Abstract
Enterococcus faecalis is a Gram-positive, commensal bacterium that lives in the gastrointestinal tracts of humans and other mammals. It causes severe infections because of high antibiotic resistance. E. faecalis can endure extremes of temperature and pH. Acyl carrier protein (ACP) is a key element in the biosynthesis of fatty acids responsible for acyl group shuttling and delivery. In this study, to understand the origin of high thermal stabilities of E. faecalis ACP (Ef-ACP), its solution structure was investigated for the first time. CD experiments showed that the melting temperature of Ef-ACP is 78.8 °C, which is much higher than that of Escherichia coli ACP (67.2 °C). The overall structure of Ef-ACP shows the common ACP folding pattern consisting of four α-helices (helix I (residues 3–17), helix II (residues 39–53), helix III (residues 60–64), and helix IV (residues 68–78)) connected by three loops. Unique Ef-ACP structural features include a hydrophobic interaction between Phe45 in helix II and Phe18 in the α1α2 loop and a hydrogen bonding between Ser15 in helix I and Ile20 in the α1α2 loop, resulting in its high thermal stability. Phe45-mediated hydrophobic packing may block acyl chain binding subpocket II entry. Furthermore, Ser58 in the α2α3 loop in Ef-ACP, which usually constitutes a proline in other ACPs, exhibited slow conformational exchanges, resulting in the movement of the helix III outside the structure to accommodate a longer acyl chain in the acyl binding cavity. These results might provide insights into the development of antibiotics against pathogenic drug-resistant E. faecalis strains.
Keywords: acyl carrier protein (ACP), biophysics, fatty acid synthase (FAS), nuclear magnetic resonance (NMR), protein structure, Enterococcus faecalis
Introduction
Fatty acids are the central building blocks of life. Fatty acid synthase (FAS)2 is an important pathway for the production of fatty acids and plays an important role in the formation of cell membranes, energy storage compounds, and messenger substances. Fatty acids also act as post-translational protein modifiers and modulate gene expression (1). In nature, there are two types of FAS systems, type I and type II. Type I FAS systems are found in mammals and fungi; this system is composed of a single polypeptide chain of covalently linked domains that forms large multienzyme complexes (2, 3). In contrast, type II FAS systems are found in bacteria and eukaryotic organelles and are dissociated so that all proteins are expressed as individual enzymes (4–7). Given the vast difference in these two systems between humans and bacteria, there has been a conscious effort to target FAS in bacteria through novel antibiotics.
Acyl carrier proteins (ACPs) are small (9-kDa) acidic proteins that are essential for numerous biosynthetic pathways that play a role in acyl group transfer. ACPs are also required for a variety of other mechanisms that require acyl group transfer steps, such as polyketide antibiotic synthesis, lipopolysaccharides, rhizobial modulation signaling factors, lipoteichoic acids, and prohemolysin toxin activation (8–11). However, the biosynthesis of fatty acids is the predominant role of ACP. ACPs are highly conserved proteins typically consisting of 70–100 residues and are essential components in type I and type II FAS systems (12, 13), which shuttle bound fatty acyl intermediates in a hydrophobic pocket to facilitate interaction with various enzyme partners (14). Therefore, ACP is believed to be a mobile protein, and its flexibility is essential for its interaction with functionally different enzyme partners.
Enterococci cause serious infections in animals and humans. Almost all enterococcal infections are caused by Enterococcus faecalis (15). E. faecalis is a bacterium that inhabits the gastrointestinal tracts of humans and other mammals. It also commonly resides in soil, water sewage, and contaminated food. Furthermore, E. faecalis can tolerate oxidative stress and dry conditions and can grow in 6.5% NaCl, 40% bile salts, and at a broad range of pH (16, 17). In particular, E. faecalis is able to grow in a high temperature range from 10 to 40 °C and can also survive at 85 °C; this thermotolerance can allow it to survive in hospital laundries (16, 18). E. faecalis also has a high resistance to salinity, detergents, antibiotics drugs, sodium azide, and bile acids, which kill most microorganisms (19, 20). E. faecalis can survive in patient clothing for over 11 days, which is a relatively long period (21). E. faecalis is able to cause life-threatening infections in humans, especially in the hospital environment (22–24). Furthermore, E. faecalis has the ability to become resistant to vancomycin, penicillins, cephalosporins, and aminoglycosides through mutation (20, 25–27). Vancomycin-resistant enterococci in particular are important etiologic agents of nosocomial infections and colonization in hospitalized patients (28). Because FAS proteins of antibiotic-resistant strains can be potential targets while designing novel antibiotics, we have developed potent inhibitors for FabH from methicillin-resistant Staphylococcus aureus and vancomycin-resistant E. faecalis, which is a condensing enzyme in bacterial FAS (29, 30).
The structures of various kinds of type II ACPs have been studied. Solution structures of ACPs from Escherichia coli (31, 32), Bacillus subtilis (33), Mycobacterium tuberculosis (34), Vibrio harveyi (32), Helicobacter pylori (35), Borrelia burgdorferi (36), and Plasmodium falciparum (37) have been determined by NMR spectroscopy. Also, NMR structures of ACP domains of various type I FAS have been studied (38–40). Structural studies of ACPs have revealed that ACPs are composed of four helix bundles and a prosthetic group attached to a conserved Ser residue located in helix II. Type II ACPs have high sequence similarity; in particular, the acidic residues in helix II that are important in interactions with FAS enzymes and DSL motifs are highly conserved (41). However, despite such considerable sequence similarity, the thermal stability of E. faecalis ACP (Ef-ACP) is much higher than that of other ACPs. In this study, to understand the origin of the thermal stability of Ef-ACP, we determined for the first time the tertiary structure of ACP from E. faecalis. In this work, we measured the melting temperatures (Tm) of wild type and mutant Ef-ACPs using CD experiments and compared these with the Tm of other ACPs to understand the origin of the high thermal stability of Ef-ACP. We also monitored amide exchange rates to understand the stability of the structural elements in Ef-ACP. To investigate the importance of Ef-ACP structural flexibility, the dynamic properties of Ef-ACP were assessed by measuring spin relaxation rates and were correlated with its thermal stability in comparison with those of other ACPs.
