Abstract
Fragments of the extracellular matrix component hyaluronan (HA) promote tissue inflammation, fibrosis and tumor progression. HA fragments act through HA receptors including CD44, LYVE1, TLR2,4 and the receptor for hyaluronan mediated motility (RHAMM/HMMR). RHAMM is a multifunctional protein with both intracellular and extracellular roles in cell motility and proliferation. Extracellular RHAMM binds directly to HA fragments while intracellular RHAMM binds directly to ERK1 and tubulin. Both HA and regions of tubulin (s-tubulin) are anionic and bind to basic amino acid-rich regions in partner proteins, such as in HA and tubulin binding regions of RHAMM. We used this as a rationale for developing bioinformatics and SPR (surface plasmon resonance) based screening to identify high affinity anionic RHAMM peptide ligands. A library of 12-mer peptides was prepared based on the carboxyl terminal tail sequence of s-tubulin isoforms and assayed for their ability to bind to the HA/tubulin binding region of recombinant RHAMM using SPR. This approach resulted in the isolation of three 12-mer peptides with nanomolar affinity for RHAMM. These peptides bound selectively to RHAMM but not to CD44 or TLR2,4 and blocked RHAMM:HA interactions. Furthermore, fluorescein-peptide uptake by PC3MLN4 prostate cancer cells was blocked by RHAMM mAb but not by CD44 mAb. These peptides also reduced the ability of prostate cancer cells to degrade collagen type I. The selectivity of these novel HA peptide mimics for RHAMM suggest their potential for development as HA mimetic imaging and therapeutic agents for HA-promoted disease.
Introduction
Hyaluronan (HA) is a polydisperse glycosaminoglycan consisting of dimeric repeats of β-glucuronic acid and N-acetylglucosamine. It performs complex structural and signaling functions required for innate and adaptive immunity, tissue organization, homeostasis and repair 1–6. These functions are mediated by interactions with specific cellular and extracellular proteins, whose mode of action are determined in part by polymer size. For example, HA oligosaccharides bind with greater affinity than high molecular weight HA to the HA receptors RHAMM and TLR2,4 resulting in activation of signaling cascades which control cell migration, survival and proliferation during response to injury processes, and during disease 7–9. In contrast, HA oligosaccharides have been reported to block signaling resulting from CD44/HA interactions 5. The molecular basis for these size dependent effects of HA is not well understood, but HA binds to its partner proteins through several distinct mechanisms which likely contribute to this size and functional specificity. HA binds to CD44 and LYVE1 via a well defined link module but to RHAMM and TLR2,4 by other mechanisms. HA:RHAMM interactions require clusters of positively charged amino acids arranged in a helix 10–12.
RHAMM is an intra- and extracellular multifunctional protein, whose expression is restricted in homeostatic tissues but transiently increased following tissue injury and chronically elevated in inflammatory and neoplastic diseases 13–15. One of its normal functions is to regulate mesenchymal/immune cell migration and differentiation during tissue repair 2, 7, 16, 17. It also promotes immune cell trafficking/invasion in inflammatory diseases 13, 18. RHAMM mRNA and protein expressions are also elevated in most human cancers (e.g. colorectal, prostate, breast, gastric, AML, MM), and have been linked to aggressive disease and poor clinical outcome 13, 14, 19, 20. Experimental evidence supports a role for extracellular RHAMM/HA interactions in response to growth factors, control of cell migration and progression through G2M 17, 21–24. Intracellular RHAMM is a tubulin and ERK1 binding protein, which decorates both interphase and mitotic spindles. Loss or gain of RHAMM proteins results in altered active ERK1,2 targeting/activation kinetics, aberrant mitotic spindle formation and unequal chromosome segregation 23, 25–28. Both extracellular and intracellular RHAMM expressions are elevated rather than decreased in the majority of human tumours and in inflammatory diseases 15, and these functions likely contribute to disease progression. These previous studies have focused attention on RHAMM as a potential therapeutic target in cancers and other diseases 13–15.Herein, we report an approach for identifying peptide ligands that bind to RHAMM, and have the potential of blocking its interaction with HA and/or tubulin. Peptides were discovered that mimic HA oligosaccharides in terms of charge, nanomolar affinity and specificity for RHAMM, and in their ability to block RHAMM:HA interactions. We further show that at least one of these peptides inhibits the invasion of an aggressive metastatic human prostate cancer cell line.
Experimental Procedures
Materials
All solvents and reagents were purchased and used without further purification, and purchased from VWR, Fisher Scientific, or Sigma Aldrich. Fmoc-Rink amide MBHA (100–200 mesh) resin, Fmoc-amino acids, Fmoc-protected aminohexanoic acid (Fmoc-Ahx) and HBTU (2-(1H-benzotriazole 1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate) coupling reagent for peptide synthesis were obtained from Peptides International. N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDAC), N-hydroxysulfosuccinimide sodium salt (sulfo-NHS), fluorescein isothiocyanate (FITC) isomer I and fetal bovine serum (FBS) were purchased from Sigma Aldrich. NHS-Biotin was obtained from Nova BioChem. Antibodies such as anti-RHAMM (Santa Cruz Biotechnology, USA), anti-CD44 (Pharmigen) and IgG ab (Santa Cruz Biotechnology USA) were obtained commercially. Hyaluronan (HA, 220 kDa) was used for all experiments and purchased from Lifecore (MN, USA). The protease inhibitor cocktail with animal-free aprotinin was purchased from Millipore (ON, CA). CD44-Fc chimera protein (54.2 kDa), which contains the HA binding region of this protein, was purchased from R&D systems.
Peptide Synthesis
Elongation of peptide chains on Rink amide MBHA resin (0.1 mmol) was performed using automated (APEX 396 auto-synthesizer) and/or manual methods using standard solid phase peptide synthesis involving Fmoc deprotection and amino acid coupling cycles, and each cycle was monitored using Kaiser test. Repeated Fmoc deprotection throughout the synthesis (15 and 20 minutes periods) was carried out using 20% piperidine solution in N,N-dimethylformide (DMF). All amino acid couplings were carried out using 0.05 M or higher concentration of Fmoc-protected amino acid and HBTU, N,N-diisopropylethylamine (DIPEA, 5 equiv.) in DMF at 30 and 90 minutes intervals. After each deprotection and coupling step, the resin was washed repeatedly with DMF (3x) and dichloromethane (DCM) (3x). Fmoc-Ahx was coupled using the same parameters. Acylation of the amino terminus was done (15 and 10 minutes) using 10% acetic anhydride in DMF following Fmoc deprotection. Fluorescein coupling was carried out by reacting the amino group of the peptide with fluorescein isothiocyanate (4 equiv.) in DMF with DIPEA (2 equiv.) for 4 hours.
Full deprotection of cysteine-containing peptides was accomplished using a solution of 94% v/v trifluoroacetic acid (TFA), 1% v/v triisopropylsilane (TIPS), 2.5% v/v H2O and 2.5% v/v 1,2-ethanedithiol 29 for 1.0–1.5 hours. Full deprotection of all other peptides was done using a solution of 88% v/v TFA, 5% v/v water, 5% m/v phenol, 2% v/v TIPS for 2–4 hours. The filtrate was collected, precipitated using cold tert-butyl methyl ether, and pelleted via centrifugation at 3000 rpm in −5°C for 10 minutes. Then, pellets were dissolved in distilled-deionized water and lyophilized yielding solid powders.
Purification of peptides was performed using gradient solvent system consisting of H2O + 0.1% TFA (solvent A) and CH3CN + 0.1% TFA (solvent B) at a flow rate of 1.5 mL/min and 20 mL/min for analytical and preparative HPLC, respectively. Analytical HPLC was performed using a Grace Vydac Protein/Peptide RP-C18 column (4.6 mm x 250 µm, 5 µm), and preparative HPLC was performed using a Grace Vydac protein/peptide RP-C18 column (22.0 mm x 250 mm, 10 µm). Absorbance was detected at wavelengths of 220 nm and 254 nm using a Waters 2998 Photodiode Array detector. During purification, fractions were collected, lyophilized, and analyzed by ESI-MS (Waters Micromass Quattro Micro™ API).
Protein Purification
Recombinant protein RHAMM-CT (aa. 706–767, sequence: RDSYAQLLGH QNLKQKIKHV VKLKDENSQL KSEVSKLRSQ LVKRKQNELR LQGELDKLQI, M.W. 7.1 kDa, pI = 10.1) was isolated from E. coli BL21 (D3) strain carrying the recombinant plasmid pPAL7-RHAMM. Bacteria were grown overnight in LB medium at 37°C containing ampicillin (100 µg/mL) and 0.5% glucose, and allowed to grow to mid-log phase. Recombinant GST-RHAMM or exact tagged-RHAMM gene expression was induced with 2 mM IPTG for 4 h at 37°C and bacterial cells were harvested by centrifugation at 10,000 x g for 20 min. Bacterial cells were re-suspended in lysis buffer (composed of 0.2 M sodium phosphate, 0.2 M potassium acetate, 1% triton X-100, and 0.1% protease inhibitors, pH 7.0), sonicated (60 s, 10 s/pulse), and centrifuged (4°C, 12000 x g, 20 min). The resulting supernatant was transferred to a clean tube and filtered (using 0.45 µm filter). GST-RHAMM was purified on a glutathione column while purification of the eXact tagged-recombinant RHAMM was conducted with Profinity eXact (Bio-Rad, USA) affinity resin, used according to manufacturer’s protocol. For this experiment, the lysate was loaded to a column packed with Profinity eXact affinity resin (4 mL resin, column 15 x 1.5 cm) equilibrated with wash buffer (0.2M sodium phosphate, pH 7.0). The column was washed with wash buffer to eliminate impurities, and recombinant RHAMM was eluted with elution buffer (composed of 0.2M sodium phosphate, 0.1M sodium fluoride, pH 7.0). Using a Millipore Filter (Millipore, USA, cut-off 3 kDa), the protein was dialyzed and concentrated in a buffer consisting of 0.2M sodium phosphate, and 0.2 M potassium acetate (pH 7.0). The purity of the isolated protein was verified on 1D SDS-PAGE, and was confirmed using Western blot analysis utilizing anti-RHAMM ab.
