Abstract
Despite much evidence of the involvement of the proteasome-ubiquitin signaling system in temperature stress response, the dynamics of the ubiquitylome during cold response has not yet been studied. Here, we have compared quantitative ubiquitylomes from a strain deficient in proteasome substrate recruitment and a reference strain during cold response. We have observed that a large group of proteins showing increased ubiquitylation in the proteasome mutant at low temperature is comprised by reverses suppressor of Ty-phenotype 5 (Rsp5)-regulated plasma membrane proteins. Analysis of internalization and degradation of plasma membrane proteins at low temperature showed that the proteasome becomes determinant for this process, whereas, at 30 °C, the proteasome is dispensable. Moreover, our observations indicate that proteasomes have increased capacity to interact with lysine 63-polyubiquitylated proteins during low temperature in vivo. These unanticipated observations indicate that, during cold response, there is a proteolytic cellular reprogramming in which the proteasome acquires a role in the endocytic-vacuolar pathway.
Keywords: endocytosis, proteasome, protein degradation, ubiquitin, vacuole, Can1, Itr1, Rad23, Rpn10, low temperature
Introduction
Proteasomal and endocytic-vacuolar pathways are responsible for the degradation of a large portion of proteins in eukaryotic cells, and their activities are fundamental for protein homeostasis. The endocytic-vacuolar pathway degrades internalized plasma membrane (PM)3 proteins, proteins trafficking from Golgi to the endosome, and proteins from autophagic processes (1). The proteasome is the fate of cytosolic and nuclear proteins, co-translationally incomplete or misfolded proteins, partially aggregated proteins, and transmembrane proteins from the endoplasmic reticulum (2, 3). Despite the functional divergence between endocytic-vacuolar and proteasomal pathways, they share a key regulatory aspect: client proteins are modified by ubiquitin, and as a consequence ubiquitin-conjugating and ubiquitin-recognizing proteins act as receptors and regulators of both processes. The involvement of distinct ubiquitin-protein ligases, deubiquitylating factors, different polyubiquitin topologies, and specific recognition of ubiquitylated substrates appear to be key factors to ensure the independence of both pathways (1, 4, 5).
In the initial steps of the endocytic pathway, yeast PM cargo proteins are mainly ubiquitylated by the Nedd4-type ubiquitin ligase Rsp5 (4, 6, 7). Ubiquitylated cargo proteins undergo endocytosis, a process orchestrated by epsins and epsin-like proteins, which possess two types of ubiquitin binding domains: ubiquitin-interacting motifs and ubiquitin-associated domains (8). Internalized proteins may exhibit monoubiquitylation or Lys-63-dependent polyubiquitylation (4, 7). In the endosome, ubiquitylated cargo is recruited to the ESCRT-0 complex constituted by Vps27 (Hrs) and Hse1 (STAM1/2). Both Vps27 and Hse1 proteins contain ubiquitin binding domains that mediate interaction with ubiquitylated cargo (9). During multivesicular body biogenesis, ubiquitin modification of cargo proteins is preserved, ensuring the subsequent recruitment of the ESCRT-I, -II, and -III complexes. Finally, cargo proteins are deubiquitylated by Doa4 (10), and intralumenal vesicles of the multivesicular body are formed, traveling to the vacuole where protein cargo is degraded.
In proteasome-dependent degradation, substrate proteins are polyubiquitylated and recognized by a set of distinct proteasomal and extraproteasomal receptors. In yeast, the receptors include the ubiquitin-interacting motif-containing protein Rpn10 (11, 12), the pleckstrin-like receptor for ubiquitin-containing Rpn13 (13), Sem1 (14), and the ubiquitin-like domain/ubiquitin-associated domain receptors Rad23, Dsk2, and Ddi2 (15). Lys-48- and Lys-11-linked polyubiquitin chains promote productive recognition and degradation of substrates, whereas Lys-63 polyubiquitin chains, even though they target substrates for proteasomal degradation in vitro (16, 17), appear not to be an efficient physiological signal under standard conditions (5, 18).
The accepted paradigm states that the endocytic-vacuolar and the proteasomal pathways operate by segregated and independent mechanisms, playing non-overlapping roles in the cell. Here, we show that during low temperature response (i) PM proteins show increased ubiquitylation in a strain deficient in proteasomal receptors Rpn10 and Rad23; (ii) these two proteasome receptors are required for the internalization, transport, and degradation of PM proteins; and (iii) elevated proteasome capacity rescues the phenotype of the ESCRT-0 VPS27 gene deletion mutant. (iv) PM proteins may interact with proteasome at low temperatures. Overall, we present a model in which the 26S proteasome pathway plays a role in the endocytic-vacuolar protein transport and degradation under a situation of cold stress.
Materials and Methods
Plasmids and strains used in this work are included in Table 1.
TABLE 1.
Strains and plasmids used in this work
a, provided by J. Vilardell; b, provided by M. Mendoza; c, provided by R. Deshaies; *, this work.
Media and Growth Conditions
Yeast extract/peptone/dextrose medium and synthetic complete medium were prepared according to standard procedures described previously (19). For SILAC quantitative ubiquitylome analysis, rad23Δrpn10Δ double mutant was grown in synthetic complete medium with heavy lysine (K8; 76 mg/liter), whereas reference cells (rad23Δ) were cultured in light (K0; 76 mg/liter) medium. To globally measure protein expression levels by tandem mass tag (TMT)-based mass spectrometry, both yeast mutants were grown in synthetic complete medium (K0; 76 mg/liter). Cultures were normally grown (30 °C) to an A600 of 1 and subjected to cold shock stress for 4 h. For plasmid selection and enhancement of the expression of the transporters, synthetic media lacking uracil and arginine (Can1) or inositol (Itr1) were prepared. In growth assays on plates (Fig. 4, C and D), cold treatments were carried out at 12 °C because it is the optimal temperature to show the phenotypes.
FIGURE 4.
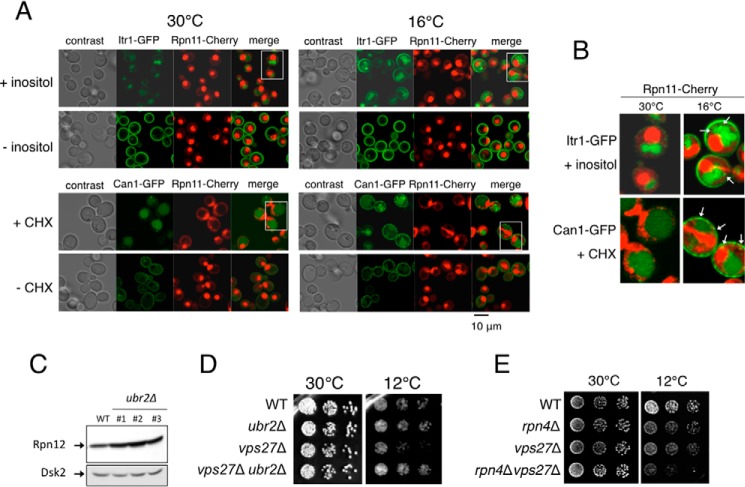
Functional links between proteasomal and endocytic-vacuolar pathways during cold response. A, internalization of Can1-GFP and Itr1-GFP, induced with cycloheximide (CHX) and inositol, respectively, in Rpn11-Cherry cells at 30 and 16 °C. Cells after 3 h of treatment are shown. B, magnified images shown in A (white rectangles). Arrows indicate aberrant distribution of Can1-GFP and Itr1-GFP signals at low temperature. C, analysis of proteasome levels in cells carrying the deletion in UBR2 gene. Wild-type and ubr2Δ strains were grown to midlog phase, and samples were taken and analyzed by immunoblotting. Rpn12 was used as an indicator of proteasome levels. Dsk2 factor was used as a control. D, increased proteasome levels rescue growth defects of vps27 mutant at low temperature. Strains were grown to midlog phase at 30 °C, and samples were taken to prepare normalized cell dilutions (⅓ dilution in successive columns). Cells were spotted on plates and incubated at 30 (2 days) and 12 °C (6 days). E, synthetic effects of the deletion of VPS27 and RPN4 genes on growth at low temperature.
