Abstract
Alternative splicing (AS) is one of the most important ways to enhance the functional diversity of genes. Huge amounts of data have been produced by microarray, expressed sequence tag, and RNA-seq, and plenty of methods have been developed specifically for this task. The most frequently asked questions in previous research were as follows. What is the content rate of AS genes among the whole gene set? How many AS types are presented in the genome, and which type is dominant? How about the conservation ability of AS among different species? Which kinds of isoforms from some genes have the environmental response to help individual adaptation? Based on this background, we collected analysis results from 17 species to try to map out the landscape of AS studies in plants. We have noted the shortages of previous results, and we appeal to all scientists working in the AS field to make a standard protocol so that analyses between different projects are comparable.
Keywords: alternative splicing, comparative analyses, conservation ability, stress response
Introduction
Alternative splicing (AS) efficiently enhances protein diversity by using limited gene loci in the formation of different isoforms. In the process of AS, particular exons of a gene are retained in the messenger RNA (mRNA), whereas some introns are erased. AS was observed first-hand in 19771,2 and in the following several years, it became one of the most active fields in biological research, especially in the search for the mechanism of AS.3,4 When we used “alternative splicing” as the keyword to search on the web, the growth of research in this area was apparent. For example, in the journal Cell, the total number of articles has doubled every 10 years. Research interests have varied greatly over time; in the early stages, focus was on AS phenomenon of one gene,5 but researchers now consider AS at the genome level6,7 or even on many diverse conditions compared tempospatially.8
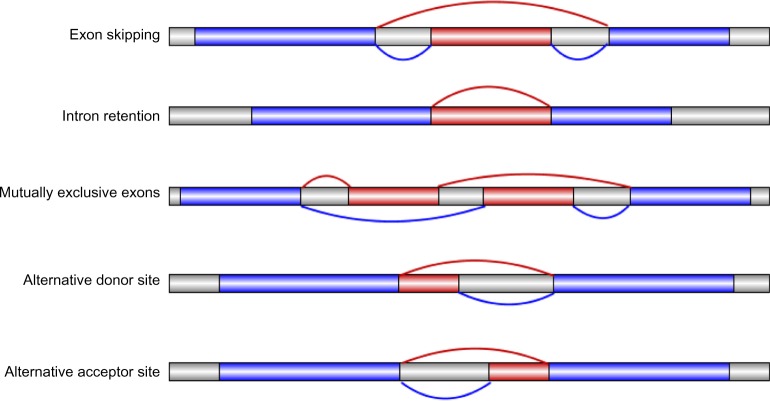
It is accepted that there are five basic modes of AS9–11: exon skipping, intron-retention, mutually exclusive exons, alternative donor site, and alternative acceptor site (Fig. 1). Exon skipping means that one exon may be spliced out to become an intron in some cases or may be retained in others; it is the most common mode of AS in mammals.9 Intron-retention means that an intron may be either spliced out or unspliced but combines with the flanking two exons to form a new, larger exon instead; intron-retention is the most common mode in plants.12 Compared with intron-retention and exon skipping, other AS modes are not dominant. Mutually, exclusive exons suggest that of two exons, only one will be displayed in the final mRNA; alternative donor site means that the 3′ boundary of the upstream exon will be changed, whereas alternative acceptor site means that the 5′ boundary of the downstream exon will be changed. Besides these five basic modes, there are several other interesting modes, such as alternative promoters and alternative poly(A).13 We present in this work a review of genome-level analyses of AS in 17 plants.
Figure 1.
Five basic modes of AS (drawing by Illustrator for Biological Sequences [IBS]59).
AS analysis at the genome level is prevalent, since microarray profiling, expressed sequence tag complementary DNA (EST-cDNA) sequence data, and RNA-seq data are easy to obtain. According to our research, AS analyses of 17 species have been released at the genome level: Oryza sativa, Arabidopsis thaliana, Glycine max, Populus euramericana, Eucalyptus grandis, Populus trichocarpa, Physcomitrella patens, Digitalis purpurea, Chlamydomonas reinhardtii, Brachypodium distachyon, Sorghum bicolor, Medicago truncatula, Lotus japonicus, Gossypium raimondii, Zea mays, Vitis vinifera, and Solanum lycopersicum. For most of these species, reference genomes have been released, which would help in AS analysis. These species include nine dicots, six monocots, one Bryopsida, and one Chlorophyceae; among them, a large number of species are from Poaceae, which indicates that crops are the most studied plants.