SPR (Surface Plasmon Resonance) Screening Assay
ProteON XPR36 system was used for the selection and ranking of different tubulin derived peptides against RHAMM. For immobilization of RHAMM, the ProteON GLC sensor chip surfaces were activated by amine coupling using 100 mM EDAC and 24 mM sulfo-NHS. RHAMM (30 µg/mL in sodium bicarbonate buffer, pH 9.7) was injected at a flow a rate of 30 µL/min. A buffer sample was injected on a different sensor plate for use as a reference. Ethanolamine HCl (1M, pH 8.5) was then injected to deactivate any remaining cross-linking groups. Peptides (10 µM in PBS-T, 2% DMSO) were injected to a RHAMM functionalized surface at 50 µL/min for 3 minutes followed by a 10 minute dissociation (i.e. injection of PBS-T buffer) period. The surfaces were regenerated using two injections of 30 µL of 1M NaCl prior to the injection of the next peptide. In all experiments, reference subtraction was performed using data obtained from reference plate (no RHAMM) and RHAMM functionalized plate.
SPR (Surface Plasmon Resonance) Binding Assays
After peptide screening, GWC SPRimager®II system was used to determine binding kinetic constants. Thiol-containing peptides at 1mM concentration in milliQ water were immobilized on a maleimide-functionalized gold-plated chip for 3 hours. Excess peptides were removed by washing with milliQ water. For binding studies, a series of concentrations (500 nM, 750 nM, and 1000 nM) of RHAMM were injected over the immobilized peptides. After a 15 min dissociation phase, the sensor chip surface was regenerated for the next peptide sample injection via treatment with two 10 min pulse injections of regenerating buffer (2 M NaCl in HBS-EP, pH 7.4) at 100 mL/min. The baseline returned to the initial value after the regeneration step, confirming the removal of all bound analytes. Data analysis and the corresponding dissociation constants (KD) were obtained via non-linear regression fitting to a Langmuir binding model. In all experiments, reference subtraction was performed using data obtained from the reference plate (no peptide) and peptide functionalized plate.
Competitive ELISA Experiments Using Fluorescein-Labelled Peptides
ELISA was carried out to test the ability of fluorescein-labelled tubulin-derived peptides to compete with HA for binding. Recombinant RHAMM (100 µL, 10 µg/mL in 0.05M PBS, pH 9) was immobilized on 96-well ELISA plates) and incubated overnight at 4°C resulting in final amount of protein of 1 ug/well. Plates were washed three times with PBS-Tween-20 buffer (5%, 200 µL/well), washed with blocking buffer (5% 200 µL/well, PBS-Tween-20 per well), and incubated for 1 hour at room temperature. Then, fluorescein-labeled peptides (final concentration of 1 µg/mL) and HA (100 µL/well, M.W. 220 kDa, 10 µg/mL in PBS, serial dilutions have been made for HA = 1:1, 1:2, 1:4, 1:8, 1:16) were added to plates and incubated overnight at 4°C. Plates were washed as described above, and absorbance was measured at 485/535 nm. Experiments were done in triplicate.
Competitive ELISA Using Alexa Fluor 647-Conjugated HA
ELISA were carried out to test the ability of non-labeled tubulin-derived peptides to compete with dye (Alexa Fluor 647)-conjugated HA for RHAMM. RHAMM (100 µL, 10 µg/mL in 0.05 M PBS, pH 9) was immobilized on 96-well ELISA plates (to achieve a final amount of 1 µg/well) and incubated overnight at 4°C. Plates were washed three times with (0.05 %) PBS-Tween-20 buffer (200 µL/well), then incubated with blocking buffer (200 µL/well, 5 % Tween-20 in PBS) for 1 hour at room temperature. Following three washes of (0.05 %) PBS-Tween-20 buffer, tubulin-derived peptides (10 µg/mL) and HA-conjugated Alexa Fluor 647 (100 µL/well, M.W. 220 kDa, 10 µg/mL in PBS, with serial dilutions of 1:1, 1:2, 1:4, 1:8, 1:16) were added to plates and incubated overnight at 4°C. Negative control plates receive no dye-conjugated HA and all experiments were done in triplicate. Plates were washed as described above and the fluorescence was measured at 650 nm.
Cellular Uptake of Alexa Fluor 647 HA
MDA-MB-231 or PC3MLN4 cells were cultured in DMEM media + 10% FBS up to 90% confluency. Then cells were seeded on glass cover slips (12 x 12 mm, coated with 50 µg/mL fibronectin) in 2 x 24-well tissue culture plates (confluency of 20,000 cells/well). Astarvation step was carried out with DMEM + 0.1% FBS overnight at 37°C. After starvation, cells were stimulated with DMEM + 10% FBS overnight at 37°C. The culture medium was aspirated and cells were rinsed with DMEM + 0.1% FBS. Then, the cells were blocked with 3% BSA in DMEM + 0.1% FCS for 1 hour at room temperature. For blocking experiments, antibodies (dilution 1:100, mouse IgG ab, goat anti-RHAMM mAb or mouse anti-CD44 mAb in DMEM + 0.1% FBS media) were added and incubated at 37°C for 1 hour. The resulting culture medium was aspirated, and the cells were washed with DMEM + 0.1% FBS at room temperature. Fluorescein-conjugated peptides (50 µg/mL) were added and incubated at 37°C for 30 minutes. Cells were washed with DMEM + 0.1% FBS, then with PBS (pH 7.6),mounted using Fluoro-gel 11 containing DAPI (Electron microscopy sciences, USA) via manufacturer’s protocol. Cells were photographed using Olympus FluoView FV1000 coupled IX81 Motorized Inverted System Microscope. Tiff images were analyzed using Image J (v1.42q) software. Each image was converted to an 8-bit format and subjected to threshold values of 20 and 255. Region of interest (ROI) were selected and mean cellular fluorescence was determined.
Growth in Methylcellulose
The importance of endogenous HA, RHAMM and CD44 expression was evaluated using methylcellulose that had been supplemented or not with high molecular weight HA (800 kDa, Life Core, Chaska, MN). For these studies PC3MLN4 cells were first suspended in 1.5% methylcellulose dissolved in growth medium. The methylcellulose/cell suspension was dispensed into 24 well culture plates (1.5 mL/well). The methylcellulose was then allowed to set at 37°C and the wells were supplemented with 0.5 mL of 1X growth medium. After 7 days the gels were solubilized in PBS, the cells were then concentrated by centrifugation and counted. For certain experiments, PC3MLN4 cells were used in which endogenous HA synthesis was inhibited using a stably expressed HAS3 antisense vector. Stably transfected cells harboring a mock vector were used as a negative control. In other experiments, the cultures included cells in which expression of RHAMM, CD44 or both were inhibited using siRNA target sequences for RHAMM (CTGGATGAGCTTGATAAATTA) or CD44 (AACTCCATCTGTGCAGCAAAC). The negative control siRNA was provided by Qiagen (Valencia, CA). Inhibition of the expression of RHAMM and CD44 was verifed using western analysis of cell extracts. A layer of 1% agarose in normal growth media (250 µL volume) was pipetted into triplicate wells/condition of a 48 well plate and allowed to solidify. PC3MLN4 cells were suspended in 2x growth media at 1x104 cells/ml and 500 µL cell suspension aliquoted into microfuge tubes for each condition. Peptide 14a or scrambled control peptide was added to the cells in the appropriate tube. 500 mL of 1.2% low melt agarose maintained at 42°C was added to the cell suspension, mixed evenly by pipetting (final concentration 0.6% agarose), and 250 µL of the mixture overlayed on the 1% agarose layer in triplicate wells. Plates were placed at 4°C for 15 minutes to facilitate rapid polymerization of the agarose, the wells were overlaid with 250 µL 1x growth media and incubated at 37°C/5% CO2 for 11 days. Colonies were counted in five random fields/well using a 10x objective, and data are shown as the average number of colonies from five fields /well
Cell invasion assay
The QCM™ Gelatin Invadopodia Assay (Millipore) was used following manufacturer’s instruction to determine cell invasion. PC3MLN4 cells were plated at 50 % confluency in Cy3-Gelatin coated 8 well chamber slides. HA mimetic and scrambled peptides were added at 10 µg/mL. 48 hrs after plating, cells were fixed, stained with FITC-Phalloidin and DAPI and images were taken by confocal microscopy.
Soft agar method
A layer of 1% agarose in normal growth media (250 µL volume) was pipetted into triplicate wells/condition of a 48 well plate and allowed to solidify. PC3MLN4 cells were suspended in 2x growth media at 1x104 cells/ml and 500 µL of the cell suspension was aliquoted into microfuge tubes for each condition. Peptide 14a or scrambled control peptide was added to the cells in the appropriate tube. 500 mL of 1.2% low melt agarose maintained at 42°C was added to the cell suspension, mixed evenly by pipetting (final concentration 0.6% agarose), and 250 µL of the mixture overlayed on the 1% agarose layer in triplicate wells. Plates were cooled to 4°C for 15 minutes to facilitate polymerization of the agarose, the wells were overlaid with 250 µL 1x growth media and incubated at 37 °C/5% CO2 for 11 days. Colonies were counted in five random fields/well using a 10x objective, and data are shown as the average number of colonies from five fields /well from triplicate wells, +/− s.e.m.
