Abstract
Long non-coding RNAs (lncRNAs) represent a class of potent regulators of gene expression that are found in a wide array of eukaryotes; however, our knowledge about these molecules in plants is still very limited. In particular, a number of model plant species still lack comprehensive data sets of lncRNAs and their annotations, and very little is known about their biological roles. To meet these shortcomings, we created an online database of lncRNAs in 10 model plant species. The lncRNAs were identified computationally using dozens of publicly available RNA sequencing (RNA-Seq) libraries. Expression values, coding potential, sequence alignments as well as other types of data provide annotation for the identified lncRNAs. In order to better characterize them, we investigated their potential roles in splicing modulation and deregulation of microRNA functions. The data are freely available for searching, browsing and downloading from an online database called CANTATAdb (http://cantata.amu.edu.pl, http://yeti.amu.edu.pl/CANTATA/).
Keywords: Database, Long-non-coding RNAs, MicroRNAs, RNA–RNA interactions, Splicing
Introduction
Long non-coding RNAs (lncRNAs) constitute untranslated RNA molecules that are at least 200 nt long. They represent a large portion of well-annotated transcriptomes; for instance there are 145,331 lncRNAs known in the human transcriptome (NONCODE v4, Xie et al. 2014), which is >6-fold more than the number of protein-coding transcripts in Ensembl 77 (Cunningham et al. 2014). In plants, the numbers of lncRNAs found are usually an order of magnitude lower than in animals, but they still constitute an important component of their transcriptomes. To date, however, the functions of very few lncRNAs have been analyzed in laboratories, while in silico analyses aiming at deciphering their functions have met a number of difficulties as they are highly heterogeneous in biogenesis, sequence, structure and function. The data accumulated so far have come mostly from animal studies, and they showed that lncRNAs participate in a variety of biological processes, including transcription, splicing, translation, protein localization, imprinting, the cell cycle and apoptosis. They are also implicated in human diseases, with much attention focused on their involvement in cancer progression and development. Plant lncRNAs have also been associated with a wide array of biological phenomena, including vernalization (Heo and Sung 2011), fertility (Ding et al. 2012), photomorphogenic processes (Wang et al. 2014), phosphate homeostasis (Jabnoune et al. 2013) and regulation of symbiosis between bacteria or fungi and leguminous plants (Kouchi et al. 1999). Plant lncRNAs can play these roles by affecting different steps of gene expression. First of all, some lncRNAs represent primary transcripts for small regulatory RNAs, such as micro RNAs (miRNAs) and small interfering RNAs (siRNAs) (Ben Amor et al. 2009). For instance, in Arabidopsis thaliana, 24 nt siRNAs were shown to originate from at least five long non-coding transcripts: npc34, npc351, npc375, npc520 and npc523. Another connection to the small RNA world is target mimicry, during which interactions between miRNAs and their targets are blocked by binding of the so-called sponge RNAs to miRNAs (Wu et al. 2013). A number of such competing endogenous RNAs (ceRNAs) have been identified in humans and some animals, suggesting that it is a widespread phenomenon (Salmena et al. 2011). Plant lncRNAs are also involved in alternative splicing regulation, e.g. ASCO-lncRNA and the nuclear speckle RNA-binding (NSR) protein that forms an alternative splicing regulatory module, which has been linked to the development of lateral roots (Bardou et al. 2014). Finally, lncRNAs participate in transcriptional regulation through chromatin remodeling (Bardou et al. 2014) as well as in mRNA translation modulation (Jabnoune et al. 2013).
There is a growing body of evidence showing that a large portion of lncRNAs may exert their functions by engaging in physical interactions with mate RNA molecules. After base-pairing with other RNAs, lncRNAs could affect their splicing by masking splice sites and splicing signals (Beltran et al. 2008). Additionally, they could mask miRNA-binding sites on mate RNAs, thus deregulating miRNA functions (Faghihi et al. 2008). lncRNA–RNA duplexes are also expected to trigger A to I editing in mRNAs (Geisler and Coller 2013). In primates, lncRNAs have been shown to guide protein-coding transcripts to the Staufen-mediated decay pathway (Gong and Maquat, 2011). Finally, mouse lncRNAs that possess SINEB2 elements were shown to increase translation efficiency in an mRNA-independent manner (Carrieri et al. 2012). These functions, relying on the formation of lncRNA–RNA duplexes, have been documented in animals. It is highly possible that, in plants, lncRNAs could also base-pair with other RNAs to affect their expression and processing.
