Abstract
The cranial neural crest (CNC) is a highly motile population of cells that is responsible for forming the face and jaw in all vertebrates and perturbing their migration can lead to craniofacial birth defects. Cell motility requires a dynamic modification of cell–cell and cell-matrix adhesion. In the CNC, cleavage of the cell adhesion molecule cadherin-11 by ADAM13 is essential for cell migration. This cleavage generates a shed extracellular fragment of cadherin-11 (EC1-3) that possesses pro-migratory activity via an unknown mechanism. Cadherin-11 plays an important role in modulating contact inhibition of locomotion (CIL) in the CNC to regulate directional cell migration. Here, we show that while the integral cadherin-11 requires the homophilic binding site to promote CNC migration in vivo, the EC1-3 fragment does not. In addition, we show that increased ADAM13 activity or expression of the EC1-3 fragment increases CNC invasiveness in vitro and blocks the repulsive CIL response in colliding cells. This activity requires the presence of an intact homophilic binding site on the EC1-3 suggesting that the cleavage fragment may function as a competitive inhibitor of cadherin-11 adhesion in CIL but not to promote cell migration in vivo.
Keywords: Cadherin, Cell adhesion, ADAM, Cranial neural crest, Xenopus
1. Introduction
Craniofacial development in all jawed vertebrates begins with the induction and migration of the cranial neural crest (CNC) cells, which are multipotent embryonic cells that later differentiate into the cartilage, bone, and ganglia of the face and head (Becker et al., 2013; LaBonne and Bronner-Fraser, 1999; Le Douarin and Kalcheim, 1999). CNC cells arise at the border of the neural plate and migrate extensive distances ventrally via defined pathways to the frontonasal prominences and pharyngeal arches. The CNC under-goes two phases of migration, starting with collective cell migration in which cell–cell adhesion maintains cohesion among the migrating group of cells, followed by a dispersion into single cell migration to reach their final locations (Alfandari et al., 2003; Alfandari et al., 2010; Sadaghiani and Thiebaud, 1987). Defects during early development that disrupt the proper induction, migration, proliferation or differentiation of CNC cells lead to an array of craniofacial abnormalities at birth. Craniofacial defects are among the most common birth defects, and many of these, including cleft lip and palate, can only be repaired by plastic surgery, while others have no treatment available. Elucidating the mechanisms by which the CNC develops is crucial to understanding how to prevent or even correct these craniofacial defects before birth.
In Xenopus, the mesenchymal cadherin-11 is an essential protein in the CNC and its levels have to be tightly regulated for proper migration. A decrease or increase of cadherin-11 protein levels caused by translation blocking morpholino oligonucleotides (MO), or overexpression leads to defects in migration (Borchers et al., 2001; Kashef et al., 2009). Cadherin-11 is a type II classical cadherin that contains five extracellular repeats (EC1-5) with immunoglobulin-like folds separated by flexible linkers, followed by a transmembrane and cytoplasmic domain (Becker et al., 2012). The membrane-distal EC1 possesses highly conserved tryptophans and the QAV motif, which function as the adhesive site for homophilic binding of cadherins on neighboring cells (Boggon et al., 2002; Patel et al., 2006). We have previously shown that ADAM13 cleaves cadherin-11 in the CNC (McCusker et al., 2009). Based on predicted molecular weights, the estimated cleavage site was determined to be between EC3 and EC4 and would therefore eliminate the homophilic binding site from the transmembrane stump. We further showed that the shed extracellular fragment of cadherin-11 containing EC1-3 retains biological function. Expression of an artificial cleavage fragment in the CNC could restore migration in cells devoid of ADAMs or overexpressing cadherin-11, suggesting that the EC1-3 fragment could promote CNC cell migration, however the mechanism by which it does so is not yet clear (McCusker et al., 2009).
Although maintenance of cell–cell adhesion is not a function that has been clearly demonstrated for cadherin-11, several pieces of evidence point towards this role. Namely, the overexpression of a cadherin-11 variant lacking 210 amino acids from the first two EC domains containing the homophilic binding site promotes CNC migration (Borchers et al., 2001), while replacing cadherin-11 with a mutant lacking the homophilic binding motif prevents CNC migration (Becker et al., 2013; Borchers et al., 2001). These variants are thought to act as dominant negatives of cell–cell adhesion. Cadherin-11 also regulates protrusion formation through its cytoplasmic domain via an interaction with the guanine nucleotide exchange factor GEF-Trio, as well as cell proliferation through binding to β-catenin (Borchers et al., 2001; Kashef et al., 2009; Koehler et al., 2013; McCusker et al., 2009). We have also recently shown that cadherin-11 plays a role in mediating cell–cell contacts during contact inhibition of locomotion (CIL; Becker et al., 2013), a mechanism that is important for the directional migration of CNC cells (Carmona-Fontaine et al., 2008). CIL is defined as the “the stopping of the continual locomotion of a cell in the same direction after collision with another cell” (Abercrombie, 1970). During CIL, cell–cell contacts are initiated by cadherins, which then modify RhoA/Rac1 activity through the non-canonical Wnt/PCP pathway (Becker et al., 2013; Carmona-Fontaine et al., 2008; Theveneau et al., 2010). This in turn affects the polarization of the cytoskeleton and leads to a change of migration direction. The process allows collectively migrating cells to define a cell-free edge at which most of the protrusive activity is concentrated, which allows the leading cells to persistently migrate ventrally away from the group and into their designated pathways. Importantly, loss of cadherin-11 in the CNC or overexpression of the homophilic binding-deficient variant causes defects in CIL (Becker et al., 2013; Kashef et al., 2009).
In this study, we demonstrate that homophilic binding is essential to the function of cadherin-11 during CNC migration. We also observe that a non-cleavable variant of cadherin-11 (Cad11-egf) cannot promote CNC migration in vivo, confirming that cleavage is essential for migration. We show that ADAM13 cleavage of cadherin-11 decreases cadherin-11-mediated cell–cell adhesion during CIL in vitro. In addition, our results reveal that the shed extracellular domain does not require homophilic binding to stimulate CNC migration in vivo indicating a novel function for EC1-3.
