Abstract
Klebsiella pneumoniae, a chief cause of nosocomial pneumonia, is a versatile and commonly multidrug-resistant human pathogen for which further insight into pathogenesis is needed. We show that the pilus regulatory gene fimK promotes the virulence of K. pneumoniae strain TOP52 in murine pneumonia. This contrasts with the attenuating effect of fimK on urinary tract virulence, illustrating that a single factor may exert opposing effects on pathogenesis in distinct host niches. Loss of fimK in TOP52 pneumonia was associated with diminished lung bacterial burden, limited innate responses within the lung, and improved host survival. FimK expression was shown to promote serum resistance, capsule production, and protection from phagocytosis by host immune cells. Finally, while the widely used K. pneumoniae model strain 43816 produces rapid dissemination and death in mice, TOP52 caused largely localized pneumonia with limited lethality, thereby providing an alternative tool for studying K. pneumoniae pathogenesis and control within the lung.
Keywords: Klebsiella pneumoniae, pneumonia, capsule, fimK, EAL domain, murine model
Klebsiella pneumoniae is a leading cause of nosocomial infections, including pneumonia, urinary tract infection (UTI), and sepsis [1, 2]. K. pneumoniae is ubiquitous in the environment and is often found in the gastrointestinal tract and on medical devices [2, 3]. The emergence of K. pneumoniae carbapenemases and extended-spectrum β-lactamases renders this pathogen increasingly difficult to treat [4–6]. Identification of critical virulence factors in K. pneumoniae will support development of new approaches to combat these daunting infections.
The virulence repertoire of K. pneumoniae is incompletely defined, although animal models and genomic studies have identified several important determinants. The capsule protects against phagocytosis, antimicrobial peptides, and serum bactericidal activity [7–10]. Adhesins, iron-scavenging systems, lipopolysaccharide, OmpA, phospholipase D1, and urease also promote successful K. pneumoniae infections [2, 11–18]. Type 1 pili, as in Escherichia coli, are critical for UTI pathogenesis; however, unlike uropathogenic E. coli, K. pneumoniae encodes a type 1 pilus regulatory gene, fimK [16, 18–21]. Deletion of fimK results in hyperfimbriate bacteria with augmented virulence in murine UTI [21]. We therefore investigated whether this conserved gene may be important for enhancing virulence in other niches.
Here, using a murine model of intratracheal inoculation, we demonstrate that FimK promotes K. pneumoniae pathogenesis in the lung. While no single approach to modeling K. pneumoniae pulmonary infection has been adopted, the most commonly used K. pneumoniae model strain, ATCC 43816, causes bacteremia and death in mice within 5 days of lung inoculation, with median lethal doses (LD50) of 101–103 colony-forming units (CFU) [11, 22–24]; capsule and O antigen are critical for 43816 dissemination and lethality [10, 17]. The present study identifies TOP52, a K. pneumoniae strain originally isolated from the human urinary tract, as also pathogenic in the murine respiratory tract. We demonstrate that loss of fimK attenuates lung infection, diminishes capsule production, and renders K. pneumoniae more susceptible to phagocytosis by host immune cells. Our work also introduces an alternative model strain of K. pneumoniae that produces infection localized to the respiratory tract.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Culture Conditions
K. pneumoniae strains included TOP52 (strain 1721), a K6 isolate from a woman with acute cystitis [19, 21]; TOP52ΔfimK, a deletion mutant that exhibits growth equivalent to the parent strain [21]; and ATCC 43816, a K2 isolate lethal in murine pneumonia models [1, 10, 17]. Plasmids included the empty arabinose-inducible expression vector pBAD33 and the pfimK vector for expression of TOP52 fimK [21]. Bacteria were grown statically in 20-mL cultures at 37°C for 16 hours in Luria-Bertani (LB) broth containing, as appropriate, 20 µg/mL chloramphenicol and 0.2% arabinose. Overnight cultures were centrifuged at 7000g for 10 minutes. Bacteria were resuspended in sterile phosphate-buffered saline (PBS) and diluted to the desired inoculum concentration according to OD600. Inocula were verified by serial dilution and plating.
Mouse Infections
Female C57BL/6J mice and complement component 3 (C3)–deficient female B6.129S4-C3tm1Crr/J mice (Jackson Laboratories, Bar Harbor, Maine) aged 7–8 weeks were used. For respiratory tract infections, an intratracheal inoculation procedure was adapted from those previously described [25]. Briefly, each mouse was anesthetized with inhaled isoflurane, and the trachea was exposed via surgical dissection. Inoculum (20 µL containing 1–2 × 107 colony-forming units [CFU] unless otherwise indicated) was injected intratracheally using a 30-gauge, caudally directed needle. Overlying tissues were replaced, and skin was closed using Vetbond (3M Animal Care Products, St. Paul, Minnesota). Control mice were inoculated with sterile PBS in identical fashion. For UTI, 50 µL of suspension (1–2 × 107 CFU) was inoculated into mouse bladders by transurethral catheterization [26]. All animal procedures were approved by the Animal Studies Committee at Washington University School of Medicine.
Mouse Survival, Weights, Organ Titers, Cytokines, and Histologic Analysis
K. pneumoniae–infected or PBS-inoculated mice were weighed daily for up to 2 weeks. Weights and organ titers were obtained only for surviving mice. Mouse organs (lungs, spleen, bladder, or kidneys) were harvested and homogenized in sterile PBS via Bullet Blender (Next Advance, Averill Park, New York) for 5 minutes. Aliquots of homogenates were serially diluted and plated on LB agar. Remaining homogenates were stored at −80°C before cytokine analysis, performed with a 23-plex magnetic bead cytokine array (Bio-Plex Pro, Mouse Group I, Bio-Rad, Hercules, California); duplicate samples were assayed at 1:20 dilution per the manufacturer's instructions. Organs removed for histologic analysis were washed in PBS, fixed in 10% neutral buffered formalin, dehydrated in ethanol, and embedded in paraffin; 5-µm sections were stained with hematoxylin and eosin. Images were obtained using an Olympus DP25 camera and BX40 light microscope.
Blood Cultures
Blood cultures were performed on a subset of mice 24 hours after infection. Mice were anesthetized with isoflurane, and cardiac puncture was performed with a 26-gauge needle. A total of 10 µL of blood was plated onto LB agar; samples yielding >100 CFU/mL (limit of detection) were considered positive.
