Abstract
Both cellular nutrient metabolism and chromatin organization are remodeled in cancer cells, and these alterations play key roles in tumor development and growth. Many chromatin modifying-enzymes utilize metabolic intermediates as cofactors or substrates, and recent studies have demonstrated that the epigenome is sensitive to cellular metabolism. The contribution of metabolic alterations to epigenetic deregulation in cancer cells is just beginning to emerge, as are the roles of the metabolism-epigenetics link in tumorigenesis. Here we review the roles of acetyl-CoA and S-adenosylmethionine (SAM), donor substrates for acetylation and methylation reactions, respectively, in regulating chromatin modifications in response to nutrient metabolism. We further discuss how oncogenic signaling, cell metabolism, and histone modifications are interconnected and how their relationship might impact tumor growth.
Graphical Abstract
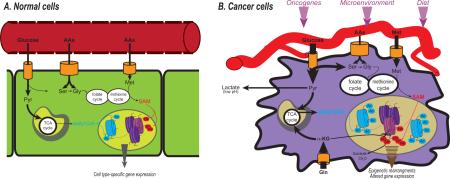
Cancer cells must survive in and adapt to a changing and often harsh microenvironment. Despite the need to adapt to the extracellular environment, cancer cells are generally more self-reliant than their normal counterparts, with weakened dependence on exogenous growth factors and cell-to-cell interaction. This outlines an apparent paradox: how can intrinsically independent cell entities also possess an enhanced ability to adapt to extracellular signals? One mechanism may be through vigilant monitoring of intracellular metabolites.
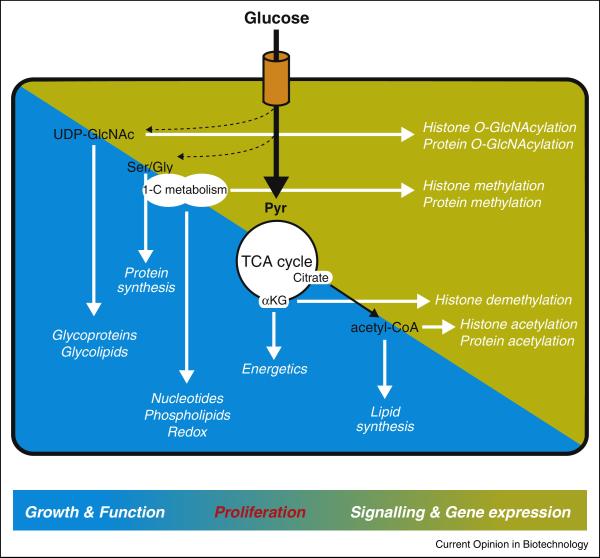
Metabolism in cancer cells is impacted both by internal stimuli such as oncogenic signal transduction and external cues such as nutrient and oxygen availability. Hence, monitoring intracellular levels of metabolites is crucial for cells to appropriately gauge their nutritional resources, taking into account both signaling cues and microenvironmental conditions. Evolutionarily conserved “nutrient-sensing” mechanisms exist to detect and respond to metabolic changes. In this respect, the role of AMP-activated protein kinase (AMPK), which is activated when the AMP:ATP ratio rises, is illustrative of the ability of mammalian cells to switch to a more catabolic state when they perceive a nutrient stress[1]. Conversely, signaling through the mechanistic target of rapamycin (mTOR) promotes growth and is active when cells sense a favorable, nutrient-replete environment[2]. Certain post-translational modifications are also sensitive to the availability of specific metabolites and thus can provide additional mechanisms for the cell to gauge its metabolic status[3-5] (Figure 1).
Figure 1.
Glucose-derived metabolic intermediates provide energy, reducing equivalents and building blocks to sustain normal cell functions and biomass accumulation (blue background). The same intermediates impact numerous signaling events, serving as substrates or cofactor for post-translational modifications of histones or proteins (yellow background). Through dual roles in the regulation of biomass generation and signal transduction, cellular metabolism is central in the decision to grow and proliferate or to remain quiescent.
Are metabolic and epigenetic alterations linked in cancer cells?