Results
BLAST search and sequence alignment
Previous studies seeking to discover ligands used unbiased screening methods to identify negatively charged peptides resembling HA, which bound to RHAMM12, 30. We therefore initially synthesized several of these and confirmed they bind to recombinant RHAMM. However, in culture comparison of the binding of these labeled peptides to RHAMM−/− fibroblasts, RHAMM-rescued fibroblast, and tumor cell lines that expressed endogenous RHAMM31 suggested high levels of non-specific peptide uptake. Nevertheless, these results predicted that negatively charged peptides which bind to RHAMM can be isolated,
The HA-binding region of RHAMM is proposed to consist of two carboxyl terminal helices enriched in hydrophobic and basic amino acids required for interaction with the HA polymer 12, 23, 30. Synthetic peptides representing this domain, but not scrambled sequences, inhibit HA binding to recombinant RHAMM 32. These results predict that the interaction of HA oligosaccharides with RHAMM protein is primarily ionic in nature, with a minor contribution from hydrophobic residues and is therefore dissimilar to the binding interactions between CD44 and link module-like sites 11, 33. RHAMM also binds directly to alpha- and beta-tubulin monomers and polymers 28 via both the N-terminal sequence 34 and a sequence that is imbedded in the carboxyl terminal HA binding region of RHAMM 25, 26, 28. Αlpha- and β-tubulin dimers contain a highly conserved helical region and a hyper-variable carboxyl terminal tail (CTT) sequence with pockets of acidic residues resembling the negative charge density of HA 35. These pockets are proteolytically released as short peptides which modulate microtubule dynamics by binding to positively charged sequences in microtubule associated proteins (MAPs), which are similar to the RHAMM HA binding sequence 29, 36–38. A query sequence corresponding to the HA-binding region (aa718–750) of RHAMM was therefore used in a basic alignment search tool to compile a list of proteins showing sequence homology to this region. Pair-wise comparisons between RHAMM and microtubule binding domains of MAPs, e.g. MAP1-4, TAU) as well as kinesins (e.g. KIF11, Klp61) revealed only moderate overall sequence homology (17%-24% as calculated using ClustalX2) to the HA binding region of RHAMM. However, RHAMM and many of these proteins share similar stretches of basic residues within helical secondary structures. Collectively, these studies predicted that RHAMM/tubulin interactions are molecularly similar to RHAMM/HA interactions and that the short CTT sequence might act as HA peptide mimics.
Screening of tubulin-derived peptides against RHAMM
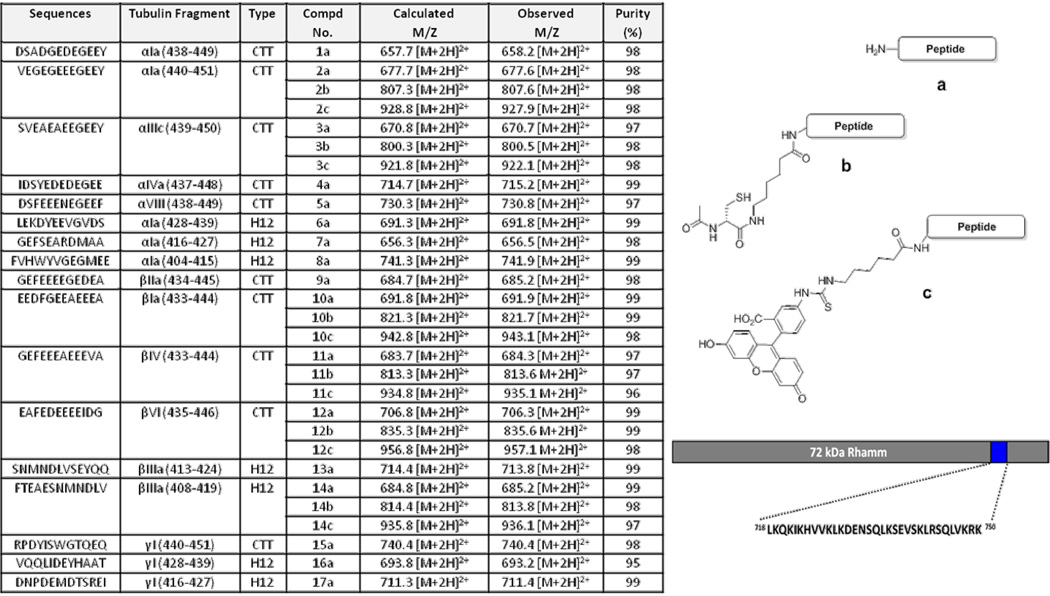
To assess this possibility, peptides corresponding to CTT regions of α- and β-tubulin sequences, which contain acidic residues and exist as helices, were synthesized on an insoluble polystyrene Rink amide resin using standard Fmoc peptide synthesis protocols (Figure 1). Most of these sequences were derived from within the CTT region of tubulin isoforms, but several contained sequences directly flanking α1a- (compounds 6, 7 and 8) and βIIIa-CTT (compounds 13 and 14). Derivatized peptides were prepared with either fluorescein or an N-acetyl cysteine modified N-terminus with the addition of an aminohexanoic acid spacer in order to increase the distance between the targeting peptide and the dye/cysteine. Based on this initial sequence analysis, seventeen tubulin-derived peptides were identified and further characterized (Figure 1).
Figure 1. Composition of the peptide ligands.
Sequences of the peptides used for evaluation (Table, left), description of the tubulin fragment being used and the compound number. Synthetic tubulin-derived peptides were analyzed using ESI-MS and RP-HPLC. The calculated and observed m/z values are based on the prominent observed signals as determined by ESI+. The percent purity was determined by RP-HPLC with detection at 220 nm. The structures (right) show: a) the general structure of unmodified tubulin-derived peptides, b) peptides conjugated to N-acetyl cysteine and c) to fluorescein isothiocyanate. A depiction of the RHAMM HA binding site is shown on bottom right.
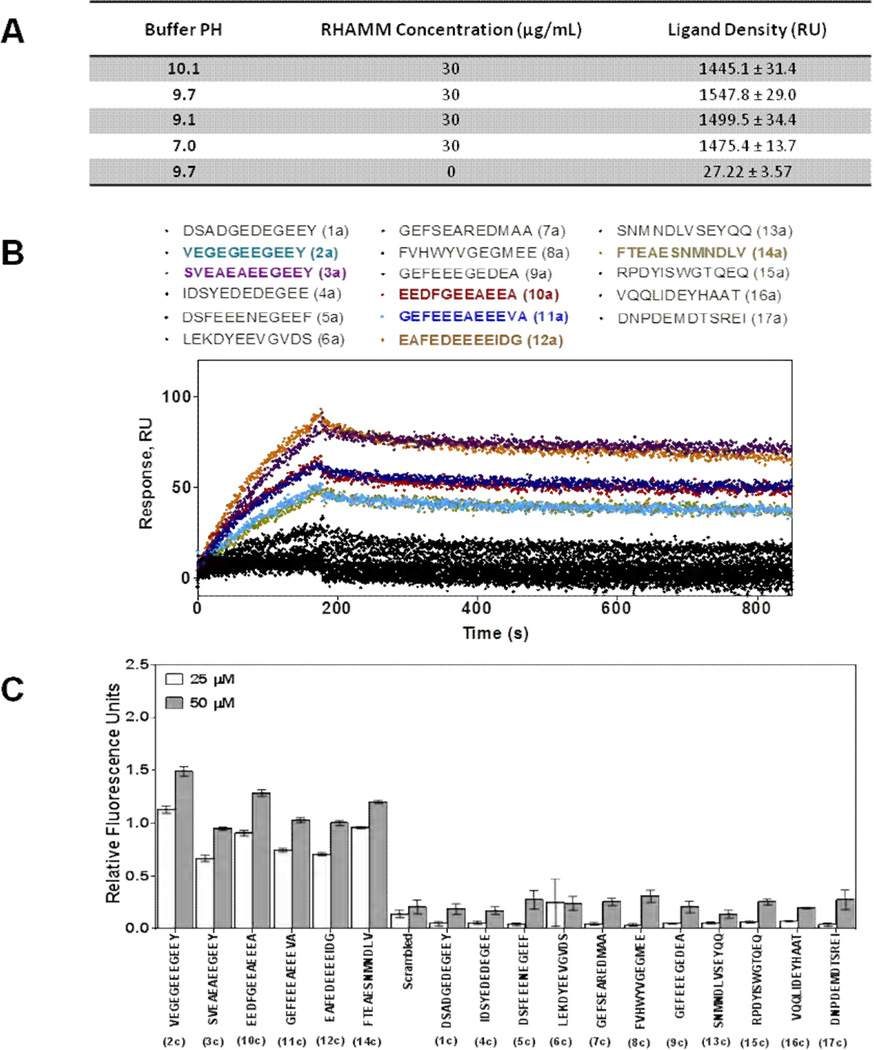
All peptides were purified by reverse-phase HPLC and analyzed by LCMS. A SPR (surface plasmon resonance)-based screening method was used to initially identify peptides with affinity for the RHAMM HA binding domain (Figure 2A,B). The optimal conditions for the immobilization of the RHAMM HA binding region to the sensor plate was determined as described in Experimental Procedures. In order to identify conditions for optimal recombinant RHAMM protein density, RHAMM was coupled to the sensor plate with varying pH. Protein density for each pH immobilization condition was determined from the average SPR response of six measurements and maximum immobilization was determined to occur at a pH of 9.7 (Figure 2A). RHAMM immobilization was slightly lower at pH 10.1 likely due in part to loss of net charge on RHAMM at its isoelectric point. For the screen, 10 µM CTT tubulin peptides were injected onto the RHAMM-covered sensor plates (Figure 2B). Sensograms with plates that did not contain immobilized RHAMM were used as negative controls and values were subtracted from those obtained with experimental sensograms. In this manner, we identified 6 out of the 17 tested peptides as potential ligands for the HA binding region of RHAMM. These are: 2a (VEGEGEEEGEEY), 3a (SVEAEAEEGEEY), 10a (EEDFGEEAEEEA), 11a (GEFEEEAEEEVA), 12a (EAFEDEEEEIDG), and 14a (FTEAESNMNDLV) (Figure 3B). Binding of these peptides to recombinant RHAMM was verified using ELISA assays and FITC-labeled peptides (Figure 2C).
