not only corrected p-eIF2α levels and the depression in global translation but also rescued the deficits in synaptic plasticity and spatial memory displayed at 10–12 months (Ma et al. 2013). In contrast, however, genetic deletion of GCN2 in the 5XFAD (five familiar AD mutations) mouse model, in which AD progresses faster than in APP/PS1 mice, did not rescue but instead aggravated the memory deficits (Devi & Ohno 2013). Thus, whether blockade of eIF2α kinases could be an effective treatment for AD remains unclear (however, see discussion below).
PKR-eIF2α signaling is also implicated in AD (Morel et al. 2009). PKR activity is upregulated in AD patients’ brains and in mouse models of AD (Chang et al. 2002b). In primary neurons, both genetic deletion and pharmacological inhibition of PKR prevented inflammation and apoptosis induced by Aβ peptides (Chang et al. 2002a; Couturier et al. 2010, 2011). Given that genetic or pharmacological inhibition of PKR enhances learning and memory in mice (Zhu et al. 2011), PKR is a promising target for AD treatment. In this regard, a recent study found that exposure of neuronal cultures to β-amyloid oligomers (AβOs) triggered PKR, but not PERK (Laurenco et al. 2013), a process that is attenuated by PKRi. Furthermore, intracerebroventricular infusion of AβOs increased eIF2α phosphorylation and resulted in cognitive impairment only in wild-type mice but not in mice lacking PKR. Hence, in future studies, it would be interesting to determine if direct inhibition of eIF2α, by other means, could also restore the memory deficits in AD mouse models.
mTORC1 Translational Control in Autism Spectrum Disorders
ASDs are a group of neurodevelopmental disorders that share common symptoms, including impaired social interactions, abnormal repetitive behavior, and often intellectual disability (Fombonne 1999, Mefford et al. 2012). Although they are genetically heterogeneous, ASDs are heritable, and some forms are linked to single gene mutations (Zoghbi & Bear 2012). In this regard, human mutations in negative regulators of mTORC1, such as PTEN or tuberous sclerosis complex (TSC) proteins (TSC1/2), are associated with ASD (O’Roak et al. 2012, Sahin 2012). Moreover, mice with mutations in Pten or Tsc1/2 causing hyperactivation of mTORC1, exhibit behaviors analogous to those of human ASD patients (see Costa-Mattioli & Monteggia 2013). Remarkably, the ASD-like phenotypes in mouse models with elevated mTORC1 signaling can be arrested or even reversed by applying rapamycin and/or its synthetic derivatives (termed rapalogs). Thus, considerable recent attention has been directed toward establishing a mechanistic understanding of how the mTORC1 signaling pathway regulates synaptic plasticity and the development of novel, mechanism-based therapies to treat mTORC1-related ASDs.
mTOR consists of two distinct complexes, mTORC1 and mTORC2, each of which critically contributes to synaptic plasticity (Stoica et al. 2011, Huang et al. 2013). Although mTORC2 activity could control some of the ASD-like behaviors in models of mTORC1 hyperactivity, here we focus on what is known about dysregulation of the mTORC1 signaling pathway in relation to disorders of synaptic dysfunction.
mTORC1 integrates a diverse set of extracellular and intracellular inputs to manage cell growth and metabolism, among other essential functions in most mammalian cell types (Laplante & Sabatini 2012). Neurons further exploit the mTORC1 pathway for the integration of activity-dependent signaling. Extracellular neurotrophin or glutamate binding to transmembrane receptors activates the PI3K-Akt-mTORC1 signaling pathway, which is outlined in Figure 2b.
Heterozygous loss-of-function mutations in either the TSC1 or TSC2 genes cause TSC, a disease that presents with neurological deficits including ASD, epilepsy, and intellectual disability in ~20–60% of patients (Fombonne 1999, Mefford et al. 2012). Like TSC patients, mice heterozygous for either Tsc1 or Tsc2 show deficits in several cognitive tasks including spatial learning in the Morris water maze, context discrimination, and fear-conditioning paradigms (Ehninger et al. 2008, Goorden et al. 2007). Tsc1+/− mice are also abnormal in their social behavior (Goorden et al. 2007). In addition, spine density and AMPA/NMDA ratios are increased in Tsc1+/− mutants while stimuli that typically induce E-LTP produce L-LTP in Tsc2+/− mutants. mGluR-LTD is also impaired in Tsc1- and Tsc2-deficient neurons (Auerbach et al. 2011, Bateup et al. 2011). At the network level, genetic deletion of Tsc1 results in chronic hyperactivity owing to a loss of inhibitory synaptic transmission (Bateup et al. 2013). These behavioral and synaptic phenotypes are thought to be caused by the hyperactivation of mTORC1 because they are restored by rapamycin.