The evidenced functionality of lncRNAs and our limited knowledge of their diversity and biology in plants motivated us to perform a large-scale prediction of novel lncRNAs and characterize them functionally. To this end, we took advantage of publicly available sets of RNA sequencing (RNA-Seq) data, and we built a computational pipeline for their prediction, which enabled us to find 45,117 lncRNAs in 10 plant species (Table 1). We annotated them, for example by estimating their expression values and identifying their homologs. We then aimed to determine their potential functions; however, there are no available tools to perform this task. For this reason, we developed a dedicated computational pipeline, focusing on one possibility, i.e. potential consequences of lncRNAs base-pairing with mate RNA molecules. We tested two scenarios: (i) modulating alternative splicing events by masking splicing signals; and (ii) deregulating miRNA functions. Altogether, we predicted 11,896 lncRNAs (26.37%) as being putatively involved in these phenomena. The computational results were made available through an online database that we called CANTATAdb (http://cantata.amu.edu.pl, http://yeti.amu.edu.pl/CANTATA/). The resource offers browsing, searching and downloading options to its users. To the best of our knowledge, the CANTATAdb database is the largest collection of plant lncRNAs.
Table 1.
A summary of long non-coding RNAs and transcripts generally identified in 10 plant species using RNA-Seq data and reference annotations from Ensembl Plants
| Species | No. of lncRNAs | No. of transcripts |
|---|---|---|
| Amborella trichopoda | 2,569 | 39,095 |
| Arabidopsis thaliana | 4,761 | 63,619 |
| Chlamydomonas reinhardtii | 2,214 | 39,716 |
| Glycine max | 3,000 | 105,001 |
| Oryza sativa | 8,594 | 131,870 |
| Physcomitrella patens | 2,711 | 9,3581 |
| Selaginella moellendorffii | 2,667 | 55,552 |
| Solanum tuberosum | 9,692 | 89,231 |
| Vitis vinifera | 4,506 | 65,213 |
| Zea mays | 4,403 | 126,734 |
Materials and Methods
Data download
Genome and transcriptome sequences as well as corresponding reference annotation data were retrieved from Ensembl Plants (Kersey et al. 2014) using BioMart and a download page. Fastq files associated with 107 RNA-Seq libraries were downloaded from the European Nucleotide Archive (Silvester et al. 2014), listed on the ‘RNA-Seq libraries’ page of CANTATAdb. Mature miRNA sequences came from miRBase Release 21 (Kozomara and Griffiths-Jones 2014).
Identification of novel transcripts and splicing isoforms using RNA-Seq data
TopHat (Trapnell et al. 2009), a splice junction mapper, was used to map RNA-Seq reads to a corresponding plant genome. Here, short reads were mapped with BowTie, a file with known transcripts from Ensembl Plants in GTF format, and the ‘mate-inner-dist’ parameter was adjusted to library characteristics. Then, StringTie (Pertea et al. 2015) was applied to assemble the obtained alignments in a BAM format into potential transcripts, using Ensembl Plants genome annotations (GTF format) as a reference. StringTie’s output files (one file per RNA-Seq library) were then processed with Cuffmerge from the Cufflinks package (Trapnell et al. 2012) to obtain a single set of genes and transcripts and compare them with reference annotations from Ensembl Plants.
lncRNA discovery
Based on the Cuffmerge results, transcripts annotated with class code ‘=‘ were removed if the reference transcript’s biotype was one of protein-coding, rRNA, tRNA, small nuclolar RNA (snoRNA) or small nuclear RNA (snRNA). Using CNCI (Sun et al. 2013) in a plant mode (‘model’ parameter set to ‘pl’), the coding potential of the transcripts was calculated, and those predicted to be protein coding were discarded. Then, a BLASTN search against sequences from the Rfam database was performed, and all sequences with hits of E-value < 1e-5 in sense orientation were removed. Additionally, transcripts originating from plastids or mitochondria were discarded. Finally, the transcripts were filtered for length to maintain only those that were at least 200 nt long.