Figure 2. Identification of RHAMM ligands from the tubulin-derived peptide library.
(A) The pH dependence of RHAMM immobilization to a surface plasmon resonance (SPR) sensor plate. Recombinant RHAMM protein was immobilized using sodium bicarbonate buffer, pH 7.0–10.1, at a flow rate of 30 µL/min. Maximum amount of immobilized RHAMM occurred at pH 9.7. The protein density was determined from the average SPR response (RU) of six measurements (± S. E.M.). (B) SPR screening of purified tubulin-derived peptides for binding to recombinant RHAMM protein. Shown are sensograms generated by the interaction of 17 tubulin-derived peptides (a versions, Figure 1) at a concentration of 10 µM to the functionalized SPR plate. Screening generated 6 peptides (colored traces; 2a, 3a, 10a, 11a, 12a and 14a), which showed high affinity to RHAMM, while black traces represent low affinity peptides. (C) The binding of fluorescein-labelled peptides (c versions) to RHAMM using ELISA binding assay. Binding studies were performed at 25 µM and 50 µM concentrations of the peptide. The negative control (no immobilized RHAMM), which showed minimal background fluorescence, was subtracted for each measurement. A scrambled peptide was also used as negative control.
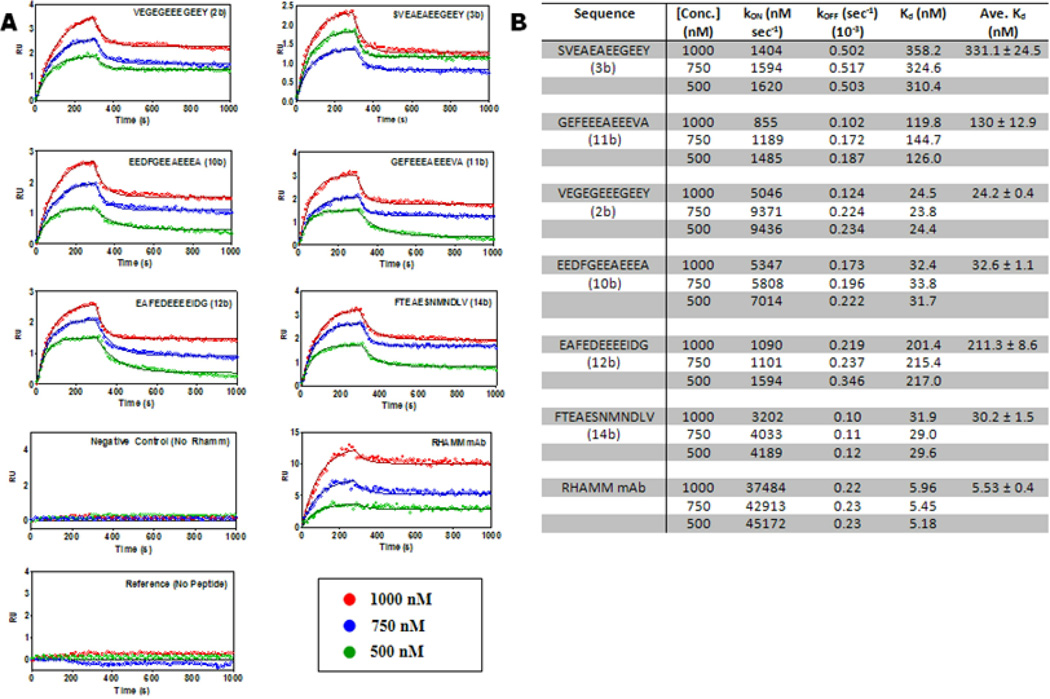
Figure 3. The kinetic profiles of peptide ligand candidates.
(A) Seven sets of sensograms showing global fits to each specific peptide-RHAMM interaction. Negative control (no RHAMM) and reference sensograms (no peptide) plot are shown. Each set of sensogram corresponds to the responses of three RHAMM concentrations (1000 nM, 750 nM and 500 nM) interacting with immobilized peptide (2b, 3b, 10b, 11b, 12b and 14b, Figure 1). The lines depict global 1:1 interaction curve-fitting models for each of the seven interactions. (B) Kinetic profiles of selected tubulin derived peptides showing calculated kON, kOFF and KD. Errors are standard deviation of the mean KD.
Affinity of CTT peptides for RHAMM
To further characterize the association of these 6 CTT peptides with the HA binding region of RHAMM, peptides were modified with a cysteine and covalently attached to the SPR sensor plate. Recombinant HA binding region of RHAMM was passed over the derivatized surface at different concentrations (Figure 3A). Peptide/RHAMM sensograms were fitted into a kinetic model for a 1:1 Langmuir binding model. The average KD for six peptides was calculated from the different RHAMM concentrations used (Figure 3B). Peptides 2b (VEGEGEEEGEEY, KD = 24 nM), 10b (EEDFGEEAEEEA, KD = 32 nM), and 14b (FTEAESNMNDLV, KD = 30 nM) showed dissociation constants in the low nanomolar range indicating strong affinity to RHAMM.
Competitive displacement of tubulin-derived peptides by HA
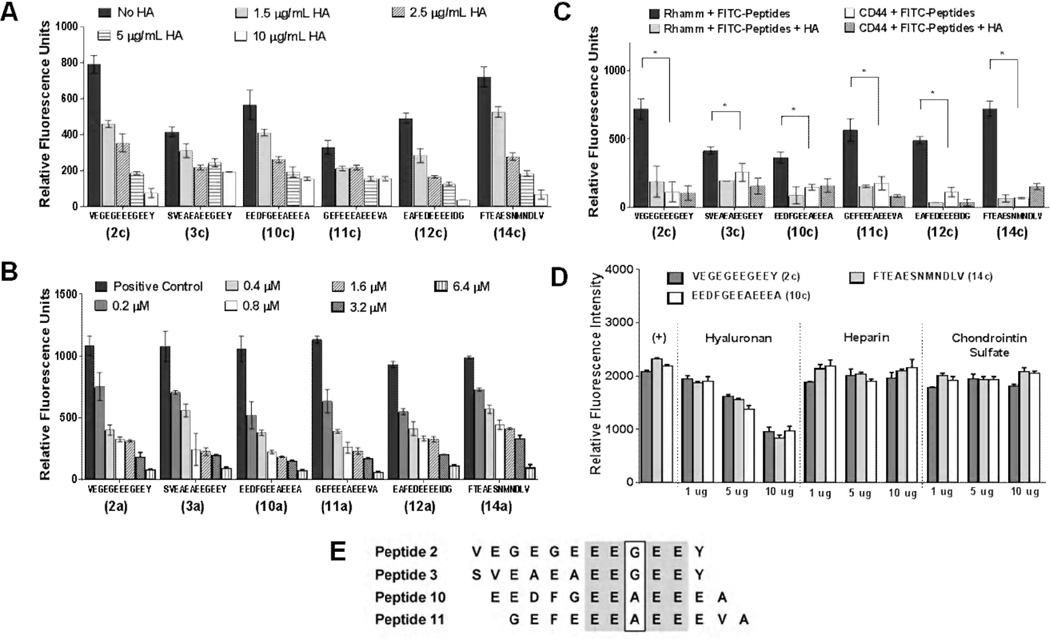
All six peptides were then tested for their ability to compete with HA for binding to RHAMM using ELISA (Figure 4). The ability of unlabeled HA to compete with fluorescein-labeled peptides for binding to RHAMM was first evaluated (Figure 4A). A concentration dependent decrease in binding of fluorescein-peptides was observed for all six candidate peptides, but HA most efficiently competed with peptides 2c, 12c and 14c for binding to RHAMM (Figure 4A). Increasing concentrations of unlabeled peptides were next used to compete with AlexaFluor 647-conjugated HA for binding to RHAMM (Figure 4B). Displacement of labeled-HA by non-fluorescent peptides was observed for all peptides so that all peptides effectively blocked labeled-HA binding to RHAMM at the highest concentration used (6.4µM). Alignment analyses (Cobalt Multiple Alignment Tool, www.ncbi.nlm.nih.gov/protein) indicate that peptides 2, 3, 10 and 11 are highly related with a conserved EEXEE sequence (Figure 4E). However peptides such as 14 do not appear to be related as determined by these types of alignment analyses. ELISA analyses also show that other glycosaminoglycans such as chondroitin sulfate and heparin do not compete with the peptides for binding to recombinant RHAMM HA binding region (Figure 4D). Overall, these results suggest that peptides 12 and 14 are most efficient at competing with HA for binding to RHAMM.
Figure 4. Binding properties of ligand candidates.
(A) Competitive displacement of six selected fluorescein-labelled peptides (2c, 3c, 10c, 11c, 12c and 14c) by HA (220 kDA) in different concentrations to immobilized recombinant RHAMM; (B) Competitive displacement of dye-labeled HA by non-labeled tubulin-derived peptides (2a, 3a, 10a, 11a, 12a and 14a) at different concentrations using ELISA. (C) ELISA binding assay of FITC-conjugated peptides (2c, 3c, 10c, 11c, 12c and 14c) to recombinant CD44 and RHAMM. The negative control (no immobilized RHAMM or CD44) was subtracted for each measurement. 10 µg/mL of the peptides and HA were used for these assays. (D) The ability of HA to compete with FITC-peptides for binding to recombinant RHAMM was compared to heparin and chondroitin sulfate. Only HA effectively competed with the peptides for binding to RHAMM. Each glycosaminoglycan was competed against FITC-peptides at three different concentrations (1 µg/mL, 5 µg/mL and 10 µg/mL). All data show the mean of three measurements ± S.E.M. in three independent experiments. Significant differences (p < 0.05) are marked by asterisks. (E) Sequence comparison of peptides 2, 3, 10 and 11 using Cobalt Multiple Alignment Tool. Identical sequences are shown in grey. A pentapeptide motif, EEXEE (where X is A or G) is present in these peptides.
