Abstract
Malaria remains one of the most prevalent infectious diseases and results in significant mortality. Isothermal amplification (loop-mediated isothermal amplification) is used to detect malarial DNA at levels of ~1 parasite/µL blood in ≤30 minutes without the isolation of parasite nucleic acid from subject’s blood or saliva. The technique targets the mitochondrial cytochrome oxidase subunit 1 gene and is capable of distinguishing Plasmodium falciparum from Plasmodium vivax. Malarial diagnosis by the gold standard microscopic examination of blood smears is generally carried out only after moderate-to-severe symptoms appear. Rapid diagnostic antigen tests are available but generally require infection levels in the range of 200–2,000 parasites/µL for a positive diagnosis and cannot distinguish if the disease has been cleared due to the persistence of circulating antigen. This study describes a rapid and simple molecular assay to detect malarial genes directly from whole blood or saliva without DNA isolation.
Keywords: malaria, point-of-care (POC), loop-mediated isothermal amplification (LAMP), molecular detection, Plasmodium falciparum, Plasmodium vivax
Introduction
In 2013, there were almost 200 million malarial infections that resulted in approximately one-half million deaths, 90% of which occurred in Africa.1 A practical alternative to the vaccination approach2 to eradicate malaria is the Test-and-Treat paradigm currently being evaluated for the eradication of HIV,3 which predicts that if all symptomatic and asymptomatic individuals in a region are tested and all of those who test positive are immediately treated, the burden of disease in that area will decrease. We believe that the same approach is feasible for malaria by deploying an inexpensive point-of-care (POC) diagnostic test for Plasmodium DNA followed by appropriate therapeutic intervention. Currently, malaria is detected by (1) microscopic examination of thick and thin blood smears, a laborious process that is time-consuming and requires trained personnel; (2) laboratory-based polymerase chain reaction (PCR) tests, which are expensive, time-consuming, and require specialized equipment and trained personnel; or (3) rapid antigen-based tests (RDTs), which are generally less sensitive.
Commercially available RDTs for malarial antigens were first introduced in 1994 and are readily available. The World Health Organization has noted the shortcomings of the available RDTs to include (1) poor quality of some products, (2) inability to detect mutant forms of plasmodia that have deleted the histidine-rich protein 2 target gene, (3) low sensitivity (500–1,000 parasites/µL), and (4) false-positive results long after the infection is cleared due to residual antigen in the bloodstream.1
Isothermal nucleic acid amplification methods such as loop-mediated isothermal amplification (LAMP) make use of a polymerase with built-in strand displacement capabilities which does not require a high-temperature denaturation step such as conventional PCR and loop primers to increase the sensitivity. This is a major advantage for POC devices since the entire amplification reaction typically occurs under isothermal conditions at 65°C. The progress of the real-time reaction can be visualized using an intercalating fluorescent dye such as SYBR Green, which preferentially binds to double-stranded DNA. Our goal was to design a POC LAMP nucleic acid test for the detection of parasites using both blood and oral fluid, and it requires no sample purification. The assay is completed in 30 minutes with a sensitivity 10–100-fold greater than conventional PCR4,5 and 500–1,000 times more sensitive than antigen detection. Other investigators have reported the detection of malarial DNA extracted from blood, saliva, or urine samples,6–9 but mostly utilized purified parasite DNA. Previous reports using LAMP targeted a conserved sequence in parasite 18S rRNA.9–11 However, we report here that targeting the mitochondrial cytochrome C oxidase subunit 1 gene improves the assay’s specificity and unlike previously described assays can discriminate between Plasmodium falciparum and Plasmodium vivax infection.12,13
Methods
Malaria-positive blood
The following reagents were obtained through the Malaria Research and Reference Reagent Resource Center (MR4) as part of the Biodefense and Emerging Infections Research Resources Repository (BEI Resources), National Institute of Allergy and Infectious Diseases (NIAID), and National Institute of Health: P. falciparum FCR-3/Gambia Subline F-86, MRA-731, deposited by W Trager; P. falciparum GB4, MRA-925, deposited by Karen Hayton, Tom Wellems (NIAID); P. falciparum 3D7, MRA-102, deposited by DJ Carucci; P. falciparum 7C46, MRA-172, deposited by TE Wellems; P. vivax Chesson, MRA-383, deposited by WE Collins; P. vivax NAM/CDC, MRA-384, deposited by WE Collins; Genomic samples—MRA 731G, MRA 925G, MRA 102G, and MRA 172; P. falciparum Genomic DNA from P. falciparum FCR-3/Gambia, MRA-731G; Genomic DNA from P. falciparum GB4, MRA-925G; P. falciparum Genomic DNA from P. falciparum 3D7, MRA-102G; and P. falciparum Genomic DNA—P. falciparum 7C46, MRA-172G.