Arabidopsis
Arabidopsis has been studied in depth; we have collected five publications relating to AS analysis at the genome level.6,7,14–16 The first technology used in genome-level analysis is ESTs and full-length cDNA. Because of the limited data generated by these two methods, the detected level of AS rate is very low, ~11.6%.7 In studies analyzing high-throughput RNA-seq data, estimates of the AS rate ranged from a little more than 40%–60%.14–16 In the Arabidopsis genome analyses, scientists also discovered some interesting results in the huge data set. For example, the main AS type in Arabidopsis is intron retention, with a high rate of ~40%, and these retained introns have a great potential to splice out as an intron in the same frame.16 Also, the alternative 3′ and 5′ splice site may introduce a frameshift mutation,15 which means that the protein diversity of the same gene is much larger than the previous expectation. Another interesting conclusion was that although the annotation of Arabidopsis was widely used in the published studies and referred to as the gold standard, novel ASs were still detected. This means that the AS rate in Arabidopsis may be much higher than what we expected, and the annotation can be improved.14–16
Rice
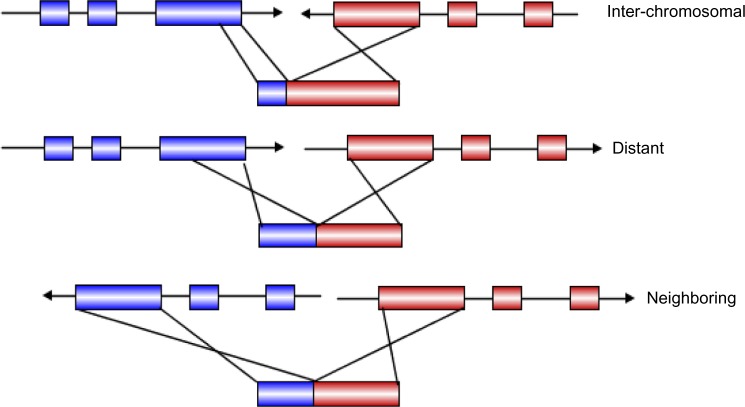
Analyses of another model plant, rice, yielded many similar conclusions. Recent genome-level analyses have suggested the AS rate in rice ranges from 33%17 to 48%.18 One interesting phenomenon that there are many chimera transcripts was also observed in rice17 – prevalent in tumor and also known as trans-splicing of RNAs or gene fusion.19 These transcripts contain the partial sequences from two genes with direct location or opposite location and also have the potential to come from two chromosomes (Fig. 2). It was suggested that AS may contribute to the fusion transcripts.17
Figure 2.
AS may contribute to the fusion transcript (according to the figure in Zhang et al17, reproduced here under the terms of a CC-BY license. Redrawing by IBS59).
Other Plants
Of other plants for which we found published genome-level AS analyses, most were crops, such as soybean,8 sorghum,20 maize,21 tomato,22 and cotton.23 The analyses from these nonmodel plants suggested that the AS events may relate to the developmental stages, with plants at a younger developmental stage possibly having a higher probability to present AS.8 Also, gene features, transcriptional level, intron length, and exon number will contribute to the AS rate.8 Analysis of cotton suggested a higher rate of AS events, of which intron-retention was the major type.23 The transposable elements (TEs) are present at a much higher rate in the retained intron (43%) compared with the rate in other introns (2.9%), and TE insertion-induced mechanisms may play an important role in the birth of new exons.24 Analyses of more than90 RNA-seq libraries of maize suggested that the majority of genotype-specific AS can be genetically mapped by cis-acting quantitative trait loci. This kind of AS plays an important role in tissue identity and genotypic variation in maize.21
Lower AS rate in plants compared to animals
With the development of improved technology and tools, as well as the transcriptome depth increase over time, it is admitted that the whole-genome-level analysis of plants in the AS-related field is in the early stage compared with that of human beings or rats, and it is one of the main reasons that the plants have a lower AS rate. However, another interesting hypothesis valuable to consider is that the AS rate is lower in plants than in animals. In 1994,25 Hughes proposed a hypothesis that after gene duplication, two AS isoforms can be fixed in two copies, which is called functional-sharing in short. Although other hypothesis, eg, duplicability-age (Age-dependent gain of alternative splice forms and biased duplication explain the relation between splicing and duplication), tries to challenge it, it is rebut by Su and Gu (Revisit on the evolutionary relationship between AS and gene duplication). Thus, as 50%–80% of angiosperms are polyploids (genome evolution in polyploids) and A. thaliana and rice are both ancient polyploids (Analysis of the genome sequence of the flowering plant A. Thaliana. Duplication and DNA segmental loss in the rice genome implications for diploidization), we can logically expect plant genes to have lower AS rate.