2. Materials and methods
2.1. Morpholinos and DNA constructs
ADAM13 morpholino antisense oligonucleotide (MO13) and morpholino against cadherin-11 (MO11) were designed as previously characterized (Kashef et al., 2009; McCusker et al., 2009) and purchased from Gene Tools, LLC (Philomath, OR, USA). Full-length ADAM13 (A13), protease-dead ADAM13-E/A, full-length cadherin-11 (C11), EC1-3-myc, GAP43-mcherry and GAP43-GFP were published previously (Alfandari et al., 2001; Kashef et al., 2009; McCusker et al., 2009). The 5′ untranslated region of cadherin-11 that is recognized by MO11 is not present in the cadherin-11 construct in pCS2+. In addition, four silent mutations were made downstream of the ATG to further prevent binding of MO11. C11-egf was engineered by introduction of a SacI site in the C11 construct immediately upstream of the transmembrane domain (QuikChange) and inserting the EGF-like domain of Xenopus ADAM13 (51 amino acids), amplified with the primers 5′-CTGAACCCCAATCCCTTAACTGTGTTTCTAAATGTAATGG-3′ and 5′-GCTCCAGTACTGAGTCCAGCAGTGACACCTACAGGGAGGT. A13-egf (containing two copies of the EGF-like domain) was made by inserting an AscI followed by a SacI site immediately upstream of the transmembrane domain, and then inserting the amplified EGF-like domain of ADAM13 into the AscI and SacI sites using the primers 5′-AGGCGCGCCTTGTGTTTCTAAATGTAATGG-3′ and 5′-CGAGCTCAGTGACACCTACAGGGAGGT-3′. The non-adhesive cadherin-11 (na-C11) and EC1-3 (na-EC1-3) each contain point mutations at W55A and W57A (W2 and W4 after prodomain removal), and A130M of the QAV motif to disrupt the homophilic binding site as previously reported (Tamura et al., 1998). EC1 and EC1-2 were made by introducing stop codons immediately upstream of EC2 (after F159) with the primers 5′-CCCGGAGTTCTAATTGCATGAAAACTACCACGCAAATGTG-3′ and 5′-TTTCATGCAATTAGAACTCCGGGGGATTATCATTTATGTC, or upstream of EC3 (after F268 with primers 5′-ACCAAAGTTTTAACCACAAAGTGCGTACCCCATGTCTGTG-3′ and 5′-CACTTTGTGGTTAAAACTTTGGTGGATTGTCATTGACATC-3′, respectively.
2.2. Cell culture and protein detection
Cos-7 cells (ATCC) were transfected according to manufacturer's instructions (FuGENE HD, Roche). After 48 h total cellular proteins were extracted with 1X TBS, 1% Triton X-100, 5 mM EDTA, 1X Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). Glycoproteins were purified from the cell extract using agarose bound Concavilin A (ConA) beads (Vector Labs) overnight and eluted in reducing Laemmli. Cadherin-11 was detected by Western blot using the mouse monoclonal antibody 1B4 (McCusker et al., 2009), and ADAM13 using the rabbit polyclonal antibody 6615 F (Alfandari et al., 1997). The monoclonal antibody 4F12 was produced against a bacterial fusion protein corresponding to the cadherin-11 EC1-3.
2.3. Embryo manipulation
Handling of Xenopus laevis embryos and CNC explants were performed as described previously (Kashef et al., 2009). All constructs were transcribed in vitro into mRNA according to manufacturer's description (Ambion Inc.). in vivo CNC migration assays by targeted injection of a fluorescent lineage tracer were performed as previously published (Abbruzzese et al., 2014; Abbruzzese et al., 2015; Cousin et al., 2011; Cousin et al., 2012). Eight-cell stage embryos were injected into the D1 blastomere with 333 pg of RFP-flag mRNA, plus 333 pg of all cadherin-11 constructs for overexpression assays. In knockdown experiments, 200 pg of RFP-flag mRNA and 5 ng of MO11 was used alone or together with 80 pg of cadherin-11 constructs. All embryos were raised at 15 °C until they reached tailbud stage and were scored for CNC migration by the presence of RFP-labeled cells in the migration pathways. Embryos were imaged using a Nikon fluorescent dissecting microscope or a Zeiss Stereo Lumar fluorescent stereoscope. For the confrontation and collision assays, 1 ng of mRNA or 2 ng of MO13 was injected into the D1 blastomere of eight-cell stage embryos.
2.4. Confrontation and collision assays
CIL assays were performed as described (Becker et al., 2013; Carmona-Fontaine et al., 2008). For the collision assay, CNC cells were dissociated for three minutes with 0.3 mM EGTA in Danilchiks buffer (lacking CaCl2) as described previously (Becker et al., 2013). Live cell images were taken with Axio Observer.Z1 spinning disc confocal microscope with 10x plan apochromate NA 0.45 air objective using AxioVision 4.8.2 software (Zeiss, Jena).