Specificity for RHAMM versus CD44
To determine if any of the peptides, which bind to RHAMM and block HA/RHAMM interactions, also bind to CD44, we performed ELISA assays with functionalized CD44 vs. RHAMM surfaces. For these assays we utilized only the HA binding regions of these proteins and quantified their binding to fluorescein-peptides (Figure 4C). As expected, fluorescein-peptides bound to RHAMM were competed for binding by HA. However, binding of fluorescein-peptides to CD44 was 6–8 fold lower than to RHAMM and HA only affected binding of two peptides to CD44 (peptides 11c and 12c). Thus, the binding of the peptides is much greater to RHAMM than to CD44.
Fluorescein-peptides bind to and are internalized by tumor cells in a RHAMM-dependent manner
To determine if the CTT-derived peptides interact with HA receptors expressed and displayed on intact cells, a cellular fluorescence assay was performed using human breast tumor (MDA-MB-231) and prostate tumor (PC3MLN4) cell lines. MDA-MB-231 cells express RHAMM and CD44 31, 39, bind and internalize dye-conjugated HA6, 40, and require HA for motility and invasion 31, 41. The prostate cancer cell line, PC3ML series (N4) was also chosen for these experiments since it expresses both CD44 and RHAMM 42, 43, and binds and internalizes Cy5 dye-conjugated HA (data not shown). Fluorescein-peptides 2c, 10c and 14c were chosen for these experiments based upon their affinity/selectivity for RHAMM, ability to compete with RHAMM/HA interactions and stability in serum (data not shown). All 3 FITC peptides bound to and were internalized by both MDA-MB-231 cells (Figure 5) and PC3MLN4 cells (Figure 6), as detected by confocal microscopy. Quantification of fluorescein-peptide uptake by image analysis confirmed these observations (Figure 5B and 6B).
Figure 5. Uptake of fluorescein-conjugated peptides in MDA-MB-231 breast cancer cells.
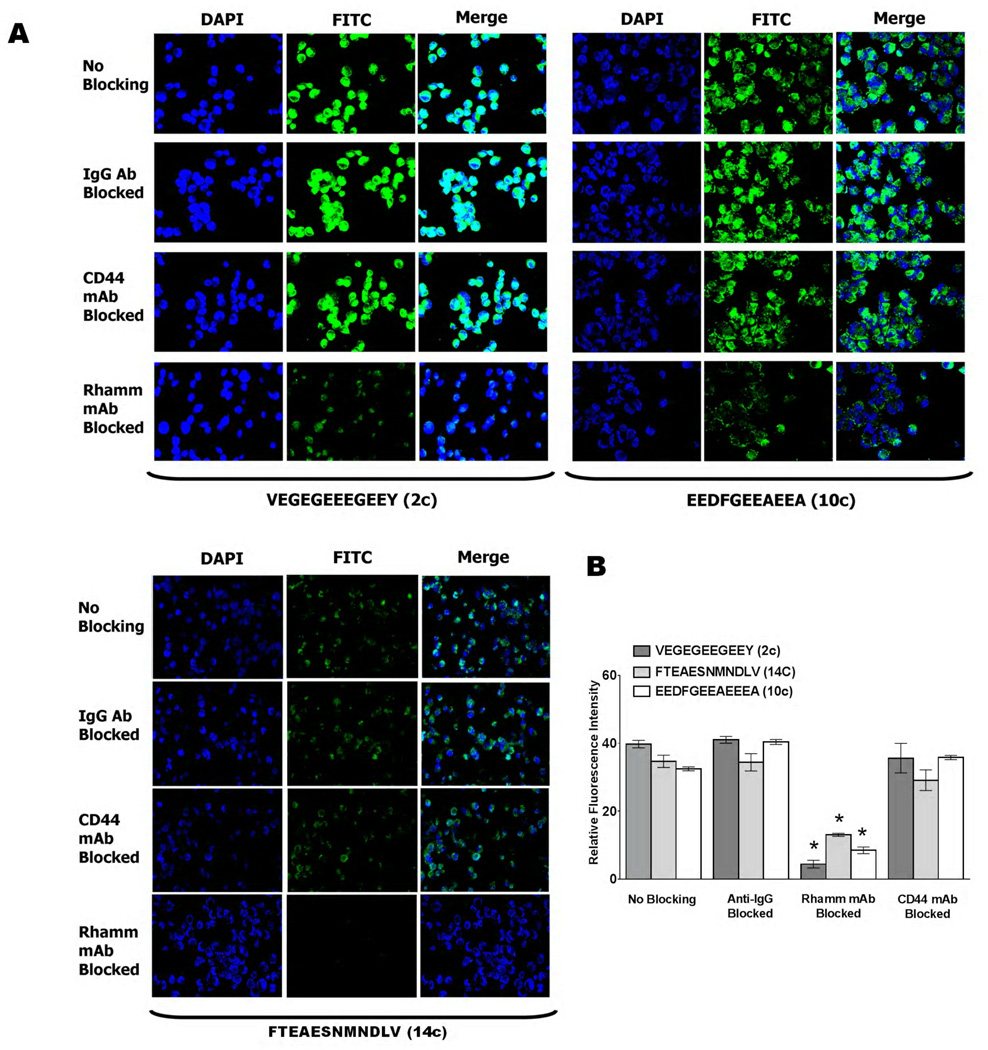
(A) Confocal microscopy of FITC-peptide uptake by MDA-MB-231 tumor cells. Images show uptake of peptides 2c, 10c and 14c. Nuclei are shown in blue (DAPI) while FITC-conjugated peptides are shown in green. Prior to addition of dye-conjugated peptides, cells were incubated with IgG, anti-RHAMM or anti-CD44 antibodies. Cells, which received no antibody treatment (no blocking) or cells treated with non-immune IgG served as controls. A reduction in fluorescence (FITC channel) was observed when cells are blocked with anti-RHAMM antibodies while no significant decrease in FITC signal was observed for cells treated with anti-CD44 antibodies. (B) Uptake in MDA-MB-231 tumor cells was quantified using ImageJ software, as described in Experimental Procedures. Values are mean fluorescence ± S.E.M., and data were analyzed using one-way ANOVA. Significant differences (p < 0.05) are marked by asterisks.
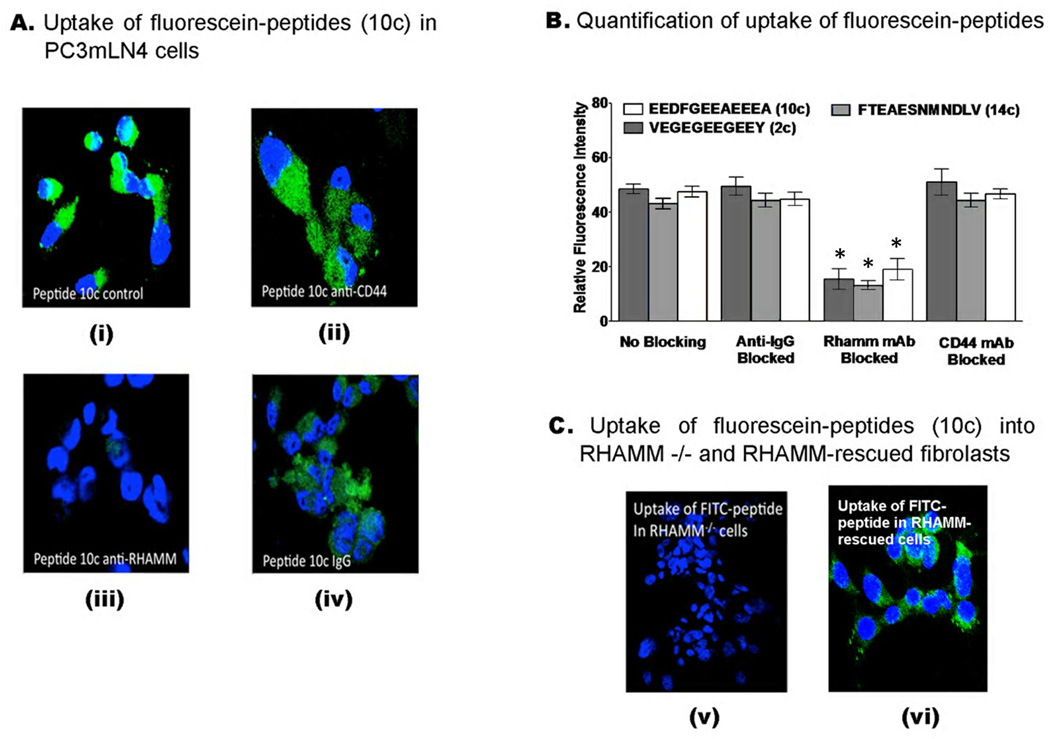
Figure 6. Uptake of FITC-peptides by PC3MLN4 prostate cancer cells.