Flow Cytometry
Lungs were harvested and placed into Incomplete Medium composed of Dulbecco's modified Eagle's medium (Gibco Life Technologies, Grand Island, New York), 10 mM HEPES buffer (Corning, Manassas, Virginia), 1 mM sodium pyruvate (Gibco), 5% MEM Nonessential Amino Acids (Corning), 1% penicillin/streptomycin (Gibco), and 2 mM l-glutamine (Corning). Single-cell suspensions were prepared by dicing lungs into Hank's balanced salt solution (Gibco) containing 1 mg/mL collagenase D (Roche, Indianapolis, Indiana), 2% fetal bovine serum (Gibco), and 25 mM HEPES buffer; incubating the material for 1 hour at room temperature with shaking; and passing the material through a 70-µM nylon cell strainer (Falcon Corning Life Sciences, Tewksbury, Massachusetts). Erythrocytes were lysed using sterile Gey's solution (500 mL sterile water, 4.15 g ammonium chloride, and 0.5 g potassium bicarbonate). Cells were counted by hemacytometer. For flow cytometry, 106 cells were incubated with 0.5 µg of Fc Block (BD Biosciences, San Jose, California) for 15 minutes at 4°C prior to staining. Surface antibody markers included GR1-PerCP-Cy5.5 (RB6-8C5; BD Biosciences) and CD11b- eFluor450 (M1/70; eBioscience, San Diego, California). Live cells were determined by forward scatter/side scatter gating. Cells were analyzed on a BD LSR II cytometer; data were collected using BD FACS Diva software and analyzed using FlowJo software (FlowJo, Ashland, Oregon).
Serum Resistance
Serum bactericidal assays were adapted from those previously described [27]. Blood specimens were collected by venipuncture from healthy adult donors (the protocol was approved by the Human Research Protection Office at Washington University). Briefly, 25 µL of a suspension of statically grown bacteria (106 CFU/mL in PBS) was mixed in 96-well microplates with 75 µL of pooled human serum and incubated for 3 hours at 37°C. Serum was either active or inactivated by preincubation at 65°C for 30 minutes. Samples were serially diluted and plated to calculate the percentage survival as a ratio of the 3-hour CFU to the input CFU.
Capsule Quantification
Capsule extraction and uronic acid quantification were performed using a modified protocol [28–30]. A total of 500 µL of overnight static LB cultures was mixed with 100 µL of 1% Zwittergent 3–14 (Sigma, St. Louis, Missouri) in 100 mM citric acid and incubated at 50°C for 20 minutes. Following centrifugation, supernatants were precipitated with cold ethanol. After recentrifugation, the pellet was dissolved in 200 µL of water, and 1200 µL of 12.5 mM tetraborate in concentrated H2SO4 was added. After vortexing, samples were boiled at 95°C for 5 minutes and then mixed with 20 µL of 0.15% 3-hydroxydiphenol (Sigma) in 0.5% NaOH. Absorbance was measured at 520 nm. The uronic acid concentration in each sample was determined from a standard curve of glucuronic acid (Sigma).
In Vivo Phagocytosis Assay
In vivo phagocytosis was assessed as previously described [31]. Briefly, bacteria were incubated with 0.2 mg/mL fluorescein 5(6)-isothiocyanate (FITC, Sigma) for 30 minutes at 37°C. Uniform FITC labeling was confirmed by fluorescence-activated cell-sorting analysis. Bronchoalveolar lavage (BAL) fluid was obtained 3 hours after infection with 1–2 × 107 CFU K. pneumoniae. Fluorescence of extracellular FITC was quenched by washing with trypan blue (Sigma). Alveolar macrophages (CD45+ CD11chi) and neutrophils (CD45+ GR-1+) were quantified by flow cytometry; phagocytosis of bacteria was measured by the shift in FITC fluorescence. Samples were blocked with 1 µg Fc block (2.4G2 hybridoma; ATCC). Surface antibody markers included CD11c-PECy7 (N418), GR1-Pacific blue (RB6-8C5), and CD45-Pacific blue (30-F11; BioLegend, San Diego, California) or CD45-BV510 (30-F11; BD Biosciences). Cells were acquired with a BD FACScan flow cytometer with DxP multicolor upgrades (Cytek Development, Woodland Park, New Jersey) and analyzed using FlowJo software.
Statistical Analysis
Comparisons between 2 groups of continuous variables were analyzed using the Mann–Whitney U test, as these values were not all normally distributed. For Kaplan–Meier survival analysis, the Mantel–Cox log-rank test was used. The Fisher exact test was used for blood culture comparisons. All tests were 2 tailed; P values of <.05 were considered statistically significant. Analyses were performed using GraphPad Prism, version 6.04.