Harvest and Cell Lysis
For the SILAC diGly analysis, equal amounts of rad23Δrpn10Δ and rad23Δ cells were mixed prior to lysis for both permissive temperature and 16 °C. For the multiplexed TMT experiment, the 10 samples (duplicates of rad23Δ at 30 °C, duplicates of rad23Δ at 16 °C, triplicates of rad23Δrpn10Δ at 30 °C, and triplicates of rad23Δrpn10Δ at 16 °C) were lysed individually. Cells were resuspended in lysis buffer (50 mm HEPES (pH 8.2), 8 m urea, 75 mm NaCl, and protease inhibitors (Roche Applied Science) and lysed by bead beating (20). Lysates were normalized by protein concentration, reduced (5 mm DTT) and alkylated (15 mm N-ethylmaleimide). Methanol-chloroform precipitation was performed prior to protease digestion. Protein extracts were resuspended in 8 m urea, 50 mm HEPES (pH 8.2) and subsequently diluted to a final concentration of 4 m urea using 50 mm HEPES (pH 8.2). Samples were digested with Lys-C protease (5 ng/μl; Wako, Japan) at room temperature for 2 h, diluted to 1 m urea with 50 mm HEPES (pH 8.2), and trypsin-digested at a 100:1 protein-to-protease ratio. Digestion proceeded overnight at 37 °C and was stopped by acidification with 0.4% TFA. Peptides were desalted using C18 solid phase extraction cartridges (Waters) and lyophilized.
Immunoprecipitation of DiGly-containing Peptides
Immunoprecipitation of diGly-containing peptides was performed as described (21). In brief, lyophilized peptides from the SILAC assay (50 mg of digested proteins) were resuspended in 1.5 ml of IAP buffer (50 mm MOPS (pH 7.4), 10 mm Na2HPO4, 50 mm NaCl), and insoluble material was removed by centrifugation. The solution was incubated with α-diGly antibody (32 μg/immunoprecipitation) coupled to protein A-agarose beads for 1 h at 4 °C with gentle end-over-end rotation. Peptides were washed with cold IAP buffer three times and twice with PBS. Peptides were eluted twice with 50 μl of 1% formic acid. Peptides were desalted using the C18 StageTip method (22), dried, and reconstituted in 4% acetonitrile, 5% formic acid for liquid chromatography (LC)-MS/MS analysis. Each lysate was immunoprecipitated sequentially two times and analyzed separately by mass spectrometry.
TMT Labeling and Basic pH Reversed Phase Fractionation
Desalted peptides were resuspended in 100 μl of 200 mm HEPES, pH 8.2. One hundred micrograms of peptides from each sample were labeled with TMT reagents (Thermo Fisher Scientific). TMT reagents (0.8 mg) were dissolved in anhydrous acetonitrile (40 μl) of which 10 μl was added to the peptides along with 30 μl of acetonitrile to achieve a final acetonitrile concentration of ∼30% (v/v). Following incubation at room temperature for 1 h, the reaction was quenched with hydroxylamine to a final concentration of 0.3% (v/v). The TMT-labeled samples were equally mixed. The sample was lyophilized and subjected to C18 solid phase extraction cartridges. The dried peptides were resuspended with 500 μl of 5% acetonitrile, 10 mm ammonium bicarbonate (pH 8) and fractionated using basic pH reversed phase HPLC. We used an Agilent 1100 pump equipped with a degasser and a photodiode array detector (set at 220- and 280-nm wavelength) from Thermo Fisher Scientific. Peptides were subjected to a 50-min linear gradient from 5 to 35% acetonitrile in 10 mm ammonium bicarbonate (pH 8) at a flow rate of 0.8 ml/min over an Agilent 300Extend C18 column (5-μm particles, 4.6-mm inner diameter, and 220 mm in length). The peptide mixture was fractionated into a total of 96 fractions, which were consolidated into 24 in a checkerboard manner. Dried samples were acidified, desalted via StageTip, dried, and reconstituted in 4% acetonitrile, 5% formic acid for LC-MS/MS processing.
Liquid Chromatography and Tandem Mass Spectrometry
The diGly-captured peptides were analyzed in a Thermo Fisher Scientific Orbitrap Elite mass spectrometer. The peptides were separated on 100-μm × 20-cm C18 reversed phase (Maccel C18, 3 μm, 200 Å; The Nest Group, Inc.) with a gradient of 6–27% acetonitrile in 0.125% formic acid over 165 min (23). The 20 most intense peaks from each full MS scan acquired in the Orbitrap were selected for MS/MS in the linear ion trap.
The TMT data were collected using an Orbitrap Fusion mass spectrometer coupled to a Proxeon EASY-nLC II LC pump (Thermo Fisher Scientific). Peptides (∼1 μg/analysis) were fractionated on a 75-μm-inner diameter microcapillary column packed with ∼0.5 cm of Magic C4 resin (5 μm, 100 Å; Michrom Bioresources) followed by ∼35 cm of GP-18 resin (1.8 μm, 200 Å; Sepax). Peptides were separated using a 2-h gradient of 6–26% acetonitrile in 0.125% formic acid at a flow rate of ∼350 nl/min. The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution, 120,000; mass range, 400–1400 m/z; automatic gain control target, 2 × 105; maximum injection time, 100 ms). Precursors for MS2/MS3 analysis were selected using a top speed of 2 s. MS2 analysis consisted of collision-induced dissociation (quadrupole ion trap analysis; automatic gain control, 4 × 103; normalized collision energy, 35; maximum injection time, 150 ms). Following acquisition of each MS2 spectrum, we collected an MS3 spectrum using a recently described method in which multiple MS2 fragment ions were captured in the MS3 precursor population using isolation waveforms with multiple frequency notches (24). MS3 precursors were fragmented by high energy collision-induced dissociation and analyzed using the Orbitrap (normalized collision energy, 55; automatic gain control, 5 × 104; maximum injection time, 150 ms; resolution, 60,000 at 400 thomsons).
Searches for DiGly-containing Peptides and Data Analysis
Mass spectra were processed using a SEQUEST-based in-house software pipeline (25). Spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included all entries from the yeast Saccharomyces Genome Database. This database was concatenated with a database composed of all protein sequences in the reversed order. Because lysines were subjected to two variable modifications, each LC-MS/MS was searched twice. First, lysine was assigned to its natural abundance value; second, +8.014199 Da was added as a static modification to reflect SILAC labeling. In both searches, diGly modification of Lys (+114.042927), methionine oxidation (+15.995 Da), and cysteine alkylation (+125.04767) were set as variables modifications. Other parameters used for database searching included: 50-ppm precursor mass tolerance; 0.9-Da product ion mass tolerance; tryptic digestion with up to three missed cleavages. The heavy and light labeled peptides from each search were then combined using custom scripts. Peptide-spectrum matches (PSMs) were adjusted to a 2% false discovery rate (26). PSM filtering was performed using a linear discriminant analysis as described (25), while considering the following parameters: XCorr (cross correlation), ΔCn (Δ correlation), missed cleavages, peptide length, charge state, and precursor mass accuracy. To score the localization of individual diGly modifications (Ascore > 13) and for their quantification, we meticulously followed methodology described previously (21).