Experimental Procedures
Cloning, Expression, and Purification of Ef-ACP
The acpP gene encoding ACP was amplified from E. faecalis genomic DNA. A restriction site (NdeI for the sense primer and BamHI for the antisense primer) was attached to the 5′-end of each primer to facilitate cloning. PCR was performed with 35 cycles of denaturation for 1 min at 95 °C, annealing for 1 min at 54 °C, and elongation for 1 min at 72 °C. The resulting product was cloned into the NdeI and BamHI sites of the pET-11a vector (Novagen, Madison, WI). The ligation mixture was transformed into E. coli DH5α competent cells. For expression of H/15N/13C-labeled ACP, plasmids encoding ACP were transformed in E. coli BL21 (DE3) cells and then grown overnight as subcultures and isolated by centrifugation. The resulting cell pellet was mixed with 100 ml of M9 minimal media containing 1 g/liter 15NH4Cl and [13C]glucose (Cambridge Isotope Laboratories, Andover, MA) and 100 mg/liter ampicillin and grown to an A600 between 0.8 and 1.0. This culture was added to 1 liter of the same minimal media, and protein expression was induced by adding isopropyl β-d-thiogalactopyranoside at an A600 of 0.8–1.0 and incubating at 25 °C for ∼24 h. Ef-ACP was purified using a HiTrapTM QFF and Hiload 16/60 Superdex 75 column. The holo-ACPs were produced by enzymatic acylation of apo-ACPs using E. coli holo-ACP synthase (AcpS) and coenzyme A (Sigma-Aldrich) (42), and the holo-ACP was separated for apo-ACP using a Resource Q column. These samples were analyzed by 20% native gel electrophoresis followed by Coomassie staining to distinguish the apo- and holo-ACPs (43).
CD Measurements of Purified ACP
CD measurements were performed in a J810 spectropolarimeter (Jasco, Tokyo, Japan) using a cell with a 1-mm path length. The CD spectra of Ef-ACP in 25 mm MES buffer, pH 6.1, containing 5 mm CaCl2 and 5 mm dithiothreitol (DTT) at 25 °C were measured at 0.1-nm intervals from 200 to 250 nm. The protein concentration was 50 μm, and data from five scans were averaged and smoothed using J810 software. CD data were shown as the mean residue ellipticity (θ) in degrees cm2 dmol−1. Protein Tm values were determined from a series of CD spectra obtained from 15 to 100 °C.
NMR Analysis and Structure Calculation
All NMR experiments were performed at 25 °C on a Bruker Avance 700-, 800-, and 900-MHz spectrometer at the Korea Basic Science Institute. All of the spectra were processed with NMRPipe (44) and analyzed using Sparky (45). The compound 2,2-dimethyl-2-silapentane-5-sulfonate was used as an internal standard. Ef-ACP samples were prepared at 1 mm concentration in 25 mm MES buffer, pH 6.1, containing 5 mm CaCl2, 5 mm DTT, 0.02% NaN3, and 10% D2O. For backbone assignments, HNCACB and CBCA(CO)NH triple resonance spectra were recorded, and assignments of aliphatic side chains were from HCCH-TOCSY and CCH-TOCSY spectra.
Anisotropic orientational restraints were obtained from 15N-labeled ACP with 40% strained polyacrylamide gel and compared with 15N-labeled ACP lacking polyacrylamide gel. 1JNH coupling measurements were performed using the IPAP-HSQC method (46) with one-bond 15N–1H residual dipolar couplings taken to be the difference between 1JNH measurements in isotropic and aligned samples. TALOS+ (47) was used in deriving backbone dihedral angle restraints (Φ and Ψ) from 1Hα, 15N, 13Cα, 13Cβ, and 13C′ chemical shift values. The NOE restraints were obtained from 15N-edited NOESY-HSQC, 13C-edited NOESY-HSQC spectra. CYANA version 3.9 (48, 49) was used for automatic NOESY peak assignments and structure calculations. NOESY peaks assigned automatically by CYANA were used as a guide to further refine the structure. Residual dipolar coupling refinement was performed using CYANA version 3.9. The analysis of Ramachandran plots for the 20 lowest energy structures was carried out using PROCHECK (50). The protein structures were represented using PyMOL and MOLMOL (51). The chemical shifts, coordinates, and NMR-derived constraints have been deposited in the Biological Magnetic Resonance Bank (accession number 25688; Protein Data Bank (PDB) entry 2N50).
Spin Relaxation Experiments
The 15N-labeled samples of ACPs contained 0.5 mm protein in 25 mm MES (pH 6.1), 5 mm DTT, 0.02% NaN3, and 10% D2O in the presence of 5 mm CaCl2 and in the absence of CaCl2. All NMR experiments were performed at 25 °C on a Bruker Advance 700-MHz spectrometer. The longitudinal relaxation rates (R1) were measured with relaxation delays of 0.002 (×2), 0.045, 0.100, 0.200, 0.315 (×2), 0.550, 0.800, and 1.000 s. The transverse relaxation rates (R2) were measured with relaxation delays of 0 (×2), 0.01696, 0.03328, 0.05088 (×2), 0.0848, 0.11872, 0.18656, and 0.28832 s (52). The heteronuclear cross-relaxation rate was measured by heteronuclear Overhauser effect (hNOE) experiments by using the interleaving pulse sequences with and without proton saturation. The recycle delay and proton saturation times in the hNOE measurement were 4.5 and 3.0 s, respectively. All relaxation spectra were acquired and analyzed as described in our previous reports (53, 54). hNOE was calculated from the ratio of peak heights with and without proton saturation pulses. R1 and R2 rates were determined by fitting the peak heights using Sparky.
Site-directed Mutagenesis of Ef-ACP
Mutagenesis of the Ef-ACP variant was performed using a Muta-DirectTM site-directed mutagenesis kit (Intron Biotechnology, Seongnam, Korea), and plasmid constructs of Ef-ACP variants containing mutations at positions His17, Phe18, Phe45, and Phe53 were cloned by PCR using the synthetic oligonucleotide primer pairs 5′-ATT TCA AAC GCG TTT GAT ATT GAA GCG-3′ (forward) and 5′-CGC TTC AAT AAA CGC GTT TGA GAT ATT-3′ (reverse) for H17A, 5′-AGC GTA ATG GAG GCG GTT TTA GAA CTT G-3′ (forward) and 5′-CAA GTT CTA AAA CCG CCT CCA TTA CGC T-3′ (reverse) for F45A, 5′-GAT TAT CTC AAA CCA CGC GGA TAT TGA AGC GGA TC-3′ (forward) and 5′-GAT CCG CTT CAA TAT CCG CGT GGT TTG AGA TAA TC-3′ (reverse) for F18A, and 5′-ACT TGA AGA TGA AGC GGG AAC AGA GAT TT-3′ (forward) and 5′-AAA TCT CTG TTC CCG CTT CAT CTT CAA GT-3′ (reverse), using Ef-ACP DNA as a template. PCR amplifications were performed using 10 fmol of template DNA, 1 μm each primer, 0.2 mm each deoxyribonucleotide triphosphate, and 2 μm Taq polymerase under the following thermocycling conditions: 25 cycles of denaturation at 95 °C (30 s), annealing at 54 °C (1 min), and primer elongation at 72 °C (1 min).