Inherited mutations in PTEN, which also increase mTORC1 activity, commonly result in multiple neurological phenotypes including ASD, intellectual disability, and macrocephaly in humans (Endersby & Baker 2008). Similarly, conditional knockout of Pten in the mouse brain causes impairments in social interaction and learning as well as epilepsy and exaggerated neuronal arborization leading to macrocephaly (Kwon et al. 2006). As in the case of Tsc2+/− mice (Bateup et al. 2013), bidirectional plasticity (LTP and LTD) is altered in Pten-deficient mice. Analysis of Nse-Cre Pten conditional knockout mice revealed that the alterations in synaptic plasticity appear prior to the onset of morphological defects and behavioral abnormalities (Takeuchi et al. 2013). Thus, synaptic dysfunction associated with mTORC1 upregulation could be causally related to both the morphological and the cognitive deficits observed in human patients with loss-of-function mutations in PTEN.
Consistent with a causal role for mTORC1 upregulation in the etiology of ASD-like phenotypes in TSC and/or PTEN-ASD mouse models, rapamycin or rapalog treatment reverses many of the synaptic and behavioral phenotypes in these models (Costa-Mattioli & Monteggia 2013, Ehninger & Silva 2011). The exact mechanisms by which rapamycin ameliorates cognitive dysfunction phenotypes remain unclear as mTORC1 regulates not only translation rates, but also lipid synthesis, autophagy, and mitochondrial function.
The most likely mechanism by which excessive mTORC1 signaling could cause ASD phenotypes is by upregulating cap-dependent translation because hyperactivation of mTORC1 promotes eIF4F complex formation (Gingras et al. 2004). Like mutant mice with hyperactive mTORC1 signaling, mice with elevated eIF4F complex in the brain (4E-BP2 knockout mice or transgenic mice overexpressing eIF4E) also show ASD-like phenotypes, including deficits in social behaviors and stereotypic repetitive behaviors (Gkogkas et al. 2013, Santini et al. 2013). Moreover, disruption of eIF4F complex formation with the protein synthesis inhibitor 4EGI-1 fully rescues the neurophysiological and autistic-like behavioral deficits in both the eIF4E-transgenic mice and the 4E-BP2 knockout mice.
Given that eIF4F complex formation is presumably elevated in TSC and PTEN-ASD mouse models, one would predict that 4EGI-1 treatment could rescue the plasticity and behavioral phenotypes in these models. Conversely, mice overexpressing eIF4E, but not 4E-BP2 knockout mice, should be sensitive to rapamycin-mediated mTORC1 inhibition. These experiments would significantly bolster the notion that the primary mechanism by which mTORC1 regulates synaptic plasticity is through eIF4F-mediated translational control. In addition, whether elevated eIF4F complex formation in the ASD brain leads to an increase in global or specific protein synthesis remains unresolved. In mice overexpressing eIF4E general translation is increased (Santini et al. 2013), whereas in 4E-BP2 knockout mice the translation of specific mRNAs encoding the ASD-associated synaptic adhesion proteins neuroligins 1–4 (Nlgn1–4) is upregulated (Gkogkas et al. 2013). As discussed above, whether, and if so how, the mRNA features that dictate mTORC1-mediated regulation of the synthesis of these synaptic proteins differ from those in nonneuronal cells are important questions that have yet to be thoroughly addressed.
Translational Control in Fragile X Syndrome
Dysregulation of translation is also implicated in the pathophysiology of FXS, the most common inherited cause of intellectual disability and ASD (for review see Bassell & Warren 2008, Bhakar et al. 2012, Darnell & Klann 2013, Nelson et al. 2013). FXS is caused by transcriptional silencing of the Fmr1 gene due to CGG triplet repeat expansion in the 5′ UTR of the mRNA (Bassell & Warren 2008, Nelson et al. 2013). Fmr1 encodes FMRP, an mRNA binding protein that represses translation (Laggerbauer et al. 2001, Li et al. 2001). Accordingly, deletion of Fmr1 leads to a region-specific increase in general translation (Qin et al. 2005). In addition, protein synthesis inhibitors rescue the LTM phenotype in Drosophila FXS mutants (Bolduc et al. 2008). The use of an in vivo UV-crosslinking procedure (CLIP) combined with high throughput sequencing (CLIP-seq) of polysome-associated FMRP showed that FMRP binds to a specific subset of mRNAs. The targets of FMRP include several mRNAs that encode proteins with significant roles in synaptic function, such as NMDAR and mGluR subunits, but also include ASD-linked mRNAs such as Pten, Tsc2, and other members of the mTOR signaling pathway (Darnell et al. 2011). In this regard, mTORC1 activity seems to be upregulated in subjects (Hoeffer et al. 2012) and mouse models of FXS (Sharma et al. 2010). Some of the phenotypes observed in Fmr1 knockout mice are restored by rapalog treatment (Busquets-Garcia et al. 2013). In addition, genetic removal of S6K1 also corrects some of the phenotypes in FXS mice (Bhattacharya et al. 2012). How S6K1 regulates translation of FMRP-target mRNAs is unclear. Given that, in one model, FMRP represses translation once associated with polysomes (Khandjian et al. 1996, Stefani et al. 2004), an interesting possibility is that S6K-mediated phosphorylation of eEF2 derepresses translation elongation.