Searches for the TMT-labeled Peptides
Searches were performed using a 50-ppm precursor ion tolerance for total protein level analysis. The product ion tolerance was set to 0.9 Da. TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+125.04767 Da) were set as static modifications, whereas oxidation of methionine residues (+15.995 Da) was set as a variable modification. For TMT-based reporter ion quantitation, we extracted the signal-to-noise ratio for each TMT channel and found the closest matching centroid to the expected mass of the TMT reporter ion. PSMs were identified, quantified, and collapsed to a 1% peptide false discovery rate and then collapsed to a final protein level false discovery rate of 1%. Proteins were quantified by summing reporter ion counts across all matching PSMs using in-house software as described previously (25). PSMs with poor quality, MS3 spectra with more than eight TMT reporter ion channels missing, MS3 spectra with a TMT reporter summed signal-to-noise ratio less than 200, or no MS3 spectra were excluded from quantitation (27). Protein quantitation values were exported for further analysis in Excel. Each reporter ion channel was summed across all quantified proteins and normalized assuming equal protein loading of all 10 samples.
Western Blot Analysis
Samples were collected, lysed with the appropriate amount of 5× SDS loading buffer (0.3 m Tris-HCl (pH 6.8), 10% SDS, 50% glycerol, 0.05% bromphenol blue, 25% β-mercaptoethanol), boiled at 95 °C for 5 min, and spun down to evaporate water in the lid. Equal amounts of protein were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). To determine protein weight, the PageRulerTM prestained protein ladder was used. The electrophoresed proteins were transferred to 0.45-μm polyvinylidene difluoride (PVDF) membranes (Immobilon-P Transfer Membrane, Millipore, Billerica, MA). HRP-conjugated secondary antibodies and ECLTM reagent (GE Healthcare) were used, and luminescence was detected by applying the membranes to an autoradiography film. Coomassie staining of the membranes was used as a loading control. Densitometry measurements were carried out with ImageJ software.
Proteasome Isolation and Immunoprecipitations
Proteasomes were affinity-purified from yeast strains carrying RPN11-TEV-PROA as described (28). Briefly, cells were harvested; resuspended in a 2-fold volume of 50 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1 mm ATP, protease inhibitor mixture tablets (one tablet/50 ml) (Roche Applied Science) buffer; and lysed. The lysate was clarified at 11,000 rpm for 45 min, filtered using cheese cloths, and incubated with IgG resin (MP Biomedicals) for 1 h at 4 °C, and the resin was washed with 30 bed volumes of 50 mm Tris-HCl (pH 7.4), 1 mm EDTA, 25 mm NaCl buffer. Proteasomes were eluted after equilibrating the IgG resin with 50 mm Tris-HCl (pH 8.0), 0.5 mm EDTA, 1 mm DTT and incubating with 1 volume of the same buffer containing 20 units/10 g TEV protease at room temperature for 1 h. TEV protease was subsequently removed from the eluate by incubation with nickel-nitrilotriacetic acid resin (Life Technologies) at 4 °C for 15 min. Methods for immunoprecipitations were adapted from Prag et al. (29) and standard protocols.
Protein Degradation Assays
For cycloheximide chase assays, exponential cultures (A600 = 1) were grown and separated at 16 and 30 °C. After 2 h of acclimation, cycloheximide (0.2 mg/ml) was added. At the indicated time points, an aliquot of each culture was removed. Samples were analyzed by immunoblot.
Proteasome Inhibition
Cells were grown to exponential phase (A600 = 1) in the synthetic complete medium described previously (30). The overnight grown culture was supplemented with 0.003% SDS and grown for 3 h at 30 °C. Cells were treated with 200 μm MG132 or dimethyl sulfoxide (DMSO) and further grown at 30 or 16 °C for 3 h. Samples were collected and analyzed by immunoblot.
Microscopy Analysis
Overnight cultures (A600 = 1) were grown at 16 and 30 °C. After 2 h of acclimation, cycloheximide (0.2 mg/ml) or inositol (60 μm) was added. At the indicated time points, an aliquot of each culture was removed and placed in an Attofluor® cell chamber (Life Technologies) on a standard (number 1.5) round coverslip 25 mm round. Cells were imaged in an UltraVIEW ERS spinning disk microscope (PerkinElmer Life Sciences) using a PLAN APOCHROMAT oil immersion 100× objective (numerical aperture, 1.4). GFP and mCherry fluorescent proteins were excited using laser lines 488 and 568 nm, respectively. The corresponding emissions were collected using band pass filter BP527 (W55) and long pass filter LP587, respectively. Single frame images or Z-stacks with a 0.5-μm step were acquired. Image processing was performed using ImageJ.
Results
Ubiquitylome Dynamics under Cold Response in Proteasome Receptor Deficiency
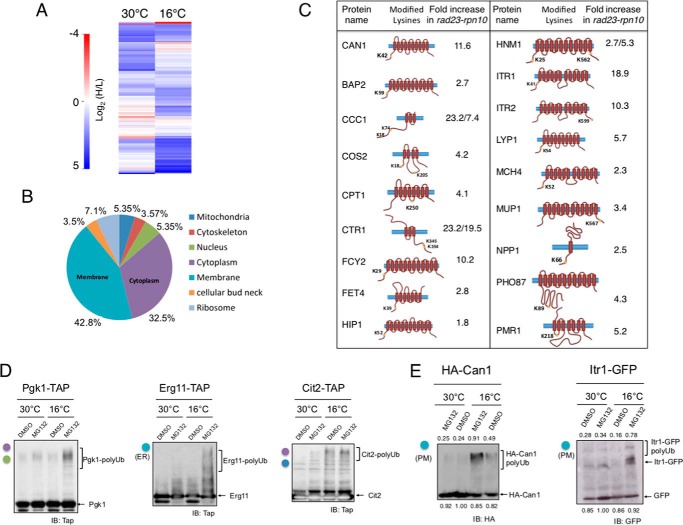
As shown in previous studies, rad23Δrpn10Δ cells show slow growth at low temperature (28, 31), suggesting a determinant role of Rpn10 and Rad23 proteins under these conditions. We reasoned that proteins showing an increased ubiquitin conjugation in rad23Δrpn10Δ after cold shock induction could be strong candidates of cold-induced proteasome substrates. Therefore, rad23Δrpn10Δ strain and the reference strain (rad23Δ) were grown in heavy isotope-labeled (K8; lysine + 8-Da shift) and standard (K0; light) media, respectively. Cells were cultured at 30 °C up to an A600 of 1 and subjected to cold shock at 16 °C for 4 h. Samples were collected before and after cold shock, and equal amounts of heavy and light cultured cells were mixed, lysed, and used for analysis. To increase the efficiency of identification and quantification of conjugated proteins, we performed an enrichment step with an antibody that specifically recognizes the diGly remnant of ubiquitylated peptides (21, 32). Signal-to-noise ratios of both heavy and light envelopes were used for site quantification, resulting in a highly confident candidate list of >300 conjugated sites quantified in both experimental conditions (supplemental Table 1).