Cancer cells undergo extensive metabolic reprogramming to sustain tumor growth[6]. Most chromatin modifying enzymes utilize metabolites as cofactors or substrates, and accumulating evidence has shown that the epigenome (and ultimately the transcriptome) is sensitive to metabolic state[5,7,8]. At the same time, it is manifest that the epigenome is reorganized in tumor cells, a feature that is now considered an enabling characteristic of cancer[9,10]. Metabolic contributions to cancer cell epigenetic alterations are, with a few notable exceptions, largely unknown, however.
A prime example in which metabolic control of the epigenome has been demonstrated is in tumors harboring isocitrate dehydrogenase (IDH1 or IDH2) mutations. In IDH mutant tumors, aberrant accumulation of the metabolite (R)-2 hydroxyglutarate competitively inhibits α-ketoglutarate-dependent JMJD histone demethylases and TET methylcytosine dioxygenases, thereby mediating a hypermethylation phenotype (reviewed in[6,11,12]). Less clear is how generalizable this paradigm will be to tumors without mutations in genes encoding metabolic enzymes. Does metabolic rewiring mediated by microenvironmental conditions or oncogenic signaling alter substrate availability to chromatin modifying enzymes to a sufficient extent to impact the cancer cell epigenome? If so, how does this impact cancer initiation, tumor growth, and treatment responses? In this Review, we will discuss the current evidence that oncogenic and microenvironment-mediated metabolic reprogramming impact tumor histone acetylation and methylation levels.
Metabolic regulation of histone acetylation
Histone acetylation participates in multiple chromatin-dependent processes, including gene regulation, DNA replication, and DNA damage repair. Acetylation is catalyzed by lysine acetyltransferases (KATs), which transfer an acetyl group from acetyl-coenzyme A (acetyl-CoA) to lysine residues (Nε), with the concomitant production of CoA[13]. Histone deacetylases (HDACs) remove the acetyl group, generating acetate (class I/II HDACs).
Acetyl-CoA levels in cells fluctuate in response to several physiological cues, including nutrient availability, circadian rhythms, and changes in metabolic state[14-17]. Several KATs have been reported to exhibit physiological regulation based on cellular levels of acetyl-CoA[15,17,18]. In yeast, Gcn5 is highly responsive to acetyl-CoA availability, and the range of acetyl-CoA reported could plausibly regulate this KAT based on KD for acetyl-CoA[17,19]. Many KATs are also inhibited by their product CoA, suggesting that the ratio of acetyl-CoA: CoA might be the physiological regulator of acetylation in response to metabolic changes [20]. Consistent with this model, the ratio of acetyl-CoA: CoA drops under conditions of glucose or growth factor deprivation, paralleling acetyl-CoA and overall histone acetylation levels[14]. Moreover, manipulation of either metabolite in isolated nuclei impacts histone acetylation levels, with high CoA suppressing histone acetylation[14].