(A) Confocal images of the uptake of FITC-conjugated peptide (10c) in prostate tumour cells using two-channel fluorescence confocal microscopy. Nuclei are shown in blue (DAPI) while FITC-conjugated peptides are shown in green. Prior to addition of dye-conjugated peptides, cells were incubated with non-immune IgG (iv), anti-RHAMM (iii) or anti-CD44 (ii) antibodies. Cells, which received no treatment (i) or were treated with non-immune IgG (iv) served as controls. A reduction in green channel fluorescence (FITC) was observed when cells are blocked with anti-RHAMM while no detectable decrease in FITC signal was observed for cells treated with anti-CD44 or IgG. (B) Uptake of 2c, 10c and 14c in PC3MLN4 tumor cells was quantified using ImageJ software, as described in Experimental Procedures. Values are mean fluorescence ± S.E.M Data were analyzed using one-way ANOVA. Significant differences (p < 0.05) are marked by asterisks. (C) FITC peptide (10c) uptake was not detectable in RHAMM−/− fibroblasts (v) but was observed when these cells were rescued by expressing a full length RHAMM cDNA (vi).
Consistent with the binding properties of these peptides, an anti-RHAMM monoclonal antibody (Clone 6B7D8, Figure 3B, KD for RHAMM = 5.5 nM) significantly blocked the binding and uptake of the fluorescein-peptides in both MDA-MB-231 and PC3MLN4 cells (Figure 5A,B and Figure 6A,B) while an anti-CD44 antibody (IM-7), did not. Non-immune IgG was used as a negative control for these experiments and did not affect uptake of fluorescein-peptides relative to controls (no blocking). To verify this result, uptake of FITC peptides 2c, 10c, and 14c were monitored in RHAMM−/− fibroblasts and compared to RHAMM-rescued fibroblasts 17. As shown in Figure 6C, FITC-labelled peptides did not bind to and were not internalized by RHAMM−/− fibroblasts (10c shown). However, expression of RHAMM in rescued fibroblasts resulted in an ability to bind and internalize fluorescein-peptides providing strong evidence that binding/internalization of these peptides is RHAMM-dependent. Collectively these results indicate a specificity of the peptides for extracellular RHAMM and further suggest that they are acting as HA oligosaccharide mimics.
Tubulin-derived peptide 14a inhibits invasion of prostate cancer cells
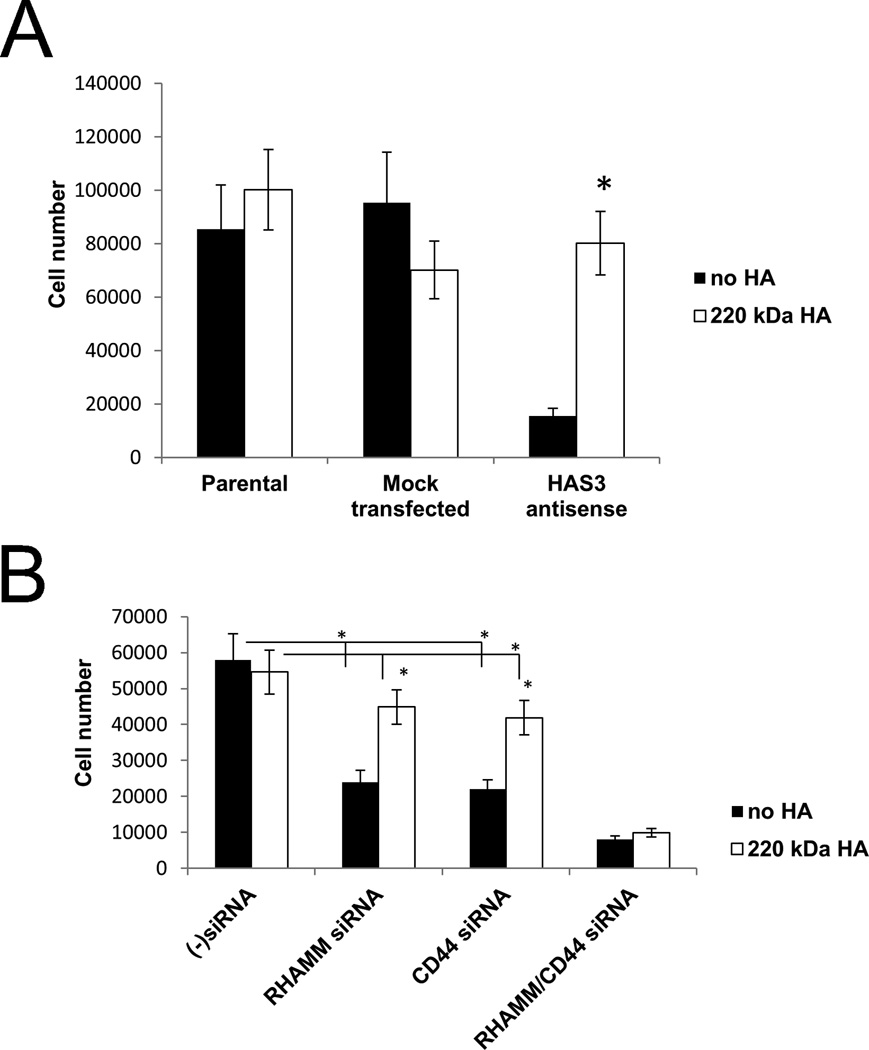
Anchorage independent survival and proliferation of PC3MLN4 cells requires HA 42–44 (Figure 7A). Thus, knockdown of HAS3 expression in PC3-MLN4 tumor cells, which is the major HAS isoform expressed by these tumor cells,44 strongly reduces HA synthesis and anchorage independent proliferation. This can be rescued by the addition of HA to the cultures (both 220 kDa Figure 7A or 800 kDa not shown). These cells express high levels of both RHAMM and CD44 by Western Analysis (not shown). To determine whether HA stimulated proliferation of PC3-MLN4 cells requires HA/RHAMM and/or HA/CD44 interactions, expression of CD44 and/or RHAMM was inhibited by siRNA. Single knockdown of either CD44 or RHAMM resulted in a partial but significant reduction in PC3MLN4 proliferation in the absence of exogenously supplied HA. This inhibition could be partially reversed by the addition of exogenous HA. By contrast, dual inhibition of RHAMM and CD44 resulted in almost complete inhibition of growth in these cultures, which could not be reversed by the addition of exogenous HA. The results indicate that both RHAMM and CD44 function as HA receptors in promoting growth of these tumor cells. Since HA can partially reverse growth inhibition observed when only one of these receptors is lost, the results suggest that the response of each receptor to HA are functionally linked in these cells. This is supported by other reports in which these two HA receptors have been functionally linked 17, 45. However, peptide 14a had marginal effects in these assays, possibly due to this peptide interacting with only cell-surface RHAMM.
Figure 7. Knock-down of HAS3, RHAMM and CD44 reduces anchorage independent growth of PC3MLN4 prostate tumor cells.
(A) PC3MLN4 cells in which HAS3 expression was inhibited using an HAS3 antisense vector or a mock vector were suspended in methylcellulose in the presence of 200 kD HA. This antisense vector inhibited HA synthesis by these cells both by ELISA and analysis of HA coat formation. After seven days, the cells were harvested by solubilizing the methylcellulose in PBS and counted. The results shown are the means of triplicate culture wells +/− S.E.M * <0.002. (B) HA stimulated growth of human prostate carcinoma cells requires both RHAMM and CD44. Expressions of CD44 and RHAMM were inhibited by transfection of specific siRNAs to PC3MLN4 cells. Twenty-four hours after transfection cells were harvested and 1.5 X 104 cells were seeded into methylcellulose cultures in the presence or absence of HA (800 kDa). The gels were solubilized using PBS seven days after plating and the cells were counted. The results shown are the means of triplicate culture wells +/− S.E.M. P < 0.01 Control siRNAs versus either RHAMM or CD44 siRNAs; P = 0.08 or 0.07 for difference between RHAMM siRNA or CD44 siRNA vs. HA addition; P < 0.002 for double siRNA versus non-transfected; P = 0.44 comparing double siRNA inhibition vs HA addition to those cells.
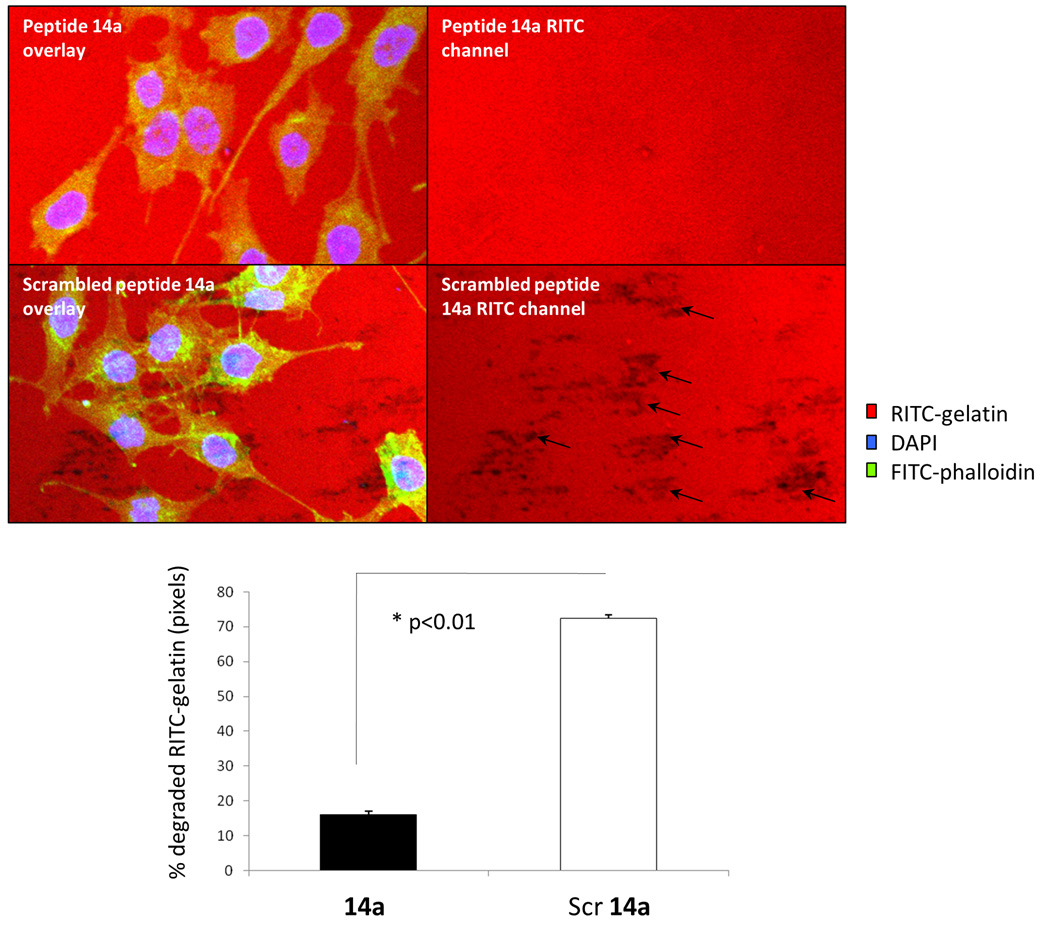
Since both cell-surface RHAMM and CD44 are strongly implicated in tumour invasion and motility46–50, we next evaluated the ability of peptide 14a to block PC3MLN4 invasion using a RITC-collagen degradation assay. Peptide 14a strongly inhibited the ability (80%) of these prostate cancer cells to degrade collagen (Figure 8, arrows indicating representative areas of degradation) when compared to the effects of the scrambled peptide control when used at 10 µg/mL. Importantly, this peptide had no observed inhibition of cell adhesion on these substrates (not shown), indicating that the inhibitory effects is likely due to specific alterations in the invasive phenotype of these cells.