Heavy/light ratios showed an increase of conjugated sites in the double mutant cells at both temperatures, consistent with a substrate recruitment deficiency in the absence of these proteasome receptors. Moreover, higher heavy/light ratios were observed at 16 °C (50% of proteins at 16 °C versus 35.5% at 30 °C with heavy/light ratios >2; Fig. 1A and supplemental Table 1). Furthermore, although the absolute amount of conjugated peptides (diGly-containing peptides) showed a 4-fold increase at 30 °C, this increase was 12-fold after 4-h incubation at 16 °C, indicating an increased cellular ubiquitylome after cold shock in the absence of Rpn10 in a RAD23 gene deletion context (Fig. 1A and supplemental Table 1). Finally, to determine whether this increase was due to a putative increase in protein levels induced during culture cold treatment, a yeast proteome analysis was performed using TMT-based proteomics. Thus, using culture conditions identical to those used in the labeled cultures, a replica of the assay was carried out, and samples were analyzed. We observed that levels of proteins showing increased conjugation remained unchanged during the assay (supplemental Tables 2 and 3).
FIGURE 1.
Ubiquitylome dynamics during low temperature response in the proteasome substrate receptor-deficient mutant rpn10Δrad23Δ. A, heat representation of GG peptides quantified in the analysis (indexed as log2(heavy/light (H/L)) where blue represents values increased in rpn10Δrad23Δ mutant and red represents decreased values). Data from cultures at 30 and 16 °C are shown. B, cellular localization of proteins showing increased conjugation at 16 °C (with respect to 30 °C). C, transmembrane proteins from “membrane” localization section in the graph shown in B. Lysine residues modified in the context of each protein and -fold increase at 16 °C with respect to 30 °C are shown. D, Western blot analysis of Pgk1, Erg11, and Cit2. Cells were permeabilized (see “Materials and Methods”) and grown to midlog phase at 30 °C, and MG132 or the vehicle dimethyl sulfoxide (DMSO) was added to cells and incubated for 3 h. Then cultures were shifted or not to 16 °C and incubated for an additional 3 h. Samples were taken and analyzed. Colors of circles correspond to subcellular localization according to B. (ER), endoplasmic reticulum localization of Erg11. E, Western blot analysis of Can1 and Itr1 in cultures grown at 30 and 16 °C in the presence or absence of MG132. Analysis was performed as in A. Densitometry values of modified forms, unmodified Can1-HA, and GFP are included. Values normalized to the band showing strongest signal are shown. (PM), plasma membrane localization of Can1 and Itr1. IB, immunoblot.
Increased Ubiquitylation of Endosome Vacuolar Substrates in Cold-induced Proteasome-deficient Cells
We performed a gene ontology enrichment analysis of proteins exhibiting an increase in conjugation in rad23Δrpn10Δ cells (log2(heavy/light) > 2), and found that a fraction showed an equivalent increase at 30 and at 16 °C. Among these, ribosomal proteins constitute the main group (Rpl20a, Rpl2a, Rps4b, and Rpl16a; supplemental Table 1). We focused our interest on the group of proteins exhibiting a conjugation increase in rad23Δrpn10Δ cells at 16 °C with respect to the permissive temperature culture (30 °C), showing a >2-fold increase at low temperature (supplemental Table 1). Interestingly, the analysis showed that 43% of the proteins exhibiting this behavior localized at the membrane (Fig. 1B). Indeed, this group contained multiple transmembrane proteins including some well characterized permeases and transporters such as the arginine transporter Can1 (Lys-42; 11-fold increase), the myo-inositol transporter Itr1 (Lys-41; 19-fold increase), the affinity methionine permease Mup1 (Lys-567; 3-fold increase), the lysine permease Lyp1 (Lys-54; 6-fold increase), and the high affinity copper transporter Ctr1 (Fig. 1C and supplemental Table 1) (7, 33, 34). The lysines modified by ubiquitin in PM proteins were exclusively located in the cytosolic loops of the proteins, and no structurally incoherent sites (located in intramembrane or extracellular loops) were identified. In addition to plasma membrane proteins, we detected other proteins that showed increased ubiquitylation at 16 °C, i.e. the prion-inducing protein Lsb2 (Pin3), Histone B2, and lanosterol 14-α-demethylase (Erg11), and 3-phosphoglycerate kinase (Pgk1) among others (supplemental Table 1).
If the increased ubiquitylation of these factors was caused by the reduced capacity of the proteasome to recruit and degrade protein substrates, chemical inhibition of the proteasome would reproduce the same pattern. We then added MG132 to cold-treated and control cultures of strains carrying C-terminally tagged versions of some of the proteins quantified in our proteomics analysis. Proteins such as Cit2, Pgk1, and Erg11 showed a pattern of reduced electrophoretic mobility consistent with an increase of ubiquitylation in cold-incubated cultures in MG132-treated cells (Fig. 1D). Moreover, PM proteins tagged with HA and expressed in a vector, such as Can1, or from its own locus, such as Itr1, also showed a pattern of reduced electrophoretic mobility at low temperature in the presence of MG132 (Fig. 1E). Thus, the behavior of increased conjugation by chemical inactivation of the proteasome is in agreement with the results obtained in our proteomics analysis of the rad23Δrpn10Δ strain, which is deficient in substrate recruitment to the proteasome.
Defects in the Internalization and Transport of Vacuolar Substrates in Cold-treated Cells
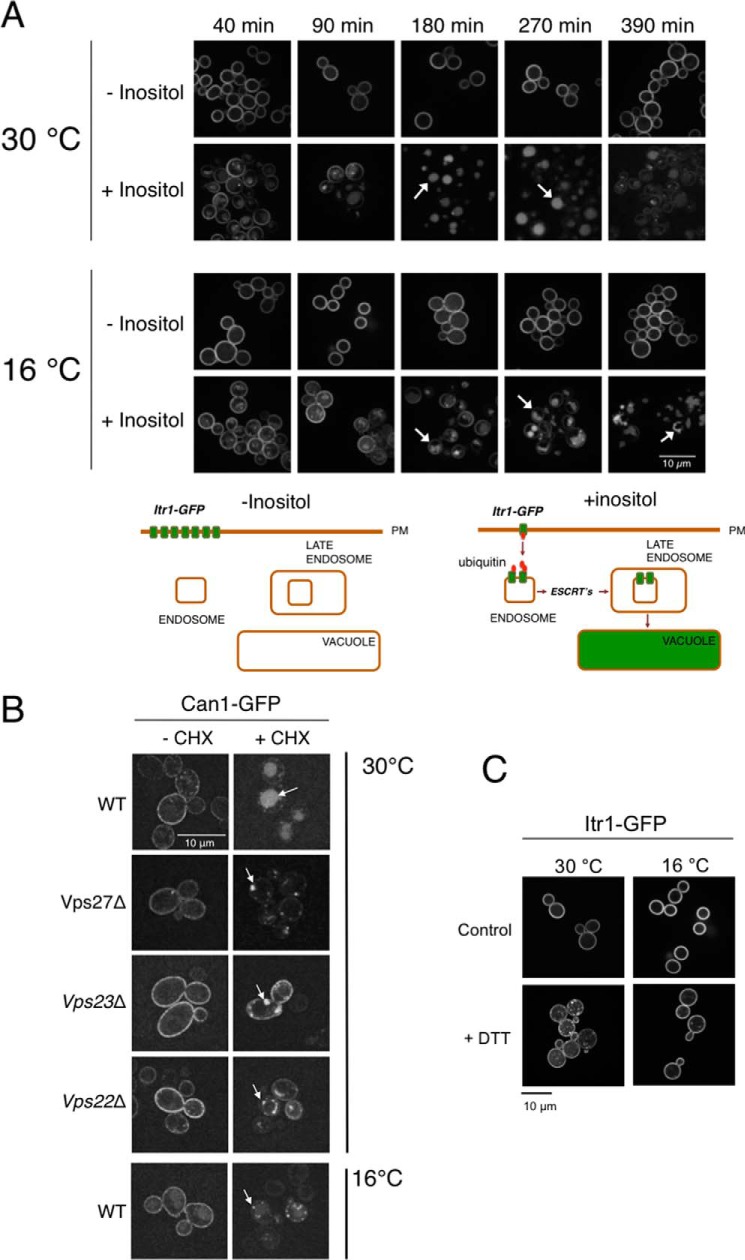