Whole mouth-stimulated saliva and negative blood samples
Negative blood samples were obtained through the New York University Medical Center, and whole mouth-stimulated saliva (WMSS) was collected14 at the Bluestone Research Center, New York University College of Dentistry, as previously described, using protocols approved by the New York University Langone School of Medicine IRB #05-51. As this research utilized existing, deidentified specimens, it was exempted from the requirement to obtain IRB approval under §46.101, b, (4).
Collectors
The following are the sample collection devices used Puritan HydraFlock (Puritan Diagnostics, Catalog #—25-3406-H), 4N6FLOQSwabs (COPAN Flock Technologies, Catalog #—4479429), and OraQuick ADVANCE HIV-1/2 kit (OraSure Technologies Inc.—Buffer and microloop).
LAMP primers
The primers specific for P. falciparum (Table 1) targeted the mitochondrial cytochrome oxidase subunit 1 gene (GenBank #AB570973.1) were optimized by varying the concentrations of each primer pair individually in the master mix, and the optimal concentration was selected based on the shortest Ct value. Primers were synthesized commercially (Sigma-Aldrich).
Table 1.
LAMP primer sequences for P. falciparum.
| P. falciparum (mitochondrial cytochrome C oxidase subunit 1 gene)13 | |
| FOP | CTCCATGTCGTCTCATCGC |
| BOP | AACATTTTTTAGTCCCATGCTAA |
| FIP | ACCCAGTATATTGATATTGCGTGACAGCCTTGCAATAAATAATATCTAGC |
| BIP | AACTCCAGGCGTTAACCTGTAATGATCTTTACGTTAAGGGC |
| LF | CGGTGTGTACAAGGCAACAA |
| LB | GTTGAGATGGAAACAGCCGG |
| P. falciparum and P. vivax (18S rRNA gene)12 | |
| FOP | GTCGATCAGGAAGGTTTCA |
| BOP | GTAACGACTTCCCCATTGT |
| FIP | CCTGAGCACCTTAACTTCCCTATCCTTAAATCTCGTAACCATGC |
| BIP | TTACCGTCGGGCCGTATGATCGCTAGTGTGAGACTCCTA |
Abbreviations: FOP, forward outer primer; BOP, back outer primer; FIP, forward inner primer; BIP, back inner primer; LF, forward loop primer; BLP, back loop primer.
Amplification facilitators/enhancers
Bovine serum albumin (BSA) 2 mg/mL—BSA Protein Assay Standard (Thermo Scientific: Catalog #23209); BSA heat shock fraction, protease free, fatty acid free, essentially globulin free, pH 7, ≥98% (10 mg/mL—Sigma Catalog #A-7030); fraction V (10 mg/mL—Sigma Catalog #A-9418); albumin, from bovine serum (10 mg/mL—Sigma Catalog #A-3294); MSD blocker A (8 mg/mL—Meso Scale Discovery Catalog #R93AA-1); ethylenediaminetetraacetic acid (EDTA) (1.5 mg/mL—Sigma Catalog #EDS-500G); sodium citrate tribasic dehydrate (3.2%—Sigma Catalog #S4641-500G); glycerol (10%—Alfa Aesar Catalog #—J16059); SuperSignal West Pico Stable Peroxide Solution (Thermo Scientific Catalog #34077); MgCl2 (2 mM—Fisher Scientific Catalog #M33-500).
DNA isolation
Dried blood spots/dried saliva spots
Spots of infected blood and saliva (Malaria Institute of Macha) were dried on Whatman filter paper. A Harris UNI-CORE punch (Catalog #WB100029) was used to cut three 2-mm-diameter disks from each dried blood spot (DBS) containing P. falciparum 3D7 at applied concentrations of 100, 103, and 106 parasites/µL and placed in 1.5-mL polyethylene microfuge tubes.15 Parasite DNA was extracted from the DBS using QIAamp DNA Mini Kit (QIAgen) (Catalog #51304) as per the manufacturer’s instructions. Briefly, lysis buffer (180 µL) was added, and the tubes were incubated with shaking (Eppendorf Thermomixer) for 60 minutes at 85°C followed by centrifugation at 14,000 × g and the addition of Proteinase K (400 µg, Ambion Catalog #AM2548) to the lysate, which was then incubated at 56°C for 60 minutes. The DNA was eluted in 150 µL of elution solution and quantified either spectrophotometrically (NanoDrop 2000 Spectrophotometer—Thermo Scientific) or fluorometrically (Quant-iT RiboGreen RNA Assay kit—Invitrogen: Catalog #R11490) as per the manufacturer’s instructions and stored at −20°C.
Malaria-positive blood DNA
A stock of malaria genomic DNA was purified from malaria-positive blood (MPB) using Dynabeads SILANE Viral NA kit (Invitrogen) (Catalog #—37011D) as per the manufacturer’s instructions to be used as an external positive control and standard.