Expression estimation
To calculate transcript expression values, RNA-Seq reads were mapped with Bowtie (Langmead et al. 2009) to all transcripts in a given species (which included our predictions enhanced with Ensembl Plants annotations) with one mismatch allowed. Reads mapping to >10 positions were discarded. Then, using in-house Python scripts, reads that mapped to more than one gene were removed and RPKM (reads per kilobase of transcript per million mapped reads) values for transcripts were calculated. The requirement for gene specificity in read mapping is expected to provide increased specificity in detecting expressed lncRNAs. With this setting, however, 18.47% of CANTATAdb lncRNAs showed no expression evidence, though they were predicted from RNA-Seq data.
Identification of lncRNA–RNA interactions
We used ‘lastal’ from the LAST package (Kielbasa et al. 2011) to identify potential lncRNA–RNA interactions. To this end, we created a custom substitution matrix that enabled us to search for G:U (wobble) pairs. The reason why we used ‘lastal’ is that other commonly used similarity search tools, such as BLAST, do not allow user-supplied scoring matrices, although BLAST has already been used in a similar task (Chen et al. 2012). In the substitution matrix, G:C, A:T and G:T matches were scored as 4, 2 and 1, respectively. These proportions have been widely used in the field of RNA–RNA interactions, e.g. in miRNA target search algorithms (Dai and Zhao 2014). Additionally, a mismatch was scored as –6, a gap opening as –20 and a gap extension as –8. ‘Lastex’, from the same package, enabled us to estimate a threshold value for alignment scores, so that there was no more than one alignment expected to occur by chance. In the interaction search procedure, predicted lncRNAs constituted a database, while Ensembl Plants transcripts were used as a query. When looking for lncRNA base pairings with pre-mRNAs, the query transcripts contained intron sequences; any intronic sequences distant by >250 bases from 3′ or 5′ splice sites were masked with N characters using Python scripts, based on exon co-ordinates from annotation files.
Retrieving interactions that could be involved in regulatory processes
To identify potential cases of splicing regulation through masking splicing signals, interactions between lncRNAs and pre-mRNAs were filtered to keep only those that spanned exon–intron borders. It was also required that the exon–intron junction was subject to alternative splicing. When looking for lncRNAs that could function as modulators of miRNA-dependent regulation, miRNA target sites were superimposed with lncRNA–RNA interaction regions. The miRNA targets were obtained using psRNAtarget (Dai and Zhao 2011) with default settings; in particular, the ‘maximum expectation’ was set to 3.0, the ‘length for complementarity scoring’ was set to 20 and the ‘ allowed maximum energy to unpair the target site’ was set to 25.0.
Further annotation of identified lncRNAs
To provide even more information about identified lncRNAs, we performed a number of computations using available software and data. First, we used the Coding Potential Calculator (CPC) (Kong et al. 2007), a Support Vector Machine-based classifier that calculates the protein-coding potential of transcripts using different sequence features from CNCI. With default settings, as many as 40,686 lncRNAs (90.18%) were predicted as non-coding. With TransDecoder (Haas et al. 2013), we identified putative proteins that could originate from the sense strand of lncRNAs. To this point, we used a –m 30 parameter, which enables identification of open reading frames (ORFs) that are at least 30 amino acids long (the default threshold is 100 amino acids). In the search procedure, ORFs were not required to possess start or stop codons. Having ORFs identified, we ran a BLASTP search against a non-redundant and manually curated set of proteins from the Swiss-Prot database (Uniprot Consortium 2015). If complete ORFs, i.e. those with a start and stop codon, showed significant similarity against annotated proteins (defined as hits with an E-value < 1e-20), a candidate was designated as a ‘low-confidence lncRNA’. We also ran a BLASTX search against Swiss-Prot proteins, with an E-value threshold set to 1e-5, to discover any lncRNA sequence similarity to known proteins, including regions outside identified ORFs.
Database implementation and testing
The database was constructed using Hypertext Markup Language (HTML), Cascading Style Sheets (CSS), PHP 5.4 (http://www.php.net/), MySQL 5.5 (http://www.mysql.com/), and Bootstrap 3 (http://getbootstrap.com/) framework. The database layout was also enhanced with the JavaScript Highcharts library (http://www.highcharts.com/) for interactive data plotting.