RESULTS
FimK Promotes Mortality and Morbidity in K. pneumoniae TOP52 Infection of the Murine Respiratory Tract
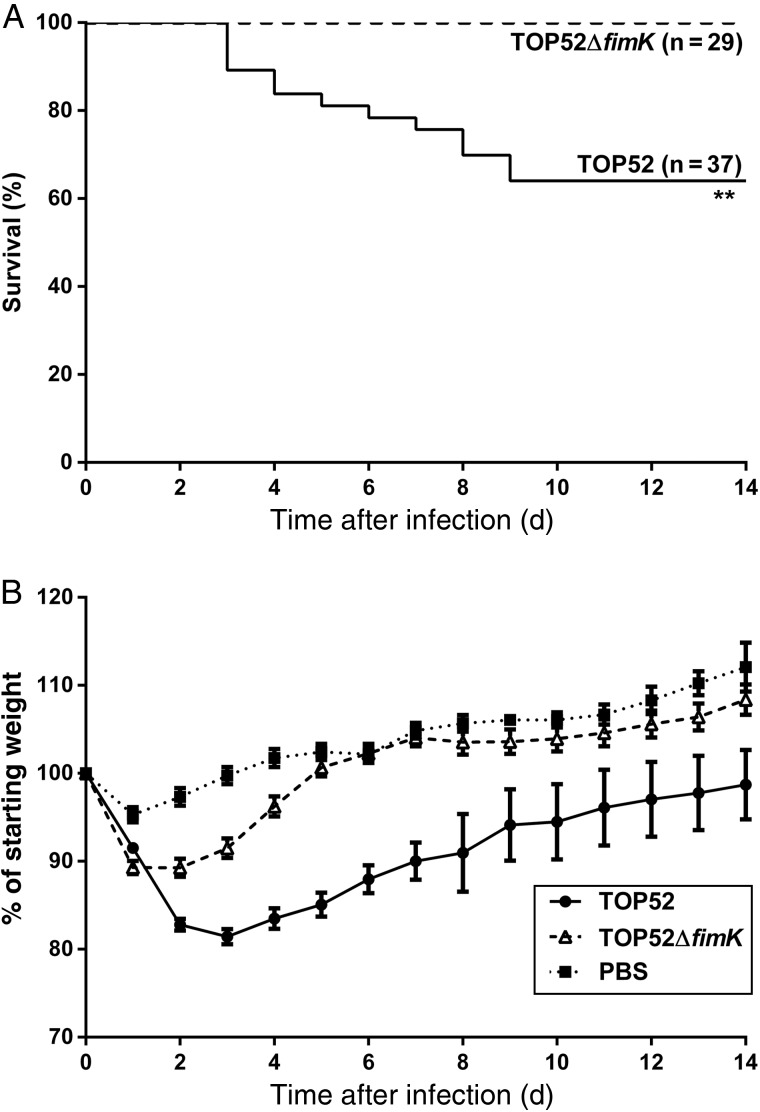
While past studies have shown that genomic carriage of fimK attenuates pathogenesis of K. pneumoniae TOP52 in the urinary tract [21], the role of fimK in other niches remains undefined. To evaluate whether fimK promotes virulence in the respiratory tract, we intratracheally inoculated C57BL/6 mice with 107 CFU of TOP52 or TOP52ΔfimK and monitored weight and survival (Figure 1). While no mice infected with TOP52ΔfimK died over 14 days, 36% of mice infected with wild-type TOP52 died between 3 and 9 days after infection (Figure 1A, P = .0013). TOP52-infected surviving mice had significantly lower weights than TOP52ΔfimK-infected mice from days 2 through 14 after infection (days 2–7, P < .0001; days 8–14, P < .01; Figure 1B). These data may understate morbidity, as only surviving mice are represented on each day. PBS–mock-infected mice lost an average of 5% body weight on day 1 after inoculation but recovered quickly and gained weight each subsequent day. TOP52ΔfimK-infected mice recovered by day 5 after infection; their weights were not statistically different from those of mock-infected mice after this interval. These data indicate that loss of fimK decreases morbidity and mortality in K. pneumoniae pneumonia.
Figure 1.
Kaplan–Meier survival and weights of mice intratracheally infected with Klebsiella pneumoniae TOP52 or TOP52ΔfimK. A, TOP52ΔfimK-infected mice exhibited a significant survival advantage relative to TOP52-infected mice (**P < .01, by the Mantel–Cox log-rank test). B, By day 2 after infection, TOP52-infected surviving mice (circles) had lost significantly more weight than TOP52ΔfimK-infected mice (triangles) and remained significantly lower in weight through the 14-day analysis. Data are shown as mean ± standard error of the mean and are combined from at least 3 independent experiments. Abbreviation: PBS, phosphate-buffered saline.
Infection With K. pneumoniae TOP52 Yields a Higher Lung Bacterial Burden Than Infection With TOP52ΔfimK
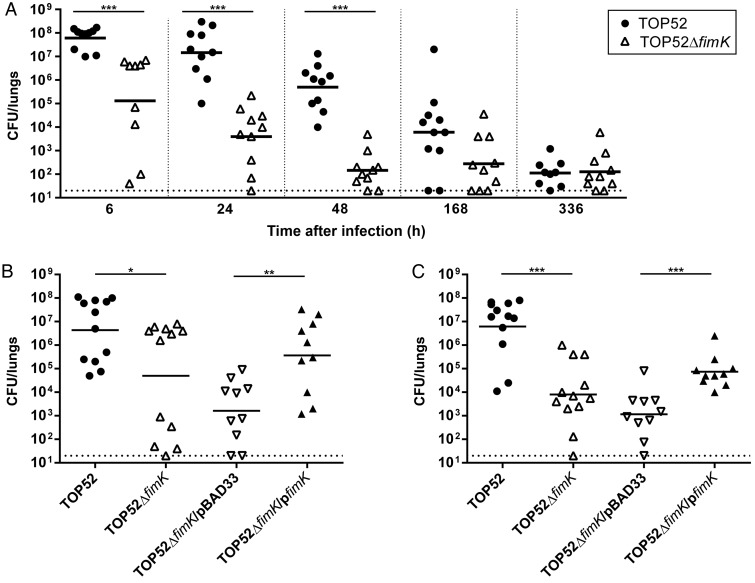
To evaluate whether fimK was important for K. pneumoniae pathogenesis within the lung itself, organs were harvested and bacterial titers quantified after infection with TOP52 or TOP52ΔfimK (Figure 2A). At 6, 24, and 48 hours after infection, TOP52ΔfimK was attenuated in the lungs (P < .0001 vs TOP52). Lung bacterial titers in surviving TOP52-infected mice continued to trend higher than those in TOP52ΔfimK-infected mice 1 week after infection (P = .0567). By 2 weeks after infection, surviving mice had substantially cleared either TOP52 or TOP52ΔfimK. Confirming that the observed differences were attributable to fimK loss, TOP52ΔfimK/pfimK yielded higher lung titers than TOP52ΔfimK/pBAD33 both 6 and 24 hours after infection (P = .0051 and .0003, respectively; Figure 2B and 2C).
Figure 2.
Time course of Klebsiella pneumoniae TOP52 and TOP52ΔfimK murine pneumonia and complementation with pfimK. A, Female C57BL/6 mice were infected with 107 TOP52 (circles) or TOP52ΔfimK (triangles) by intratracheal inoculation. Lung bacterial loads were significantly higher in TOP52-infected mice than in TOP52ΔfimK-infected mice at 6, 24, and 48 hours after infection. B and C, This phenotype was complemented with fimK in trans at both 6 hours after infection (B) and 24 hours after infection (C). Data are combined from at least 3 independent experiments. Each symbol represents 1 animal, short bars represent geometric means of each group, and full dotted horizontal lines represent limits of detection (*P < .05, **P < .01, and ***P < .001, by the Mann–Whitney U test). Abbreviation: CFU, colony-forming units.
To verify that the increased urovirulence of TOP52ΔfimK previously demonstrated in C3H/HeN mice [21] was not specific to that murine background, we transurethrally infected C57BL/6 female mice with TOP52 or TOP52ΔfimK. Indeed, C57BL/6 bladders infected with TOP52ΔfimK bore significantly higher bacterial burdens 6 and 24 hours after infection than TOP52-infected bladders (Supplementary Figure 1). Taken together, these data indicate that, while fimK diminishes TOP52 virulence in the urinary tract, it is required for optimal virulence in K. pneumoniae murine pneumonia.