Hydrogen/Deuterium Exchange Experiments
Ef-ACP samples were prepared at a concentration of 0.5 mm protein in 25 mm MES buffer, pD 6.1, 5 mm DTT, and 0.02% NaN3 containing 5 mm CaCl2 and in the absence of CaCl2. Data acquisition was started ∼5 min after the addition of D2O to the protein. A series of heteronuclear single quantum coherence spectroscopy (HSQC) spectra were acquired every 10 min using two scans and 1024 × 256 data points for 10 h; after 4 days, additional HSQC spectra were acquired using the same conditions. Decay of amide proton signal as a function of time was measured by the decrease in peak height. To determine the exchange rate constant of protein, decay curves were fit using a single-exponential function (I = I0 exp(−kext) + C), where I represents the NH resonance intensities as a function time (t), I0 is initial intensity, and kex is exchange rate of protein. The protection factor was calculated by the comparison of measured hydrogen/deuterium exchange rates for protein (kprot) with the rates for the random coil conformation (krc) (55).
Results
Thermal Stability of the Ef-ACP
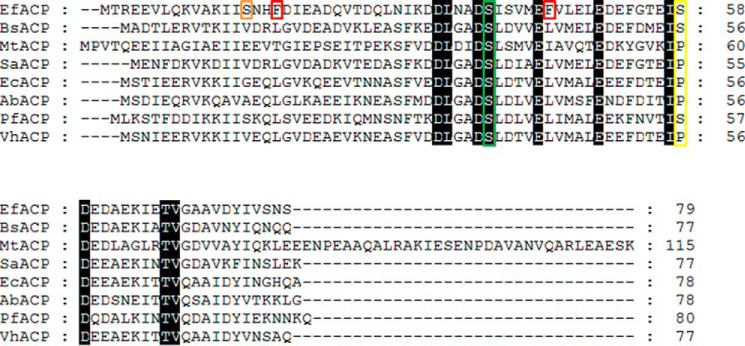
Ef-ACP is a highly acidic protein with a pI value of 4.2 with 21 acidic residues. As shown in Fig. 1, sequence alignments revealed that type II bacterial ACPs have high sequence similarities with Ef-ACP. ACPs have a strictly conserved serine residue where the 4′-phosphopantetheine prosthetic group is attached (marked with a green box in Fig. 1). Some distinct features were noted in the sequence of Ef-ACP. All ACPs have a conserved aromatic residue at the end of helix II, whereas Ef-ACP has an additional Phe (Phe45) in the middle of helix II. In addition, Ef-ACP has Phe18 at the beginning of loop I, resulting in stable hydrophobic interactions with Phe45 (marked with a red box in Fig. 1). There are proline residues in the α2α3 loops connecting helix II and helix III in most ACPs, including Ec-ACP (Pro55), whereas in Ef-ACP, this is replaced with a serine residue (Ser58), providing more flexibility (marked with a yellow box in Fig. 1). As shown in Fig. 1, all ACPs are highly acidic; divalent cations reduce the severe electrostatic repulsions between the acidic residues in ACP. Ef-ACP has 11 Glu and 10 Asp residues, whereas Ec-ACP has 14 Glu and 6 Asp. It has been reported previously that Ec-ACP has two metal binding sites: site A consisting of Glu30, Asp35, and Asp38 and site B consisting of Glu47, Asp51, Glu53, and Asp56 (56).
FIGURE 1.
Sequence alignment of ACP. Sequence alignment of the Ef-ACP protein with three Gram-positive homologues (B. subtilis ACP (BsACP), M. tuberculosis ACP (MtACP), and S. aureus-ACP (SaACP)) and four Gram-negative bacterial homologues (E. coli ACP (EcACP), A. baumanni ACP (AbACP), P. falciparum ACP (PfACP), and V. harveyi ACP (VhACP)). The conserved residues with 100% identity are shown in black. The prosthetic group attachment site, which is highly conserved (Ser39 in Ef-ACP) is marked by a green box. Unique features of Ef-ACP are marked with orange (Ser15), red (Phe18 and Phe45), and yellow boxes (Ser58). The residues marked with a red box form hydrophobic interactions with each other and are found only in Ef-ACP. The residues in the yellow box are composed of a proline in the α2α3 loop of most ACPs, but Ef-ACP carries a serine residue in the α2α3 loop.
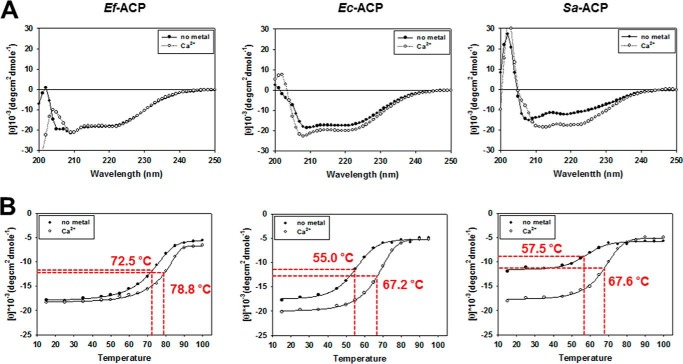
Despite high similarities between Ec-ACP and Ef-ACP, the melting temperature of Ef-ACP was markedly different from that of Ec-ACP. The CD spectrum of Ef-ACP showed double minima at 205 and 222 nm, which are characteristic of an α-helical structure (Fig. 2A). Analysis of the thermal denaturation curves obtained from 15 to 100 °C showed that the Tm of Ef-ACP with CaCl2 is 6.3 °C higher than that in the absence of Ca2+ ions (Fig. 2B). Table 1 lists the Tm values of various ACPs. We measured the Tm values of Ec-ACP and S. aureus ACP, and compared these with that of Ef-ACP by CD experiments. The Tm of Ec-ACP in the presence of Ca2+ was 67.2 °C, much higher than 55.0 °C, the Tm of Ec-ACP in the absence of metal ions. The Tm of S. aureus ACP in the presence of Ca2+ was 67.6 °C, which was relatively lower than those of other ACPs, whereas the Tm of S. aureus ACP in the absence of metal ions was 57.5 °C, an additional 10.1 °C lower than that with Ca2+. For Ec-ACP, as listed in Table 1, we compared our data with the previously reported Tm measured by differential scanning calorimetry and found that the buffer condition as well as the measurement methods resulted in a small difference in Tm (57). However, both results confirmed that the Tm of Ec-ACP in the presence of Ca2+ was >12 °C higher than that of Ec-ACP in the absence of metal ions. Also, it has been reported that V. harveyi ACP has an α-helical structure with a Tm value of 66.4 °C in the presence of Ca2+, whereas this protein is unfolded in the absence of Ca2+ (58). The measurement of melting temperatures implied that Ef-ACP has much higher thermal stability than does other ACPs. Therefore, in this study, we investigated the key factors that stabilize the specific conformations of Ef-ACP.