Isothermal amplification (LAMP)
Infected whole blood was heated at ~90°C for five minutes to lyse blood cells and parasites, releasing target DNA. LAMP reactions contained 17 µL of a complete master mix (OptiGene: Catalog #ISO-004), 4 µL of primers, and 4 µL of sample DNA in a total volume of 25 µL. OptiGene Isothermal master mix was used for all experiments and required only the addition of primers and target nucleic acid. The P. falciparum-specific LAMP reaction mix contained final concentrations of 0.2 µM for outer primers, 1.6 µM for inner primers, and 0.8 µM for the loop primers. The P. vivax-specific LAMP reaction mix contained final concentrations of 1.5 µM for outer primers and 2.5 µM for inner primers and did not require loop primers. Reactions were followed in real time at 65°C for 30 minutes using either a Genie II or III instrument (OptiGene/Pro-Lab Diagnostics) and analyzed using Genie Explorer Software (OptiGene). Amplification was confirmed by observing the banding patterns after electrophoresis on 2% agarose gels stained with ethidium bromide.
Results
Detection using dried blood spots
DNA isolated from DBS spotted with 100, 103, or 106 parasites/µL yielded concentrations of 1.7–2.0 ng/µL, respectively. The P. falciparum mitochondrial cytochrome oxidase subunit 1 gene was readily detected using LAMP (Fig. 1).
Figure 1.
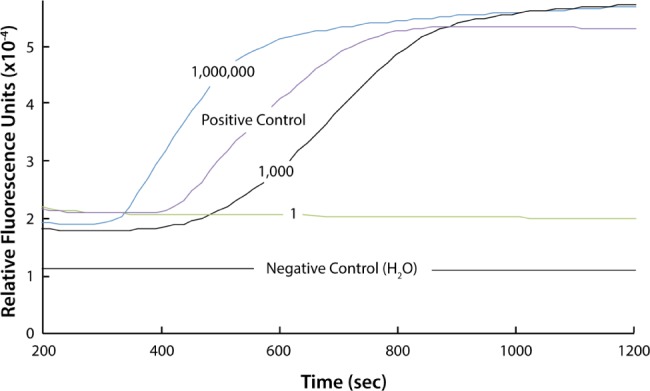
LAMP of DNA isolated from DBS showing real-time amplification in the positive control (genomic Plasmodium DNA) and amplification from DNA isolated from DBS containing 103 and 106 parasites/µL.
Amplification without DNA isolation
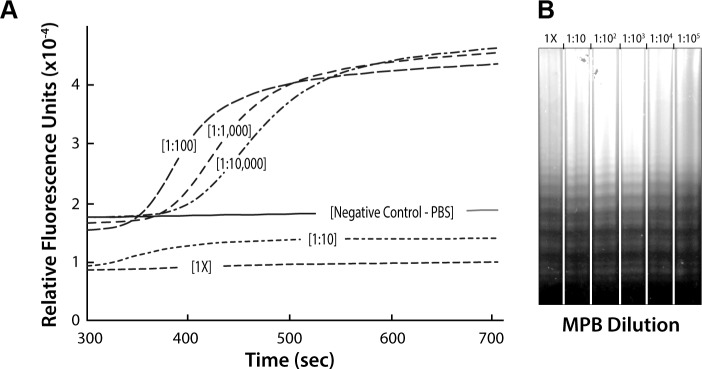
Experiments using MPB directly for LAMP were carried out to test the possibility of eliminating a DNA isolation step. Our studies indicated that a 1:1,000 dilution of blood allowed nucleic acid amplification with detection in real time (Fig. 2A). Although undiluted (1×) and 1:10 dilutions of blood were not visualized by real-time fluorescence, amplification did occur as verified by agarose gel electrophoresis of the amplification reaction products (Fig. 2B). The apparent inhibition of real-time amplification was attributed to fluorescence quenching by blood proteins.
Figure 2.
(A) Direct LAMP of MPB without prior DNA isolation. Serial dilution of blood samples from 1:1,000 to 10,000 dilutions allowed following the amplification with real-time detection of fluorescence. However, smaller dilutions of blood (ie, 1× and 1:10) showed an apparent lack of amplification in real time. It should be noted that a dilution of 1:100 was not consistently sufficient to allow real-time detection depending on the blood sample. (B) Image of the agarose gel electrophoresis of the samples amplified in (A). All of the samples showed the characteristic ladder pattern for LAMP after isothermal amplification.