The web interface and functionalities of the database were tested on Windows (XP, 7, 8 and 8.1) and Mac OS X (Snow Leopard, Lion, Mountain Lion, Mavericks and Yosemite) operating systems using Internet Explorer (versions 6, 7, 8, 9, 10 and 11), Mozilla Firefox (versions 37 and 40), Chrome (version 42), Opera (versions 12.15 and 12.16) and Safari (versions 5.1, 6, 6.1, 7 and 8) web browsers. Only in the case of Internet Explorer versions 6 and 7 was the web interface visualized improperly without affecting the functionality of CANTATAdb.
CANTATAdb: Database Composition and Usage
Search page
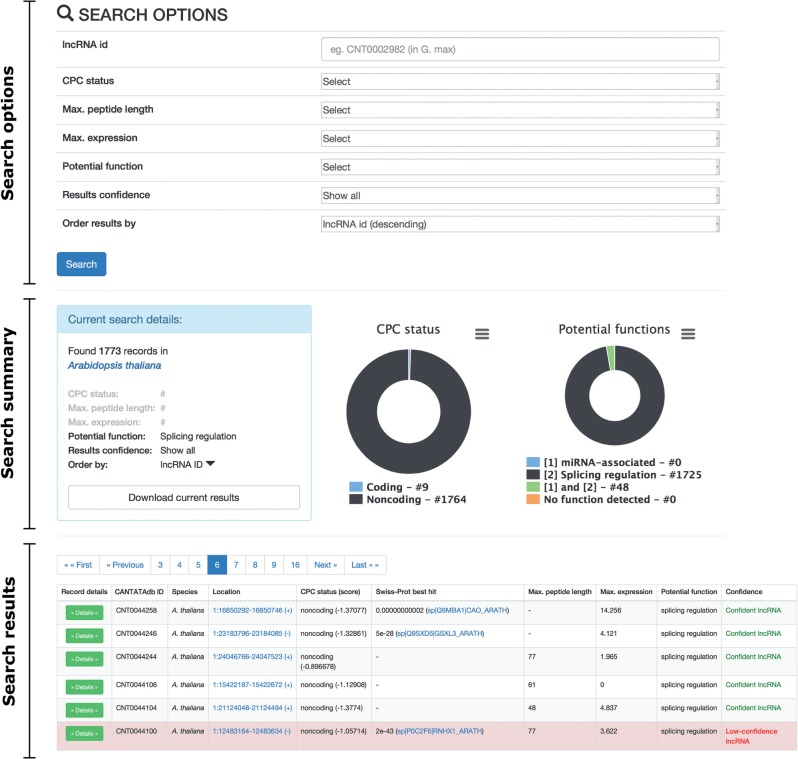
To access the data stored at CANTATAdb, the user is required to select a species from the drop-down list in the main menu. A search page will appear with the following components (Fig. 1).
Fig. 1.
A view of the search page, with search options, search summary and search results components marked.
Search options. Here, the user can select criteria for data search and filtering: CANTATAdb lncRNA id, CPC status (coding or non-coding), length of the longest peptide found in an lncRNA, maximal expression value, potential function (miRNA-associated or splicing regulation), lncRNA confidence and options for ordering of the results.
Search summary. In this section, user-selected search parameters are provided, and the number of records found is reported. To download the filtered data into a tab-delimited text file, one needs to press the ‘Download current results’ button. Additionally, two pie charts summarize the CPC status and potential functions for filtered lncRNAs.