Intratracheal Inoculation With TOP52 Results in Limited Bacteremia and Dissemination
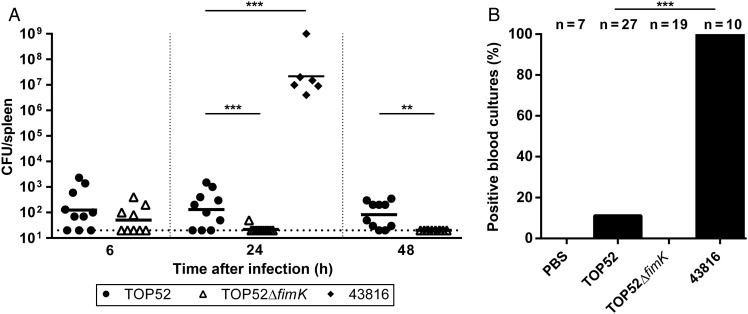
To test whether TOP52 murine pneumonia results in dissemination of infection beyond the respiratory tract, we first evaluated spleen bacterial titers. Spleen titers in TOP52-infected mice were very low but significantly higher than in TOP52ΔfimK-infected mice 24 and 48 hours after infection (P = .0027 and P = .0007, respectively; Figure 3A). At 1 and 2 weeks after intratracheal infection with TOP52 or TOP52ΔfimK, spleen titers were extremely low or absent and did not differ significantly (data not shown). In contrast, intratracheal inoculation of an equivalent inoculum (107 CFU) of the commonly used K. pneumoniae model strain 43816 yielded markedly higher spleen titers 24 hours after infection (P = .0002; Figure 3A). Intratracheal inoculation with 105 CFU of 43816, a typical experimental dose of this strain, similarly resulted in uniformly high (>104 CFU) spleen titers 24 hours after infection (data not shown). Cultures of blood specimens obtained 24 hours after infection were uniformly negative in mock-infected and TOP52ΔfimK-infected mice (Figure 3B). A total of 11.1% of TOP52-infected mice exhibited positive blood cultures, compared with 100% of 43816-infected mice (P < .0001). Intratracheal infection with the lower inoculum (105 CFU) of 43816 also yielded 100% bacteremia 24 hours after infection (data not shown). These data demonstrate that, while respiratory tract infection with TOP52 may be associated with bacteremia more often than its isogenic fimK mutant, TOP52 has substantially less propensity to disseminate than 43816.
Figure 3.
Klebsiella pneumoniae TOP52 and TOP52ΔfimK display limited dissemination and bacteremia. A, Female C57BL/6 mice were inoculated intratracheally with 107 TOP52 (circles), TOP52ΔfimK (triangles), or ATCC 43816 (diamonds). Spleen bacterial loads were measured at indicated time points. Data are combined from at least 3 independent experiments. Each symbol represents 1 animal, short bars represent geometric means of each group, and full dotted horizontal lines represent limits of detection (**P < .01 and ***P < .001, by the Mann–Whitney U test). B, Blood cultures at 24 hours after infection were positive in only 11.1% of TOP52-infected mice and 0% of TOP52ΔfimK-infected mice, compared with 100% of ATCC 43816–infected mice (***P < .001, by the Fisher exact test). Abbreviations: CFU, colony-forming units; PBS, phosphate-buffered saline.
Murine TOP52 Pneumonia Features Increased Inflammation Compared With TOP52ΔfimK
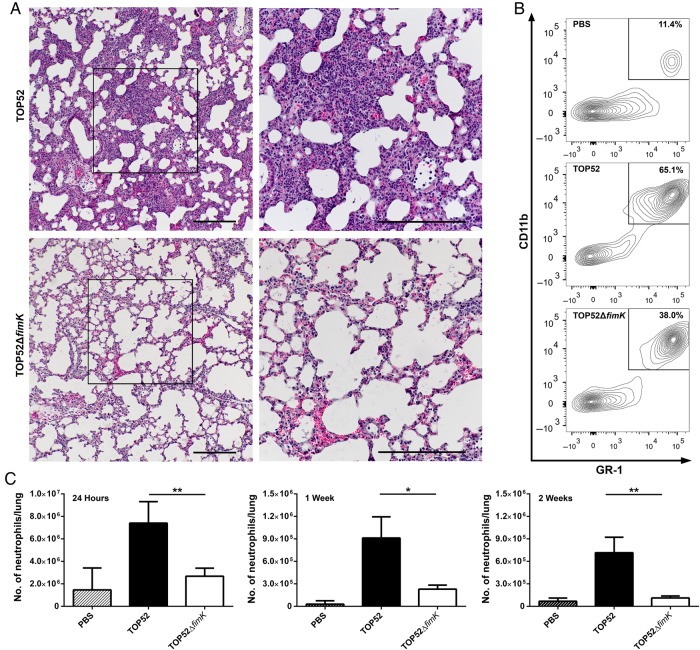
To gain insight into host response during TOP52 versus TOP52ΔfimK pneumonia, histologic analysis was performed on lung sections prepared 24 hours after infection. Mouse lungs infected with TOP52 demonstrated significant consolidation throughout, with few remaining morphologically normal alveoli (Figure 4A). A marked acute inflammatory infiltrate (neutrophils and macrophages) and areas of hemorrhage were evident. In contrast, TOP52ΔfimK-infected lungs exhibited only sporadic areas of acute inflammation and mild hemorrhage, with sparing of most distal airways. These differences in lung inflammation between TOP52-infected mice and TOP52ΔfimK-infected mice were amplified 48 hours after infection; TOP52-infected lungs were completely consolidated at this time point, while TOP52ΔfimK histologic findings were nearly normal (data not shown).
Figure 4.
Klebsiella pneumoniae TOP52 elicits a more robust innate immune response in the lungs at 24 hours after infection than TOP52ΔfimK. A, Representative histologic staining of murine lungs at 24 hours after infection with TOP52 (top panels) or TOP52ΔfimK (bottom panels) reveals marked differences in consolidation and acute inflammatory infiltrate. Boxes indicate areas of increased magnification in right panels (scale bars, 200 µm). B, Representative flow cytometry shows a higher proportion of neutrophils in lungs after TOP52 infection, compared with TOP52ΔfimK infection. One representative plot shown for each group, with mean neutrophil percentages indicated from 33 mice across 3 independent experiments. C, Flow cytometric quantification demonstrates significantly higher absolute numbers of neutrophils in TOP52-infected mouse lungs (solid bars) than in TOP52ΔfimK-infected lungs (open bars) at 24 hours, 1 week, and 2 weeks after infection. Data are shown as mean ± standard error of the mean (*P < .05 and **P < .01, by the Mann–Whitney U test). Abbreviation: PBS, phosphate-buffered saline.