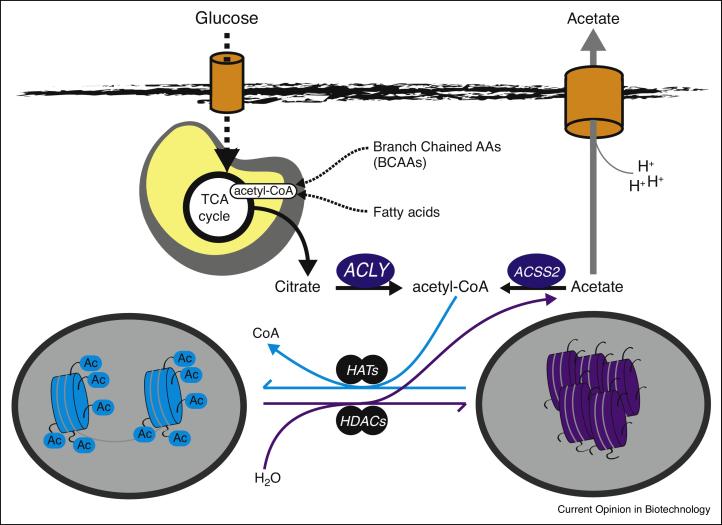
In mammals, the nucleo-cytoplasmic pool of acetyl-CoA is produced largely by 2 enzymes; ATP-Citrate Lyase (ACLY), which produces acetyl-CoA from citrate, and acyl-CoA synthetase short-chain family member 2 (ACSS2, also known as AceCS1), which ligates coenzyme A to acetate to generate acetyl-CoA[4]. Generation of nuclear-cytoplasmic acetyl-CoA from breakdown of glucose, fatty acids, and amino acids relies on export of mitochondrial citrate to the cytoplasm and its subsequent cleavage by ACLY. Given the rapid turnover of histone acetylation (minutes to hours)[21,22], a close interplay between ACLY and ACSS2 likely occurs to maintain the nuclear-cytoplasmic acetyl-CoA pools, with ACLY responsible for net input of acetyl-CoA from nutrients, and ACSS2 playing a key role in acetyl group recycling after generation of acetate from deacetylation reactions (Figure 2). In addition to ACLY and ACSS2, production of nuclear acetyl-CoA has very recently also been attributed to the Pyruvate Dehydrogenase Complex (PDC), which normally functions to generate acetyl-CoA from pyruvate in mitochondria for TCA cycle entry, but has now been shown to also be present and enzymatically active within the nucleus[23]. All three enzymes, ACLY, ACSS2, and PDC, have been shown to contribute to histone acetylation[23-25].
Figure 2.
Acetyl-CoA production and utilization for histone acetylation. ATP-citrate lyase (ACLY) generates nuclear-cytoplasmic acetyl-CoA from nutrients such as glucose, amino acids, and fatty acids. Each of these nutrients generates acetyl-CoA within mitochondria. Transfer of these acetyl units from mitochondria to cytoplasm involves mitochondrial synthesis of citrate from oxaloacetate and acetyl-CoA, then subsequent mitochondrial export and then cleavage of citrate into acetyl-CoA and oxaloacetate by ACLY. Acetyl-CoA serves as the substrate for lysine acetyltransferases (KATs). Histone deacetylases (HDACs) remove acetyl moieties from histones, leading to local production of acetate, which in turn can be recycled back into acetyl-CoA by ACSS2 or exported in a proton-coupled manner to facilitate pH balance. Not shown, the pyruvate dehydrogenase complex has also recently been shown to be present within the nucleus and to provide an additional source of nuclear acetyl-CoA for histone acetylation.
Oncogenic control of acetyl-CoA metabolism and histone acetylation
Acetyl-CoA plays crucial roles in energy generation, as well as in lipid and amino acid metabolism[4]. Hence, it is not surprising that production and utilization of this central metabolite are impacted by oncogenic signal transduction. Not all oncogenic stimuli influence acetyl-CoA metabolism identically; for example, in a direct comparison of metabolic flux changes mediated by myr-AKT or H-RASV12G, AKT stimulated citrate to lipid conversion (which requires ACLY-dependent acetyl-CoA production), while H-RAS suppressed this flux[26].
AKT has long been recognized as a major regulator of glucose uptake and metabolism[6]. AKT also promotes the phosphorylation of ACLY on Ser455, thereby increasing its activity[27,28]. We have recently found that levels of AKT phosphorylation in human gliomas and prostate tumors correlate significantly with global levels of histone acetylation[14]. In support of a metabolic mechanism underlying this relationship, expression of myr-AKT or an ACLY phosphomimetic (S44D) facilitated sustained acetyl-CoA and histone acetylation levels during glucose limitation. Reciprocally, Akt inhibition suppressed acetyl-CoA and histone acetylation levels, which were partially restored upon acetate supplementation[14]. Importantly, elevated histone acetylation is also detectable upon AKT activation in vivo[14].