Because eIF4F complex formation seems to be increased in the FXS mouse model (Sharma et al. 2010), FMRP could also repress translation at the initiation level. In this model, FMRP binds to its partner CYFIP1, which acts as a 4E-BP, binding eIF4E through a noncanonical eIF4E motif, thus repressing cap-dependent translation initiation (Napoli et al. 2008).
Several important questions remain regarding the role of translational control in FXS pathophysiology. First, does FMRP inhibit polypeptide elongation or translation initiation? If so, is this a dynamic and reversible process? Second, does disruption of eIF4F complex formation rescue some of the FXS phenotypes? Indeed, would this be specific to eIF4F, or could a more general translation inhibitor, like in FXS Drosophila experiments (Bolduc et al. 2008), also correct the FXS phenotypes in mice and/or humans? In this regard, crossing Tsc2 mice—which are expected to have increased eIF4F but show reduced overall translation rates and several behavioral deficits similar to the Fmr1 knockout mice—with Fmr1 knockout mice rescued the mutant phenotypes (Auerbach et al. 2011). Third, if FMRP binds to specific mRNAs, why is general translation increased in Fmr1 knockout hippocampus? Furthermore, given that modulation of actin polymerization through PAK also corrects the behavioral deficits in the FXS mouse (Dolan et al. 2013, Hayashi et al. 2004), it will be interesting to determine the link between translation and cytoskeletal dynamics in FXS. Recent data demonstrating that, through interaction with Rac1, CYFIP1 also contributes to cytoskeletal remodeling at dendritic spines represent a promising first step in this direction (De Rubeis et al. 2013).
FUTURE PERSPECTIVE
Regulation of protein synthesis is crucial for long-lasting changes in synaptic efficacy and long-term memory storage. Over the past two decades, significant progress has been made in understanding how translation is modulated by synaptic activity. However, the identities of the specific mRNAs translated during L-LTP or LTD remain unknown. One also wonders whether these plasticity-related mRNAs can be exclusively translated at dendrites but not in the soma. If so, what would be the mechanism underlying this process? Furthermore, how many mRNAs can be locally translated in a given spine?
Dysregulated protein synthesis control has been associated with several cognitive disorders, including ASD and Alzheimer’s disease. Recent studies targeting different signaling pathways that modulate translation, such as S6K, Tsc, and CPEB, have led to the amelioration of some of the symptoms associated with FXS (Auerbach et al. 2011, Bhattacharya et al. 2012, Udagawa et al. 2013). Although one could therefore envision the dream of personalized medicine for some of these disorders, we should also understand our limitations. For instance, S6K, Tsc, and CPEB each control a different translational program, yet genetic deletion of any one of these in the Fmr1 knockout rescues some of the FXS symptoms. Thus, in addition to moving toward a much-needed application of these findings in translational medicine, we should also pursue a more thorough understanding of the basic principles of translational control in the nervous system. For instance, how is eIF4F complex formation regulated in the brain? Is neuroguidin, CYFIP, or 4E-BP the primary regulator of cap-dependent translation initiation in neurons? Do CPEB1–4 assume an activity-dependent prion-like conformation in the mammalian brain? If so, how does the aggregated isoform regulate translation rates? Is translation initiation or elongation the principal translational control mechanism in neurons? Are plasticity-related mRNAs degraded or recycled following translation at dendrites? If so, does the mRNA circularize in neurons? Could internal ribosome entry site (IRES)-dependent translation be an alternative mechanism for translational control of some forms of synaptic plasticity (Dyer et al. 2003)? Finally, how does translational control at the pre- and postsynaptic domains, respectively, contribute to the protein synthesis dependence of long-term synaptic plasticity? The next few years are likely to see significant progress toward many of these goals, as well as the development of new treatments for cognitive disorders that target specific translational control mechanisms.
Acknowledgments
The authors thank Drs. K. Krnjević and D. Nelson for insightful comments and discussion and apologize to those whose work was not discussed owing to space limitations. This work was supported by the Whitehall Foundation, the Searle Scholars Program, the U.S. Department of Defense, the National Institute of Neurological Disorders and Stroke, and the National Institute of Mental Health.