We quantified neutrophilic inflammation by flow cytometry in the lungs of mice inoculated with TOP52, TOP52ΔfimK, or PBS. In mock-infected lungs, neutrophils (Gr-1+, CD11b+) comprised on average 11.4% of total live cells 24 hours after infection (Figure 4B). TOP52-infected lungs harbored a greater mean percentage of neutrophils 24 hours after infection, compared with TOP52ΔfimK-infected lungs (65% vs 38%; P = .0005). In addition, absolute neutrophil numbers in TOP52-infected mouse lungs were >2-fold higher than those in TOP52ΔfimK-infected lungs 24 hours after infection (P = .0016) and >3-fold higher at 1 and 2 weeks after infection (P = .0207 and P = .0022, respectively; Figure 4C).
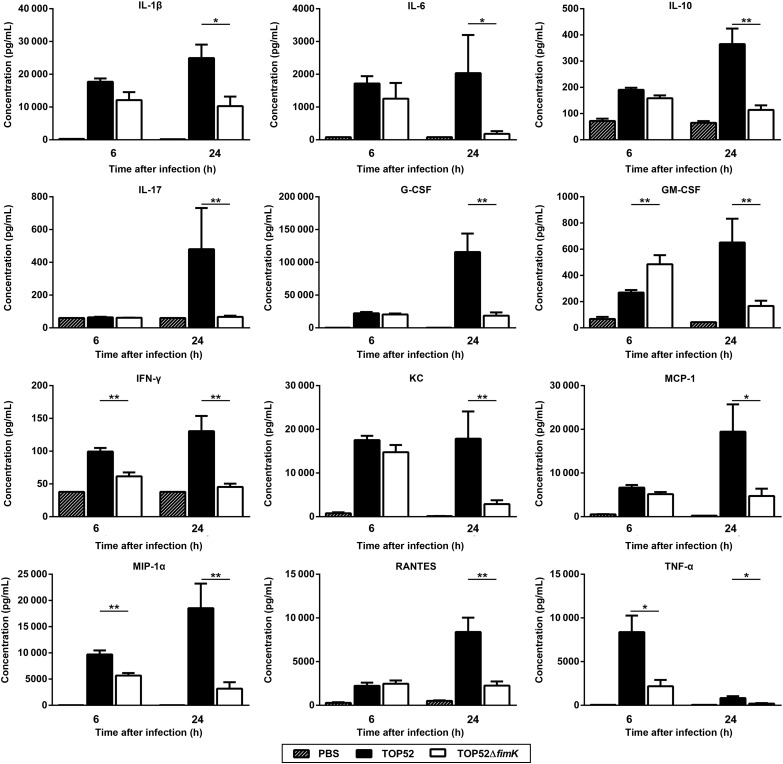
Quantification of 23 cytokines in lung homogenates was performed 6 and 24 hours after infection (Figure 5). There were few significant differences in lung cytokine levels 6 hours after infection with TOP52 or TOP52ΔfimK, notably including higher IFN-γ and TNF-α levels in TOP52-infected samples (P = .0022 and P = .0152, respectively). However, TOP52-infected lungs 24 hours after infection contained significantly higher levels of many proinflammatory cytokines (IL-1β, IL-6, IL-17, granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, IFN-γ, and TNF-α) and chemokines (KC, monocyte chemotactic protein 1, macrophage inflammatory protein 1α [MIP-1α], and RANTES) than TOP52ΔfimK-infected lungs. Together, these data indicate that the higher-titer infection achieved by TOP52 is accompanied by a more robust immune response (involving inflammatory mediators and immune cells), compared with TOP52ΔfimK.
Figure 5.
Klebsiella pneumoniae TOP52–infected lungs have an increased proinflammatory cytokine response at 24 hours after infection, relative to TOP52ΔfimK-infected lungs. TOP52-infected (solid bars), TOP52ΔfimK-infected (open bars), or phosphate-buffered saline (PBS) mock-infected (hatched bars) mouse lungs were harvested at 6 or 24 hours after infection, and homogenates were analyzed by multiplex bead array for cytokine content. Data were collected using organ homogenates from at least 3 independent mouse experiments and are shown as mean ± standard error of the mean (*P < .05 and **P < .01, by the Mann–Whitney U test). Abbreviations: G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte macrophage colony-stimulating factor; IFN-γ, interferon γ; IL-1β, interleukin 1β; IL-6, interleukin 6; IL-10, interleukin 10; IL-17, interleukin 17; MCP-1, monocyte chemotactic protein 1; MIP-1α, macrophage inflammatory protein 1α; TNF-α, tumor necrosis factor α.