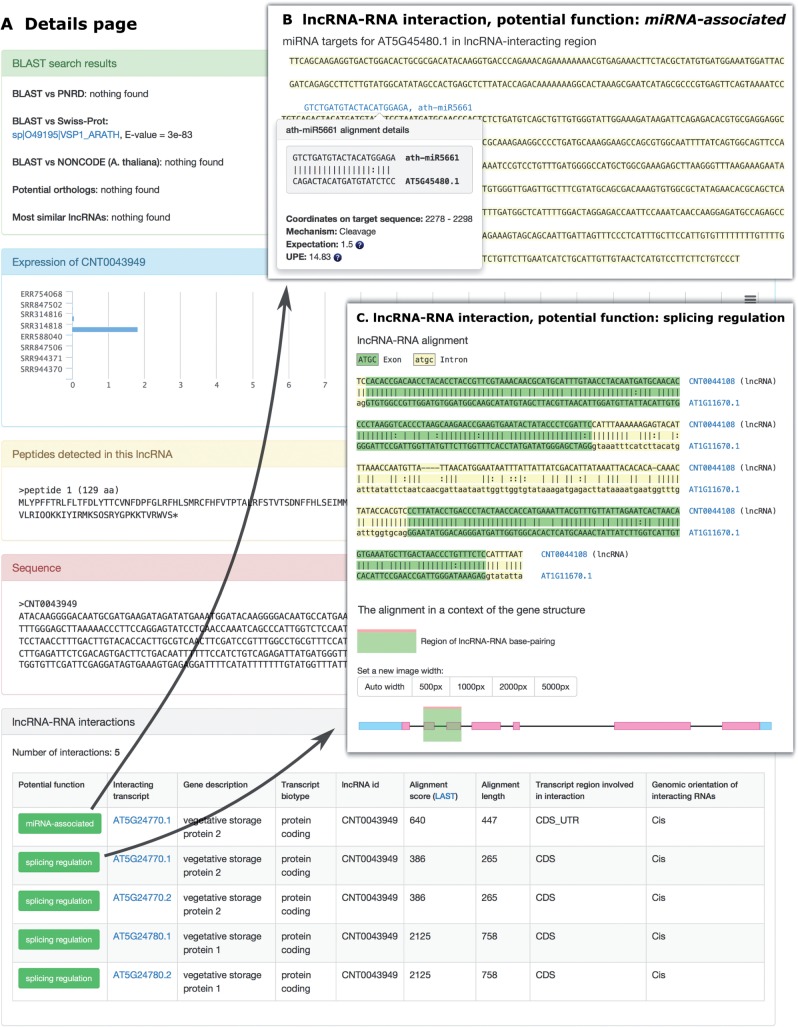
Search results. The search results are displayed in a table with one row per lncRNA. The presented data include: lncRNA id, species, genomic location, best hit against Swiss-Prot proteins (if any), maximal length of all peptides found in a lncRNA (if any), maximal expression value across used RNA-Seq libraries, potential function (either miRNA-associated or splicing regulation), and confidence. Low-confidence records are highlighted with a pale red color. There is also a ‘Details’ button—upon clicking this, the user gets access to more information on selected lncRNA (Fig. 2A), which includes the following
The lncRNA sequence.
BLAST search results against the Plant Non-coding RNA Database (PNRD), Swiss-Prot and A. thaliana lncRNAs from NONCODE. Additionally, most similar lncRNAs are presented there, including potential orthologs.
The lncRNA expression profile across analyzed RNA-Seq libraries is shown in the form of a bar chart
Information about peptides detected in the lncRNAs. In the case of ‘low-confidence lncRNAs’, there is also a red button on top of the page; upon clicking, the best five peptide alignments against Swiss-Prot proteins are visualized.
A list of identified lncRNA–RNA interactions, including potential function predictions (either splicing regulation or miRNA-associated) with a link to detailed information on the base-pairing (Fig. 2B, C).
Fig. 2.
An example of the ‘Details page’ (A) and information regarding potential roles of a lncRNA in deregulation of miRNA functions (B) and in modulation of alternative splicing (C).
From this details page, the user can return to the search page without losing the search parameters by clicking a button on the top of the page.
BLAST page
Here, one can perform a sequence-based search of data stored in CANTATAdb using BLAST version 2.2.26. A user-submitted nucleotide sequence should be in FASTA format. It is possible to select between two tools, i.e. BLASTN and MEGABLAST. Except for the expectation value (E-value) and maximal number of found hits, the search parameters are left as the default, and they are provided for user reference on the same page.
Download page
Although the search results can be easily downloaded from the search page, we also enabled a bulk data download option. All lncRNAs can be downloaded in FASTA format for each species separately. There are also files with genomic co-ordinates of lncRNAs, their potential function and annotation data, including database id, species and expression values. Finally, the best BLAST hits against selected databases of lncRNAs and Swiss-Prot proteins are available there.