Comparative Analysis in Plants
Comparative analyses of AS patterns in multiple plants are prevalent, including analysis of pairs such as rice and Arabidopsis,12,26,27 Populus and Eucalyptus,28 Brassica and Arabidopsis,29 Populus and Arabidopsis,30 and tomato and Arabidopsis.31 The analysis between Populus and Arabidopsis indicates that the isochorismate synthase gene in Populus has extensive AS, while it is rare in Arabidopsis.30 Through analysis of AS events in developing xylem of two trees, Xu et al reveal that the wood-forming tissues of Populus and Eucalyptus have one-third of their genes related to AS, and among these AS genes, ~42% of AS events result in changes to the original reading frame and about one-third cause protein domain modification.28
Variable rate of AS events in plants
From analysis of microarray profiling, EST-cDNA sequence data, and RNA-seq data, it has been estimated that more than 95% of human genes and 60% of Drosophila multiexon genes are alternatively spliced.9,32 In plants, this content rate is much lower; 61% of intron-containing genes in Arabidopsis undergo AS and 48% of genes in rice are affected by AS.16,18 More than 63% of soybean multiexon genes undergo AS.8 The AS ratio data in plants and animals are provided in Table 1. Recent studies in plants have found AS rates of 50%–60%.14,16,18 The most common type of AS in Drosophila and humans is exon skipping, which occurs when an exon is spliced out along with its flanking introns (Fig. 1), accounting for ~40% of AS events.13 Even though the AS rates are lower in plants, they have a higher percentage of intron-retention AS, where an intron remains in the mature RNA transcript as part of the exon (Fig. 1); intron-retention accounts for ~45.1%–55% of the AS events in rice and 30%–64.1% of those in Arabidopsis (Table 1) but occurs at a much lower frequency in Drosophila and humans, with rates ~5%–15% (Table 1). Unlike the model organisms, other plants have a much lower AS ratio, eg, the AS ratios in S. lycopersicum, L. Japonicus, and M. truncatula are less than 10%.22,31,33
Table 1.
The AS rate and the dominant AS types in species (updated from Zhang et al.44).