It is well described that the levels of PM proteins are actively regulated by a turnover mechanism orchestrated by the endocytic pathway. In this process, internalized proteins that are not recycled to the membrane are targeted to vacuolar degradation (1). This complex process is controlled by ubiquitylation, which triggers internalization of cargo proteins and the sequential recruitment of ESCRT complexes (1). To assess the influence of low temperature in the internalization and degradation of PM proteins, we monitored the process of endocytosis and vacuolar transport at permissive temperature (30 °C) and 16 °C. We analyzed the behavior of Can1 and Itr1, two well characterized vacuolar substrates (7, 34), in these conditions. To induce Itr1 or Can1 endocytosis, we added inositol (the substrate of Itr1 transporter) or cycloheximide (triggers internalization of Can1 and other PM proteins) to midlog phase growing cultures (7, 35) (schematically shown in Fig. 2A). We observed that the localization of Itr1-GFP at the PM shifted to the vacuole upon inositol addition at standard temperature in agreement with Nikko and Pelham (34) (Fig. 2A). This relocalization effect was also observed for Can1-GFP after cycloheximide addition (7) (Fig. 2B). However, in cold-shocked cultures, the stimulus-dependent vacuolar localization of Can1-GFP and Itr1-GFP was delayed or partially impaired, and both proteins were observed not only in the vacuole but also in the PM and in endocytic intermediates (Fig. 2, A and B). Moreover, in these conditions, the vacuole adopted an abnormal morphology, observed after 3–6.5 h of internalization stimuli (Fig. 2, A and B). The absence of rapid vacuolar localization of Can1-GFP and Itr1-GFP in cold-shocked cells suggests a slowdown or partial dysfunction of endocytic transport under these conditions. For a further characterization of the PM protein internalization defects at 16 °C, we analyzed the internalization of Can1 in a set of mutant strains including genes of components of ESCRT complexes (VPS22, VPS23, and VPS27) at permissive temperature. We observed that vps22, vps23, and vps27 mutants exhibited slight defects in the process of internalization and vacuolar transport of Can1-GFP, showing a phenotype similar to that observed in cold-shocked wild-type cells (Fig. 2B). Therefore, in wild-type cells, a sudden decrease in temperature causes a partial loss of function of the endocytic-vacuolar pathway, resulting in a delayed or incomplete delivery of PM proteins to the vacuole, which adopts an abnormal morphology.
FIGURE 2.
Defects of internalization and vacuolar transport of Itr1 and Can1 at low temperature. A, inositol-induced internalization and vacuolar localization of Itr1in cultures grown at 30 and 16 °C. Cells were grown to midlog phase at 30 °C and shifted or not to 16 °C for 3 h. Then inositol was added or not to cultures, and samples were taken as shown. Panels showing samples of cultures incubated in the absence of inositol are included. Bottom, scheme showing the transport of Itr1from the PM to the vacuole in the presence of inositol. B, cycloheximide (CHX)-induced internalization and vacuolar localization of Can1 in WT, vps22Δ, vps23Δ, and vps27Δ strains. Cultures were grown at 30 °C to midlog phase, and cycloheximide was added. Then cultures were grown for 3 h more at 30 or 16 °C as shown. Arrows depict normal and aberrant vacuolar compartments in A and B. C, effect of treatment with DTT for 1.5 h in Itr1 localization. The effect of cold shock to the localization of the same protein is also shown.
In cold-treated cells and in the absence of internalizing stimuli, the localization of Itr1-GFP or Can1-GFP was peripheral. Moreover, the signal levels of these proteins were identical to those observed in cell treated at permissive temperature (Fig. 2, A and B). Thus, the partial functional loss at low temperature does not alter the process of synthesis and localization of these PM proteins (Fig. 2, A and B), suggesting that, in these conditions, PM proteins are not specifically targeted to the endoplasmic reticulum-associated protein degradation pathway during protein synthesis. To corroborate this idea, we treated cells expressing Itr1-GFP with DTT to promote endoplasmic reticulum-associated protein degradation (36), observing an altered pattern of localization, not observed at 16 °C, in the absence of inositol (Fig. 2C).
Severe Defects in Internalization and Degradation of Can1-GFP in Proteasome-deficient Cells during Low Temperature Response
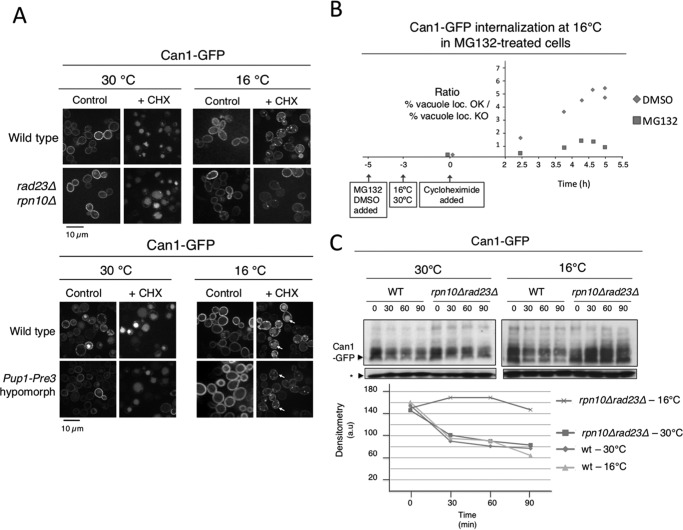
We analyzed Can1-GFP internalization and vacuolar transport in rad23Δrpn10Δ cells at 16 and 30 °C. Can1-GFP exhibited rapid internalization in rad23Δrpn10Δ cells at permissive temperature, similar to that observed in WT cells (Fig. 3A). At low temperature, Can1-GFP showed a moderately delayed internalization in WT cells; however, in rad23Δrpn10Δ cells, it exhibited severely impaired internalization and mainly a PM signal (Fig. 3A, top panels), suggesting a role for these proteasome receptors in transport and degradation of PM proteins during cold response. Moreover, mutations in the catalytic active sites of the proteasome (PUP1-PRE3 hypomorphic strain) exhibited a similar behavior (Fig. 3A, bottom panels). We further analyzed the involvement of the proteasome in the regulation of PM protein transport and turnover by quantifying the effect of MG132 in cycloheximide-induced Can1-GFP vacuolar transport at 16 and 30 °C. In this analysis, Can1-GFP showed a substantial increase of aberrant localization in MG132-treated cells at 16 °C as compared with dimethyl sulfoxide-treated cells (Fig. 3B). At 30 °C, internalization and transport of Can1 were successfully promoted in both wild-type and proteasome-deficient conditions (see Fig. 3A, wild-type panels, as representative results).
FIGURE 3.
Internalization, vacuolar localization, and turnover of Can1 in proteasome-deficient conditions at permissive and low temperatures. A, localization of Can1-GFP at 30 and 16 °C in the presence or absence of internalizing stimulus (cycloheximide (CHX)) in wild-type, rpn10Δrad23Δ, and Pup1-Pre3 mutant cells. Cells were grown to midlog phase, then temperatures were shifted accordingly, cultures were incubated for an additional 3 h, and cycloheximide was added. Finally, upon 2 h of treatment, samples were analyzed. B, graphical representation of the effect of proteasome inhibition using MG132 on Can1 internalization in wild-type cells at 16 °C. Fields of ∼300 cells were counted at the time points indicated. Ratios of cells showing vacuolar Can1-GFP signal (vacuole loc. OK) versus PM, intermediate, or aberrant signal (vacuole loc. KO) are shown. The same experiment was performed at 30 °C, and vacuolar localization was as efficient as all controls performed in this work with no differences between wild-type and proteasome-deficient cells (e.g. A at 30 °C). C, turnover of Can1-GFP in wild-type and rpn10Δrad23Δ cells at 30 and 16 °C. The assay was performed using the experimental design shown in B. Samples were taken as shown and analyzed by immunoblotting using anti-GFP. *, levels of free GFP.