RNA-Seq libraries
On this page, we summarize the RNA-Seq libraries used to estimate the lncRNA expression values. The library name with a direct link to the SRA (Short Read Archive) database is provided as well as the source of the sequenced material.
Discussion and Conclusions
Confidence of found lncRNAs
Determining the protein-coding ability for a transcript is a critical point in the identification of lncRNAs, yet it represents quite a challenging task. First of all, algorithms designed for coding potential calculations generate a significant number of false positives and false negatives. For example, CNCI showed 97.3% accuracy when tested on human protein-coding and long non-coding transcripts (Sun et al. 2013). Additionally, lncRNAs are likely to possess ORFs purely by chance (Dinger et al. 2008), so discarding any transcripts containing ORFs would result in losing a substantial number of true lncRNAs. For example, we were able to detect peptides >30 amino acids long in 74.63% of the lncRNAs stored at CANTATAdb and in 80.24% of the PNRD lncRNAs. Regarding peptides that do not exceed 100 amino acids, the proportions are 82.53% and 76.63% in our database and PNRD, respectively, while long peptides (exceeding 200 amino acids) are contained in 8.19% of CANTATAdb lncRNAs, compared with 6.35% in PNRD. This problem might be mitigated by exploiting protein sequence conservation, with the assumption that lncRNA-derived ORFs should not be conserved unless they are translated and their products are functional. We noticed that 8.23% of our lncRNAs contained ORFs with start and stop codons and showed significant similarity to known proteins from Swiss-Prot (E-value threshold of 1e-20). The same was true for only 4.72% of the lncRNAs in PNRD and 5.40% of plant lncRNAs in NONCODE. What is more, only 43.33% were predicted by CPC as non-coding, compared with 90.18% in the case of all the CANTATAdb lncRNAs. We dubbed them ‘low-confidence lncRNAs’ as their non-coding status was questionable. To discriminate them from other candidates, they are marked with a pale red color in the database.
Comparison with existing databases of lncRNAs
There are already several databases that store plant lncRNAs, yet they are much less comprehensive and store fewer data than CANTATAdb. The biggest of them, i.e. the PNRD (Yi et al. 2015), contains fasta sequences of lncRNAs from four plant species: A. thaliana (2,577 records), Oryza sativa (752 records), Populus trichocarpa (538 records) and Zea mays (1,704 records). NONCODE v4 stores lncRNAs for eight animals, yeast and 3,853 lncRNAs from A. thaliana. LncRNAdb (Quek et al. 2015) collects experimentally tested lncRNAs from nine plant species: seven from A. thaliana and between one and three from other species. The recently released PLNlncRbase (Xuan et al. 2015) contains manually collected data from nearly 200 publications, covering a total of 1,187 plant lncRNAs in 43 plant species. Finally, PLncDB (Jin et al. 2013) stores 13,230 novel transcription units from A. thaliana, including long intergenic RNAs (lincRNAs), identified using high-throughput technologies, such as TILING arrays and RNA-Seq sequencing. We found that 53.22% of CANTATAdb lncRNAs for A. thaliana are contained in PlncDB. A detailed comparison between the two resources in provided in Supplementary Table S1.
A distinguishing feature of CANTATAdb is annotation data, including predicted functions in the context of lncRNA–RNA interactions. Altogether, 11,896 lncRNAs have assigned functions, including 440 lncRNAs that are thought to take part in deregulation of miRNA functions and 11,659 lncRNAs that could function as splicing modulators through masking splicing signals. Notably, only 4.35% of them were predicted as coding by the CPC, and 2.97% of them are classified as ‘low-confidence lncRNAs’. For all CANTATAdb lncRNAs, these numbers were 9.82% and 8.23%, respectively. Moreover, in the case of 66.65% of lncRNA–RNA interactions, both RNA components are co-expressed in at least one of the libraries used in this study, which involved 73.50% of all unique lncRNAs with predicted functions. These calculations provide support for predicted functions and the non-coding status of particular lncRNAs, but extensive laboratory tests are required to check the predictions experimentally. Moreover, there are a number of other possibilities regarding roles of plant lncRNAs. With this in mind, we are planning further studies to enhance the annotation of lncRNAs stored at CANTATAdb. Still, we believe that the provided lncRNA sequences and their annotation, including bona fide functions playing a role in the context of lncRNA–RNA interactions, will contribute significantly to future efforts aimed at deciphering the biology of plant lncRNAs.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the National Science Centre [grant No. 2014/15/D/NZ2/00525 to M.W.S]; the Foundation for Polish Science [a START Scholarship grant editions 2014/2015 and 2015/2016 to M.W.S.]; the KNOW Poznan RNA Centre [grant No. 01/KNOW2/2014].