| SPECIES | AS RATE (%) | INTRON RETENTION RATE (%) | EXON SKIPPING RATE (%) | REF |
|---|---|---|---|---|
| Fly | 40 | – | – | 50 |
| 18.6 | 30.8 | 13.6 | 51 | |
| – | ~10 | ~32 | 52 | |
| 60.8 | ~11 | ~11.4 | 32 | |
| Human | 63 | 36 | – | 53 |
| – | 14.8 | 52 | 54 | |
| 32.1 | 15.8 | 28.8 | 51 | |
| – | <10 | ~42 | 52 | |
| 94 | ~1 | ~35 | 33 | |
| 95 | – | – | 9 | |
| 88 | 41 | – | 55 | |
| Rice | 8.1 | 55 | 55.0 | 51 |
| 21.2 | 53.5 | 13.8 | 12 | |
| 32.5 | 45.1 | 12.8 | 26 | |
| 48 | – | – | 34 | |
| Arabidopsis | 1.2 | – | 6.4 | 6 |
| 7~10 | 30.5 | 3.2 | 56 | |
| 11.6 | 44.8 | 15.5 | 7 | |
| 14.1 | 42.8 | 42.8 | 51 | |
| 21.8 | 56.1 | 8 | 12 | |
| 23.5 | 47.9 | 6.8 | 26 | |
| 42 | 64.1 | – | 14 | |
| – | ~30 | ~5 | 52 | |
| 61 | 40 | ~6.8 | 16 | |
| Soybean | 63 | 26.74 | 8.89 | 8 |
| Populous | 28.3 | 45 | 8.6 | 28 |
| 36 | 40 | 8 | 57 | |
| Eucalyptus | 20.7 | 33.9 | 10.5 | 28 |
| Physcomitrella patens | 47 | – | – | 42 |
| 49 | 49 | 1 | 41 | |
| Brachypodium distachyon | 6.3 | 55.5 | 5 | 58 |
| 42 | 36 | 9 | 59 | |
| Sorghum Bicolor | – | 41 | 2 | 20 |
| Lotus japonicus | 2.8 | 41.7 | 8.7 | 35 |
| Zea maysa | 40 | 58 | 39 | 21 |
| Vitis vinifera | 30 | 51 | 4.1 | 60 |
| Solanum lycopersicum | 10 | 55 | – | 22 |
| 10 | 53 | 3 | 31 | |
| Gossypium raimondii | 32 | 40 | 10 | 23 |
| Medicago truncatula | 9.6 | 51.3 | 10.7 | 35 |
Conservation ability of AS in plants
The evolutionary conservation of plant AS has been studied across many species, and the results indicate that the conservation ratio is not high. Genome-wide analysis has identified that 56 out of 380 AS events (14.7%) are conserved between Arabidopsis and rice and 49 out of 298 AS events (16.4%) are conserved between rice and maize.27 In closely related species of Brassica and Arabidopsis, 537 out of 9878 AS events (5.4%) were reported as conserved events, and the results indicated that intron-retention and exon skipping were underrepresented.29 In L. japonicus, 22 out of 115 AS events (19.1%) were conserved AS events between two or more legume species33 and 71 out of 716 orthologous groups (9.9%) have conserved AS events between Populus and Eucalyptus.28 AS between paralogous genes in two independently synthesized allotetraploid Brassica napus lines showed parallel loss of AS events after polyploidy, and 26%–30% of genes showed changes in AS compared with the parents.34
Call AS junctions from RNA sequencing data
The AS data now available, even for the human genome, are not enough to support a perfect or complete annotation for genes at the genome-wide level through the whole life cycle. The reality is that plenty of RNA sequencing data are produced and sleeping in the NCBI’s SRA database. The number of RNA sequencing data sets in NCBI is now ~1,70,000. The most prevalent organisms are Homo sapiens and Mus musculus, with more than 40,000 total data sets. The most represented data sets in plants, A. thaliana, Z. Mays, and O. sativa, number far less than for animals, ranging from 887 to 3646. These data may partially account for why estimates of the AS content are as high as 95% in humans, whereas they are only ~60% in plants. Additionally, a large number of RNA sequencing data were not used to call the AS junction.
Useful tools for AS analysis in plants
Most software for transcriptome analysis, such as Cufflinks, could be used in AS analysis. However, the so-called gold standard for isoform annotations is far from impeccable. Even in such exhaustive annotations as human and mouse, the new gene annotations are sometimes inconsistent.35 Previous analysis used eight popular publicly available software packages to reveal that, even using the exact same data, the detected isoforms using different tools can be distinctly variable.36 These results have indicated that no significant evidence supports the notion that any one tool has the best performance, and the overlap of detected isoforms from different tools is low. In light of this, we suggest people use as many tools as they can and transfer each result into tracks that are suitable for display in a genome browser (eg, GBrowse) to provide convenient access and use for others.
Distinctive AS analysis in plants
Unlike animals, plants cannot move, run, or hide, so they need to put up with some difficult environments, such as the dry weather, floods and waterlog, salts, and many kinds of passive damage. Environmental stresses will lead to specific changes in the AS analysis of plants.