FIGURE 2.
CD spectra of Ef-ACP, Ec-ACP, and S. aureus ACP (Sa-ACP). A, CD spectrum of ACPs in the presence of Ca2+ (○) and in the absence of Ca2+ (●) at 25 °C. B, temperature-induced folding change of ACPs as monitored by changes of ellipticity at 222 nm. The red dotted line indicates the Tm point of each graph.
TABLE 1.
Melting temperatures of ACPs
| Buffer conditions | Metal | Tm | |
|---|---|---|---|
| °C | |||
| Ef-ACP | 25 mm MES (pH 6.1), 5 mm DTT | Nonea | 72.5b |
| 5 mm CaCl2 | 78.8b | ||
| Ec-ACP | 25 mm MES (pH 6.1), 5 mm DTT | Nonea | 55.0b |
| 5 mm CaCl2 | 67.2b | ||
| S. aureus ACP | 25 mm MES (pH 6.1), 5 mm DTT | Nonea | 57.5b |
| 5 mm CaCl2 | 67.6b | ||
| Ec-ACP | 50 mm Sodium acetate (pH 6.1), 0.5 mm DTT | Nonea | 52.7 (57)c |
| 8.4 mm CaCl2 | 64.3 (57)c | ||
| V. harveyi ACP | 5 mm Hepes (pH 6.5) | Nonea | Unfolded (58) |
| 2 mm CaCl2 | 66.4 (58)c |
a Tm was measured in the absence of metal ion.
b Tm was measured by CD spectroscopy.
c Tm was measured by differential scanning calorimetry.
Chemical Shift Perturbations of Apo- and Holo-Ef-ACP
To investigate the effect of the prosthetic group binding in Ef-ACP, chemical shift perturbations upon conversion of the apo- to the holo-form were studied by comparing the 1H-15N HSQC spectra of holo- and apo-ACPs (Fig. 3A). This analysis showed that the chemical shifts of apo- and holo-Ef-ACPs are very similar. Upon this conversion by covalent modification of Ser39 by a phosphopantetheine prosthetic group, large chemical shift changes (>0.1 ppm) were observed near the prosthetic group binding site at Ser39 and Ile40. The results are in reasonable agreement with those of the most perturbed residues reported for phosphopantetheinylation of B. subtilis ACP, Ec-ACP, and M. tuberculosis ACP (33, 34, 59). In addition, the residues Ala37, Asp38, Ser41, Val42, Met43, and Glu44 near the site of prosthetic group attachment undergo chemical shift changes (0.03 < ppm < 0.1), as indicated with different colors on the ribbon structure of holo-Ef-ACP (Fig. 3B). Furthermore, residues such as Ile57, Ser58, Ile65, and Ala70, which form a hydrophobic cleft and are affected by the prosthetic group, show chemical shift perturbations (0.03 < ppm < 0.05) between the apo- and holo-forms.
FIGURE 3.
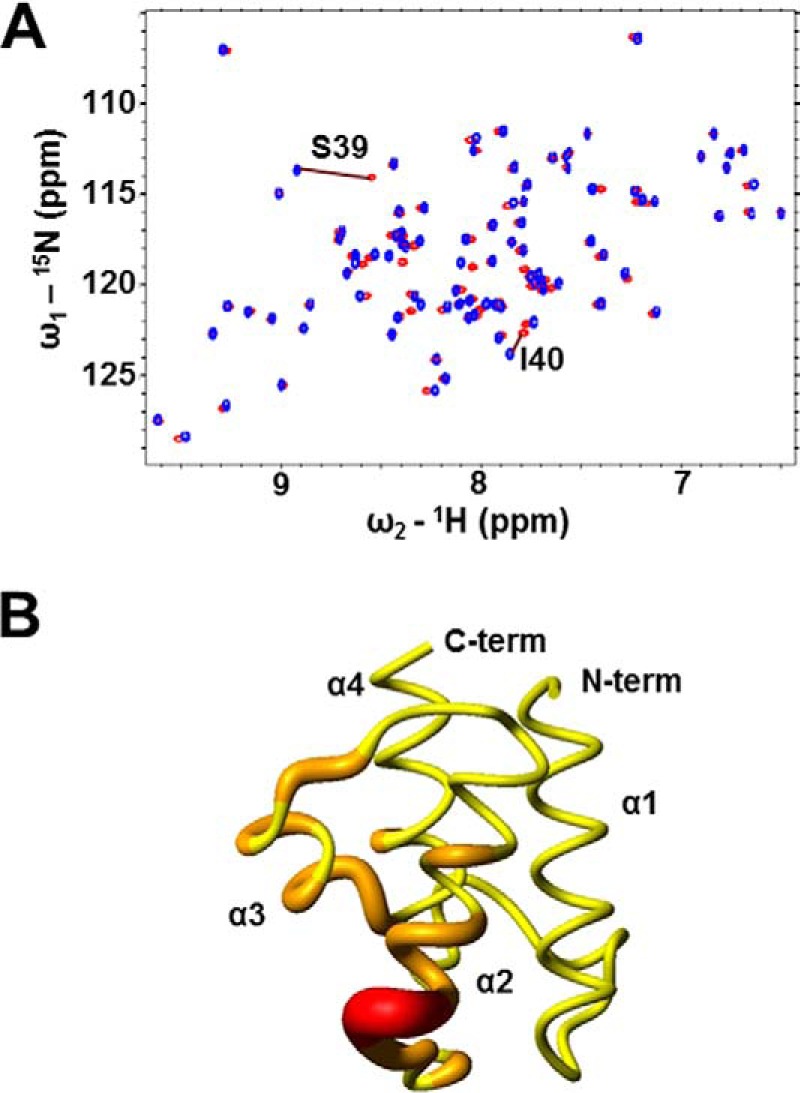
NMR data of apo- and holo-ACP. A, overlay of the 1H-15N HSQC spectra of apo- (red) and holo-ACP (blue). B, chemical shift change value ranges induced by 4′-phosphopantetheine prosthetic group modification of apo-ACP are indicated in various colors in tubular drawings. Residues with ppm >0.03 are shown with graduated larger ribbon diameters. N-term and C-term, N and C termini, respectively. Red, >0.1 ppm; orange, 0.03 < ppm < 0.1; yellow, <0.03 ppm. Chemical shift perturbations were calculated using the equation, Δδav = (0.5(Δδ(1HN)2 + (0.2Δδ(15N))2))½.