FimK Promotes K. pneumoniae Serum Resistance, Capsule Production, and Resistance to Phagocytosis
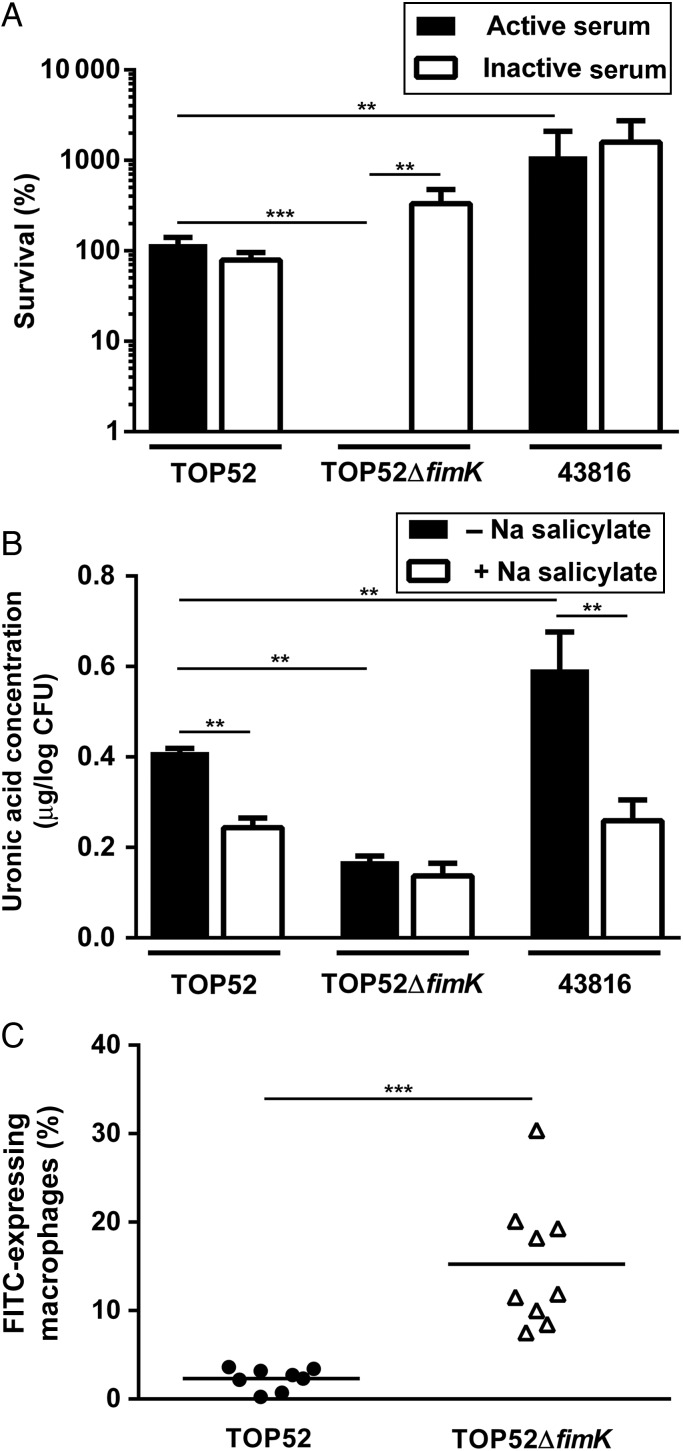
To test the hypothesis that differences in pathogenesis of TOP52 and TOP52ΔfimK reflect an influence of FimK on the expression of virulence factors important in resisting innate defenses, we first performed serum resistance assays (Figure 6A). The serum-resistant, heavily encapsulated K. pneumoniae strain 43816 served as a comparator. TOP52 titers were approximately 100% of input CFU after 3-hour incubation in active serum, while TOP52ΔfimK showed <1% survival (P = .0006). Meanwhile, 43816 exhibited 10-fold higher titers than TOP52 after 3 hours in active serum (P = .0041). Interestingly, while inactivation of serum did not alter TOP52 and 43816 survival, TOP52ΔfimK grew to 300% of input in inactivated serum (vs <1% survival in active serum; P = .0012), indicating that lack of FimK confers marked susceptibility to complement-mediated killing in vitro.
Figure 6.
Klebsiella pneumoniae fimK promotes serum resistance, capsule production, and resistance to phagocytosis. A, In serum resistance assays, bacteria were incubated with pooled human active (solid bars) or inactivated (open bars) serum for 3 hours. Bacterial survival is expressed as a percentage of input colony-forming units (CFU). B, Capsule extraction and quantification was performed on bacteria grown with (open bars) and without (solid bars) the capsule inhibitor sodium (Na) salicylate. C, Mice were intratracheally inoculated with fluorescein isothiocyanate (FITC)–labeled bacteria, and bronchoalveolar lavage was performed at 3 hours after infection. The percentage of macrophages expressing FITC was determined by flow cytometry after infection with TOP52 (circles) or TOP52ΔfimK (triangles). All data are combined from 2 independent experiments and shown as mean or mean ± standard error of the mean (**P < .01 and ***P < .001, by the Mann–Whitney U test).
To test whether increased susceptibility to complement was responsible for the attenuated in vivo virulence of TOP52ΔfimK, we intratracheally inoculated mice lacking the complement component 3 (C3) gene. Lungs and spleens of C3-deficient mice infected with TOP52 harbored increased bacterial loads relative to those infected with TOP52ΔfimK (Supplementary Figure 2A). Additionally, a significantly higher rate of bacteremia (80%) in C3−/− mice 24 hours after infection was noted after infection with TOP52, while TOP52ΔfimK blood cultures were all sterile (Supplementary Figure 2B). Thus, increased susceptibility of TOP52ΔfimK to complement activity in vitro does not account for its diminished virulence within the murine lung.
Beyond its role in serum resistance, capsule mediates interactions between bacteria and host epithelial and immune cells [8, 9, 32]. To test for alterations in capsule production, TOP52, TOP52ΔfimK, and 43816 were grown with and without the capsule inhibitor sodium salicylate [29], and uronic acid was extracted and quantified (Figure 6B). TOP52 produced significantly more capsule than TOP52ΔfimK and significantly less than 43816 (P = .0022 for each comparison). Both TOP52 and 43816 exhibited lower capsule production in the presence of sodium salicylate, as expected (P = .0022). However, sodium salicylate induced no further decrement in capsule production by TOP52ΔfimK.
To determine whether decreased capsule in TOP52ΔfimK might allow early immune clearance in vivo, we quantified phagocytosis by host leukocytes in BAL fluid of mice 3 hours after infection with FITC-labeled TOP52 or TOP52ΔfimK (Figure 6C). Only 2.3% of alveolar macrophages isolated from TOP52 infections were FITC positive (indicating uptake of bacteria), compared with 15.3% of macrophages from TOP52ΔfimK infections (P < .0001). A similar disparity was observed in neutrophils (1.8% vs 9.6%; P = .0159; data not shown). These data indicate that FimK positively influences production of capsule and impedes phagocytosis of K. pneumoniae in the lung.
DISCUSSION
K. pneumoniae is a versatile pathogen able to infect multiple host sites. K. pneumoniae as a species harbors substantial genomic diversity, but in contrast to E. coli, genomic content in most K. pneumoniae strains does not reliably predict host tissue tropism [33]. Although pyogenic liver abscesses with systemic sequelae have been associated with K. pneumoniae expressing magA, rmpA, and other capsular determinants [34, 35], the genomic attributes required for virulence in other sites or for gastrointestinal tract colonization remain largely unknown. Hyperexpression of type 1 pili by E. coli may reduce bowel colonization [36], but this has not been shown for K. pneumoniae. In addition, K. pneumoniae environmental isolates may colonize the murine gastrointestinal tract and infect the urinary tract just as effectively as clinical isolates [37]. The present study demonstrates that a strain derived from the human urinary tract can cause murine pneumonia. Many K. pneumoniae isolates likely retain the genetic machinery required to thrive in multiple environments into which they may be introduced. Past work demonstrated that the terminal gene in the type 1 pilus operon, fimK, reduces the virulence of K. pneumoniae in UTI [21]. Here we show that, in contrast, fimK carriage is imperative for virulence in the respiratory tract. Thus, fimK is not broadly deleterious and, in fact, confers a niche-specific survival advantage in the lung.