3. Results
3.1. Cadherin-11 cleavage is required for CNC migration
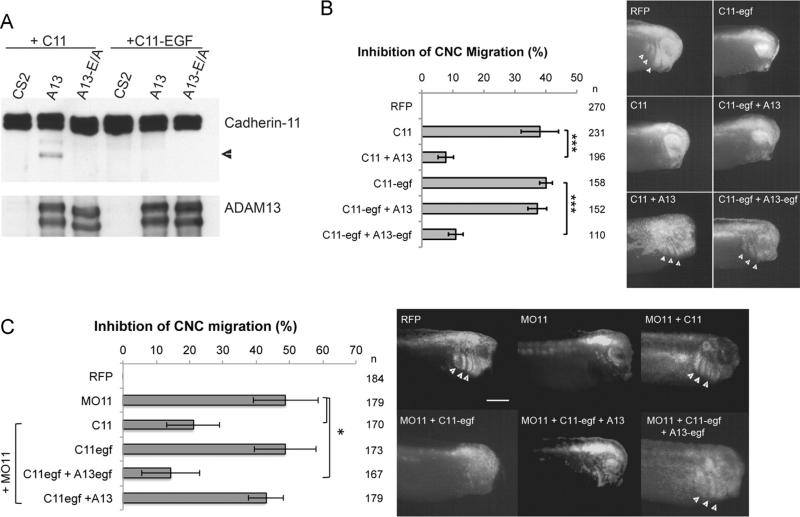
We have previously shown that cleavage of cadherin-11 by ADAM13 stimulates CNC migration. ADAM13 cleaves cadherin-11 in vitro and in vivo. In the CNC, overexpression of cadherin-11 blocks migration, and this can be rescued by increasing the levels of ADAM13 or by expressing an artificial extracellular cleavage fragment of cadherin-11 (EC1-3), which is normally generated by ADAM13. Furthermore, the triple knockdown of ADAM9, 13 and 19 can also be rescued by expressing EC1-3 (McCusker et al., 2009), suggesting that cleavage is important to generate an active soluble cadherin-11 fragment. However, attempts to prevent cleavage by mutating the predicted cleavage site in the linker region between extracellular repeats 3 and 4 were unsuccessful (data not shown). Based on the hypothesis that cleavage specificity was structural rather than sequence-specific, we used an alternative strategy of misaligning the protease active site of ADAM13 and the putative extracellular cadherin-11 cleavage site. Using protein modeling, we predicted the extracellular repeats of cadherin-11 to be approximately 50 Å and an EGF-like repeat to be 30 Å. Thus, by inserting the ADAM13 EGF-like repeat immediately N-terminal to the cadherin-11 transmembrane domain (C11-egf), the linker region between EC3 and EC4 would presumably be raised by 30 Å relative to the plasma membrane and the protease active site would then instead be in line with the globular domain of EC4. When transfected into Cos-7 cells, wild-type cadherin-11 is cleaved by ADAM13 but not by the inactive ADAM13-E/A mutant (Fig. 1A). However, when C11-egf is co-expressed with ADAM13, cleavage does not occur, as shown by the absence of the cleaved protein fragment that remains in the membrane (Fig. 1A).
Fig. 1.
Cleavage of cadherin-11 is essential for CNC migration in vivo. (A) Western blot from showing cleavage of C11 by ADAM13 but not by the inactive ADAM13-E/A containing a mutation in the active site. Glycoproteins were purified from transfected Cos-7 cells with ConA-agarose beads and detected using the antibody to the cadherin-11 cytoplasmic domain 1B4. Cleavage is observed by the presence of a shorter membrane-bound C11 fragment at 65 kDa (arrowhead). This cleavage fragment is absent when C11-egf is co-transfected with ADAM13. ADAM13 was detected using the antibody to the ADAM13 cytoplasmic domain 6615 F. The 120 kDa Pro-form and the 100 kDa mature form are shown. B–D) Targeted injection assays testing non-cleavable C11-egf in CNC migration in vivo. Histograms represent the percentage of embryos with no CNC migration, normalized to injection of RFP alone, for the overexpression of C11 or C11-egf with A13 or A13-egf (B) or the replacement of endogenous C11 with wildtype or non-cleavable C11-egf (C). Representative images of embryos in (C) are shown in (D) with arrowheads pointing to RFP-labeled cells that successfully migrated into the branchial and hyoid arches. n=number of embryos scored from three or more independent experiments. Error bars represent standard deviation to the mean. Student's t-test was performed to determine statistical significance. *p < 0.05, ***p < 0.005. Scale bar, 500 μm.
Overexpression of cadherin-11 inhibits CNC cell migration due to both increase in cell adhesion and association with β-catenin (Borchers et al., 2001). To test if the C11-egf construct was functional in the CNC, we first performed a targeted injection in which a single CNC precursor (dorsal animal blastomere) is injected at the 8-cell stage with a lineage tracer, red fluorescent protein (RFP), to follow the migration of CNC cells (Abbruzzese et al., 2014; Cousin et al., 2011, 2012). At the tailbud stage, embryos are scored for the presence of fluorescent cells in the CNC migration pathways, and for each condition the percentage of embryos without migration is normalized to the injection of RFP alone (% inhibition of CNC migration). Overexpression of either wild type cadherin-11 or C11-egf inhibits migration in 38% and 40% of embryos, respectively (Fig. 1B). The inhibition observed for C11-egf suggests that the protein is still functional as it can block CNC cell migration as well as the wild-type protein. While migration defects due to overexpression of wild-type cadherin-11 can be rescued by co-expressing ADAM13, overexpression of the non-cleavable C11-egf could not be rescued by ADAM13 (Fig. 1B). We then designed a new variant of ADAM13 that contains one extra copy of its EGF-like domain (A13-egf), so that the extracellular domains of C11-egf and A13-egf are each raised from the membrane by a similar distance. With this construct, we do observe a significant rescue of migration when expressed with C11-egf in the CNC, suggesting that A13-egf is able to cleave C11-egf. In embryos, expression of A13-egf but not A13 reduces the level of full-length C11-egf suggesting that A13-egf can indeed cleave C11-egf (Supplemental Fig. 1A). This also appears to be the case in Hek293T cells but is not as clear as these cells possess another protease that cleaves both C11 and C11-egf in the absence of ADAM13 (Supplemental Fig. 1B).
Next we sought to determine if cleavage of cadherin-11 is essential for CNC cell migration by replacing the endogenous cadherin-11 with the non-cleavable C11-egf. In a targeted injection assay, we injected a morpholino oligonucleotide against cadherin-11 (MO11) to prevent translation of the endogenous protein, which blocked migration in 40% of the embryos (Fig. 1C and D), consistent with previous results (Kashef et al., 2009). CNC migration is significantly restored by co-injecting MO11 with mRNA encoding a morpholino-resistant cadherin-11. However, expression of C11-egf in the MO11 background leads to migration defects similar to MO11 alone, which can be rescued by co-expressing ADAM13-egf but not by wild type ADAM13 (Fig. 1C). Taken together, these results demonstrate that cleavage of cadherin-11 by ADAM13 is essential for its function during CNC cell migration.