Elucidating the functional roles of metabolic regulation of histone acetylation will require further investigation. We speculate that oncogenes may exploit acetyl-CoA metabolism in part to modify chromatin in such a way to promote transformation and growth. In support of this possibility, work in yeast has shown that high acetyl-CoA stimulates promoter histone acetylation and expression of genes involved in cell growth and division[17,29]. Analogously, acetyl-CoA availability in glioblastoma cells regulates a gene expression signature enriched for genes involved in DNA replication and cell cycle[14]. While enabling acetyl-CoA production in glucose-limited conditions by providing cells with acetate does not per se accelerate proliferation, it does appear to poise cells for growth, potentially through gene regulation [14].
The proto-oncogene MYC has also been identified as a key regulator of acetyl-CoA metabolism and histone acetylation. Specifically, c-Myc was found to determine an increase in histone H4 acetylation in fibroblasts, an event that is coincident with and likely propaedeutic to cell cycle entry[30,31]. Myc-mediated histone acetylation relies on acetyl-CoA derived from mitochondria[31], presumably via citrate export and ACLY activity. In the absence of Myc, acetyl-CoA levels fall, despite compensatory increases in fatty acid oxidation[32]. Thus, both AKT and MYC have roles in promoting acetyl-CoA production and provision for histone acetylation.
Microenvironmental control of acetyl-CoA metabolism and histone acetylation
In addition to oncogenic controls, tumor microenvironmental conditions such as hypoxia, glucose deprivation, and pH changes also alter cellular metabolism and metabolite levels. Solid tumors expand and frequent incur a state of chronic hypoxia, due to poor or abnormal vascularization [33]. This activates a hypoxic response program coordinated by the hypoxia inducible factor (HIF) transcription factors, that includes enhanced glycolysis, reduced glucose entry into the TCA cycle, and upregulation of glutamine-dependent reductive carboxylation to produce citrate and lipogenic acetyl-CoA[34-36]. Hypoxia also increases the expression of ACSS2[37]. How hypoxia impacts acetyl-CoA availability for histone acetylation is not yet clear, but this could conceivably affect cellular responses and adaptability to a hypoxic environment.
Another distinguishing feature of the tumor microenvironment is the presence of a local acidic pH[38], due in part to elevated secretion of lactic acid by glycolytic cells. Extracellular acidosis also lowers intracellular pH (pHi), which promotes histone deacetylation [39]. Acidic pHi favors histone deacetylation as a mechanism to extrude protons from the cell; acetate is transported in a proton-coupled manner across the plasma membrane through monocarboxylate transporters. Chromatin has thus been proposed to function as a capacitor to ensure pHi balance[40]. Interestingly, hypoxia- and low pH-activated genes generally favor an aggressive, pro-metastatic phenotype [38], and likewise low histone acetylation levels are associated with poor patient prognosis [41]. This at first may seem at odds with the notion discussed above that nutrient-regulated histone acetylation appears to promote growth. One potential explanation is that these divergent effects may dominate at different disease stages. For example, histone acetylation changes are detected early in disease progression as a consequence of oncogene activation and may serve to initially facilitate growth. As disease progresses and pH in the microenvironment drops, acetate is mobilized from chromatin. Hence, the low histone acetylation that correlates with poor prognosis may be a consequence rather than a mediator of disease progression.
Tissue-specific environmental conditions can also play a major role in acetyl-CoA metabolism. This is exemplified by the role of butyrate, which is produced by commensal microorganisms in the gut, through fermentation of diet-derived fibers [42]. A short chain fatty acid, butyrate is a major energy source of normal colonocytes. Butyrate contributes to the mitochondrial acetyl-CoA pool after undergoing β-oxidation; consistently, in normal colonocytes butyrate promotes ACLY-dependent increase in histone acetylation and cell proliferation[43]. On the other hand, in cancer cells, butyrate is not oxidized but instead enters the nucleus and functions as an HDAC inhibitor, underlying the selective toxicity of butyrate to cancer cells. Notably, gene expression profiles mediated by butyrate are strikingly different whether elevated histone acetylation results from increased acetyl-CoA production or from HDAC inhibition[43], highlighting that the consequences of shifts in overall histone acetylation levels can be markedly different depending on the underlying mechanisms. Importantly, high fiber diet was shown to suppress colorectal tumorigenesis in mice, in a manner dependent on the presence of butyrate-producing gut bacteria[42]. Moreover, both butyrate and histone acetylation levels are elevated in human colon tumors compared to surrounding normal tissue[42].