Figure 8. HA mimetic peptide 14a reduces gelatin degradation by PC3MLN4 cells.
Cells were plated at subconfluency on fluorescently labeled gelatin (red) as described in Experimental Procedures. Cultures were incubated in growth medium containing either 10 µg/mL peptide 14a or 10 µg/mL scrambled control peptide. After 48 hrs, cells were fixed and stained with DAPI (blue) and fluorescently labeled phalloidin (green). Confocal images were taken and used to count number of degraded areas per cell (arrows indicate representative areas of degradation). Graph represent mean +/− SE of 5 images.
Discussion
RHAMM is an oncogene that is commonly over-expressed in human cancers and this over-expression is associated with poor outcome in breast, gastric, colorectal and other cancers 1, 13, 14. Increased accumulation of HA, the extracellular ligand for RHAMM, is also a prognostic factor for poor outcome in breast and prostate cancers 6, 51. Thus, peptide ligands, which mimic HA in that they compete with HA for binding to RHAMM, appear to act as antagonists for RHAMM and as such may be useful for both diagnostic and therapeutic purposes in cancers and other diseases.
In this study we developed peptides, rather than previously reported polysaccharide ligands 5, 43, to target the HA binding domain of RHAMM. We reasoned that dual binding might occur because the microtubule and HA binding region in the carboxyl terminus of RHAMM overlap 15, and because binding to both HA and tubulin is largely based upon ionic interactions. Therefore we predicted that the negatively charged CTT tubulin sequences would associate with the HA binding region of RHAMM and be susceptible to competition for this interaction by HA. Intriguingly, recent studies suggest that cell surface proteins can bind to exposed cytoskeleton proteins in damaged or apoptosing cells and that this interaction is specific, of high affinity, and essential for detecting damaged cells 52, 53. Thus, our method for identifying ligands that bind to “intracellular” cytoskeleton proteins and to “extracellular” RHAMM may have an important physiological counterpart and, besides offering a novel therapeutic approach, could also help to dissect the novel inside/outside functions of both RHAMM and HA.
Database searches and pair wise comparisons between RHAMM and tubulin associated proteins (e.g. MAPs) that bind to CTT tubulin sequences were used to identify sequences that also have affinity for RHAMM. Although specific interactions between RHAMM and novel ligands were thus deduced, our ability to identify high affinity RHAMM-specific peptides that mimic HA was greatly aided by the use of cell based assays where the involvement of RHAMM was queried with function blocking antibodies or genetic deletion of the target protein. Initially 17 peptides of twelve amino acid length were evaluated using SPR (Figure 3B) and six of these were identified as interacting with RHAMM (Figure 3C). Surprisingly, CTT peptides only competed with HA and not with other anionic glycosaminoglycans, thus further conferring specificity to the HA binding site (Figure 4D). These CTT peptides showed moderate stability in bovine serum (approximately 110–250 minutes half-life), which is predicted to be sufficiently long for in vivo imaging. Further analyses indicated that three of these ligands interact with KD values of 24–32 nM, which is within an acceptable affinity range for in vivo analyses. We demonstrated that the three HA peptide mimics with the highest affinity for RHAMM, bound to and were taken up by breast and prostate cancer cells in a RHAMM dependent manner. The relative lack of CD44 involvement, as detected by an inability to block peptide uptake with anti-CD44 antibodies, is consistent with major differences in the structure and mechanisms, in particular the lower reliance on ionic interactions, by which CD44 binds to HA in comparison to RHAMM 54. Intriguingly, the results of this study predict an ability of MAPs that resemble RHAMM to bind to HA. The existence of intracellular HA has been reported by a number of groups 55–57 and careful analysis has also demonstrated its presence on microtubules. Whether or not the RHAMM-like sequences in MAPs bind to HA remains to be further investigated. A recurring pentapeptide motif was found in most of the CTT peptides identified in the present study. The motif, EEXEE (X = G or A) is present in the peptides with the highest affinity for RHAMM. Our present results are in agreement with previous reports that the short sequence EEGEE 58–60 could be involved in tubulin and MAP binding, a family of proteins that shares sequence homology to RHAMM. Despite this role of acidic functional groups in HA:RHAMM interactions, our results also indicate additional, possibly conformational influences. For example it is surprising that peptides containing DEXEEZ (as seen in peptides 1 and 4) and EEXEDZ (e.g. peptide 9) motifs failed the initial screening, suggesting that Asp residues within the first and fifth sequence cannot substitute the acidic Glu residue within this motif. This predicts that the CTT interaction with RHAMM is mediated by both electrostatic forces and conformational effects.
Conclusion
In this study we describe the discovery of novel ligands, which interact with the HA binding domain of RHAMM but not CD44 and which can be competed in this binding by HA, collectively suggesting that they are functioning as HA oligosaccharides peptide mimetics specific for RHAMM. RHAMM plays a role in a number of diseases including cancers, diabetes and arthritis. Therefore, these peptides may serve as antagonists that could block the RHAMM-HA interaction, thus limiting the transforming potential of RHAMM. For example, these HA mimetic peptides inhibit the invasion of these highly metastatic PC3MLN4 prostate cancer cells, which is a property associated with malignant tumor progression.
Supplementary Material
Acknowledgments
This work is supported by the Ontario Institute for Cancer Research (OICR, to LL), the Canadian Breast Cancer Foundation (CBCF, to LL) and the Pamela Greenaway-Kohlmeier Translational Breast Cancer Research Unit (LL, ET) and the NIH (#1RO1CA119092) to JBM and ET.