Deletion of fimK from K. pneumoniae TOP52 resulted in decreased bacterial loads, lower morbidity and mortality, and attenuated lung pathology and cytokine response during murine pneumonia. TOP52ΔfimK produced less capsule, which rendered it more susceptible to phagocytosis in vivo. This likely favors earlier clearance and diminished bacterial loads and, in turn, a less dramatic cellular inflammatory response. In contrast, lung titers of TOP52 at early time points were modestly higher than in the inoculum, reflecting replication (as opposed to simple persistence) of TOP52 in the lung. IFN-γ and TNF-α, levels of which were elevated in TOP52-infected lungs, represent important stimulators of activated proinflammatory macrophages [38, 39], which subsequently may fuel the globally increased proinflammatory milieu observed 24 hours after infection with TOP52. In turn, several locally produced chemokines promote neutrophil recruitment to the lungs (eg, KC and MIP-2) [40–42]. The amplified cellular response elicited by TOP52 persisted through 2 weeks, at which time it likely contributes to tissue repair.
The mechanisms by which FimK promotes pathogenesis in the lungs are not yet fully elucidated. FimK enhances production of capsule, an important virulence determinant in the lung [8–10]. As host C3 deficiency did not rescue TOP52ΔfimK, the contribution of capsule within the lung is likely in resistance to phagocytosis rather than to complement activity. Of note, previous work has proposed that fimbria and capsule may be counterregulated or exhibit steric interference in K. pneumoniae [43, 44]. FimK decreases expression of type 1 pili in TOP52 [21], which are critical for uropathogenesis but dispensable for virulence in the lung [18]. Wang et al showed that FimK in K. pneumoniae CG43S3 may instead increase expression of type 1 pili; however, fimB was overexpressed in this strain [45], which would bypass the known ability of FimK to influence orientation of the fimS promoter [21]. In our new studies, we confirmed that fimK deletion yields increased FimA expression (by Western blot; data not shown) in TOP52, as was shown previously [21]. FimK might also regulate other virulence factors important for pneumonia pathogenesis. FimK contains a predicted N-terminal helix-turn-helix domain, which may bind regions of DNA, and a C-terminal EAL domain, characteristic of phosphodiesterases that cleave the second messenger cyclic di-GMP [45, 46]. Ongoing work continues to investigate the broader range of K. pneumoniae virulence properties influenced by FimK.
Murine pulmonary infection with TOP52 provides an alternative preclinical model organism that contrasts with 43816. First, the histologic features of TOP52-infected lungs 24 hours after infection parallel those observed in human disease [47, 48]. Additionally, infection after intratracheal inoculation with TOP52 is largely survivable and contained to the respiratory tract; conversely, inoculation with 43816 (which causes mortality likely due to sepsis rather than respiratory failure) may be more valuable in studying disseminated K. pneumoniae infection. Compared with 100% bacteremia 24 hours after infection with 43816, only 11% of TOP52-infected mice were bacteremic, a proportion similar to that observed in human pneumonia (7%–15%) [49, 50]. Of note, the TOP52 genome was recently sequenced, facilitating additional genetic manipulation and genomic comparisons [19]. TOP52 may thus prove a more useful model strain for studying specific aspects of pneumonia pathogenesis (eg, adaptive immune responses). Continued work will provide an improved understanding of Klebsiella virulence mechanisms and avenues for inhibition, informing efforts to combat this versatile and increasingly antibiotic-resistant pathogen.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank C. Stallings, for critical review of the manuscript; E. Crouch, for assistance with pathology review; and S. Fritz, for statistical advice.
Financial support. This work was supported by Novartis Vaccines and Diagnostics (Pediatric Infectious Diseases Fellowship Award to D. A. R.); the National Institutes of Health (grants R01-DK080752, P50-DK064540, T32-AI106688, and R01AI104732), and the Child Health Research Center at Washington University School of Medicine (K12-HD076224 to S. C. M.).
Potential conflicts of interest. D. A. R. is partially supported by a grant funded by Novartis Vaccines and Diagnostics. D. A. H. serves on the scientific advisory board of BioVersys. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bakker-Woudenberg IA, van den Berg JC, Vree TB et al. Relevance of serum protein binding of cefoxitin and cefazolin to their activities against Klebsiella pneumoniae pneumonia in rats. Antimicrob Agents Chemother 1985; 28:654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Micro Rev 1998; 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rock C, Thom KA, Masnick M et al. Frequency of Klebsiella pneumoniae carbapenemase (KPC)-producing and non-KPC-producing Klebsiella species contamination of healthcare workers and the environment. Infect Cont Hosp Epidemiol 2014; 35:426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz-Price LS, Poirel L, Bonomo RA et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paterson DL, Ko WC, Von Gottberg A et al. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum beta-lactamases. Clin Infect Dis 2004; 39:31–7. [DOI] [PubMed] [Google Scholar]
- 6.Santino I, Bono S, Nuccitelli A et al. Microbiological and molecular characterization of extreme drug-resistant carbapenemase-producing Klebsiella pneumoniae isolates. Int J Immunopathol Pharmacol 2013; 26:785–90. [DOI] [PubMed] [Google Scholar]
- 7.Campos MA, Vargas MA, Regueiro V et al. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun 2004; 72:7107–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomas JM, Benedi VJ, Ciurana B et al. Role of capsule and O antigen in resistance of Klebsiella pneumoniae to serum bactericidal activity. Infect Immun 1986; 54:85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabha K, Nissimov L, Athamna A et al. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect Immun 1995; 63:847–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawlor MS, Handley SA, Miller VL. Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect Immun 2006; 74:5402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fodah RA, Scott JB, Tam HH et al. Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS One 2014; 9:e107394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lery LM, Frangeul L, Tomas A et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol 2014; 12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llobet E, March C, Gimenez P et al. Klebsiella pneumoniae OmpA confers resistance to antimicrobial peptides. Antimicrob Agents Chemother 2009; 53:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maroncle N, Rich C, Forestier C. The role of Klebsiella pneumoniae urease in intestinal colonization and resistance to gastrointestinal stress. Res Microbiol 2006; 157:184–93. [DOI] [PubMed] [Google Scholar]