Glossary
- STM
short-term memory
- LTM
long-term memory
- LTP
long-term potentiation
- LTD
long-term depression
- mGluR-LTD
a protein synthesis–dependent form of LTD induced by activation of group 1 mGluRs
- NMDAR-LTD
a protein synthesis–independent form of LTD induced by NMDAR activation
- Ternary complex
composed of the initiator methionyl-tRNA (Met-tRNAiMet), eIF2, and GTP
- Upstream open reading frame (uORF)
a short open reading frame found in the 5′ UTR that serves as a regulatory unit for expression of its downstream gene
- Rapamycin
an immunosuppressant and antiproliferative macrolide
- Reconsolidation
stabilization of an existing memory following a retrieval period during which it becomes labile and subject to modifications
- Ribosome profiling
a technique using deep sequencing of ribosome-protected mRNA fragments to quantitatively assess genome-wide translational activity with subcodon resolution
- Homeostatic plasticity
plasticity initiated at the single cell or network level as a compensatory adaptive response to moderate network excitability
- Translating ribosome affinity purification (TRAP)
a biochemical method to isolate polysomal mRNAs from specific cell populations
- Rapalog
a synthetic analogue of rapamycin
- mTOR
mechanistic target of rapamycin
Footnotes
NOTE ADDED IN PROOF
After this manuscript was accepted for publication, Viana Di Prisco et al. (2014) reported a new mechanism of translational control of hippocampal mGluR-dependent LTD. The authors provide data highly relevant to the content of this review by identifying neuronal mRNAs that are translationally upregulated by mGluR activation as well as the translational program underlying mGluR-LTD.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Shelly A. Buffington, Email: shelly.buffington@bcm.edu.
Wei Huang, Email: wei.huang@bcm.edu.
Mauro Costa-Mattioli, Email: costamat@bcm.edu.
LITERATURE CITED
- Alarcon JM, Hodgman R, Theis M, Huang Y-S, Kandel ER, Richter JD. Selective modulation of some forms of Schaffer Collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem. 2004;11:318–27. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antion MD, Hou L, Wong H, Hoeffer CA, Klann E. mGluR-dependent long-term depression is associated with increased phosphorylation of S6 and synthesis of elongation factor 1A but remains expressed in S6K-deficient mice. Mol Cell Biol. 2008;28:2996–3007. doi: 10.1128/MCB.00201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Tassone F, Neri G, Hagerman R. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J Clin Investig. 2012;122:4314–22. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko JL, Merhav M, Stern E, Sonenberg N, Rosenblum K, Klann E. Behavioral alterations in mice lacking the translation repressor 4E-BP2. Neurobiol Learn Mem. 2007;87:248–56. doi: 10.1016/j.nlm.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Banko JL, Poulin F, Hou L, DeMaria CT, Sonenberg N, Klann E. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci. 2005;25:9581–90. doi: 10.1523/JNEUROSCI.2423-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak S, Liu F, Hamida SB, Yowell QV, Neasta J, et al. Disruption of alcohol-related memories by mTORC1 inhibition prevents relapse. Nat Neurosci. 2013;16:1111–17. doi: 10.1038/nn.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–14. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Johnson CA, Denefrio CL, Saulnier JL, Kornacker K, Sabatini BL. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78:510–22. doi: 10.1016/j.neuron.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hip-pocampal mGluR-LTD and increases excitatory synaptic function. J Neurosci. 2011;31:8862–69. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH. Persistence of long-term memory storage requires a late protein synthesis- and BDNF- dependent phase in the hippocampus. Neuron. 2007;53:261–77. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Belelovsky K, Elkobi A, Kaphzan H, Nairn AC, Rosenblum K. A molecular switch for translational control in taste memory consolidation. Eur J Neurosci. 2005;22:2560–68. doi: 10.1111/j.1460-9568.2005.04428.x. [DOI] [PubMed] [Google Scholar]
- Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn Mem. 2006;13:4–7. doi: 10.1101/lm.73706. [DOI] [PubMed] [Google Scholar]
- Berlanga JJ, Ventoso I, Harding HP, Deng J, Ron D, et al. Antiviral effect of the mammalian translation initiation factor 2 α kinase GCN2 against RNA viruses. EMBO J. 2006;25:1730–40. doi: 10.1038/sj.emboj.7601073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakar AL, Dölen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–43. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Kaphzan H, Alvarez-Dieppa AC, Murphy JP, Pierre P, Klann E. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76:325–37. doi: 10.1016/j.neuron.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidinosti M, Ran I, Sanchez-Carbente MR, Martineau Y, Gingras AC, et al. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol Cell. 2010;37:797–808. doi: 10.1016/j.molcel.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Lømo T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J Physiol. 1973;232:381–89. doi: 10.1113/jphysiol.1973.sp010273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Kouser M, Powell CM. Systemic inhibition of mammalian target of rapamycin inhibits fear memory reconsolidation. Neurobiol Learn Mem. 2008;90:28–35. doi: 10.1016/j.nlm.