3.2. Increase of ADAM13 expression leads to higher CNC invasiveness and blocks repulsive response in single colliding CNC cells
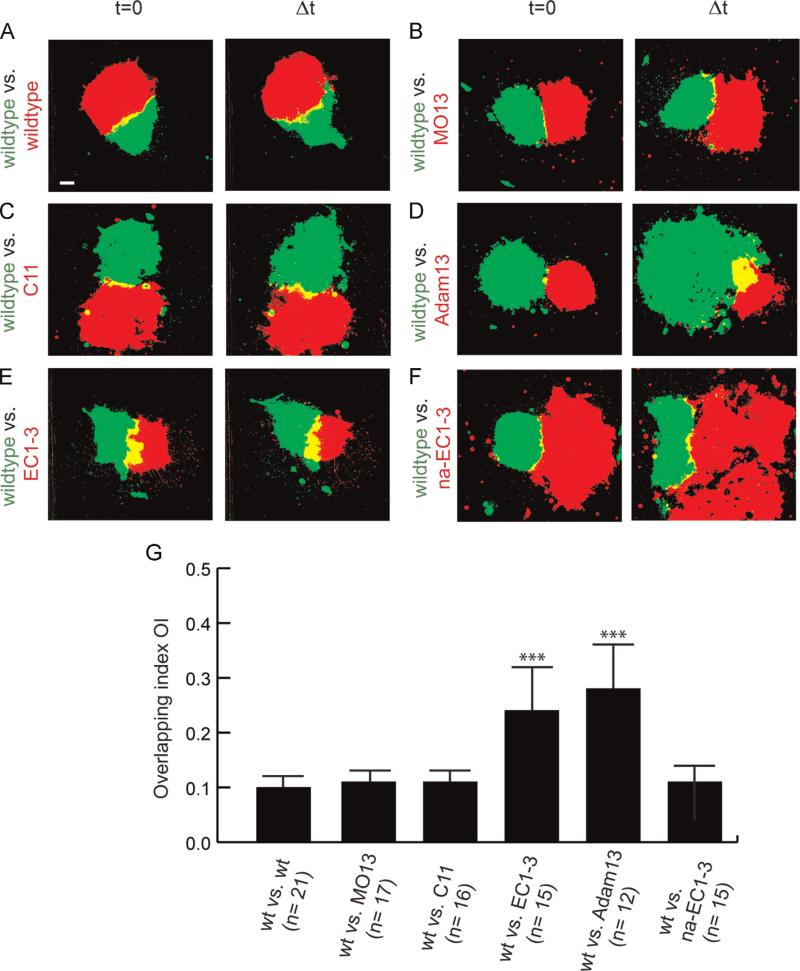
As cadherin-11 mediated cell–cell adhesion promotes CIL and is important for the directional migration of CNC in vivo (Becker et al., 2013), we hypothesized that ADAM13 might play a role in CIL. To test this hypothesis we performed explant confrontation assays in vitro (Becker et al., 2013; Carmona-Fontaine et al., 2008). In wild type explants, CIL prevents the mutual invasion of CNC cells (Carmona-Fontaine et al., 2008). By confronting explants injected with two different lineage tracers (RFP and GFP), it is possible to quantify the degree of overlap of the two explants (overlapping index, OI, defined as the quotient of the explant overlapping area (yellow) and the normalized area of one explant (red or green) at the time point of highest invasion Δt (Becker et al., 2013). Inhibition of CIL increases the invasiveness of the explants resulting in an increase of the OI.
Fluorescently labeled wild-type CNC explants show characteristic CIL with a small OI (10.4%, Fig. 2A and G; Movie S1). Interestingly both ADAM13 KD (Fig. 2B, OI= 11.4%) and cadherin-11 overexpression (Fig. 2C, OI=11%) were not significantly different from the wild-type CNC (Movies S2 and S3). In contrast, over-expression of ADAM13 (Fig. 2D, OI=24%) or the cadherin-11 cleavage fragment EC1-3 (Fig. 2E, OI = 28.5%) showed significantly higher invasion suggesting that CIL was inhibited in these explants (Movies S4 and S5). We then tested whether this inhibition of CIL depends on the presence of a functional homophilic binding site on the EC1-3 fragment. The homophilic interaction occurs by a “β-strand swapping” mechanism where the two amino-terminal tryptophan residues from each cadherin-11 molecule extend into the large hydrophobic QAV pockets of the partner molecule (Patel et al., 2006). Mutating the two tryptophan residues to alanine and replacing the alanine of the QAV motif with methionine (QMV) is sufficient to disrupt these interactions (Tamura et al., 1998). In the confrontation assay, expression of the non-adhesive mutant of EC1-3 (na-EC1-3) in one of the explants did not increase cell invasion (Fig. 2F, OI of 11.3%).
Fig. 2.
Increase of ADAM13 expression leads to higher CNC invasiveness in vitro. Confrontation assay. Left column: Confronted explants at time point t=0. Right column: Confronted CNC explants at time point of highest invasion Δt. Confrontations of wildtype explants with (A) wildtype CNC explants, (B) explants of ADAM13-depleted CNC (MO13) or (C) CNC explants overexpressing C11 show only small overlapping explant borders with low invasiveness. In contrast, yellow overlapping area increases significantly in confrontations with CNC cells overexpressing (D) ADAM13 or (E) cleavage product EC1-3. (F) Overexpression of the non-adhesive extracellular fragment na-EC1-3 in CNC explants leads to small overlap when confronted with wild-type explants. Average Overlapping Index (OI) is given in (G) with n=number of confrontations. wt: wildtype. Error bar shows standard error. (***) Significance to wildtype vs. wildtype with p < 0.005 after Student's t-test. Scale bar, 50 μm.