- 15.Murphy CN, Mortensen MS, Krogfelt KA et al. Role of Klebsiella pneumoniae type 1 and type 3 fimbriae in colonizing silicone tubes implanted into the bladders of mice as a model of catheter-associated urinary tract infections. Infect Immun 2013; 81:3009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosen DA, Pinkner JS, Walker JN et al. Molecular variations in Klebsiella pneumoniae and Escherichia coli FimH affect function and pathogenesis in the urinary tract. Infect Immun 2008; 76:3346–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shankar-Sinha S, Valencia GA, Janes BK et al. The Klebsiella pneumoniae O antigen contributes to bacteremia and lethality during murine pneumonia. Infect Immun 2004; 72:1423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Struve C, Bojer M, Krogfelt KA. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect Immun 2008; 76:4055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson JG, Spurbeck RR, Sandhu SK et al. Genome sequence of Klebsiella pneumoniae urinary tract isolate Top52. Genome Announc 2014; 2:e00668–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosen DA, Hung CS, Kline KA et al. Streptozocin-induced diabetic mouse model of urinary tract infection. Infect Immun 2008; 76:4290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen DA, Pinkner JS, Jones JM et al. Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infect Immun 2008; 76:3337–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau HY, Clegg S, Moore TA. Identification of Klebsiella pneumoniae genes uniquely expressed in a strain virulent using a murine model of bacterial pneumonia. Microb Pathog 2007; 42:148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavender HF, Jagnow JR, Clegg S. Biofilm formation in vitro and virulence in vivo of mutants of Klebsiella pneumoniae. Infect Immun 2004; 72:4888–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawlor MS, Hsu J, Rick PD et al. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Micro 2005; 58:1054–73. [DOI] [PubMed] [Google Scholar]
- 25.Deng JC, Zeng X, Newstead M et al. STAT4 is a critical mediator of early innate immune responses against pulmonary Klebsiella infection. J Immunol 2004; 173:4075–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung CS, Dodson KW, Hultgren SJ. A murine model of urinary tract infection. Nat Protoc 2009; 4:1230–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podschun R, Sievers D, Fischer A et al. Serotypes, hemagglutinins, siderophore synthesis, and serum resistance of Klebsiella isolates causing human urinary tract infections. J Infect Dis 1993; 168:1415–21. [DOI] [PubMed] [Google Scholar]
- 28.Blumenkrantz N, Asboe-Hansen G. New method for quantitative determination of uronic acids. Anal Biochem 1973; 54:484–9. [DOI] [PubMed] [Google Scholar]
- 29.Domenico P, Schwartz S, Cunha BA. Reduction of capsular polysaccharide production in Klebsiella pneumoniae by sodium salicylate. Infect Immun 1989; 57:3778–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin TL, Yang FL, Yang AS et al. Amino acid substitutions of MagA in Klebsiella pneumoniae affect the biosynthesis of the capsular polysaccharide. PLoS One 2012; 7:e46783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deady LE, Todd EM, Davis CG et al. L-plastin is essential for alveolar macrophage production and control of pulmonary pneumococcal infection. Infect Immun 2014; 82:1982–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Favre-Bonte S, Joly B, Forestier C. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect Immun 1999; 67:554–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar V, Sun P, Vamathevan J et al. Comparative genomics of Klebsiella pneumoniae strains with different antibiotic resistance profiles. Antimicrob Agents Chemother 2011; 55:4267–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang CT, Chuang YP, Shun CT et al. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 2004; 199:697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu WL, Ko WC, Cheng KC et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 2006; 42:1351–8. [DOI] [PubMed] [Google Scholar]
- 36.McCormick BA, Klemm P, Krogfelt KA et al. Escherichia coli F-18 phase locked ‘on’ for expression of type 1 fimbriae is a poor colonizer of the streptomycin-treated mouse large intestine. Microb Pathog 1993; 14:33–43. [DOI] [PubMed] [Google Scholar]
- 37.Struve C, Krogfelt KA. Pathogenic potential of environmental Klebsiella pneumoniae isolates. Environ Microbiol 2004; 6:584–90. [DOI] [PubMed] [Google Scholar]
- 38.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity 2004; 21:467–76. [DOI] [PubMed] [Google Scholar]
- 39.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008; 8:958–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craig A, Mai J, Cai S, Jeyaseelan S. Neutrophil recruitment to the lungs during bacterial pneumonia. Infect Immun 2009; 77:568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Driscoll KE, Hassenbein DG, Howard BW et al. Cloning, expression, and functional characterization of rat MIP-2: a neutrophil chemoattractant and epithelial cell mitogen. J Leukoc Biol 1995; 58:359–64. [DOI] [PubMed] [Google Scholar]
- 42.Wolpe SD, Sherry B, Juers D et al. Identification and characterization of macrophage inflammatory protein 2. Proc Natl Acad Sci USA 1989; 86:612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schembri MA, Blom J, Krogfelt KA et al. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect Immun 2005; 73:4626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matatov R, Goldhar J, Skutelsky E et al. Inability of encapsulated Klebsiella pneumoniae to assemble functional type 1 fimbriae on their surface. FEMS Microbiol Lett 1999; 179:123–30. [DOI] [PubMed] [Google Scholar]
- 45.Wang ZC, Huang CJ, Huang YJ et al. FimK regulation on the expression of type 1 fimbriae in Klebsiella pneumoniae CG43S3. Microbiol 2013; 159:1402–15. [DOI] [PubMed] [Google Scholar]
- 46.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 2009; 7:263–73. [DOI] [PubMed] [Google Scholar]
- 47.Okada F, Ando Y, Honda K et al. Clinical and pulmonary thin-section CT findings in acute Klebsiella pneumoniae pneumonia. Eur Radiol 2009; 19:809–15. [DOI] [PubMed] [Google Scholar]
- 48.Bullowa JGM, Chess J, Friedman NB. Pneumonia due to Bacillus friedlanderi. Arch Intern Med 1937; 60:735–52. [Google Scholar]
- 49.Metersky ML, Ma A, Bratzler DW et al. Predicting bacteremia in patients with community-acquired pneumonia. Am J Resp Crit Care 2004; 169:342–7. [DOI] [PubMed] [Google Scholar]
- 50.Waterer GW, Wunderink RG. The influence of the severity of community-acquired pneumonia on the usefulness of blood cultures. Resp Med 2001; 95:78–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.