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila Fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11:1143–45. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busquets-Garcia A, Gomis-González M, Guegan T, Agustín-Pavón C, Pastor A, et al. Targeting the endocannabinoid system in the treatment of fragile X syndrome. Nat Med. 2013;19:603–7. doi: 10.1038/nm.3127. [DOI] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, Tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–66. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammalleri M, Lütjens R, Berton F, King AR, Simpson C, et al. Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci USA. 2003;100:14368–73. doi: 10.1073/pnas.2336098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, et al. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron. 2013;79:712–24. doi: 10.1016/j.neuron.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio A, Martin KC, Giustetto M, Zhu H, Chen M, et al. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell. 1999;99:221–37. doi: 10.1016/s0092-8674(00)81653-0. [DOI] [PubMed] [Google Scholar]
- Chang RC, Suen KC, Ma CH, Elyaman W, Ng HK, Hugon J. Involvement of double-stranded RNA-dependent protein kinase and phosphorylation of eukaryotic initiation factor-2alpha in neuronal degeneration. J Neurochem. 2002a;83:1215–25. doi: 10.1046/j.1471-4159.2002.01237.x. [DOI] [PubMed] [Google Scholar]
- Chang RC, Wong AK, Ng HK, Hugon J. Phosphorylation of eukaryotic initiation factor-2α(eIF2α) is associated with neuronal degeneration in Alzheimer’s disease. Neuroreport. 2002b;13:2429–32. doi: 10.1097/00001756-200212200-00011. [DOI] [PubMed] [Google Scholar]
- Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, et al. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron. 2003;39:655–69. doi: 10.1016/s0896-6273(03)00501-4. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Harding H, Herdy B, Azzi M, et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2α kinase, GCN2. Nature. 2005;436:1166–73. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, et al. eIF2α phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Monteggia LM. mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat Neurosci. 2013;16:1537–43. doi: 10.1038/nn.3546. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Morel M, Pontcharraud R, Gontier V, Fauconneau B, et al. Interaction of double-stranded RNA-dependent protein kinase (PKR) with the death receptor signaling pathway in amyloid β (Aβ)-treated cells and in APPSLPS1 knock-in mice. J Biol Chem. 2010;285:1272–82. doi: 10.1074/jbc.M109.041954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Paccalin M, Morel M, Terro F, Milin S, et al. Prevention of the β-amyloid peptide-induced inflammatory process by inhibition of double-stranded RNA-dependent protein kinase in primary murine mixed co-cultures. J Neuroinflammation. 2011;8:72. doi: 10.1186/1742-2094-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–36. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidkova G, Carroll RC. Characterization of the role of microtubule-associated protein 1B in metabotropic glutamate receptor-mediated endocytosis of AMPA receptors in hippocampus. J Neurosci. 2007;27:13273–78. doi: 10.1523/JNEUROSCI.3334-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, Pasciuto E, Li KW, Fernández E, Di Marino D, et al. CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron. 2013;79:1169–82. doi: 10.1016/j.neuron.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi CR, Ohno M. Deletion of the eIF2α kinase GCN2 fails to rescue the memory decline associated with Alzheimer’s disease. PLoS ONE. 2013;8:e77335. doi: 10.1371/journal.pone.0077335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan BM, Duron SG, Campbell DA, Vollrath B, Shankaranarayana Rao BS, et al. Rescue of fragile X syndrome phenotypes in Fmr1 KO mice by the small-molecule PAK inhibitor FRAX486. Proc Natl Acad Sci USA. 2013;110:5671–76. doi: 10.1073/pnas.1219383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–62. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudai Y. The restless engram: consolidations never end. Annu Rev Neurosci. 2012;35:227–47. doi: 10.1146/annurev-neuro-062111-150500. [DOI] [PubMed] [Google Scholar]
- Dyer JR, Michel S, Lee W, Castellucci VF, Wayne NL, Sossin WS. An activity-dependent switch to cap-independent translation triggered by eIF4E dephosphorylation. Nat Neurosci. 2003;6:219–20. doi: 10.1038/nn1018. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem Res. 2002;27:1065–77. doi: 10.1023/a:1020956805307. [DOI] [PubMed] [Google Scholar]
- Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, et al. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14:843–48. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Silva AJ. Rapamycin for treating tuberous sclerosis and autism spectrum disorders. Trends Mol Med. 2011;17:78–87. doi: 10.1016/j.molmed.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endersby R, Baker SJ. PTEN signaling in brain: neuropathology and tumorigenesis. Oncogene. 2008;27:5416–30. doi: 10.1038/onc.2008.239. [DOI] [PubMed] [Google Scholar]
- Fombonne E. The epidemiology of autism: a review. Psychol Med. 1999;29:769–86. doi: 10.1017/s0033291799008508. [DOI] [PubMed] [Google Scholar]
- Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–36. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- García MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Gildish I, Manor D, David O, Sharma V, Williams D, et al. Impaired associative taste learning and abnormal brain activation in kinase-defective eEF2K mice. Learn Mem. 2012;19:116–25. doi: 10.1101/lm.023937.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. mTOR signaling to translation. Curr Top Microbiol Immunol. 2004;279:169–97. doi: 10.1007/978-3-642-18930-2_11. [DOI] [PubMed] [Google Scholar]
- Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, et al. Autism-related deficits via dys-regulated eIF4E-dependent translational control. Nature. 2013;493:371–77. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62:648–55. doi: 10.1002/ana.21317. [DOI] [PubMed] [Google Scholar]
- Govindarajan A, Israely I, Huang S-Y, Tonegawa S. The dendritic branch is the preferred integrative unit for protein synthesis-dependent LTP. Neuron. 2011;69:132–46. doi: 10.1016/j.neuron.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hayashi ML, Choi SY, Rao BS, Jung HY, Lee HK, et al. Altered cortical synaptic morphology and impaired memory consolidation in forebrain-specific dominant-negative PAK transgenic mice. Neuron. 2004;42:773–87. doi: 10.1016/j.neuron.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Henry FE, McCartney AJ, Neely R, Perez AS, Carruthers CJ, et al. Retrograde changes in presynaptic function driven by dendritic mTORC1. J Neurosci. 2012;32:17128–42. doi: 10.1523/JNEUROSCI.2149-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–50. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hoeffer CA, Cowansage KK, Arnold EC, Banko JL, Moerke NJ, et al. Inhibition of the interactions between eukaryotic initiation factors 4E and 4G impairs long-term associative memory consolidation but not reconsolidation. Proc Natl Acad Sci USA. 2011;108:3383–88. doi: 10.1073/pnas.1013063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Sanchez E, Hagerman RJ, Mu Y, Nguyen DV, et al. Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes Brain Behav. 2012;11:332–41. doi: 10.1111/j.1601-183X.2012.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Santini E, Ma T, Arnold EC, Whelan AM, et al. Multiple components of eIF4F are required for protein synthesis-dependent hippocampal long-term potentiation. J Neurophysiol. 2013;109:68–76. doi: 10.1152/jn.00342.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Klann E. Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2004;24:6352–61. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Chotiner JK, Steward O. The mRNA for elongation factor 1α is localized in dendrites and translated in response to treatments that induce long-term depression. J Neurosci. 2005;25:7199–209. doi: 10.1523/JNEUROSCI.1779-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhu PJ, Zhang S, Zhou H, Stoica L, et al. mTORC2 controls actin polymerization required for consolidation of memory. Nat Neurosci. 2013;16:441–48. doi: 10.1038/nn.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–48. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–76. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–57. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–27. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- Jakawich SK, Nasser HB, Strong MJ, McCartney AJ, Perez AS, et al. Local presynaptic activity gates homeostatic changes in presynaptic function driven by dendritic BDNF synthesis. Neuron. 2010;68:1143–58. doi: 10.1016/j.neuron.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Belforte JE, Lu Y, Yabe Y, Pickel J, et al. eIF2α phosphorylation-dependent translation in CA1 pyramidal cells impairs hippocampal memory consolidation without affecting general translation. J Neurosci. 2010;30:2582–94. doi: 10.1523/JNEUROSCI.3971-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–38. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–6. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Karim MM, Svitkin YV, Kahvejian A, De Crescenzo G, Costa-Mattioli M, Sonenberg N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci USA. 2006;103:9494–99. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin YV, et al. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol. 2001;21:5200–13. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- Khoutorsky A, Yanagiya A, Gkogkas CG, Fabian MR, Prager-Khoutorsky M, et al. Control of synaptic plasticity and memory via suppression of poly(A)-binding protein. Neuron. 2013;78:298–311. doi: 10.1016/j.neuron.2013.02.025. [DOI] [PubMed] [Google Scholar]
- Krüttner S, Stepien B, Noordermeer JN, Mommaas MA, Mechtler K, et al. Drosophila CPEB Orb2A mediates memory independent of its RNA-binding domain. Neuron. 2012;76:383–95. doi: 10.1016/j.neuron.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–88. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–38. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Langstrom NS, Anderson JP, Lindroos HG, Winblad B, Wallace WC. Alzheimer’s disease-associated reduction of polysomal mRNA translation. Brain Res Mol Brain Res. 1989;5:259–69. doi: 10.1016/0169-328x(89)90060-0. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenco MV, Clarke JR, Frozza RL, Bomfim TR, Forny-Germano L, et al. TNF-α mediates PKR-dependent memory impairment and brain IRS-1 inhibition induced by Alzheimer’s β-amyloid oligomers in mice and monkeys. Cell Metab. 