Finally, the capacity of cold-shocked wild-type cells to degrade Can1-GFP was analyzed. Although cold-shocked wild-type cells showed slight defects in the morphology of the vacuole during its transport and delivery (Fig. 2), they exhibited the same capacity to degrade Can1-GFP as cells growing at permissive temperature (Fig. 3C). However, in rad23Δrpn10Δ, after induction of cold shock, Can1-GFP degradation rates were remarkably decreased, whereas at 30 °C they were identical to those observed in wild-type cells (Fig. 3C). This observation supports the idea that Rpn10 and Rad23 proteasomal receptors play a key role in the turnover of Can1-GFP at low temperature and suggests a basis for the detection of increased ubiquitylation of multiple PM proteins in our proteomics study (Fig. 1 and supplemental Table 1).
Functional Links between Proteasomal and Endocytic-Vacuolar Pathways during Cold Response
Our results indicate an involvement of the proteasome in the process of PM internalization, transport, and degradation during cold response. These data are supported by an increased ubiquitylation and half-life, delayed internalization and aberrant localization of PM proteins, and cells showing reduced proteasome activity (MG132-treated or rad23Δrpn10Δ cells) under low temperature conditions. We were interested in analyzing the cellular localization of the proteasome and PM proteins during internalization at low temperature. Thus, we analyzed cells carrying Rpn11-Cherry together with either Can1-GFP or Itr1-GFP fusion protein. Rpn11 is an integral subunit of the proteasome that shows polyubiquitin processing activity coupled with protein degradation (37, 38). As such, this protein is a suitable marker of proteasomes in the cell. We observed cytosolic and strong nuclear detection of Rpn11 in our analysis at both permissive and low temperatures (Fig. 4, A and B). When Itr1-GFP, Can1-GFP, and Rpn11-Cherry subcellular localization were analyzed simultaneously, two different scenarios arose. At permissive temperature, Can1 and Itr1 internalization resulted in a rapid transport to the vacuole. In contrast, at low temperature, internalization was delayed, showing Can1-GFP and Itr1-GFP at PM and endocytic vesicles and exhibiting potential contacts with cytosolic Rpn11-Cherry signal (Fig. 4B, white arrows). This scenario of slow transport of PM proteins favors the functional interaction between endocytic-vacuolar transport of PM proteins and the proteasome during cold response.
Therefore, if the proteasome is involved in the regulation of PM protein turnover specifically during cold shock response, cells with increased proteasome capacity should show a compensatory effect of phenotypes. To approach this question, we deleted the ubiquitin ligase ubr2 in our strains. Ubr2 is involved in the turnover of the proteasome transcription factor Rpn4 (39), and in its absence, Rpn4 cellular levels are higher than in wild-type cells (39, 40), causing an increase of proteasome abundance (Fig. 4C). We observed that the strain carrying the deletion of VPS27 gene, which showed slow growth at low temperature, exhibited a growth rescue when combined with the deletion of ubr2 (Fig. 4D). This rescue was not clearly observed in the absence of VPS22 or VPS23 (not shown). Moreover, deletion of RPN4 gene in vps27 caused an inverse effect in colony growth at low temperature (Fig. 4E).
Putative Interaction of Lysine 63-polyubiquitylated Proteins with the Proteasome during Cold Response
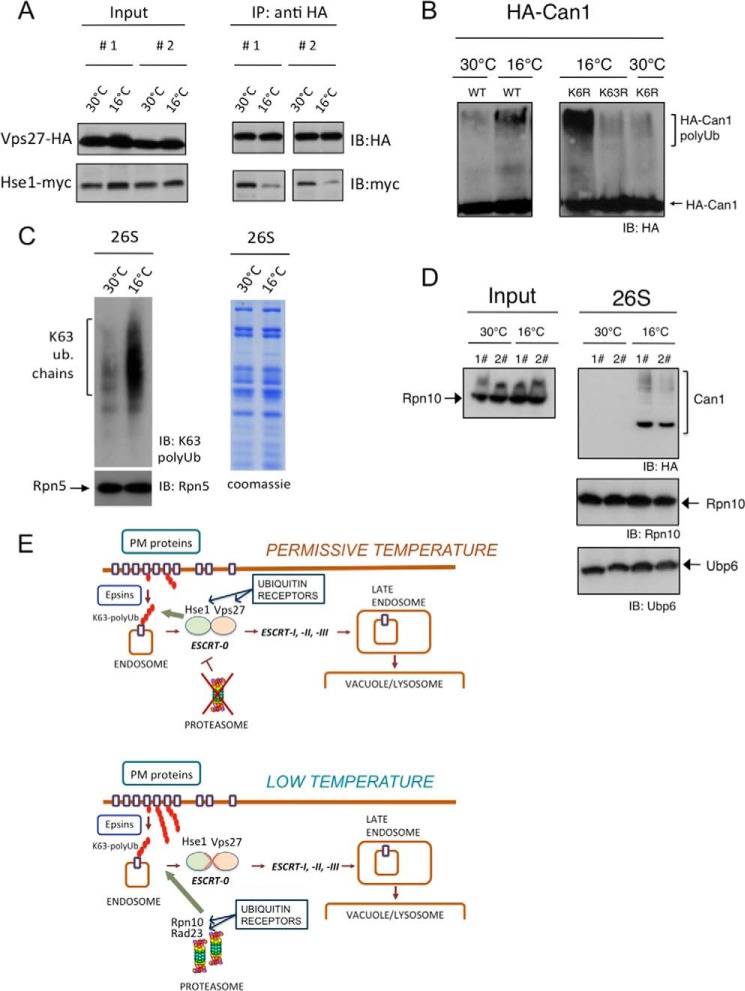
Our observations suggest that the partial loss of function of the endocytic-vacuolar pathway during cold response promotes the involvement of the proteasome in the pathway, which has an effect on the turnover of internalized PM proteins. Moreover, our genetic data reveal an interaction between Vps27 and proteasome regulation at low temperature. To further understand the mechanisms underlying this process, we focused on the role of the Vps27-Hse1 (ESCRT-0) complex. This complex interacts with internalized ubiquitylated cargo proteins and facilitates the initial steps of cargo protein transport to the vacuole (9, 29, 41). Moreover, it has also been proposed that the ESCRT-0 complex prevents Lys-63-polyubiquitylated proteins from interacting with the proteasome in vivo (5). We wondered whether, at low temperature conditions, the status of the Vps27-Hse1 complex could be affected. We co-expressed Vps27-HA and Hse1-Myc and immunoprecipitated the complex. We observed that, after low temperature treatment, the levels of Hse1 co-precipitated with Vps27 decreased significantly as assessed by immunoblot analysis (Fig. 5A). This observation suggested a decreased association of the ESCRT-0 complex during cold response.
FIGURE 5.