The Structure of Ef-ACP
To understand the relationship between the structure and the thermal stability of Ef-ACP, we determined the tertiary structure of Ef-ACP. The three-dimensional structure of Ef-ACP was calculated based on NMR constraints. A total of 1510 NOE distance restraints, 148 backbone torsion angle restraints, and 72 orientational restraints were identified for Ef-ACP (Table 2). The patterns of short and medium range NOEs observed in the 15N and 13C HSQC-NOESY spectra of Ef-ACP indicate that Ef-ACP possesses three relatively long helices and one short helix. In the final structural calculations, a total of 100 structures for ACP were calculated using the program CYANA, and 20 structures with the lowest target functions were selected and superimposed (Fig. 4A). The root mean square deviations of Ef-ACP were 0.18 ± 0.02 Å for backbone (N, Cα, and C′) atoms, and 0.66 ± 0.05 Å for all heavy atoms of residues 2–79, respectively. The solution structures of Ef-ACPs consisted of four α-helices where α1 (Arg3–His17), α2 (Ser39–Phe53), α3 (Glu60–Lys64), and α4 (Val68–Asn78) were connected by three loops (Fig. 4B). Helix III was shorter relative to the other helices. A shorter α2α3 loop (Gly54–Asp59) connected helix II to III, and a 3-residue α3α4 loop (Ile65–Thr67) linked helices III and IV.
TABLE 2.
Structural statistics and mean pairwise root mean squared deviations for the 20 lowest energy structures of Ef-ACP
| Distance restraints | Ef-ACP |
|---|---|
| Total | 1510 |
| Intraresidual and sequential | 752 |
| Medium range | 461 |
| Long range | 297 |
| Dihedral angle restraints | 148 |
| Hydrogen bonds | 44 |
| Orientational restraints | 72 |
| Mean Cyana target function (+) | 4.01 ± 0.03 |
| Coordinate precisiona | |
| Backbone (+) | 0.18 ± 0.02 |
| Heavy atom (Å) | 0.66 ± 0.05 |
| Ramachandran plot statistics (%)b | |
| Residues in most favored regions | 82.9 |
| Residues in additional allowed regions | 17.1 |
| Residues in generously allowed regions | 0.0 |
| Residues in disallowed regions | 0.0 |
a The average root mean square deviation for the overall structure between the 20 structures calculated for Ef-ACP.
b Determined using the program PROCHECK (50).
FIGURE 4.
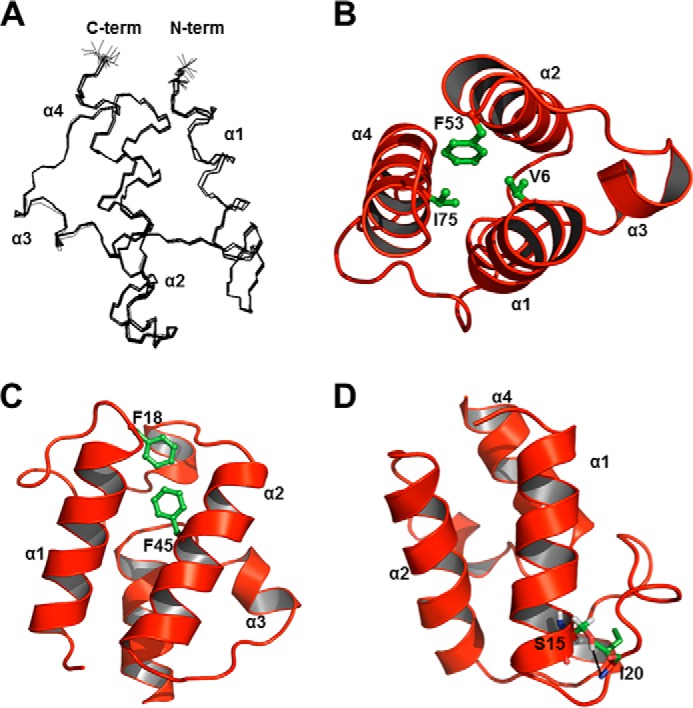
Structure of Ef-ACP. A, superposition of backbone (nitrogen, Cα, and C′) atoms of the 20 lowest energy structures of holo-Ef-ACP. All residues (positions 1–79) are shown, and N and C termini are labeled as N-term and C-term, respectively. B, hydrophobic contacts of Val6, Phe53, and Ile75 in holo-Ef-ACP. C, hydrophobic interactions between Phe18 and Phe45. D, hydrogen bond interactions between Ser15 and Ile20 are depicted with black dotted lines. In B–D, all structures are represented as schematic diagrams, and each helix is labeled from α1 to α4. Each residue is depicted as green sticks.
The α-helices of Ef-ACP have amphipathic character. Many of the long range NOEs involved in packing of the helical bundle arose from the contacts between the hydrophobic side chains, corresponding to residues Val6, Val10, and Ile13 in helix I; Phe18 and Ile31 in the α1α2 loop; Phe45, Val46, Leu49, and Phe53 in helix II; Ile57 in the α2α3 loop; Ala61 in helix III; and Ala71, Tyr74, and Ile75 in helix IV. These residues form the hydrophobic cavity of Ef-ACP to accommodate the growing acyl chain, in which hydrophobic interactions between these residues stabilize the global fold of Ef-ACP. Unusually, Ef-ACP has three phenylalanines, Phe18, Phe45, and Phe53, which play an important role in hydrophobic packing of the helical bundle, as shown in Fig. 4, B and C, whereas Ec-ACP has only two phenylalanines, Phe28 and Phe50. As commonly shown in most ACP structures, Val6, Phe53, and Ile75 at the top end of the hydrophobic cavity of ACP have close hydrophobic contacts with each other (Fig. 4B). Phe18 is located at the beginning of the first long loop and forms a hydrophobic interaction with Phe45 in helix II (Fig. 4C). We also noted a unique structural feature that stabilizes the flexible long α1α2 loop of Ef-ACP; the side chain oxygen of Ser15 and the backbone amide proton of Ile20 form a hydrogen bonding interaction (Fig. 4D). In addition, the backbone carboxyl group of Glu50 and the backbone amide proton of Thr55 form a hydrogen bonding interaction.