AS patterns under stress
The patterns of AS in plants are not only variable along the life cycle and in different tissues but also subject to variation in diverse conditions, such as salt stress, cold stress, strong light, and diseases. High-coverage RNA-seq data of seedlings treated with different concentrations of NaCl indicated that AS increases significantly under salt stress compared with unstressed conditions; most differential AS genes may not be regulated by salt stress but associated with specific functional pathways linked to stress responses and RNA splicing, like serine/arginine-rich splicing factors.37 Analysis of AS under cold conditions indicated that the process of cold regulation may introduce a premature termination codon, and the resulting transcripts could be potential targets for degradation by the nonsense-mediated mRNA decay process, thus protecting the normal transcripts.38 Analysis based on P. patens indicated that AS was related to light exposure and that during photomorphogenesis, light regulated AS, with intron-retention occurring preferentially in transcripts involved in photosynthesis and translation.39 Another analysis based on P. patens indicated that heat shock treatment also leads to novel AS events, and ~50% of genes are alternatively spliced.40 Analysis of B. distachyon with the Panicum mosaic virus and its satellite virus reveals that ~30% more transcripts were detected, and the infected plant was enriched in defense-related genes, such as Nucleotide-binding site, Leucine-rich repeat (NB-LRR)-resistance proteins.41 In addition, in a more specific group of genes, such as clock genes, the analysis suggested that the structure of ELF3 (Early Flowering) gene and ZTL (Zeitlupe) gene varied significantly during photoperiod, temperature, and salt stress.42 Extensive AS in clock genes including LHY (Late Elongated Hypocoyl) pseudo-response regulator 7.43
Intron-retention may contribute to very complex gene structures in plants
Since intron-retention is a dominant mode of AS in the majority of plants, our recent research has focused on the structural evolution related to AS. We have found that it is possible for intron-retention isoforms from the ancestor genes coupled with RNA to reverse to cDNA and then be recruited into the genome and become a new gene. When compared with the parental gene, the new gene has lost some introns and also kept some introns, thus making the gene structure look very complex among these homologs. In the new gene, the lost intron was due to the RNA splicing, whereas the retained intron was kept in the RNA due to the intron-retention. This hypothesis was proved using 25 pairs of such complex genes to show that the ratio of complex gene pairs in plants is much higher than that in animals.44 This hypothesis could help to explain the recurrent loss of introns in orthologous gene families in plants.45
Conclusion and Future Perspectives
Shortage of current studies in AS analysis
Although a lot of AS analyses were conducted in the last few decades and hundreds of thousands of RNA-seq data were produced, we are still far away from fullness of knowledge. The results of different comparisons among species are biased because the AS was affected by the depth of coverage, the stage of the sample, the environment of time, and the tools used. Since these elements have too much variability, the results may lead to wrong conclusions. Besides, the conservation ratios of different analyses now are not comparable, because of the unclear methods. We propose the following method for measuring AS conservation: (1) find the orthologous gene pairs between two species (eg, L pair); (2) find the AS events among these gene pairs, respectively (eg, Ma and Mb); and (3) find the perfect CDS match pairs with no gaps and no extension (eg, N); thus, the conservation ratio can be presented as N/L or 2N/(Ma + Mb). To compare different groups, dividing the divergence time of two species would be a choice.
AS in the developmental stage with the single cell
In recent years, more and more single cell RNA sequencing analyses have been done, opening a new chapter of AS research focused on more specific targets. Zhang et al profiled the mRNA in 10 types of cells, including five major cell types of the differentiating endosperm, and in the embryo and four maternal compartments of the maize kernel.46 The results suggest that even in the small seed, the mRNA population has diverged gene expression profiles.46 Adrian et al quantified the gene expression profile using RNA sequencing data of stomata lineage cells marked by fluorescence and showed the regulatory modules that have different developmental decisions compared to previous expectation.47
This review has discussed AS analysis in the plant field as well as the tools used in such research. Although plenty of analyses of AS in plants have been presented and a huge quantity of data produced, it is necessary to think about the most efficient way to display these data. An AS database, eg, Alternative Splicing in Plants (ASIP),12 and genome browser tracks that are open source and easy to download may be the best choice.
Also, since AS was affected by many factors, such as time, stress, and tissues, a professional standard may also be essential in the future. In this standard, the species name, the sampling time, the environment, the tissues, the analysis tools, and the reference genome should be recorded as indispensable.