Parasite concentration determination
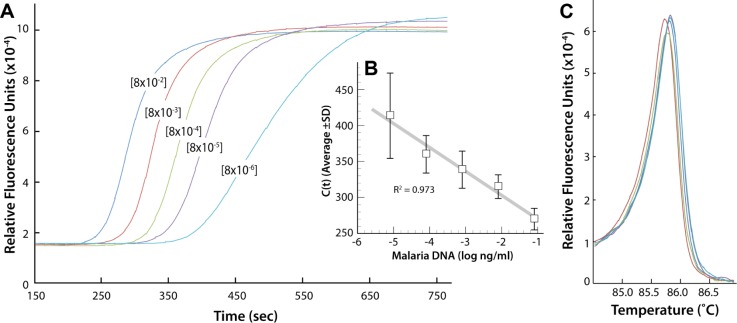
The gold standard used for diagnosing malaria is microscopic observation of thin and thick blood-smeared slides, where the number of parasites per microliter is counted. The samples obtained from BEI were not characterized with a parasite titer. DNA from the genomic sample MRA 731G, as a representative MPB, was isolated and used to create a standard curve of DNA concentration as a function of the respective real-time amplification cross-over thresholds (Ct) values observed from real-time amplification (Fig. 3). This allowed calculating the DNA concentration and estimating the parasite count. We calculated that the MRA 731 sample contained ~54 ng of parasite genomic DNA/µL blood.
Figure 3.
(A) LAMP as a function of concentration of genomic DNA sample MRA-731G. (B) Graph of the resulting Ct values as a function of MRA-731 DNA concentration (ng/µL) (n = 7). (C) Annealing curves of the genomic sample MRA-731 for each dilution yielded a single amplicon product.
Mu et al reported that the P. falciparum genome consists of ~23 × 106 bp.16 Dolezel et al17 calculated the parasite’s DNA content in picograms by the following formula:
Thus, 1 parasite = 23 × 106 bp = 0.0235 pg. This formula and the concentration were determined from the amplification standard curve (Fig. 3); 1 ng of DNA contains ~42,553 parasites and 1 µL of MRA 731 blood sample containŝ2,297,862 parasites.
The number of parasites in 1 µL of MPB was also determined by the isolation of DNA from all the MPB samples using the Dynabeads SILANE viral NA kit. The isolated DNA was quantified using the Quant-iT RiboGreen assay kit, and the number of parasites was calculated (Supplementary Table 1). The values obtained by the isolation of DNA from MPB (MRA 731) using Dynabeads (~36 ng/µL) and from the standard curve (Fig. 3B, ~54 ng/µL) were considered similar when taking into account the efficiency of extraction and recovery by the Dynabeads SILANE Viral NA kit.
Real-time LAMP
Blood samples were diluted to eliminate fluorescence quenching resulting from blood proteins in order to follow the amplification in real time. Blood was diluted 1,000-fold in PBS and then serially diluted 10-fold up to 1:108 to determine the lowest dilution detectable by LAMP. The lowest dilution detected was 1 × 106 of MPB in water (Fig. 4), which is equivalent to 1.5 parasites/µL of MPB or 0.01 parasites/µL in the 25 µL of reaction mix. This experiment supports the high sensitivity of LAMP, and its use for diagnosing recently infected asymptomatic patients.
Figure 4.
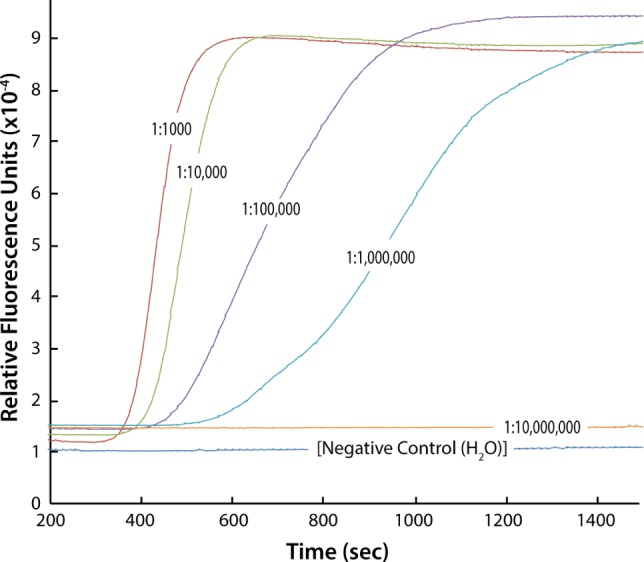
LAMP curves for a serial dilution of MPB blood (MRA-731) from 1:1,000 to 10,000,000.