Dynamics of Ef-ACP
Fast backbone dynamics on a picosecond-to-nanosecond time scale were studied by measuring the R1, R2, and hNOEs of Ef-ACP (Fig. 5). The average R1, R2, and hNOE values of Ef-ACP were 1.76 ± 0.027, 7.38 ± 0.24, and 0.79 ± 0.007 s−1. The average R2/R1 ratio for Ef-ACP was 3.93, implying that Ef-ACP exists as monomer. Relaxation rates showed relatively uniform behavior across the protein sequence with the exception of the loop regions and the residues at the N and C termini. The R2 values were uniform in all regions except for Ile57 and Ser58 in the α2α3 loop region. Ser58 in Ef-ACP corresponds to Pro55 in Ec-ACP, which forms the rigid bent structure right before helix III. Ile57 (10.51 ± 0.76 s−1) and Ser58 (39.86 ± 1.04 s−1) have high R2 values, implying that they have conformational exchange caused by the relatively more flexible loop motions compared with those of Ile54 and Pro55 in Ec-ACP (59). As shown in Fig. 3B, these two residues show chemical shift perturbations (0.042 ppm in Ile57 and 0.037 ppm in Ser58) induced by phosphopantetheine modification. These results imply that the Ile57 and Ser58 loop region might have a conformational exchange to generate enough room to accommodate the longer acyl chains.
FIGURE 5.
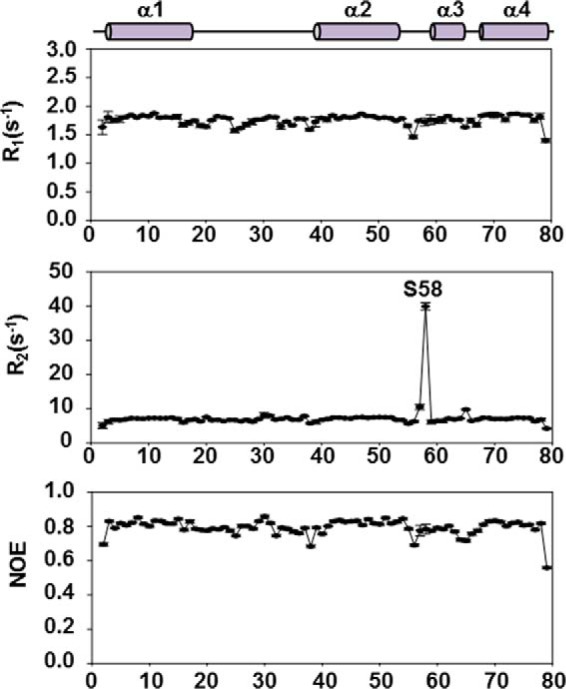
Spin relaxation rates of Ef-ACP in the presence of Ca2+. Relaxation data of Ef-ACP were measured at 298 K. The values of R1, R2, and the steady-state heteronuclear NOEs are plotted as a function of the residue sequence number for Ef-ACP. Above the plots, the solid and filled bars represent the four helices of Ef-ACP.
The average hNOE values in the helix regions, α1 (Arg3–His17), α2 (Ser39–Phe53), α3 (Glu60–Lys64), and α4 (Val68–Asn78) were high, which implied structural rigidity: 0.81, 0.82, 0.77, and 0.81. Residues at each end of helix II, such as Asp38 and Glu56, exhibited relatively lower hNOE values. Asp38 is located at the end of the long α1α2 loop region (Phe18–Asp38) near the prosthetic group binding sites, and Glu56 is at the flexible α2α3 loop region of Ef-ACP. Significantly reduced hNOE values were observed for the N and C termini because of the flexibilities of the terminal regions in protein structure. The hNOE value of Ser79 at the C terminus was not negative but was low because the peak for Val25 was overlapped with the peak for Ser79. The hNOE values for the α1α2 loop are relatively high in consideration of its length, resulting from hydrogen bonds between Ser15 and Ile20. Therefore, the structural rigidity and thermal stability of Ef-ACP resulted from the hydrophobic interactions between the residues forming the hydrophobic cleft of Ef-ACP.
Hydrogen/Deuterium Exchange Experiments
Amide hydrogen/deuterium exchange rates provide information about the amides buried in stable protein structures, which result in slow exchange rates at a second-level time scale. To understand how the dynamic behaviors in slow time scales are related to the structural stability of Ef-ACP, hydrogen/deuterium exchange experiments were performed, and the protection factors were calculated to obtain information about the degree of protection of protein amide protons against hydrogen/deuterium exchange. The first HSQC spectrum was acquired 5 min after the addition of D2O to the protein. Many residues showed rapid exchange, and only 53 amide peaks among 78 remained after 10 min in the first HSQC spectrum. The helical regions of Ef-ACP with the exception of the short helix III had high protection factor values in the presence of Ca2+ (Fig. 6A). This implies that the residues in helices I, II, and IV were involved in stable folds unlike the residues in the other regions, and this result agrees well with the hNOE data. Although some residues, such as Phe18, Ile20, Gln24, Leu29, Ile31, Lys32, Asp34, Leu35, and Thr55, were in the loop region, they showed high protection factors (logP > 2). Fig. 6B shows the examples of fitting results of the slow hydrogen/deuterium-exchanging residues (Ser15, Phe18, Phe45, and Phe53). The exchange rates shown in Fig. 6B are 1.58 × 10−3 min−1 (Ser15), 5.82 × 10−3 min−1 (Phe18), 4.74 × 10−3 min−1 (Phe45), and 1.15 × 10−3 min−1 (Phe53). Although Phe18 is located in the α1α2 loop, the exchange rate is slow, resulting from a stable hydrophobic interaction with Phe45. Phe53 plays important roles in hydrophobic interactions in the hydrophobic cleft of Ef-ACP, and its exchange rate (1.15 × 10−3 min−1) is similar to the previously reported value of Phe50, the corresponding residue in Ec-ACP (4.61 × 10−3 min−1) (60). Hydrogen bonds between Ser15 and Ile20 result in slow exchange rate of the amide proton of Ile20 in the α1α2 loop as well. Amide protons of Gln24 and Leu29 form hydrogen bonds with Glu21 and Thr29, respectively, and demonstrate slow hydrogen/deuterium exchange rates. Ef-ACP contains one turn from Ile31 to Leu35 in the α1α2 loop, resulting in the decrease of exchange rates of Ile31, Lys32, Asp34, and Leu35. Previous work on Ec-ACP has also reported that residues in the C-terminal portion from 26 to 34 in the α1α2 loop also exhibit greater protection factors than do residues at the N-terminal region of this loop (60). Hydrogen bonding interaction between Thr55 and Glu50 decreased the exchange rate of the amide proton of Thr55. Even after 4 days, the Ef-ACP spectrum still showed many peaks for the residues located in helices I, II, and IV. These residues are Val6, Leu7, Val10, Ala11, Ile13, and Ile14 in helix I; Val46, Leu47, Glu48, Leu49, Glu50, and Phe53 in helix II; and Val68, Ala71, Val72, Asp73, Tyr74, Ile75, and Val76 in helix IV.