Putative interaction of lysine 63-polyubiquitylated proteins with the proteasome during cold response. A, analysis of the association of the Vps27-Hse1 complex at permissive and low temperature. A strain expressing Vps27-HA and Hse1-Myc was used to carry out immunoprecipitations (IP) of Vps27-HA from cultures at 30 or 16 °C (4-h treatment). Immunoblots (IB) of inputs and immunoprecipitated material using anti-HA and anti-Myc antibodies are shown. B, Western blot analysis of Can1 in cultures grown at 30 and 16 °C in the presence or absence of MG132 in strains that express ubiquitin K63R or ubiquitin K6R as indicated. The procedure was the same as that used in Fig. 1A. C, affinity-purified proteasomes from cultures at 30 or 16 °C (4-h treatment) were analyzed by immunoblot using anti-Lys-63 polyubiquitin and anti-Rpn5 antibodies. Proteasomes were analyzed by SDS-PAGE followed by Coomassie staining. D, affinity-purified proteasomes in the presence of MG132 from cultures expressing Can1-HA at 30 or 16 °C were analyzed by immunoblot using anti-HA, anti-Rpn10, and anti-Ubp6 antibodies. The input of extracts prior to proteasome purification is shown (left). E, scheme of the endocytic-vacuolar pathway and the proteasome involvement during cold response. Top, under permissive temperature, PM proteins are internalized and transported through the endosomal system to the vacuole, a process orchestrated by the ESCRT pathway. The role of ESCRT-0 (Hse1-Vps27) is depicted. Bottom, in a situation of cold response, the proteasome is involved at the first steps of the transport of PM proteins. The proteasomal ubiquitin receptors Rpn10 and Rad23 are shown.
Cargo proteins internalized from the PM to the vacuole undergo monoubiquitylation or Lys-63-dependent polyubiquitylation (4). To assess which type of lysine linkage is produced in cold-induced Can1 polyubiquitylation, Can1-HA ubiquitylation was analyzed in a strain that uniquely expresses ubiquitin K63R (UbK63R) (42). After MG132 and low temperature induction, we observed that Can1-HA conjugation was strongly diminished in the presence of UbK63R. This decrease in conjugation was not observed when a strain expressing UbK6R mutant was used (Fig. 5B). This result indicates that Can1 polyubiquitylation mainly involves Lys-63 linkage. Is Lys-63-polyubiquitylated Can1 interacting with the proteasome during low temperature response? We analyzed the content of polyubiquitin associated to affinity-purified proteasomes after treatment at low temperature. We observed that Lys-63 polyubiquitin-immunoreactive species increased substantially in proteasomes isolated from cold-treated cultures (Fig. 5C). Importantly, polyubiquitylated Can1 could be detected in affinity-purified proteasomes, and this detection could only be observed after cold treatment (Fig. 5D).
Therefore, we propose that during cold response, internalized ubiquitylated cargo proteins, such as Can1, can transiently interact with the proteasome. Our data also suggest that this unconventional interactive capacity of Lys-63 polyubiquitin with the proteasome may be promoted by a functional failure of the Vps27-Hse1 complex under low temperature conditions.
Discussion
The process of internalization, transport, sorting, and lysosomal/vacuolar degradation of PM proteins is fundamental to regulate PM composition, cell physiology, and response to environmental changes (1, 43). This complex pathway, which is conserved in all eukaryotes, requires a ubiquitin signal at multiple stages including the first step of endocytosis and the successive steps of endosome transport controlled by ESCRT complexes (1, 43). It is widely accepted that the endocytic-vacuolar pathway does not require the 26S proteasome and that these two essential proteolytic systems are functionally segregated in eukaryotic cells. In the current work, we demonstrate that, during cold response, 26S activity becomes essential for the regulation of a large set of PM proteins. Using as initial evidence a diGly quantitative proteomics analysis, we focused on Can1 and Itr1, two well characterized RSP5-regulated PM proteins. These two factors, which show a remarkable increase in ubiquitylation in the absence of Rpn10 and Rad23 at 16 °C, localize normally to PM at this temperature in the absence of internalizing stimuli. However, when their endocytosis and turnover are triggered at 16 °C, the process appears to be severely impaired in the absence of a fully functional 26S proteasome. The involvement of the proteasome cannot be explained by endoplasmic reticulum-associated protein degradation activation as shown by the correct synthesis and localization of PM proteins during cold response in the absence of internalizing stimuli and by the fact that endoplasmic reticulum-associated protein degradation induction by DTT caused abnormal subcellular localization of Itr1-GFP in the absence of internalizing stimuli (Fig. 2C).
These observations represent an important change in our understanding of how eukaryotic proteolytic systems function and cooperate. Moreover, they uncover a novel role for 26S proteasome (depicted in Fig. 5D): the assistance of the endocytic-vacuolar pathway under a low temperature response. This new cross-talk between distinct protein degradation systems has very important implications to further understand how cells react under certain types of stress not only during temperature fluctuation but also in situations of disease, such as protein turnover failure in neuropathology, or other environmental changes that challenge basic cellular functions.
The confluence of endocytic-vacuolar and 26S pathways in low temperature response has a molecular link: ubiquitin signaling. Both pathways use ubiquitylation of substrates and, importantly, ubiquitin recognition by a diverse set of ubiquitin-interacting receptors. Namely, proteasome components Rpn10, Rpn13, and Sem1 and proteasome shuttling receptors such as Rad23, Dsk2, and Ddi1 that interact with ubiquitin through specific motifs play important roles in substrate delivery to the proteasome. Similarly, the endocytic-vacuolar pathway contains factors such as epsin proteins Ent1/2 and Ede1 and the ESCRT components Vps27, Hse1, Vps23, and Vps36, which are ubiquitin receptors, that play pivotal roles in protein cargo internalization, transport, and sorting. A substantial difference is that 26S pathway targeting involves mainly Lys-48 and Lys-11 polyubiquitin and endocytic-vacuolar signaling involves Lys-63 polyubiquitin or monoubiquitin with the corresponding recognition specificities of the receptors (5, 9, 17, 39).
How does the 26S proteasome play a role in PM protein turnover at low temperature? Our data support the idea of a direct involvement of the 26S proteasome in the degradation of PM proteins during cold response. This would imply the formation of Lys-48 polyubiquitin in targeted PM proteins or, alternatively, the capacity of the proteasome to recognize and process Lys-63-polyubiquitylated substrates. Both scenarios are novel but conceivable if additional specific factors are involved. For example, the recruitment of ubiquitin-ligating factors with E4 activity playing a role in temperature response, such as Hul5 (44, 45), could be postulated. Conversely, it has been shown that the 26S proteasome is mechanistically capable of degrading Lys-63-polyubiquitylated proteins in vitro (17). Furthermore, the capacity of Vps27-Hse1 (ESCRT-0) complex in preventing Lys-63 polyubiquitin-proteasome interaction in vivo (5) suggests a hypothetical regulatory checkpoint in the cross-talk between 26S and endocytic systems. We have observed that Lys-63 is crucial in the induction of Can1 polyubiquitylation at low temperature (Fig. 5B) and that proteasomes show increased content of associated Lys-63-polyubiquitulated species (Fig. 5, B and C). Therefore, it is possible that Lys-63-polyubiquitylated cargo proteins, on their way from the PM to the vacuole, interact with proteasomes. In this scenario, a decreased functionality of the Vps27-Hse1 complex could elicit the adaptive mechanism (Fig. 5D).
The capacity of cells to generate the appropriate response under temperature fluctuation is a key aspect in biology. Budding yeast tolerate changes in temperature by stress response programs at multiple levels that include metabolic, transcriptional, and degradation pathways. Low temperature promotes a decrease in the transcription levels of multiple genes involved in plasma membrane transport as well as a specific metabolic response (46, 47). We propose that, in the same way, proteolytic systems tune their roles to produce the appropriate protein homeostasis response at low temperature.