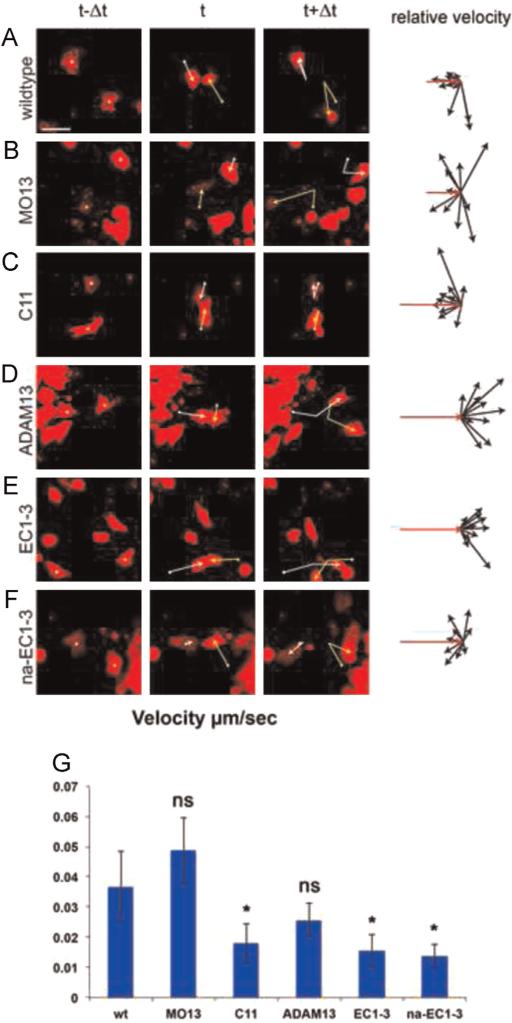
To confirm our results in single CNC cells we used a collision analysis assay (Becker et al., 2013). Wild-type CNC cells change direction after a collision with another CNC cell as demonstrated by relative velocity analyses (Fig. 3A, original vector in red). Although depletion of ADAM13 (Fig. 3B) or overexpression of cadherin-11 (Fig. 3C) leads to formation of larger and longer lasting CNC cell clusters in vitro, single cells show typical CIL response and change of migration direction after the collision. In contrast, collisions of two CNC cells overexpressing either ADAM13 or EC1-3 lead to minimal change of direction upon contact (Fig. 3D and E, respectively). Again, this suggests that increased ADAM13 expression or expression of the cleavage fragment of cadherin-11 interferes with CIL in single CNC cells. Expression of the non-adhesive fragment na-EC1-3, however, did not perturb single cell repulsion (Fig. 3F). Interestingly, over-expression of cadherin-11, the EC1-3 or na-EC1-3 significantly reduced cell velocity by more than two fold (Fig. 3G). It is interesting that the velocity of these cells is similar to cells during the first phase of collective cell migration (50–60 μm/h), while both the wild type and cells lacking ADAM13 have the typical velocity of single CNC cells migrating during the second phase of migration (130–175 μm/h) (Alfandari et al., 2003).
Fig. 3.
Increase of ADAM13 expression blocks the repulsive response in colliding single CNC cells in vitro. Collision assay. First three columns: Single CNC cells before (t-Δt), during (t) and after (t+Δt) mutual contact. Yellow and white vectors give change of cell location. Fourth column: Relative velocity vectors with initial velocity vector (red, n=10 collisions). Mutual collisions of (A) wildtype CNC cells, (B) ADAM13-depleted CNC cells (MO13) and (C) CNC cells overexpressing C11 lead to repulsive response of the cells and CIL. In contrast, CNC cells overexpressing (D) ADAM13 or (E) the cleavage product EC1-3 show no change of direction. (F) Repulsive response and CIL are observed in collisions in single CNC cells over-expressing the non-adhesive fragment na-EC1-3. Scale bar, 50 μm. (G) Average velocity of single cells in the various conditions. Overexpression of cadherin-11 and expression of EC1-3 or na-EC1-3 significantly reduce the cell velocity.
Taken together, these results demonstrate that ADAM13 can modulate CIL. Given that the proteolytic fragment of cadherin-11 (EC1-3) produces the same effect as ADAM13 overexpression it is likely that ADAM13 decreases CIL by cleaving cadherin-11 and producing the EC1-3 fragment. The presence of the homophilic site on the EC1-3 is critical suggesting that it may act as a competitive inhibitor for cadherin-11 mediated cell–cell adhesion during CIL. Interestingly, the loss of ADAM13 function leads to normal CIL in vitro indicating that migration defects due to higher levels of cadherin-11 or loss of ADAM13 in vivo are not due to a defect in CIL.
3.3. EC1-3 promotes CNC migration independently of the homophilic binding site
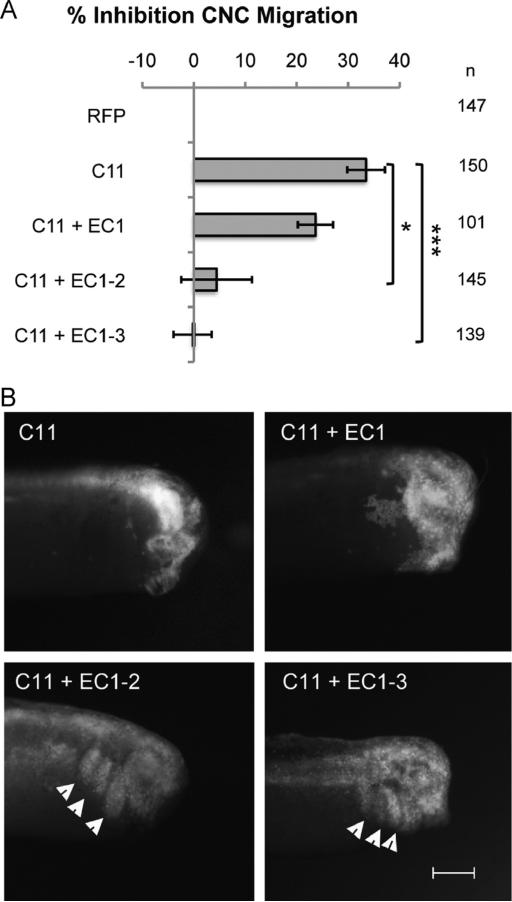
The previous results suggest that EC1-3 can act as a competitive inhibitor of cadherin-11 in CIL and that this function requires the homophilic binding site. We next tested if the same site was also critical to promote CNC migration in vivo. Using the targeted injection assay, we first investigated which EC repeats of the shed cadherin-11 fragment are required to promote CNC migration. Thus, we generated truncations of EC1-3 that included either only EC1, or EC1 and EC2 (EC1-2). We found that EC1 alone was not sufficient to rescue CNC migration defects that are due to overexpression of cadherin-11 (23.7% inhibition compared to 33.5% inhibition for overexpression of cadherin-11 alone; Fig. 4). However, EC1-2 was able to rescue the migration defects similar to EC1-3 (4.4% and −0.2% inhibition, respectively; Fig. 4). It is important to note that a negative inhibition number for EC1-3 shows that the protein tested can not only rescue the overexpression of full-length cadherin-11 but also actually increase CNC migration, something that was also seen previously (McCusker et al., 2009).