2013;18:831–43. doi: 10.1016/j.cmet.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–83. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher C, Huber KM. Group 1 mGluR-dependent synaptic long-term depression: mechanisms and implications for circuitry and disease. Neuron. 2010;65:445–59. doi: 10.1016/j.neuron.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, et al. Suppression of eIF2α kinases alleviates Alzheimer’s disease-related plasticity and memory deficits. Nat Neurosci. 2013;16:1299–305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–29. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, et al. Synapse-specific, long-term facilitation of Aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–38. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–30. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh JL. Memory–a century of consolidation. Science. 2000;287:248–51. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Batshaw ML, Hoffman EP. Genomics, intellectual disability, and autism. N Engl J Med. 2012;366:733–43. doi: 10.1056/NEJMra1114194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milekic MH, Alberini CM. Temporally graded requirement for protein synthesis following memory reactivation. Neuron. 2002;36:521–25. doi: 10.1016/s0896-6273(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Miniaci MC, Kim JH, Puthanveettil SV, Si K, Zhu H, et al. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59:1024–36. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel M, Couturier J, Lafay-Chebassier C, Paccalin M, Page G. PKR, the double stranded RNA-dependent protein kinase as a critical target in Alzheimer’s disease. J Cell Mol Med. 2009;13:1476–88. doi: 10.1111/j.1582-4934.2009.00849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Kelić S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–48. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader K, Schafe GE, Le Doux JE. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature. 2000;406:722–26. doi: 10.1038/35021052. [DOI] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–54. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Orr HT, Warren ST. The unstable repeats—three evolving faces of neurological disease. Neuron. 2013;77:825–43. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat Rev Neurosci. 2008;9:65–75. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- O’Connor T, Sadleir KR, Maus E, Velliquette RA, Zhao J, et al. Phosphorylation of the translation initiation factor eIF2α increases BACE1 levels and promotes amyloidogenesis. Neuron. 2008;60:988–1009. doi: 10.1016/j.neuron.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuki R, Bando Y, Suyama E, Katayama T, Kawasaki H, et al. An RNA-dependent protein kinase is involved in tunicamycin-induced apoptosis and Alzheimer’s disease. EMBO J. 2004;23:959–68. doi: 10.1038/sj.emboj.7600049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page G, Rioux Bilan A, Ingrand S, Lafay-Chebassier C, Pain S, et al. Activated double-stranded RNA-dependent protein kinase and neuronal death in models of Alzheimer’s disease. Neuroscience. 2006;139:1343–54. doi: 10.1016/j.neuroscience.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Park S, Park JM, Kim S, Kim JA, Shepherd JD, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–26. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney J, Tsurudome K, Liao EH, Elazzouzi F, Livingstone M, et al. TOR is required for the retrograde regulation of synaptic homeostasis at the Drosophila neuromuscular junction. Neuron. 2012;74:166–78. doi: 10.1016/j.neuron.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–99. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005;25:5087–95. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran I, Gkogkas CG, Vasuta C, Tartas M, Khoutorsky A, et al. Selective regulation of GluA subunit synthesis and AMPA receptor-mediated synaptic function and plasticity by the translation repressor 4E-BP2 in hippocampal pyramidal cells. J Neurosci. 2013;33:1872–86. doi: 10.1523/JNEUROSCI.3264-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo RL, Morris RGM. Making memories last: the synaptic tagging and capture hypothesis. Nat Rev Neurosci. 2011;12:17–30. doi: 10.1038/nrn2963. [DOI] [PubMed] [Google Scholar]
- Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- Ron D, Harding HP. eIF2α phosphorylation in cellular stress responses and disease. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2007. pp. 345–68. [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin M. Targeted treatment trials for tuberous sclerosis and autism: no longer a dream. Curr Opin Neurobiol. 2012;22:895–901. doi: 10.1016/j.conb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–15. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shires KL, Da Silva BM, Hawthorne JP, Morris RG, Martin SJ. Synaptic tagging and capture in the living rat. Nat Commun. 2012;3:1246. doi: 10.1038/ncomms2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Choi Y-B, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–35. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in Aplysia. Cell. 2003a;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell. 2003b;115:879–91. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992a;257:206–11. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992b;257:201–6. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–76. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern E, Chinnakkaruppan A, David O, Sonenberg N, Rosenblum K. Blocking the eIF2α kinase (PKR) enhances positive and negative forms of cortex-dependent taste memory. J Neurosci. 2013;33:2517–25. doi: 10.1523/JNEUROSCI.2322-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O. mRNA localization in neurons: a multipurpose mechanism? Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–91. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- Stoica L, Zhu PJ, Huang W, Zhou H, Kozma SC, Costa-Mattioli M. Selective pharmacogenetic inhibition of mammalian target of Rapamycin complex I (mTORC1) blocks long-term synaptic plasticity and memory storage. Proc Natl Acad Sci USA. 2011;108:3791–96. doi: 10.1073/pnas.1014715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Ito HT, Cressy P, Kempf C, Woo JC, Schuman EM. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–99. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–61. doi: 10.1016/j.neuron.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Wall NR, Aakalu GN, Schuman EM. Regulation of dendritic protein synthesis by miniature synaptic events. Science. 2004;304:1979–83. doi: 10.1126/science.1096202. [DOI] [PubMed] [Google Scholar]
- Swanger SA, Bassell GJ. Making and breaking synapses through local mRNA regulation. Curr Opin Genet Dev. 2011;21:414–21. doi: 10.1016/j.gde.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha E, Gildish I, Gal-Ben-Ari S, Rosenblum K. The role of eEF2 pathway in learning and synaptic plasticity. Neurobiol Learn Mem. 2013;105:100–6. doi: 10.1016/j.nlm.2013.04.015. [DOI] [PubMed] [Google Scholar]
- Takei N, Kawamura M, Hara K, Yonezawa K, Nawa H. Brain-derived neurotrophic factor enhances neuronal translation by activating multiple initiation processes: comparison with the effects of insulin. J Biol Chem. 2001;276:42818–25. doi: 10.1074/jbc.M103237200. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Gertner MJ, Zhou J, Parada LF, Bennett MV, Zukin RS. Dysregulation of synaptic plasticity precedes appearance of morphological defects in a Pten conditional knockout mouse model of autism. Proc Natl Acad Sci USA. 2013;110:4738–43. doi: 10.1073/pnas.1222803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA. 2002;99:467–72. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–13. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh MA, Kaphzan H, Wek RC, Pierre P, Cavener DR, Klann E. Brain-specific disruption of the eIF2α kinase PERK decreases ATF4 expression and impairs behavioral flexibility. Cell Rep. 2012;1:676–88. doi: 10.1016/j.celrep.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P, Grace EA, Chan P, Ma T, Sealfon SC, et al. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J Neurosci. 2005;25:5833–43. doi: 10.1523/JNEUROSCI.0599-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P, Ma T, Iyengar R, Landau EM, Blitzer RD. Mitogen-activated protein kinase upregulates the dendritic translation machinery in long-term potentiation by controlling the mammalian target of rapamycin pathway. J Neurosci. 2007;27:5885–94. doi: 10.1523/JNEUROSCI.4548-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med. 2001;7:1128–32. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- Udagawa T, Farney NG, Jakovcevski M, Kaphzan H, Alarcon JM, et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–77. doi: 10.1038/nm.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, et al. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell. 2012;47:253–66. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–74. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana Di, Prisco G, Huang W, Buffington SA, Hsu C-C, Bonnen P, et al. Translational control of mGluR-dependent long-term depression and object-place learning by eIF2α. Nat Neurosci. 2014 doi: 10.1038/nn.3754. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Martin KC, Zukin RS. Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 2010a;33:173–82. doi: 10.1016/j.tins.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Luo YX, He YY, Li FQ, Shi HS, et al. Nucleus accumbens core mammalian target of rapamycin signaling pathway is critical for cue-induced reinstatement of cocaine seeking in rats. J Neurosci. 2010b;30:12632–41. doi: 10.1523/JNEUROSCI.1264-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherill DB, Dyer J, Sossin WS. Ribosomal protein S6 kinase is a critical downstream effector of the target of rapamycin complex 1 for long-term facilitation in Aplysia. J Biol Chem. 2010;285:12255–67. doi: 10.1074/jbc.M109.071142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherill DB, McCamphill PK, Pethoukov E, Dunn TW, Fan X, Sossin WS. Compartment-specific, differential regulation of eukaryotic elongation factor 2 and its kinase within Aplysia sensory neurons. J Neurochem. 2011;117:841–55. doi: 10.1111/j.1471-4159.2011.07251.x. [DOI] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, et al. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–39. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Zhu PJ, Huang W, Kalikulov D, Yoo JW, Placzek AN, et al. Suppression of PKR promotes network excitability and enhanced cognition by interferon-γ-mediated disinhibition. Cell. 2011;147:1384–96. doi: 10.1016/j.cell.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4(3):a009886. doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]