Enhancers
Methods to reduce or eliminate the apparent fluorescence quenching of blood in real-time LAMP besides dilution were explored. Previous reports indicated that a buffer composed of 320 nM of saccharose, 5 nM MgCl2, 1% Triton X-100, 10 mM Tris–HCl (pH 7.5) was successful in using a whole blood sample for amplification by PCR in the diagnosis of brucellosis.18 Hypotonic solution, increased pH, and MgCl2 concentration have also been successfully used in the genotypic identification of closely related alleles.19 The addition of anticoagulants was shown to be helpful in the amplification of human hemochromatosis and apoE genes. Na-heparin, a commonly used anticoagulant for the collection of blood from humans, was cited to be better than using either EDTA or sodium citrate.20
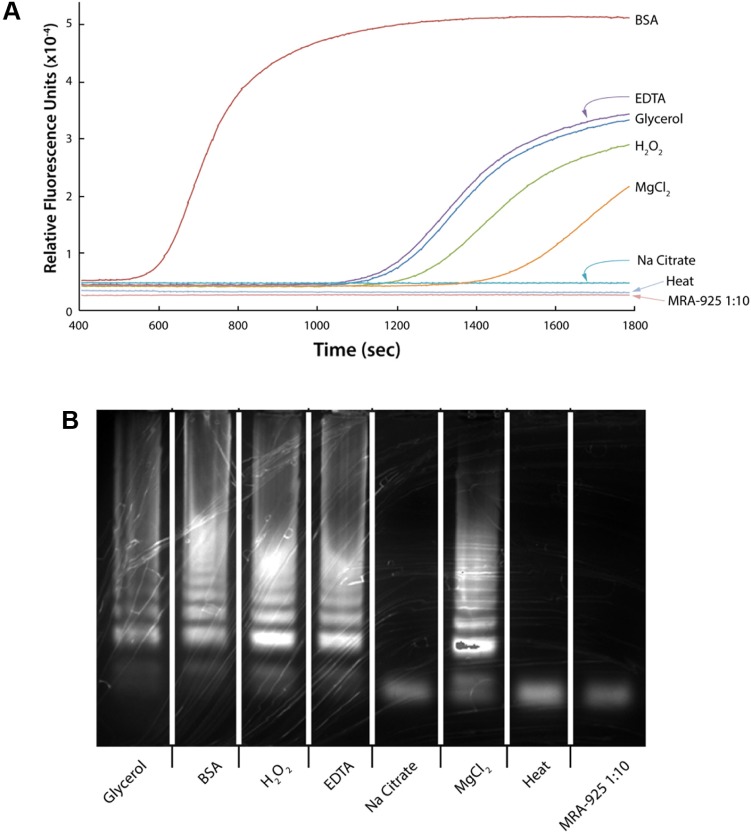
The addition of BSA (0.24 µM) or glycerol (0.87 µM) was confirmed in our system to facilitate DNA amplification.21–24 Heating whole blood samples in a microwave25 or at 99°C for 10 minutes were also reported to be successful for LAMP of the P. falciparum 18S ribosomal RNA gene.26 Other reagents used as enhancers included EDTA (410 µM), Na citrate (0.01 µM), MgCl2 (0.08 µM), and H2O2 (0.04%) and observed the following: Ct’s: BSA—630 seconds, glycerol solution—1,230 seconds, EDTA – 1,200 seconds, hydrogen peroxide solution—1,305 seconds, and MgCl2—1,560 seconds (Fig. 5A). BSA was prepared by initial fractionation by heat shock. The amplification of 0.8% MPB (50 µL of the 1:10 diluted MPB) was performed after incubation with shaking at 70°C for 15 minutes with the enhancer tested. BSA was considered the best facilitator based on amplification starting earlier than for the other compounds tested (Fig. 5). The observed annealing temperature was consistently 87.7°C for all samples, suggesting that no alternative products were produced (Fig. 5B).
Figure 5.
(A) LAMP of MPB treated with agents indicating that BSA facilitates amplification more efficiently than other agents when heated at 70°C for 15 minutes. (B) Image of the electrophoresis gel of the LAMP amplified treated MPB.
Heat lysis
Heat treatment of blood was found to facilitate LAMP without the need for isolating parasite DNA.25,26 We examined the temperature range from 75°C to 95°C and show that dilution with PBS of MPB (1:40), followed by heating for five minutes at 90°C, gave reproducible amplification (Fig. 6).
Figure 6.
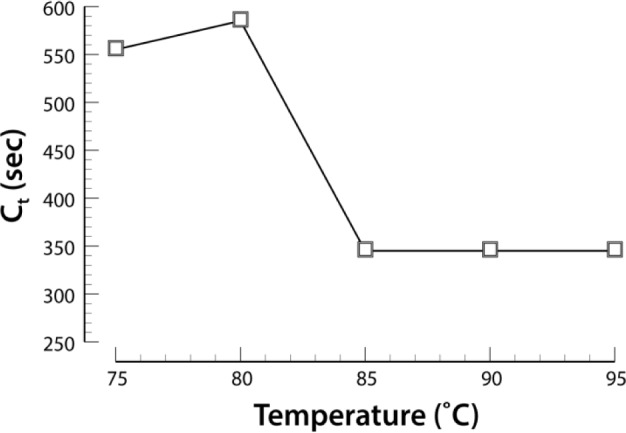
Effect of temperature on the LAMP cross-over threshold (Ct). All incubations were for 5 minutes at the indicated temperature, followed by a 4-µL aliquot amplified using LAMP.