Most importantly, in the plant field, more appropriate tools or suitable pipelines need to be developed according to gene structure characters.
Footnotes
ACADEMIC EDITOR: J. T. Efird, Associate Editor
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1216 words, excluding any confidential comments to the academic editor
FUNDING: This study was funded by Hundred Talents Program (Chinese Academy of Sciences). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Hong Yang is responsible for data collection and writing. Huizhao Yang and Chengjun Zhang are responsible for writing. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 2.Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977;74:3171–5. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogers J, Wall R. A mechanism for RNA splicing. Proc Natl Acad Sci U S A. 1980;77:1877–9. doi: 10.1073/pnas.77.4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi E, Kuehl M, Wall R. RNA splicing generates a variant light chain from an aberrantly rearranged kappa gene. Nature. 1980;286:776–9. doi: 10.1038/286776a0. [DOI] [PubMed] [Google Scholar]
- 5.Werneke JM, Chatfield JM, Ogren WL. Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell. 1989;1:815–25. doi: 10.1105/tpc.1.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W, Schlueter SD, Brendel V. Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol. 2003;132:469–84. doi: 10.1104/pp.102.018101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iida K, Seki M, Sakurai T, et al. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 2004;32:5096–103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Y, Zhou Z, Wang Z, et al. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell. 2014;26:996–1008. doi: 10.1105/tpc.114.122739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 10.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–98. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 11.Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–82. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 12.Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci U S A. 2006;103:7175–80. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–55. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 14.Filichkin SA, Priest HD, Givan SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard BE, Hu Q, Babaoglu AC, et al. High-throughput RNA sequencing of pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS One. 2013;8:e74183. doi: 10.1371/journal.pone.0074183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012;22:1184–95. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang G, Guo G, Hu X, et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 2010;20:646–54. doi: 10.1101/gr.100677.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu T, Lu G, Fan D, et al. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010;20:1238–49. doi: 10.1101/gr.106120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–61. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 20.Panahi B, Abbaszadeh B, Taghizadeghan M, Ebrahimie E. Genome-wide survey of alternative splicing in Sorghum bicolor. Physiol Mol Biol Plants. 2014;20:323–9. doi: 10.1007/s12298-014-0245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thatcher SR, Zhou W, Leonard A, et al. Genome-wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell. 2014;26:3472–87. doi: 10.1105/tpc.114.130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aoki K, Yano K, Suzuki A, et al. Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics. 2010;11:210. doi: 10.1186/1471-2164-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Xiao G, Zhu YX. Single-nucleotide resolution mapping of the Gossypium raimondii transcriptome reveals a new mechanism for alternative splicing of introns. Mol Plant. 2014;7:829–40. doi: 10.1093/mp/sst175. [DOI] [PubMed] [Google Scholar]
- 24.Chen SD, Krinsky BH, Long MY. New genes as drivers of phenotypic evolution. Nature Reviews Genetics. 2013;14:645–60. doi: 10.1038/nrg3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes AL. The Evolution of Functionally Novel Proteins after Gene Duplication. Proceedings of the Royal Society B-Biological Sciences. 1994;256:119–24. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 26.Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Severing EI, van Dijk AD, Stiekema WJ, van Ham RC. Comparative analysis indicates that alternative splicing in plants has a limited role in functional expansion of the proteome. BMC Genomics. 2009;10:154. doi: 10.1186/1471-2164-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu P, Kong Y, Song D, Huang C, Li X, Li L. Conservation and functional influence of alternative splicing in wood formation of Populus and Eucalyptus. BMC Genomics. 2014;15:780. doi: 10.1186/1471-2164-15-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darracq A, Adams KL. Features of evolutionarily conserved alternative splicing events between Brassica and Arabidopsis. New Phytol. 2013;199:252–63. doi: 10.1111/nph.12238. [DOI] [PubMed] [Google Scholar]