Footnotes
These might include comments relevant to but not central to the matter under discussion, limited experimental and spectral data, and crystallographic data.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
References
- 1.Jiang D, Liang J, Noble PW. Physiol Rev. 2011;91:221–264. doi: 10.1152/physrev.00052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lennon FE, Singleton PA. Am J Physiol Lung Cell Mol Physiol. 2011;301:L137–L147. doi: 10.1152/ajplung.00071.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misra S, Heldin P, Hascall VC, Karamanos NK, Skandalis SS, Markwald RR, Ghatak S. FEBS J. 2011;278:1429–1443. doi: 10.1111/j.1742-4658.2011.08071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murano E, Perin D, Khan R, Bergamin M. Nat Prod Commun. 2011;6:555–572. [PubMed] [Google Scholar]
- 5.Toole BP, Slomiany MG. Semin Cancer Biol. 2008;18:244–250. doi: 10.1016/j.semcancer.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veiseh M, Turley EA. Integr Biol (Camb) 2011;3:304–315. doi: 10.1039/c0ib00096e. [DOI] [PubMed] [Google Scholar]
- 7.Gao F, Yang CX, Mo W, Liu YW, He YQ. Clin Invest Med. 2008;31:E106–E116. doi: 10.25011/cim.v31i3.3467. [DOI] [PubMed] [Google Scholar]
- 8.Kouvidi K, Berdiaki A, Nikitovic D, Katonis P, Afratis N, Hascall VC, Karamanos NK, Tzanakakis GN. J Biol Chem. 2011;286:38509–38520. doi: 10.1074/jbc.M111.275875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manzanares D, Monzon ME, Savani RC, Salathe M. Am J Respir Cell Mol Biol. 2007;37:160–168. doi: 10.1165/rcmb.2006-0413OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson DG. Immunol Rev. 2009;230:216–231. doi: 10.1111/j.1600-065X.2009.00803.x. [DOI] [PubMed] [Google Scholar]
- 11.Milner CM, Higman VA, Day AJ. Biochem Soc Trans. 2006;34:446–450. doi: 10.1042/BST0340446. [DOI] [PubMed] [Google Scholar]
- 12.Ziebell MR, Prestwich GD. J Comput Aided Mol Des. 2004;18:597–614. doi: 10.1007/s10822-004-5433-8. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, Casalegno-Garduno R, Chen H, Schmitt A, Schmitt M, Maxwell CA. ScientificWorldJournal. 2010;10:1244–1257. doi: 10.1100/tsw.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell CA, McCarthy J, Turley E. J Cell Sci. 2008;121:925–932. doi: 10.1242/jcs.022038. [DOI] [PubMed] [Google Scholar]
- 15.Turley EA, Naor D. Front Biosci. 2012;17:1775–1794. doi: 10.2741/4018. [DOI] [PubMed] [Google Scholar]
- 16.Slevin M, Krupinski J, Gaffney J, Matou S, West D, Delisser H, Savani RC, Kumar S. Matrix Biol. 2007;26:58–68. doi: 10.1016/j.matbio.2006.08.261. [DOI] [PubMed] [Google Scholar]
- 17.Tolg C, Hamilton SR, Nakrieko KA, Kooshesh F, Walton P, McCarthy JB, Bissell MJ, Turley EA. J Cell Biol. 2006;175:1017–1028. doi: 10.1083/jcb.200511027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naor D, Nedvetzki S, Walmsley M, Yayon A, Turley EA, Golan I, Caspi D, Sebban LE, Zick Y, Garin T, Karussis D, Assayag-Asherie N, Raz I, Weiss L, Slavin S. Ann N Y Acad Sci. 2007;1110:233–247. doi: 10.1196/annals.1423.025. [DOI] [PubMed] [Google Scholar]
- 19.Casalegno-Garduno R, Schmitt A, Schmitt M. Expert Rev Vaccines. 2011;10:785–799. doi: 10.1586/erv.11.56. [DOI] [PubMed] [Google Scholar]
- 20.Tabarkiewicz J, Giannopoulos K. Transplant Proc. 2010;42:3293–3296. doi: 10.1016/j.transproceed.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 21.Du YC, Chou CK, Klimstra DS, Varmus H. Proc Natl Acad Sci U S A. 2011;108:16753–16758. doi: 10.1073/pnas.1114022108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall CL, Yang B, Yang X, Zhang S, Turley M, Samuel S, Lange LA, Wang C, Curpen GD, Savani RC, Greenberg AH, Turley EA. Cell. 1995;82:19–26. doi: 10.1016/0092-8674(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Chang MC, Zylka D, Turley S, Harrison R, Turley EA. J Biol Chem. 1998;273:11342–11348. doi: 10.1074/jbc.273.18.11342. [DOI] [PubMed] [Google Scholar]
- 24.Mohapatra S, Yang X, Wright JA, Turley EA, Greenberg AH. J Exp Med. 1996;183:1663–1668. doi: 10.1084/jem.183.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. Curr Biol. 2004;14:1801–1811. doi: 10.1016/j.cub.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, Walter JC, Livingston DM. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell CA, Keats JJ, Crainie M, Sun X, Yen T, Shibuya E, Hendzel M, Chan G, Pilarski LM. Mol Biol Cell. 2003;14:2262–2276. doi: 10.1091/mbc.E02-07-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolg C, Hamilton SR, Morningstar L, Zhang J, Zhang S, Esguerra KV, Telmer PG, Luyt LG, Harrison R, McCarthy JB, Turley EA. J Biol Chem. 2010;285:26461–26474. doi: 10.1074/jbc.M110.121491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chau MF, Radeke MJ, de Ines C, Barasoain I, Kohlstaedt LA, Feinstein SC. Biochemistry. 1998;37:17692–17703. doi: 10.1021/bi9812118. [DOI] [PubMed] [Google Scholar]
- 30.Ziebell MR, Zhao ZG, Luo B, Luo Y, Turley EA, Prestwich GD. Chem Biol. 2001;8:1081–1094. doi: 10.1016/s1074-5521(01)00078-3. [DOI] [PubMed] [Google Scholar]
- 31.Hamilton SR, Fard SF, Paiwand FF, Tolg C, Veiseh M, Wang C, McCarthy JB, Bissell MJ, Koropatnick J, Turley EA. J Biol Chem. 2007;282:16667–16680. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang B, Yang BL, Savani RC, Turley EA. EMBO J. 1994;13:286–296. doi: 10.1002/j.1460-2075.1994.tb06261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milner CM, Tongsoongnoen W, Rugg MS, Day AJ. Biochem Soc Trans. 2007;35:672–676. doi: 10.1042/BST0350672. [DOI] [PubMed] [Google Scholar]
- 34.Assmann V, Marshall JF, Fieber C, Hofmann M, Hart IR. J Cell Sci. 1998;111(Pt 12):1685–1694. doi: 10.1242/jcs.111.12.1685. [DOI] [PubMed] [Google Scholar]
- 35.Nogales E. Annu Rev Biophys Biomol Struct. 2001;30:397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- 36.Downing KH, Nogales E. Cell Struct Funct. 1999;24:269–275. doi: 10.1247/csf.24.269. [DOI] [PubMed] [Google Scholar]
- 37.Manning J, Kumar S. Int J Biochem Cell Biol. 2007;39:7–11. doi: 10.1016/j.biocel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Sackett DL, Bhattacharyya B, Wolff J. J Biol Chem. 1985;260:43–45. [PubMed] [Google Scholar]
- 39.Bourguignon LY, Wong G, Earle C, Krueger K, Spevak CC. J Biol Chem. 2010;285:36721–36735. doi: 10.1074/jbc.M110.162305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veiseh M, Breadner D, Ma J, Akentieva N, Savani RC, Harrison R, Mikilus D, Collis L, Gustafson S, Lee TY, Koropatnick J, Luyt LG, Bissell MJ, Turley EA. Biomacromolecules. 2012;13:12–22. doi: 10.1021/bm201143c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montgomery N, Hill A, McFarlane S, Neisen J, O’Grady A, Conlon S, Jirstrom K, Kay EW, Waugh D. Breast Cancer Res. 2012;14:R84. doi: 10.1186/bcr3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lokeshwar VB, Lopez LE, Munoz D, Chi A, Shirodkar SP, Lokeshwar SD, Escudero DO, Dhir N, Altman N. Cancer Res. 2010;70:2613–2623. doi: 10.1158/0008-5472.CAN-09-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benitez A, Yates TJ, Lopez LE, Cerwinka WH, Bakkar A, Lokeshwar VB. Cancer Res. 2011;71:4085–4095. doi: 10.1158/0008-5472.CAN-10-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson MA, Wilson CM, McCarthy JB. Am J Pathol. 2002;161:849–857. doi: 10.1016/S0002-9440(10)64245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikitovic D, Kouvidi K, Karamanos NK, Tzanakakis GN. Biomed Research International. 2013 doi: 10.1155/2013/929531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tolg C, McCarthy JB, Yazdani A, Turley EA. Biomed Res Int. 2014:103923. doi: 10.1155/2014/103923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heldin P, Basu K, Olofsson B, Porsch H, Kozlova I, Kahata K. J Biochemistry. 2013;154:395–408. doi: 10.1093/jb/mvt085. [DOI] [PubMed] [Google Scholar]
- 48.Meier C, Spitschak A, Abshagen K, Gupta S, Mor JM, Wolkenhauer O, Haier J, Vollmar B, Alla V, Putzer BM. J Pathol. 2014;234:351–64. doi: 10.1002/path.4400. [DOI] [PubMed] [Google Scholar]
- 49.Du YC, Chou CK, Klimstra DS, Varmus H. Proc Natl Acad Sci U S A. 2011;108:16753–8. doi: 10.1073/pnas.1114022108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Wu Y, Wang H, Zhang Y, Mei L, Fang X, Zhang X, Zhang F, Chen H, Liu Y, Jiang Y, Sun S, Zheng Y, Li N, Huang L. Proc Natl Acad Sci U S A. 2014;111:E89–E98. doi: 10.1073/pnas.1319190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tammi RH, Kultti A, Kosma VM, Pirinen R, Auvinen P, Tammi MI. Semin Cancer Biol. 2008;18:288–295. doi: 10.1016/j.semcancer.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Podor TJ, Singh D, Chindemi P, Foulon DM, McKelvie R, Weitz JI, Austin R, Boudreau G, Davies R. J Biol Chem. 2002;277:7529–7539. doi: 10.1074/jbc.M109675200. [DOI] [PubMed] [Google Scholar]
- 53.Zhang JG, Czabotar PE, Policheni AN, Caminschi I, Wan SS, Kitsoulis S, Tullett KM, Robin AY, Brammananth R, van Delft MF, Lu J, O’Reilly LA, Josefsson EC, Kile BT, Chin WJ, Mintern JD, Olshina MA, Wong W, Baum J, Wright MD, Huang DC, Mohandas N, Coppel RL, Colman PM, Nicola NA, Shortman K, Lahoud MH. Immunity. 2012;36:646–657. doi: 10.1016/j.immuni.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Day AJ, Prestwich GD. J Biol Chem. 2002;277:4585–4588. doi: 10.1074/jbc.R100036200. [DOI] [PubMed] [Google Scholar]
- 55.Collis L, Hall C, Lange L, Ziebell M, Prestwich R, Turley EA. FEBS Lett. 1998;440:444–449. doi: 10.1016/s0014-5793(98)01505-1. [DOI] [PubMed] [Google Scholar]
- 56.Evanko SP, Parks WT, Wight TN. J Histochem Cytochem. 2004;52:1525–1535. doi: 10.1369/jhc.4A6356.2004. [DOI] [PubMed] [Google Scholar]
- 57.Wang A, de la Motte C, Lauer M, Hascall V. FEBS J. 2011;278:1412–1418. doi: 10.1111/j.1742-4658.2011.08069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Devred F, Barbier P, Lafitte D, Landrieu I, Lippens G, Peyrot V. Methods Cell Biol. 2010;95:449–480. doi: 10.1016/S0091-679X(10)95023-1. [DOI] [PubMed] [Google Scholar]
- 59.Mukhopadhyay R, Kumar S, Hoh JH. Bioessays. 2004;26:1017–1025. doi: 10.1002/bies.20088. [DOI] [PubMed] [Google Scholar]
- 60.Paschal BM, Obar RA, Vallee RB. Nature. 1989;342:569–572. doi: 10.1038/342569a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.