Detection of parasites from saliva
WMSS samples were spiked with the malarial genomic sample MRA-172, and Hydraflock and Copan collectors were submerged in 500 µL of 0.1 ng/µL (~4,255 parasites/µL) of spiked saliva for one minute, transferred to a Mobicol column (Fig. 7A), inserted in a microfuge tube, and centrifuged at 2,400 rpm for five minutes, and 4 µL of eluted DNA was (Fig. 7B) taken for amplification. Both the collectors transferred a volume of ~150 µL with an average of 96% volume recovery. Both collectors picked up ~50% of the genomic sample, though only 8% and 0.02% of the total sample were recovered from the Hydraflock and Copan collectors, respectively (Figs. 7 and 8).
Figure 7.
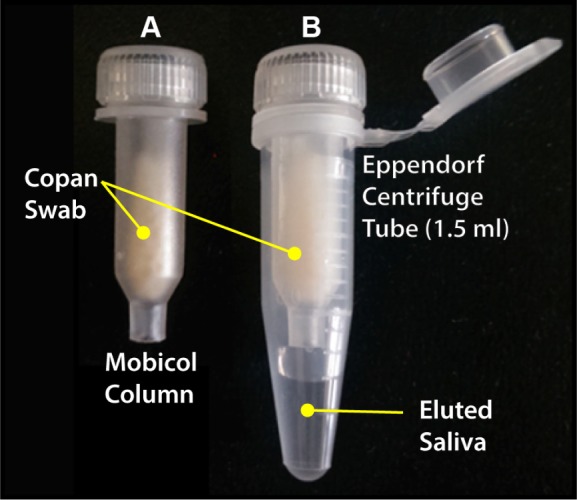
(A) Mobicol column with loaded Copan collector head. (B) Mobicol assembly post-centrifugation with eluted sample after centrifugation from the Copan collector head suspended in Mobicol column inserted in the 1.5-mL Eppendorf centrifuge tube.
Figure 8.
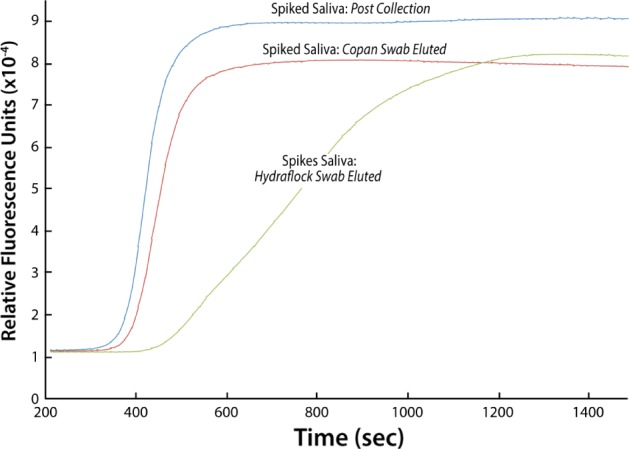
LAMP of spiked WMSS samples recovered from Hydraflock and Copan collectors.
P. falciparum detection specificity
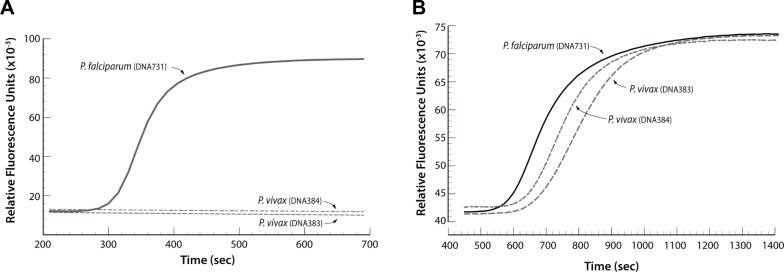
We targeted the P. falciparum mitochondrial cytochrome oxidase subunit 1 gene, which is specific for P. falciparum and does not amplify P. vivax genomic DNA (MRA 383 and MRA 384; Fig. 9). Primers that target and amplify the 18S rRNA gene present in all Plasmodium species (falciparum, vivax, malaria, and ovale) have been reported.12,27
Figure 9.
(A) LAMP detection of P. falciparum DNA using LAMP primers targeted to the parasite’s mitochondrial cytochrome oxidase subunit 1 gene. The P. falciparum-specific primers did not amplify two different samples of P. vivax DNA. (B) LAMP detects both P. falciparum and P. vivax DNA when targeting the 18S rRNA gene present in all Plasmodium species.