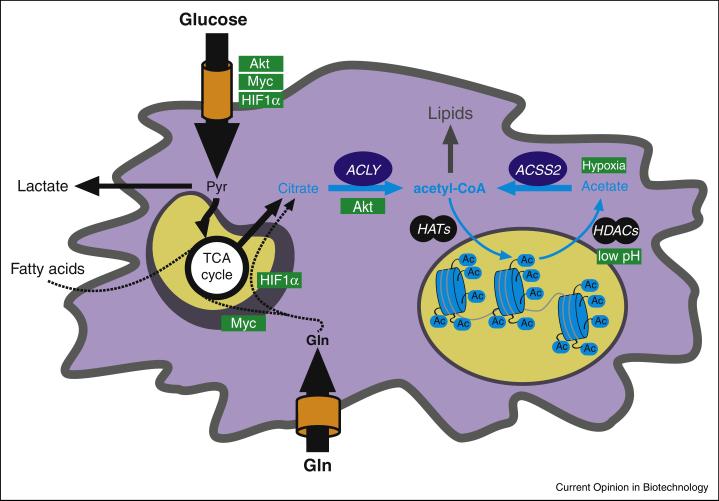
These data underscore the concept that acetyl-CoA serves not only as a metabolic intermediate, but also as signaling molecule that confers metabolic information to chromatin (Figure 3). Beyond chromatin and gene regulation, recent studies have also shown that acetyl-CoA levels impact acetylation of many other proteins, influencing autophagy and multiple metabolic processes[44]. How oncogenic control of acetyl-CoA metabolism and acetylation influence gene expression programs, differentiation state, and stress responses in cancer will be exciting areas for further investigation.
Figure 3.
Acetyl-CoA metabolism is impacted by oncogenes and microenvironmental cues (positive regulators indicated in green boxes).
Metabolic control of histone methylation
Histone lysines can be mono-, di- or tri-methylated and are recognized by particular chromatin-binding proteins, which locally orchestrate the gene expression machinery to either repress or activate neighboring genes. The universal methyl donor in mammals is S-adenosylmethionine (SAM), synthesized from methionine and ATP. SAM is an intermediate of one-carbon metabolism, constituted of two interconnected cycles of reactions, the folate and methionine cycles, which have metabolic inputs from methionine, serine and glycine metabolism, and folate[45,46]. Transfer of the methyl unit from SAM results in the production of S-adenosylhomocysteine (SAH), which is then recycled in the methionine cycle, but also acts as competitive inhibitor of methyltransferases. Hence, the SAM: SAH ratio is considered an indicator of the “methylation potential” of a cell [47].
Metabolic alterations, SAM levels, and histone methylation
Like acetyl-CoA, 1-carbon metabolism also plays key roles in biosynthetic reactions, including nucleotide and phospholipid synthesis[45,46]. Moreover, regulation of the availability of 1-carbon units is influenced by at least two major control points that are altered frequently in cancer cells. First, cellular uptake of methionine occurs through solute transporters such as Lat1 (Slc7a5), which is overexpressed in many tumors[48]. Secondly, the serine-glycine biosynthesis pathway, which donates 1-carbon units to regenerate methionine from homocysteine, is also frequently enhanced in cancer cells via amplification of 3-Phospoglycerate Dehydrogenase (PHGDH), a gatekeeper enzyme of the pathway[49,50]. Suppression of PHGDH in cancer cells with PHGDH amplification impairs proliferation[49,50], although its impact on histone methylation has not yet been addressed. Interestingly, overexpression of PHGDH in normal breast acini causes disruption of acinar organization, hinting at possible chromatin rearrangements[50].
Alterations in SAM and histone methylation levels have been described in cancer cells overexpressing nicotinamide N-methyltransferase (NNMT), an enzyme that catabolizes SAM to 1-Methyl Nicotinamide (1MNA). The NNMT reaction drains SAM and traps it into 1MNA, a highly stable metabolite that is not known to be further utilized by the cell. Hence, increased NNMT activity in cancer cells lowers SAM and histone methylation levels, and this is associated with the acquisition of a more aggressive phenotype[47].