FIGURE 6.
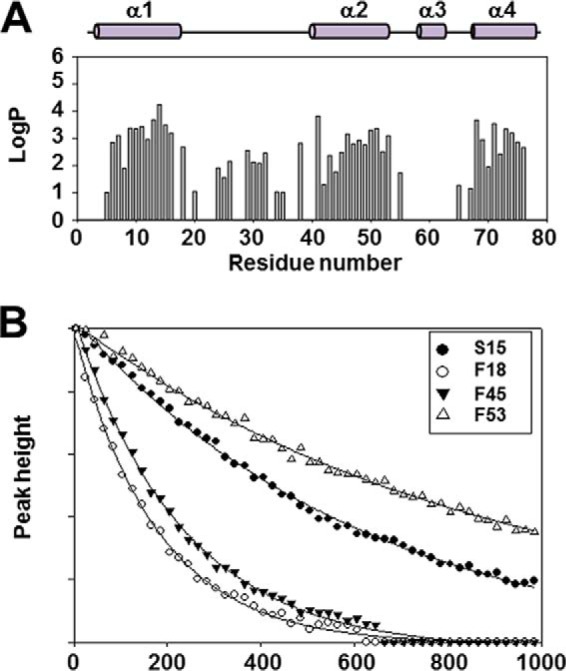
Hydrogen/deuterium exchange experiment of Ef-ACP in the presence of Ca2+. A, the protection factor of Ef-ACP is depicted by a bar graph as a function of the residue sequence number of Ef-ACP. The solid and filled bars above the plots represent the four helices of Ef-ACP. B, time course of exchange showing signal loss due to hydrogen/deuterium exchange as a function of time after the addition of D2O for each of the four residues. Peak height is in arbitrary units.
Effect of Ca2+ on Ef-ACP
To investigate the effect of Ca2+, fast backbone dynamics were studied by measuring the R1, R2, and hNOEs of Ef-ACP in the absence of Ca2+ (Fig. 7, A–C). In the absence of Ca2+, the average R1, R2, and hNOE values of Ef-ACP were 1.76 ± 0.021, 7.66 ± 0.15, and 0.81 ± 0.020 s−1, which are very similar to those in the presence of Ca2+. Similar to the data in the presence of Ca2+, Ser58 in the α2α3 loop region showed high R2 values, 38.49 ± 2.16 s−1 in the absence of Ca2+, implying that they have conformational exchange in both conditions. The hydrogen/deuterium exchange rates in both conditions were also very similar (Figs. 6A and 7D), except over the short helix III region. In the case of metal-free conditions, helix III of Ef-ACP has higher protection compared with in conditions with Ca2+ ions. This may be because Asp61 and Glu63 in this helix may be the Ca2+ binding site and must be exposed to bind to Ca2+ ions, whereas in the absence of Ca2+, they are buried inside and protected. Overall, the protection factors of Ef-ACP in both conditions are very similar. These results agree well with the thermal stability of Ef-ACP because the difference between Tm of Ef-ACP in the calcium-bound form and in the calcium-free form is only 6 °C.
FIGURE 7.
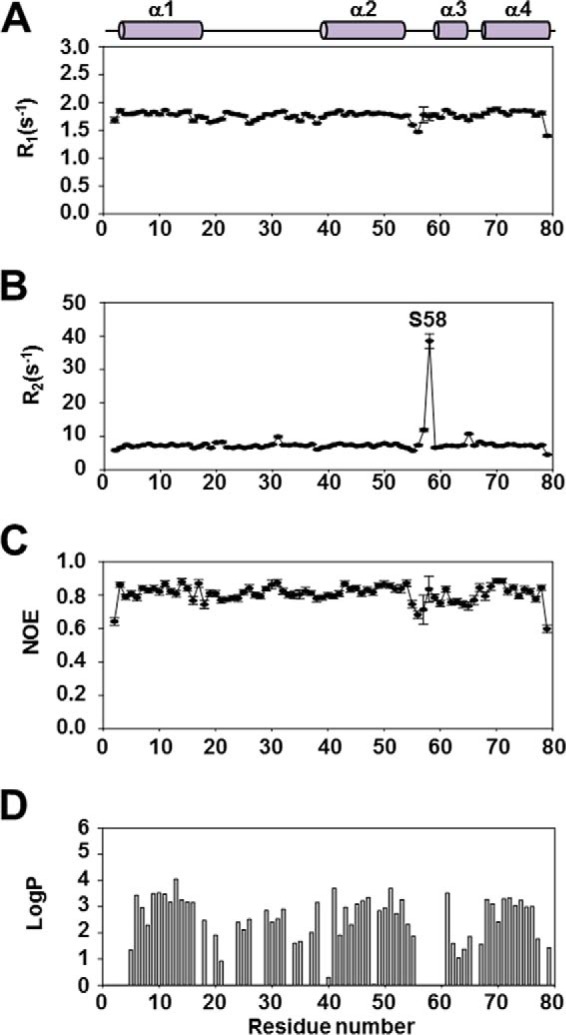
Spin relaxation rates and Protection factor of Ef-ACP in the absence of Ca2+. The values of R1 (A), R2 (B), and the steady-state heteronuclear NOEs (C) are plotted as a function of the residue sequence number for Ef-ACP. Above the plots, the solid and filled bars represent the four helices of Ef-ACP. D, protection factor of Ef-ACP in the absence of Ca2+ depicted by a bar graph as a function of the residue sequence number of Ef-ACP. The solid and filled bars above the plots represent the four helices of Ef-ACP. All NMR data of Ef-ACP were measured at 298 K.