Fig. 4.
Cadherin-11 extracellular repeats 1 and 2 are sufficient to stimulate CNC migration in vivo. (A) Histogram representing the percentage of embryos with no migration due to the overexpression of C11, normalized to RFP. Rescue of migration requires co-expression of the artificial C11 cleavage fragment containing at least extracellular repeats 1 and 2 (EC1-2), while EC1 alone is not sufficient. (B) Representative images of embryos in (A) are shown with arrowheads pointing to RFP-labeled cells that successfully migrated into the branchial and hyoid arches. n=number of embryos scored from three independent experiments. Error bars are standard deviation to the mean. A Student's t-test was performed for significance to C11 overexpression. * < 0.05, *** < 0.005. Scale bar, 500 μm.
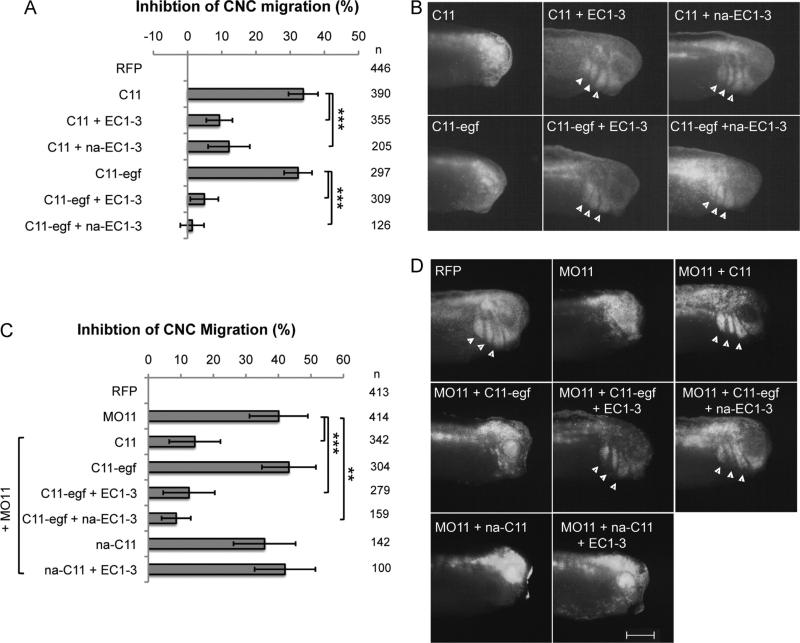
We then asked whether homophilic interactions of cadherin-11 are required to promote CNC cell migration. Our results show that in embryos overexpressing cadherin-11 in the CNC (33.8% inhibition), migration can be significantly recovered by co-expressing either wild type EC1-3 (9.3% inhibition) or the non-adhesive mutant, na-EC1-3 (12% inhibition; Fig. 5A and B). This was also true for the overexpression of C11-egf, where the inhibition of CNC migration was restored to 4.9% with co-expression of EC1-3, and 1.4% with na-EC1-3, compared to 32.3% inhibition for overexpression of C11-egf alone (Fig. 5A and B).
Fig. 5.
EC1-3 does not promote migration in vivo through the homophilic site. (A–D) Targeted injection assays to measure CNC migration with histograms showing the percentage of embryos with inhibited migration normalized to RFP (A, C) and representative images of each condition with arrowheads pointing to RFP-positive cells within the branchial and hyoid arches (B, D). na-EC1-3 rescues the overexpression of C11 or C11-egf as well as EC1-3 (A, B), and also rescues migration when expressed along with MO11 and C11-egf (C, D). Full-length na-C11 cannot rescue migration when expressed with MO11, and cannot be rescued by wildtype EC1-3 in the MO11 background (C, D). n=number of embryos scored from at least three independent experiments. Error bars are standard deviation to the mean and a Student's t-test was performed to determine statistical significance. ** < 0.01, *** < 0.005. Scale bar, 500 μm.
We then used the non-cleavable cadherin-11 (C11-egf) to test if the homophilic binding site was required both in the transmembrane and the extracellular fragment. For this we used the morpholino to cadherin-11 with various combination of the rescue mRNA. As seen previously, C11-egf cannot rescue CNC migration by itself but can when co-injected with the EC1-3 (Fig. 5C and D). Surprisingly, this rescue is also achieved by the na-EC1-3 showing that the homophilic binding site is not required once cadherin-11 is cleaved. In contrast, the same mutation present on the full-length cadherin-11 (na-C11) renders the protein incapable of rescuing migration whether the EC1-3 is provided or not (Fig. 5C and D).
Taken together these results show that cadherin-11-mediated cell–cell adhesion supported by the homophilic binding site is essential for CNC migration in vivo. However, once cleaved by ADAM13, the extracellular fragment of cadherin-11 does not rely on homophilic binding for its activity. Additionally, our results suggest that the target receptor of EC1-3 is not cadherin-11 or at least not the classical binding site.