Disclosures
The authors have no conflicts of interest to declare.
Supplementary Material
Glossary
Abbreviations
- CPC
Coding Potential Calculator
- lncRNA
long non-coding RNA
- miRNA
microRNA
- ORF
open reading frame
- PNRD
Plant Non-coding RNA Database
- RNA-Seq
RNA sequencing
References
- Bardou F., Ariel F., Simpson C.G., Romero-Barrios N., Laporte P., Balzergue S., et al. (2014) Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 30166–30176. [DOI] [PubMed] [Google Scholar]
- Beltran M., Puig I., Peña C., García J.M., Alvarez A.B., Peña R., et al. (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial–mesenchymal transition. Genes Dev. 22: 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor B., Wirth S., Merchan F., Laporte P., d’Aubenton-Carafa Y., Hirsch J., et al. (2009) Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 19: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., et al. (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491: 454–457. [DOI] [PubMed] [Google Scholar]
- Chen D., Yuan C., Zhang J., Zhang Z., Bai L., Meng Y., et al. (2012) PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Res. 40: D1187–D1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., et al. Ensembl (2015). Nucleic Acids Res. 2014; Jan; 43(Database issue): D662–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhao P.X. (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39: W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Lu Q., Ouyang Y., Mao H., Zhang P., Yao J., et al. (2012) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 109: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger M.E., Pang K.C., Mercer T.R., Mattick J.S. (2008) Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., et al. (2008) Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14: 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Coller J. (2013) RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Maquat L.E. (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J.B., Sung S. (2011) Encoding memory of winter by noncoding RNAs. Epigenetics 6: 544–547. [DOI] [PubMed] [Google Scholar]
- Jabnoune M., Secco D., Lecampion C., Robaglia C., Shu Q.Y., Poirier Y. (2013) A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Liu J., Wang H., Wong L., Chua N.H. (2013) PLncDB: plant long non-coding RNA database. Bioinformatics 29: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P.J., Allen J.E., Christensen M., Davis P., Falin L.J., Grabmueller C., et al. (2014) Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 42: D546–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa S.M., Wan R., Sato K., Horton P., Frith M.C. (2011) Adaptive seeds tame genomic sequence comparison. Genome Res. 21: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Zhang Y., Ye Z.Q., Liu X.Q., Zhao S.Q., Wei L., et al. (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35: W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., Takane K., So R.B., Ladha J.K., Reddy P.M. (1999) Rice ENOD40: isolation and expression analysis in rice and transgenic soybean root nodules. Plant J. 18: 121–129. [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel J.F., Goodrich J.A. (2012) Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 37: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek X.C., Thomson D.W., Maag J.L., Bartonicek N., Signal B., Clark M.B., et al. (2015) lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 43: D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. (2011) A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 146: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvester N., Alako B., Amid C., Cerdeño-Tárraga A., Cleland I., Gibson R., et al. (2014) Content discovery and retrieval services at the European Nucleotide Archive. Nucleic Acids Res. 43: D23–D29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., et al. (2013) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 41: e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., et al. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43: D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fan X., Lin F., He G., Terzaghi W., Zhu D., et al. (2014) Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. USA 111: 10359–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.J., Wang Z.M., Wang M., Wang X.J. (2013) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 161: 1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Yuan J., Li H., Li M., Zhao G., Bu D., et al. (2014) NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 42: D98–D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan H., Zhang L., Liu X., Han G., Li J., Li X., et al. (2015) PLNlncRbase: a resource for experimentally identified lncRNAs in plants. Gene 573: 328–332. [DOI] [PubMed] [Google Scholar]
- Yi X., Zhang Z., Ling Y., Xu W., Su Z. (2015) PNRD: a plant non-coding RNA database. Nucleic Acids Res. 43: D982–D989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.