Another striking example of metabolic control of histone methylation is in the context of stem cell pluripotency. SAM-dependent regulation of histone methylation is required for maintenance of pluripotency. In mouse ES cells, this occurs through threonine catabolism into glycine to generate 1-carbon units[51]. In human ES cells, methionine consumption is crucial for sustaining histone methylation and pluripotency[52]. It is not yet known whether SAM-dependent control of histone methylation regulates cellular plasticity or differentiation in the context of tumor development.
Perspectives
Metabolites such as acetyl-CoA and SAM connect nutritional status to signaling and gene expression. This connection is impacted by oncogenic and microenvironmental stimuli, thereby contributing to chromatin organization in cancer cells. The biological programs regulated by metabolic control of the epigenome are still largely elusive. Contributions to growth and proliferation, cellular plasticity, and differentiation will be intriguing areas for future investigation. Also poorly understood is whether diet induces changes in cellular metabolite levels that could help explain the increasingly clear link between overnutrition and cancer. Further elucidation of the connections between metabolism, oncogenic signaling and chromatin organization is expected to enhance our understanding of the basic biology of cancer and to potentially point towards new therapeutic approaches.
Highlights.
Oncogenic and microenvironmental stimuli rewire metabolism in cancer cells.
Many chromatin-modifying enzymes rely on metabolites as cofactors or substrates.
Histone acetylation is sensitive to acetyl-CoA levels, and linked to growth and proliferation.
Histone methylation is sensitive to the methyl donor S-adenosylmethionine.
Acknowledgements
Work in the Wellen lab is currently funded by the NCI, the American Diabetes Association, Pew Charitable Trusts and AACR/Pancreatic Cancer Action Network. We apologize to authors whose work we did not cite due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Hardie DG, Ross FA, Hawley SA. Ampk: A nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laplante M, Sabatini DM. Mtor signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metallo CM, Vander Heiden MG. Metabolism strikes back: Metabolic flux regulates cell signaling. Genes Dev. 2010;24(24):2717–2722. doi: 10.1101/gad.2010510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellen KE, Thompson CB. A two-way street: Reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13(4):270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 5.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16(1):9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward PS, Thompson CB. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaelin WG, Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153(1):56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaochar S, Tu BP. Gatekeepers of chromatin: Small metabolites elicit big changes in gene expression. Trends Biochem Sci. 2012;37(11):477–483. doi: 10.1016/j.tibs.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153(1):38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339(6127):1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: Mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 12.Losman JA, Kaelin WG., Jr. What a difference a hydroxyl makes: Mutant idh, (r)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27(8):836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 14**.Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, Worth AJ, Yuan ZF, Lim HW, Liu S, Jackson E, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20(2):306–319. doi: 10.1016/j.cmet.2014.06.004. [The first study to link oncogenic metabolic rewiring to changes in tumor histone acetylation levels. Authors demonstrate that AKT activation positively regulates ACLY-dependent acetyl-CoA production, both in vitro and in vivo. Augmenting acetyl-CoA availability ultimately leads to enhanced histone acetylation and increase expression of genes involved in gene proliferation. AKT activation was found to correlate with histone acetylation marks in human gliomas and prostate cancers.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, Zamzami N, et al. Regulation of autophagy by cytosolic acetyl-coenzyme a. Mol Cell. 2014;53(5):710–725. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 16.Sahar S, Masubuchi S, Eckel-Mahan K, Vollmer S, Galla L, Ceglia N, Masri S, Barth TK, Grimaldi B, Oluyemi O, Astarita G, et al. Circadian control of fatty acid elongation by sirt1 protein-mediated deacetylation of acetyl-coenzyme a synthetase 1. J Biol Chem. 2014;289(9):6091–6097. doi: 10.1074/jbc.M113.537191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Cai L, Sutter BM, Li B, Tu BP. Acetyl-coa induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42(4):426–437. doi: 10.1016/j.molcel.2011.05.004. [Authors found that fueling acetyl-CoA pool in yeast (with either glucose and galactose or end products of glycolytic metabolism such as ethanol or acetate) promotes entry into growth phase and eventually cell division. Unbiased screening for acetylated proteins indicated that Gcn5/SAGA-mediated histone acetylation is responsible for cell cycle entry through activation of proliferative genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. A glycolytic burst drives glucose induction of global histone acetylation by picnua4 and saga. Nucleic Acids Res. 2009;37(12):3969–3980. doi: 10.1093/nar/gkp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langer MR, Fry CJ, Peterson CL, Denu JM. Modulating acetyl-coa binding in the gcn5 family of histone acetyltransferases. J Biol Chem. 2002;277(30):27337–27344. doi: 10.1074/jbc.M203251200. [DOI] [PubMed] [Google Scholar]
- 20.Albaugh BN, Arnold KM, Denu JM. Kat(ching) metabolism by the tail: Insight into the links between lysine acetyltransferases and metabolism. Chembiochem. 2011;12(2):290–298. doi: 10.1002/cbic.201000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Evertts AG, Zee BM, Dimaggio PA, Gonzales-Cope M, Coller HA, Garcia BA. Quantitative dynamics of the link between cellular metabolism and histone acetylation. J Biol Chem. 2013;288(17):12142–12151. doi: 10.1074/jbc.M112.428318. [Authors investigate dynamics of histone acetylation tracing the incorporation of labelled nutrients into histones. Glucose-derived acetyl groups were detected in acetylated histones within 10 minutes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterborg JH. Dynamics of histone acetylation in saccharomyces cerevisiae. Biochemistry. 2001;40(8):2599–2605. doi: 10.1021/bi002480c. [DOI] [PubMed] [Google Scholar]
- 23.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-coa and histone acetylation. Cell. 2014;158(1):84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 24.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. Atp-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23(2):207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 26.Fan J, Kamphorst JJ, Mathew R, Chung MK, White E, Shlomi T, Rabinowitz JD. Glutamine-driven oxidative phosphorylation is a major atp source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol. 2013;9:712. doi: 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of atp-citrate lyase as a protein kinase b (akt) substrate in primary adipocytes. J Biol Chem. 2002;277(37):33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- 28.Potapova IA, El-Maghrabi MR, Doronin SV, Benjamin WB. Phosphorylation of recombinant human atp:Citrate lyase by camp-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of atp:Citrate lyase by phosphorylated sugars. Biochemistry. 2000;39(5):1169–1179. doi: 10.1021/bi992159y. [DOI] [PubMed] [Google Scholar]
- 29.Shi L, Tu BP. Acetyl-coa induces transcription of the key g1 cyclin cln3 to promote entry into the cell division cycle in saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2013;110(18):7318–7323. doi: 10.1073/pnas.1302490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7(8):1054–1066. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrish F, Noonan J, Perez-Olsen C, Gafken PR, Fitzgibbon M, Kelleher J, VanGilst M, Hockenbery D. Myc-dependent mitochondrial generation of acetyl-coa contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J Biol Chem. 2010;285(47):36267–36274. doi: 10.1074/jbc.M110.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Edmunds LR, Sharma L, Kang A, Lu J, Vockley J, Basu S, Uppala R, Goetzman ES, Beck ME, Scott D, Prochownik EV. C-myc programs fatty acid metabolism and dictates acetyl-coa abundance and fate. J Biol Chem. 2014;289(36):25382–25392. doi: 10.1074/jbc.M114.580662. [Authors performed metabolic tracing of long and medium-chain fatty acids in c-Myc KO fibroblasts. Compared to WT cells, Myc-deficient cells showed increased fatty acids oxidation and increased utilization of acetyl-CoA for TCA cycle-mediated energy production, and decreased channeling to anabolic processes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keith B, Johnson RS, Simon MC. Hif1alpha and hif2alpha: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, et al. Reductive glutamine metabolism by idh1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, Osterman AL, Smith JW. Comparative metabolic flux profiling of melanoma cell lines: Beyond the warburg effect. J Biol Chem. 2011;286(49):42626–42634. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshii Y, Furukawa T, Yoshii H, Mori T, Kiyono Y, Waki A, Kobayashi M, Tsujikawa T, Kudo T, Okazawa H, Yonekura Y, et al. Cytosolic acetyl-coa synthetase affected tumor cell survival under hypoxia: The possible function in tumor acetyl-coa/acetate metabolism. Cancer Sci. 2009;100(5):821–827. doi: 10.1111/j.1349-7006.2009.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K, Hong CS, Christofk HR, Vogelauer M, Seligson DB, Kurdistani SK. Histone acetylation regulates intracellular ph. Mol Cell. 2013;49(2):310–321. doi: 10.1016/j.molcel.2012.10.025. [Authors found that low intracellular pH forces the cell to extrude protons using acetate as co-transporter. To mediate this, HDACs are hyperactivated and acetate is mobilized from the chromatin. The effect of acidosis is independent on nutrient availability and restricted to histones.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurdistani SK. Chromatin: A capacitor of acetate for integrated regulation of gene expression and cell physiology. Curr Opin Genet Dev. 2014;26C:53–58. doi: 10.1016/j.gde.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurdistani SK. Histone modifications in cancer biology and prognosis. Prog Drug Res. 2011;67:91–106. doi: 10.1007/978-3-7643-8989-5_5. [DOI] [PubMed] [Google Scholar]
- 42**.Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A, Curry KP, Renner SW, Greenwalt A, Ryan EP, Godfrey V, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-14-0501. [Authors demonstrated that a fiber-rich diet protects against chemical-induced colorectal carcinoma in mice. The anti-tumor effect was found to be mediated by increased production of butyrate from gut microbiota (B. fibrisolvens). Importantly, high fiber diet fails to reduce tumor burden in the absence of commensal bacteria or in presence of mutant B. fibrisolvens, unable to generate butyrate. Authors also found that tumor masses display a highr level of histone acetylation compared to proximal normal tissue.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48(4):612–626. doi: 10.1016/j.molcel.2012.08.033. [Authors found that normal colonocytes consume extremely high levels of butyrate, which is incorporated into the TCA cycle to produce energy but also significantly contribute to the acetyl-CoA pool utilized for anabolic purposes. Hence, in normal colonocytes butyrate triggers histone acetylation and determine a cell-specific gene expression profile. Conversely, malignant colonocytes utilize mostly glucose as energy source, driving the accumulation of butyrate in the nucleus, where it acts as HDAC inhibitor, with significantly different effects on gene expression.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15(8):536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 45.Locasale JW. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39(4):191–198. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.Ulanovskaya OA, Zuhl AM, Cravatt BF. Nnmt promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9(5):300–306. doi: 10.1038/nchembio.1204. [One of the first studies to demonstrate cancer cell metabolic alterations that influence histone acetylation levels through regulation of SAM metabolism. Authors demonstrated that overexpression of NNMT in cancer cells reduces the availability of SAM, transfering a carbon unit to a stable metabolite (1NMA), resulting in histone hypomethylation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuchs BC, Bode BP. Amino acid transporters asct2 and lat1 in cancer: Partners in crime? Semin Cancer Biol. 2005;15(4):254–266. doi: 10.1016/j.semcancer.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51**.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, Asara JM, et al. Influence of threonine metabolism on s-adenosylmethionine and histone methylation. Science. 2013;339(6116):222–226. doi: 10.1126/science.1226603. [First paper to demonstrate that SAM-dependent histone methylation plays a role in stem cell pluripotency. In different models of mouse stem cells, radiotype labeling demonstrated that threonine is metabolized to SAM via Threonine dehydrogenase (Tdh). Threonine-dependent SAM production contributes to histone H3K4 trimethylation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19(5):780–794. doi: 10.1016/j.cmet.2014.03.017. [Authors demonstrate that SAM-dependent histone methylation plays a role in human stem cell pluripotency.] [DOI] [PubMed] [Google Scholar]