4. Discussion
Increased levels of cadherin-11 are implicated in both natural and disease-related cell migration and invasion. For example it plays a critical role in trophoblast cell invasion during implantation (Getsios and MacCalman, 2003; Peng et al., 2015) as well as during cancer cell migration and invasive metastasis (Huang et al., 2010). Cadherin-11 is a multifaceted regulator of CNC cell migration. Its cytoplasmic domain regulates both filopodia formation by binding to the guanine nucleotide exchange factor (GEF) Trio (Kashef et al., 2009), and signaling by physically interacting with β-catenin to regulate its transcriptional activity resulting in the regulation of both CNC cell proliferation and cell specification (Koehler et al., 2013). On the extracellular side, cleavage of the extracellular domain of cadherin-11 by ADAM13 promotes CNC cell migration via an unknown mechanism (McCusker et al., 2009). Here we investigated the domains of cadherin-11 that are critical for its function when inserted into the plasma membrane or after shedding by ADAM13 to provide new insight about the mechanism by which the extracellular fragment of cadherin-11 can promote cellular migration, a role that is likely conserved in other invasive cells.
We originally proposed that cleavage of cadherin-11 by ADAM13 could promote migration in multiple ways. First, by removing the homophilic domain (EC1) from the plasma membrane it should decrease cell–cell adhesion. Second, the released EC1-3 fragment could also act as a competitive inhibitor binding intact cadherin-11 and further decreasing the cell–cell adhesion. Finally, the released EC1-3 could bind to a signaling receptor on the surface of CNC to promote cell migration. We engineered a noncleavable form of cadherin-11 in addition to mutating the homophilic site exclusively on the transmembrane or the shed form of cadherin-11, to test these various possibilities.
By replacing endogenous cadherin-11 by the non-cleavable cadherin-11 (C11-egf) we demonstrated that cleavage is absolutely essential for CNC migration, confirming our previous findings (McCusker et al., 2009). Expressing the EC1-3 together with the non-cleavable cadherin-11 fully rescued migration even when the homophilic binding site was mutated (na-EC1-3), demonstrating that once shed, the cadherin-11 fragment promotes migration by a mechanism independent of homophilic competition. On the other hand, exchanging the endogenous cadherin-11 with full-length cadherin-11 containing a mutated homophilic domain did not rescue migration even in the presence of a wild type EC1-3, clearly demonstrating the absolute requirement of the homophilic binding on the integral cadherin-11 form. While this confirms previous results showing that non-adhesive cadherin-11 could not substitute for the endogenous protein (Becker et al., 2013), our results further define which form (integral) requires the homo-philic binding domain. These results point to a model in which the EC1-3 binds to a unknown receptor at the surface of the CNC to promote cell migration. Other cadherins have also been shown to be cleaved and promote cell migration. For example, ADAM10 cleaves N-cadherin and promotes glioblastoma cell migration (Kohutek et al., 2009). In addition the cleavage of N-cadherin releases a pool of β-catenin from the membrane to the cytoplasm increasing β-catenin downstream target genes (Reiss et al., 2005). Furthermore, cleavage of E-cadherin by ADAM15 produces an extracellular fragment that binds to the EGF receptor Her2 and Her3 and can promote signaling throught the Erk pathway (Najy et al., 2008). In squamous cell carcinoma the E-cadherin extracellular fragment binding to EGF- and IGF1- receptors, stimulates both MAPK and AKT pathway promoting cell migration and invasion (Brouxhon et al., 2014). In these studies the role of the homophilic binding site was not tested. Thus, it seems that shed cadherin fragments are widely used to promote cell migration and invasion in both natural (neural crest) and pathological (cancer) processes. Further work will be required to define the signaling pathway stimulated by the cadherin-11 extracellular fragment in the CNC in vivo.
Cadherin-11 has also been shown to mediate contact inhibition of locomotion (CIL) during CNC migration (Becker et al., 2013). Together with N-cadherin, cadherin-11 engagment at the surface of CNC prevents cell invasion and promotes a change in direction when two neural crest cells interact. Our results show that ADAM13 activity can reduce CIL. Given that the cleavage product of cadherin-11 by ADAM13 (EC1-3) also decreases CIL, it is likely that the observed decrease of CIL by ADAM13 is due to ADAM13 cleavage of cadherin-11. Obviously we cannot eliminate the possibility that ADAM13 overexpression also affects other cell adhesion molecules that could account for the inhibition of CIL. In particular N-cadherin has been shown to play an essential role in CIL (Theveneau et al., 2010) and Xenopus ADAM13 can cleave N-cadherin in vitro (unpublished observation). It is also possible that ADAM13-induced decrease of CIL is due to a protease independent function, for example a role of the adhesive disintegrin and cysteine-rich or the cytoplasmic domains, but this is less likely given the ability of EC1-3 to produce the same phenotype. In addition these results suggests that the cleaved EC1-3 fragment can indeed act as a homophilic competitor for the integral cadherin-11 and that ADAM13 can decrease cell adhesion by reducing intact cadherin-11 and producing the EC1-3 fragment. The fact that the homophilic site on EC1-3 is important to interfere with CIL suggests that it is not the mechanism that allows EC1-3 to promote CNC migration in vivo. In addition our result showing that knockdown of ADAM13 does not affect CIL suggests that the integral cadherin-11, rather than the cleavage fragment, is involved in this process. On the other hand, our result also suggests that when CIL needs to be reduced, for example to invade tissues that may express cadherin-11, ADAM13 can perform this function rapidly. Given that the proteolytic activity of ADAM13 is inhibited by Fz4 in the CNC (Abbruzzese et al., 2015), this protein could act as a switch to control cadherin-11 cleavage at various phases of CNC development and as such finely tune the intensity of CIL.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health, U.S. Public Health Service, Grants RO1-DE016289 to D.A and F31-DE023275 to G.A. G.A. and S.F.B. were supported by the KHYS visiting Grant from Karlsruhe Institute of Technology. S.F.B was supported by the Deutsche Forschungsgemeinschaft (DFG) WE-1208-13-1. J.K.s' Young Investigator Group received financial support from the “Concept for the Future” of the Karlsruhe Institute of Technology within the framework of the German Excellence Initiative. J.K. is further supported by the Deutsche Forschungsgemeinschaft (DFG) DFG-FOR 1756 (www.dfg.de).
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2015.07.018.