Author Contributions
M. I., C. S., M. D., P. P.-S., A. Z., and A. B. performed the experiments. E. R. supervised the microscopy. S. P. G. supervised the proteomics. B. C. designed and supervised the project. M. I., C. S., and B. C. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Josep Vilardell, Dr. Jim Hurley, Dr. Ray Deshaies, Dr. Mark Hochstrasser, Dr. Shahri Raasi, Dr. Marion Schmidt, and Dr. Daniel Finley for strains. We thank Dr. Scott Emr for plasmids.
This work was supported by Spanish Government (Ministry of Economy and Competitiveness) Grants BFU2009-06985 and BFU2012-35716, Agència de Gestió d'Ajuts Universitaris i de Recerca Grants 2009SGR-1482 and FI-DGR 2013, and European Molecular Biology Organization Fellowship ASTF 363.00-2010. The authors declare no conflict of interest.

This article contains supplemental Tables 1–3.
- PM
- plasma membrane
- Rsp5
- reverses suppressor of Ty-phenotype 5
- ESCRT
- endosomal sorting complexes required for transport
- TMT
- tandem mass tag
- SILAC
- stable isotope labeling using amino acids in cell culture
- PSM
- peptide-spectrum match
- TEV
- tobacco etch virus
- Ub
- ubiquitin.
References
- 1. MacGurn J. A., Hsu P. C., and Emr S. D. (2012) Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81, 231–259 [DOI] [PubMed] [Google Scholar]
- 2. Schmidt M., and Finley D. (2014) Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thibault G., and Ng D. T. (2012) The endoplasmic reticulum-associated degradation pathways of budding yeast. Cold Spring Harb. Perspect. Biol. 4, a013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Erpapazoglou Z., Dhaoui M., Pantazopoulou M., Giordano F., Mari M., Léon S., Raposo G., Reggiori F., and Haguenauer-Tsapis R. (2012) A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting. Mol. Biol. Cell 23, 2170–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nathan J. A., Kim H. T., Ting L., Gygi S. P., and Goldberg A. L. (2013) Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 32, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horák J. (2003) The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: insights from yeast. Biochim. Biophys. Acta 1614, 139–155 [DOI] [PubMed] [Google Scholar]
- 7. Lin C. H., MacGurn J. A., Chu T., Stefan C. J., and Emr S. D. (2008) Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725 [DOI] [PubMed] [Google Scholar]
- 8. Shih S. C., Katzmann D. J., Schnell J. D., Sutanto M., Emr S. D., and Hicke L. (2002) Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4, 389–393 [DOI] [PubMed] [Google Scholar]
- 9. Bilodeau P. S., Urbanowski J. L., Winistorfer S. C., and Piper R. C. (2002) The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4, 534–539 [DOI] [PubMed] [Google Scholar]
- 10. Amerik A. Y., Nowak J., Swaminathan S., and Hochstrasser M. (2000) The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11, 3365–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Nocker S., Sadis S., Rubin D. M., Glickman M., Fu H., Coux O., Wefes I., Finley D., and Vierstra R. D. (1996) The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 16, 6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fu H., Sadis S., Rubin D. M., Glickman M., van Nocker S., Finley D., and Vierstra R. D. (1998) Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J. Biol. Chem. 273, 1970–1981 [DOI] [PubMed] [Google Scholar]
- 13. Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., and Dikic I. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paraskevopoulos K., Kriegenburg F., Tatham M. H., Rösner H. I., Medina B., Larsen I. B., Brandstrup R., Hardwick K. G., Hay R. T., Kragelund B. B., Hartmann-Petersen R., and Gordon C. (2014) Dss1 is a 26S proteasome ubiquitin receptor. Mol. Cell 56, 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Müller B., Feng M. T., Tübing F., Dittmar G. A., and Finley D. (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4, 725–730 [DOI] [PubMed] [Google Scholar]
- 16. Elsasser S., Chandler-Militello D., Müller B., Hanna J., and Finley D. (2004) Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279, 26817–26822 [DOI] [PubMed] [Google Scholar]
- 17. Saeki Y., Kudo T., Sone T., Kikuchi Y., Yokosawa H., Toh-e A., and Tanaka K. (2009) Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 28, 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., and Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 20. Villén J., and Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rappsilber J., Ishihama Y., and Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 23. Haas W., Faherty B. K., Gerber S. A., Elias J. E., Beausoleil S. A., Bakalarski C. E., Li X., Villén J., and Gygi S. P. (2006) Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell Proteomics 5, 1326–1337 [DOI] [PubMed] [Google Scholar]
- 24. McAlister G. C., Nusinow D. P., Jedrychowski M. P., Wühr M., Huttlin E. L., Erickson B. K., Rad R., Haas W., and Gygi S. P. (2014) MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 27. McAlister G. C., Huttlin E. L., Haas W., Ting L., Jedrychowski M. P., Rogers J. C., Kuhn K., Pike I., Grothe R. A., Blethrow J. D., and Gygi S. P. (2012) Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 84, 7469–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Isasa M., Katz E. J., Kim W., Yugo V., González S., Kirkpatrick D. S., Thomson T. M., Finley D., Gygi S. P., and Crosas B. (2010) Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol. Cell 38, 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prag G., Watson H., Kim Y. C., Beach B. M., Ghirlando R., Hummer G., Bonifacino J. S., and Hurley J. H. (2007) The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev. Cell 12, 973–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J., Sitaram A., and Burd C. G. (2007) Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase. Traffic 8, 1375–1384 [DOI] [PubMed] [Google Scholar]
- 31. Lambertson D., Chen L., and Madura K. (1999) Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 153, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Udeshi N. D., Mertins P., Svinkina T., and Carr S. A. (2013) Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 8, 1950–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Butler P. L., and Mallampalli R. K. (2010) Cross-talk between remodeling and de novo pathways maintains phospholipid balance through ubiquitination. J. Biol. Chem. 285, 6246–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nikko E., and Pelham H. R. (2009) Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Galan J. M., and Haguenauer-Tsapis R. (1997) Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16, 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ellgaard L., and Helenius A. (2003) Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 37. Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R. 3rd, Koonin E. V., and Deshaies R. J. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 38. Yao T., and Cohen R. E. (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 39. Wang L., Mao X., Ju D., and Xie Y. (2004) Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J. Biol. Chem. 279, 55218–55223 [DOI] [PubMed] [Google Scholar]
- 40. Kruegel U., Robison B., Dange T., Kahlert G., Delaney J. R., Kotireddy S., Tsuchiya M., Tsuchiyama S., Murakami C. J., Schleit J., Sutphin G., Carr D., Tar K., Dittmar G., Kaeberlein M., Kennedy B. K., and Schmidt M. (2011) Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 7, e1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hurley J. H. (2008) ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 20, 4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spence J., Sadis S., Haas A. L., and Finley D. (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15, 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raiborg C., and Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 44. Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., and Finley D. (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 45. Fang N. N., Chan G. T., Zhu M., Comyn S. A., Persaud A., Deshaies R. J., Rotin D., Gsponer J., and Mayor T. (2014) Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress. Nat. Cell Biol. 16, 1227–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tai S. L., Daran-Lapujade P., Walsh M. C., Pronk J. T., and Daran J. M. (2007) Acclimation of Saccharomyces cerevisiae to low temperature: a chemostat-based transcriptome analysis. Mol. Biol. Cell 18, 5100–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walther D., Strassburg K., Durek P., and Kopka J. (2010) Metabolic pathway relationships revealed by an integrative analysis of the transcriptional and metabolic temperature stress-response dynamics in yeast. Omics 14, 261–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., and Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 49. Arendt C.S., and Hochstrasser M. (1999) Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. EMBO J. 18, 3575–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.