Discussion
These results suggest that the diagnosis of malaria can be achieved by using LAMP of DBS isolated from infected individuals (Fig. 1). MPB was used to demonstrate that nucleic acid isolation was not required for amplification, although the dilution of parasite-infected blood needs to be diluted to prevent quenching or inhibition of the amplification reaction (Fig. 2A and B). A 1,000 fold dilution of the blood samples was required to consistently detect the Plasmodium target with real-time detection without first isolating the parasite DNA (Fig. 4). BSA was found to be the best agent (Fig. 5), and together with preparing the blood/parasite sample for nucleic acid amplification by dilution and heat lysis were the best for facilitating amplification (Fig. 6). Target specificity was demonstrated for P. falciparum. A proof-of-concept study showed that malaria could be diagnosed from oral samples collected by cheek-swab collectors (Figs. 7 and 8).
Primers that target highly conserved mitochondrial sequences in Plasmodium species13,28 and targeting the highly conserved Pvr64 family of genes and the alpha-tubulin gene in P. vivax29,30 have also been reported. These are being explored for their ability to differentiate species using LAMP (Fig. 9). The advantage of the mitochondrial sequences is that they are highly conserved, which facilitates the distinguishing of falciparum in a mixture of genetic variation.
We designed a rapid LAMP-based assay for the detection of Plasmodium parasites using both blood and oral fluid without sample purification. The optimized protocol is suitable for POC diagnosis of malaria infection by P. falciparum. Heat lysis coupled with dilution allows the amplification of parasites without the need to first purify target DNA with both high sensitivity and specificity from a subject’s blood or saliva samples. The technique is rapid (ie, ~30 minutes to result) with a detection limit of ~1–2 parasites/µL and can detect either P. falciparum alone or differentially with P. vivax using primers directed against ribosomal RNA. BSA was used as an amplification enhancer, but further study needs to be done to optimize its use.
Supplementary Material
Supplementary Table 1.
Calculation of the number of parasites present in a microliter of blood.
| MPB | CONCENTRATION (ng/µL) | NUMBER OF PARASITES/µL |
|---|---|---|
| MRA-731 | 36 | 15,31,908 |
| MRA-925 | 10 | 4,25,530 |
| MRA-102 | 23 | 9,78,719 |
| MRA-172 | 2 | 85,106 |
| MRA-383 | 43 | 18,29,779 |
| MRA-384 | 62 | 26,38,286 |
Acknowledgments
Dried blood and saliva spots were generously provided by Dr. Sungano Mharakurwa, Malaria Institute of Macha, Choma. Jane Carlton, New York University, New York, NY, USA, is acknowledged for her generous contribution of parasite blood samples.
Footnotes
ACADEMIC EDITOR: Douglas MacPherson, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1739 words, excluding any confidential comments to the academic editor.
FUNDING: Funding for this project was received from the National Institute of Dental and Craniofacial Research Grants U01DE017855 and R44DE024456. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE). Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived and designed the experiments: SSM, EG, DM, and YSYO. Analyzed the data: SSM, CAB, and WRA. Wrote the first draft of the manuscript: SSM. Contributed to the writing of the manuscript: SSM, CAB, EG, WRA, DM, and YSYO. Agreed with the manuscript results and conclusions: SSM, CAB, EG, WRA, DM, and YSYO. Jointly developed the structure and arguments for the paper: SSM, CAB, EG, WRA, DM, and YSYO. Made critical revisions and approved the final version: DM and WRA. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.World Health Organization . World Malaria Report 2014. Geneva, Switzerland: World Health Organization; 2014. [Google Scholar]
- 2.Snounou G, Renia L. The vaccine is dead—long live the vaccine. Trends Parasitol. 2007;23(4):129–132. doi: 10.1016/j.pt.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Walensky RP, Paltiel AD, Losina E, et al. Test and treat DC: forecasting the impact of a comprehensive HIV strategy in Washington DC. Clin Infect Dis. 2010;51(4):392–400. doi: 10.1086/655130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505(7481):50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D, Mauk M, Qiu X, et al. An integrated, self-contained microfluidic cassette for isolation, amplification, and detection of nucleic acids. Biomed Microdevices. 2010;12(4):705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Byun D, Mauk MG, et al. A disposable, self-contained PCR chip. Lab Chip. 2009;9(4):606–612. doi: 10.1039/b807915c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Mauk MG, Hart R, et al. A self-heating cartridge for molecular diagnostics. Lab Chip. 2011;11(16):2686–2692. doi: 10.1039/c1lc20345b. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Qiu X, Ongagna S, et al. A timer-actuated immunoassay cassette for detecting molecular markers in oral fluids. Lab Chip. 2009;9(6):768–776. doi: 10.1039/b814322f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paris DH, Imwong M, Faiz AM, et al. Loop-mediated isothermal PCR (LAMP) for the diagnosis of falciparum malaria. Am J Trop Med Hyg. 2007;77(5):972–976. [PubMed] [Google Scholar]