- 30.Yuan Y, Chung JD, Fu X, et al. Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in Populus and Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:22020–5. doi: 10.1073/pnas.0906869106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ner-Gaon H, Leviatan N, Rubin E, Fluhr R. Comparative cross-species alternative splicing in plants. Plant Physiol. 2007;144:1632–41. doi: 10.1104/pp.107.098640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graveley BR, Brooks AN, Carlson JW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–9. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang BB, O’Toole M, Brendel V, Young ND. Cross-species EST alignments reveal novel and conserved alternative splicing events in legumes. BMC Plant Biol. 2008;8:17. doi: 10.1186/1471-2229-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou R, Moshgabadi N, Adams KL. Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc Natl Acad Sci U S A. 2011;108:16122–7. doi: 10.1073/pnas.1109551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang YE, Landback P, Vibranovski M, Long M. New genes expressed in human brains: implications for annotating evolving genomes. Bioessays. 2012;34:982–91. doi: 10.1002/bies.201200008. [DOI] [PubMed] [Google Scholar]
- 36.Liu R, Loraine AE, Dickerson JA. Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems. BMC Bioinformatics. 2014;15:364. doi: 10.1186/s12859-014-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding F, Cui P, Wang Z, Zhang S, Ali S, Xiong L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics. 2014;15:431. doi: 10.1186/1471-2164-15-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leviatan N, Alkan N, Leshkowitz D, Fluhr R. Genome-wide survey of cold stress regulated alternative splicing in Arabidopsis thaliana with tiling microarray. PLoS One. 2013;8:e66511. doi: 10.1371/journal.pone.0066511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu HP, Su YS, Chen HC, et al. Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 2014;15:R10. doi: 10.1186/gb-2014-15-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang CY, Lin WD, Tu SL. Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 2014;165:826–40. doi: 10.1104/pp.113.230540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandadi KK, Scholthof KB. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell. 2015;27:71–85. doi: 10.1105/tpc.114.133991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon YJ, Park MJ, Kim SG, Baldwin IT, Park CM. Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 2014;14:136. doi: 10.1186/1471-2229-14-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.James AB, Syed NH, Bordage S, et al. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24:961–81. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C, Gschwend AR, Ouyang Y, Long M. Evolution of gene structural complexity: an alternative-splicing-based model accounts for intron-containing retrogenes. Plant Physiol. 2014;165:412–23. doi: 10.1104/pp.113.231696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, Devos KM, Bennetzen JL. Recurrent loss of specific introns during angiosperm evolution. PLoS Genet. 2014;10:e1004843. doi: 10.1371/journal.pgen.1004843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhan J, Thakare D, Ma C, et al. RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell. 2015;27:513–31. doi: 10.1105/tpc.114.135657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adrian J, Chang J, Ballenger CE, et al. Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev Cell. 2015;33:107–18. doi: 10.1016/j.devcel.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stolc V, Gauhar Z, Mason C, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–60. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- 49.Nagasaki H, Arita M, Nishizawa T, Suwa M, Gotoh O. Species-specific variation of alternative splicing and transcriptional initiation in six eukaryotes. Gene. 2005;364:53–62. doi: 10.1016/j.gene.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 50.Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–31. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kan Z, States D, Gish W. Selecting for functional alternative splices in ESTs. Genome Res. 2002;12:1837–45. doi: 10.1101/gr.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–65. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mollet IG, Ben-Dov C, Felício-Silva D, et al. Unconstrained mining of transcript data reveals increased alternative splicing complexity in the human transcriptome. Nucleic Acids Res. 2010;38:4740–54. doi: 10.1093/nar/gkq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–85. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 56.Bao H, Li E, Mansfield SD, Cronk QC, El-Kassaby YA, Douglas CJ. The developing xylem transcriptome and genome-wide analysis of alternative splicing in Populus trichocarpa (black cottonwood) populations. BMC Genomics. 2013;14:359. doi: 10.1186/1471-2164-14-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walters B, Lum G, Sablok G, Min XJ. Genome-wide landscape of alternative splicing events in Brachypodium distachyon. DNA Res. 2013;20:163–71. doi: 10.1093/dnares/dss041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vitulo N, Forcato C, Carpinelli EC, et al. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014;14:99. doi: 10.1186/1471-2229-14-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu W, Xie Y, Ma J, et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015;31(20):3359–61. doi: 10.1093/bioinformatics/btv362. [DOI] [PMC free article] [PubMed] [Google Scholar]