References
- Abbruzzese G, Cousin H, Salicioni AM, Alfandari D. GSK3 and Polo-like kinase regulate ADAM13 function during cranial neural crest cell migration. Mol. Biol. Cell. 2014 doi: 10.1091/mbc.E14-05-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbruzzese G, Gorny AK, Kaufmann LT, Cousin H, Kleino I, Steinbeisser H, Alfandari D. The Wnt receptor Frizzled-4 modulates ADAM13 metalloprotease activity. J. Cell Sci. 2015;128:1139–1149. doi: 10.1242/jcs.163063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abercrombie M. Contact inhibition in tissue culture. In vitro. 1970;6:128–142. doi: 10.1007/BF02616114. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Gaultier A, Hoffstrom BG, DeSimone DW. Integrin alpha5beta1 supports the migration of Xenopus cranial neural crest on fibronectin. Dev. Biol. 2003;260:449–464. doi: 10.1016/s0012-1606(03)00277-x. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, DeSimone DW. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr. Biol. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Marsden M. Mechanism of Xenopus cranial neural crest cell migration. Cell Adhes. Migr. 2010;4:553–560. doi: 10.4161/cam.4.4.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfandari D, Wolfsberg TG, White JM, DeSimone DW. ADAM 13: a novel ADAM expressed in somitic mesoderm and neural crest cells during Xenopus laevis development. Dev. Biol. 1997;182:314–330. doi: 10.1006/dbio.1996.8458. [DOI] [PubMed] [Google Scholar]
- Becker SF, Langhe R, Huang C, Wedlich D, Kashef J. Giving the right tug for migration: cadherins in tissue movements. Arch. Biochem. Biophys. 2012;524:30–42. doi: 10.1016/j.abb.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Becker SF, Mayor R, Kashef J. Cadherin-11 mediates contact inhibition of locomotion during Xenopus neural crest cell migration. PLoS One. 2013;8:e85717. doi: 10.1371/journal.pone.0085717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Borchers A, David R, Wedlich D. Xenopus cadherin-11 restrains cranial neural crest migration and influences neural crest specification. Development. 2001;128:3049–3060. doi: 10.1242/dev.128.16.3049. [DOI] [PubMed] [Google Scholar]
- Brouxhon SM, Kyrkanides S, Teng X, Athar M, Ghazizadeh S, Simon M, O'Banion MK, Ma L. Soluble E-cadherin: a critical oncogene modulating receptor tyrosine kinases, MAPK and PI3K/Akt/mTOR signaling. Oncogene. 2014;33:225–235. doi: 10.1038/onc.2012.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin H, Abbruzzese G, Kerdavid E, Gaultier A, Alfandari D. Translocation of the cytoplasmic domain of ADAM13 to the nucleus is essential for Calpain8-a expression and cranial neural crest cell migration. Dev. Cell. 2011;20:256–263. doi: 10.1016/j.devcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin H, Abbruzzese G, McCusker C, Alfandari D. ADAM13 function is required in the 3 dimensional context of the embryo during cranial neural crest cell migration in Xenopus laevis. Dev. Biol. 2012 doi: 10.1016/j.ydbio.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getsios S, MacCalman CD. Cadherin-11 modulates the terminal differentiation and fusion of human trophoblastic cells in vitro. Dev. Biol. 2003;257:41–54. doi: 10.1016/s0012-1606(03)00041-1. [DOI] [PubMed] [Google Scholar]
- Huang CF, Lira C, Chu K, Bilen MA, Lee YC, Ye X, Kim SM, Ortiz A, Wu FL, Logothetis CJ, Yu-Lee LY, Lin SH. Cadherin-11 increases migration and invasion of prostate cancer cells and enhances their interaction with osteo-blasts. Cancer Res. 2010;70:4580–4589. doi: 10.1158/0008-5472.CAN-09-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashef J, Kohler A, Kuriyama S, Alfandari D, Mayor R, Wedlich D. Cadherin-11 regulates protrusive activity in Xenopus cranial neural crest cells upstream of Trio and the small GTPases. Genes Dev. 2009;23:1393–1398. doi: 10.1101/gad.519409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler A, Schlupf J, Schneider M, Kraft B, Winter C, Kashef J. Loss of Xenopus cadherin-11 leads to increased Wnt/beta-catenin signaling and up-regulation of target genes c-myc and cyclin D1 in neural crest. Dev. Biol. 2013;383:132–145. doi: 10.1016/j.ydbio.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Kohutek ZA, diPierro CG, Redpath GT, Hussaini IM. ADAM-10-mediated N-cadherin cleavage is protein kinase C-alpha dependent and promotes glioblastoma cell migration. J. Neurosci.: Off. J. Soc. Neurosci. 2009;29:4605–4615. doi: 10.1523/JNEUROSCI.5126-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Molecular mechanisms of neural crest formation. Annu. Rev. Cell Dev. Biol. 1999;15:81–112. doi: 10.1146/annurev.cellbio.15.1.81. [DOI] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The neural crest. 2nd ed. Cambridge University Press; Cambridge, UK; New York, NY, USA.: 1999. [Google Scholar]
- McCusker C, Cousin H, Neuner R, Alfandari D. Extracellular cleavage of cadherin-11 by ADAM metalloproteases is essential for Xenopus cranial neural crest cell migration. Mol. Biol. Cell. 2009;20:78–89. doi: 10.1091/mbc.E08-05-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najy AJ, Day KC, Day ML. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. J. Biol. Chem. 2008;283:18393–18401. doi: 10.1074/jbc.M801329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, Shapiro L. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell. 2006;124:1255–1268. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Peng B, Zhu H, Ma L, Wang YL, Klausen C, Leung PC. AP-1 transcription factors c-FOS and c-JUN mediate GnRH-induced cadherin-11 expression and trophoblast cell invasion. Endocrinology. 2015;156:2269–2277. doi: 10.1210/en.2014-1871. [DOI] [PubMed] [Google Scholar]
- Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell–cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani B, Thiebaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev. Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L. Structure-function analysis of cell adhesion by neural (N−) cadherin. Neuron. 1998;20:1153–1163. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.