- 10.Ghayour Najafabadi Z, Oormazdi H, Akhlaghi L, et al. Detection of Plasmodium vivax and Plasmodium falciparum DNA in human saliva and urine: loop-mediated isothermal amplification for malaria diagnosis. Acta Trop. 2014;136:44–49. doi: 10.1016/j.actatropica.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Mohon AN, Elahi R, Khan WA, et al. A new visually improved and sensitive loop mediated isothermal amplification (LAMP) for diagnosis of symptomatic falciparum malaria. Acta Trop. 2014;134:52–57. doi: 10.1016/j.actatropica.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han ET, Watanabe R, Sattabongkot J, et al. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol. 2007;45(8):2521–2528. doi: 10.1128/JCM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polley SD, Mori Y, Watson J, et al. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol. 2010;48(8):2866–2871. doi: 10.1128/JCM.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phelan JA, Abrams WR, Norman RG, et al. Design aspects of a case-control clinical investigation of the effect of HIV on oral and gastrointestinal soluble innate factors and microbes. PLoS One. 2014;9(11):e112901. doi: 10.1371/journal.pone.0112901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ongagna-Yhombi SY, Corstjens P, Geva E, et al. Improved assay to detect Plasmodium falciparum using an uninterrupted, semi-nested PCR and quantitative lateral flow analysis. Malar J. 2013;12:74. doi: 10.1186/1475-2875-12-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mu J, Seydel KB, Bates A, et al. Recent progress in functional genomic research in Plasmodium falciparum. Curr Genomics. 2010;11(4):279–286. doi: 10.2174/138920210791233081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolezel J, Bartos J, Voglmayr H, et al. Nuclear DNA content and genome size of trout and human. Cytometry A. 2003;51(2):127–128. doi: 10.1002/cyto.a.10013. Author reply 129. [DOI] [PubMed] [Google Scholar]
- 18.Morata P, Queipo-Ortuno MI, de Dios Colmenero J. Strategy for optimizing DNA amplification in a peripheral blood PCR assay used for diagnosis of human brucellosis. J Clin Microbiol. 1998;36(9):2443–2446. doi: 10.1128/jcm.36.9.2443-2446.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J, Chen L, Mao Y, et al. Multiplex allele-specific amplification from whole blood for detecting multiple polymorphisms simultaneously. Genet Test Mol Biomarkers. 2013;17(1):10–15. doi: 10.1089/gtmb.2012.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunenwald H. Direct PCR from whole blood. Epicentre. 2013;7(4):10–12. [Google Scholar]
- 21.Abu Al-Soud W, Radstrom P. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J Clin Microbiol. 2000;38(12):4463–4470. doi: 10.1128/jcm.38.12.4463-4470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62(3):1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagai M, Yoshida A, Sato N. Additive effects of bovine serum albumin, dithiothreitol, and glycerol on PCR. Biochem Mol Biol Int. 1998;44(1):157–163. doi: 10.1080/15216549800201172. [DOI] [PubMed] [Google Scholar]
- 24.Farell EM, Alexandre G. Bovine serum albumin further enhances the effects of organic solvents on increased yield of polymerase chain reaction of GC-rich templates. BMC Res Notes. 2012;5:257. doi: 10.1186/1756-0500-5-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandford AJ, Pare PD. Direct PCR of small genomic DNA fragments from serum. Biotechniques. 1997;23(5):890–892. doi: 10.2144/97235st05. [DOI] [PubMed] [Google Scholar]
- 26.Poon LL, Wong BW, Ma EH, et al. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem. 2006;52(2):303–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- 27.Aydin-Schmidt B, Xu W, Gonzalez IJ, et al. Loop mediated isothermal amplification (LAMP) accurately detects malaria DNA from filter paper blood samples of low density parasitaemias. PLoS One. 2014;9(8):e103905. doi: 10.1371/journal.pone.0103905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao ZY, Zhou HY, Xia H, et al. Adaptation of a visualized loop-mediated isothermal amplification technique for field detection of Plasmodium vivax infection. Parasit Vectors. 2011;4:115. doi: 10.1186/1756-3305-4-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel JC, Oberstaller J, Xayavong M, et al. Real-time loop-mediated isothermal amplification (RealAmp) for the species-specific identification of Plasmodium vivax. PLoS One. 2013;8(1):e54986. doi: 10.1371/journal.pone.0054986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinzouna-Boutamba SD, Yang HW, Joo SY, et al. The development of loop-mediated isothermal amplification targeting alpha-tubulin DNA for the rapid detection of Plasmodium vivax. Malar J. 2014;13:248. doi: 10.1186/1475-2875-13-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Calculation of the number of parasites present in a microliter of blood.
| MPB | CONCENTRATION (ng/µL) | NUMBER OF PARASITES/µL |
|---|---|---|
| MRA-731 | 36 | 15,31,908 |
| MRA-925 | 10 | 4,25,530 |
| MRA-102 | 23 | 9,78,719 |
| MRA-172 | 2 | 85,106 |
| MRA-383 | 43 | 18,29,779 |
| MRA-384 | 62 | 26,38,286 |