Mutagenesis Studies of Ef-ACP
To assess the effects of specific residues on the thermal stability of Ef-ACP, site-directed mutagenesis was performed to generate five single-point mutations of Ef-ACP, S15G, H17A, F18A, F45A, and F53A, and their Tm values were determined using CD spectroscopy, as shown in Table 3. The Tm of the S15G mutant of Ef-ACP was 60.1 °C, 18.7 °C lower than that of the wild type protein, and the helical content was also much lower than that of the wild type Ef-ACP, implying that the hydrogen bond between Ser15 and Ile20 in the long α1α2 loop might be the key factor in stabilizing the conformation. The Tm of the H17A mutant was 62.3 °C, 16.5 °C lower than that of the wild type, whereas its overall secondary structure was similar to that of wild type Ef-ACP. This implies that His17 might play an important role in conformational stabilization by reducing the electrostatic repulsion between the negatively charged residues but that mutation of His17 does not induce substantial conformational change. The Tm of F18A was also 9.3 °C lower than that of the wild type, and the helical content was a little lower than that of wild type ACP. In contrast, the Tm of the F45A mutant was 52.9 °C, 25.9 °C lower than that of the wild type, and the helical content of F45A mutant Ef-ACP was much lower than that of wild type Ef-ACP. The HSQC spectrum showed that F45A mutation caused partial unfolding of protein structure (data not shown). The mutations of Phe53 with alanine at F53A also exhibited a big influence (24.1 °C) on stability and on the Ef-ACP structure as well. These results imply that the hydrophobic interactions between Phe45 and Phe18, as well as other hydrophobic interactions in the hydrophobic cleft, including those between Val6, Phe53, and Ile75, play critical roles in the proper folding and structural stability of Ef-ACP. These results demonstrate that electrostatic as well as hydrophobic interactions in Ef-ACP are key factors in thermal and structural stability in Ef-ACP.
TABLE 3.
Melting temperatures of mutant Ef-ACPs in 25 mm MES buffer, pH 6.1, containing 5 mm CaCl2
| Wild type | S15G | H17A | F18A | F45A | F53A | |
|---|---|---|---|---|---|---|
| Melting temperaturea | 78.8 °C | 60.1 °C | 62.3 °C | 69.5 °C | 52.9 °C | 54.7 °C |
a Melting temperature was measured by CD spectroscopy.
Discussion
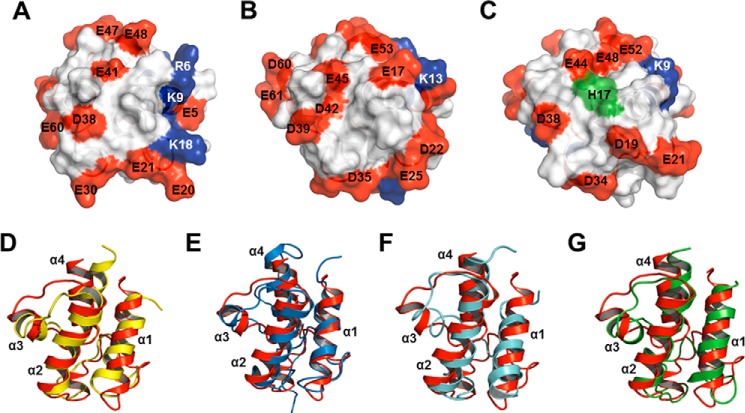
To provide insights into understanding the thermal stability of Ef-ACP, we identify the key factors in its stability by investigating its structure, dynamics, and melting temperatures of mutants. In previous studies, Ec-ACP has been reported to exhibit higher melting temperatures (64.3 °C) in the presence of calcium ion compared with Ec-ACP in the absence of calcium ion (52.7 °C) (57). In addition, in the presence of calcium ion, V. harveyi ACP has an α-helical structure with a Tm of 66.4 °C; however, in the absence of calcium ion, this protein is unfolded (58). As shown in Fig. 9 (A–C), severe electrostatic repulsions exist in helix II and in the α1α2 loop near the metal binding site A in both Ec-ACP and V. harveyi ACP. The low thermal stability of Ec-ACP and V. harveyi ACP in conditions of the absence of calcium results from the severe electrostatic repulsion between negatively charged residues, which reduces the stability of Ec-ACP. In previous reports, the neutralization of acidic residues in helix II was shown to stabilize V. harveyi ACP conformation (12). In the case of Ec-ACP, His75 at helix IV retained a stable native conformation by shielding the electrostatic repulsion between negatively charged residues (59). Furthermore, V. harveyi ACP, which has an alanine instead of histidine at the C terminus, becomes unfolded, whereas the A75H mutation restores proper folding of the α-helical structures by reducing the electrostatic repulsion (58, 61, 62).
FIGURE 9.
Electrostatic potential surface structure and structural comparison of ACPs. A–C, electrostatic potential of Ec-ACP (PDB entry 2K93) (A), V. harveyi ACP (PDB entry 2L0Q) (B), and Ef-ACP (PDB entry 2N50) (C). The residue numbers are labeled. Positive and negative charge residues are colored in blue and red, respectively. The His17 residue is colored in green. This surface structure is rotated about the y axis by 90° compared with that shown in Fig. 4A. D–G, structural comparison of Ef-ACP with other ACPs; overlay of structures of Ef-ACP (red; PDB entry 2N50) and Ec-ACP (yellow; PDB entry 2K93) (D), Ef-ACP and M. tuberculosis ACP (blue; PDB entry 1KIP) (E), Ef-ACP and P. falciparum ACP (cyan; PDB entry 2QF0) (F), and Ef-ACP and B. subtilis ACP (green; PDB entry 1HY8) (G). Each helix is labeled from α1 to α4, and this overlay structure has the same orientation as the structure shown in Fig. 4A.
As shown in this study, Ef-ACP is more thermally stable than are other ACPs in the absence of calcium, and the thermal stabilities of Ef-ACP in calcium-bound (78.8 °C) and calcium-free conditions (72.5 °C) are only 6.3 °C different, which is a relatively small difference compared with other ACPs, as shown in Table 1. Also, the relaxation rates as well as hydrogen/deuterium exchange data of Ef-ACP are similar for both conditions. This may result from His17, which is unique in Ef-ACP, at the end of the first helix. As shown in Fig. 9C, His17 in Ef-ACP reduced the electrostatic repulsion between negatively charged residues in the α1α2 loop and helix II. The H17A mutant was identified to have a 12 °C decrease in melting temperature. Because distribution of negatively charged residues in Ef-ACP is different from those of Ec-ACP and V. harveyi ACP, the metal binding sites of Ef-ACP might differ from those of other ACPs, which needs to be investigated in further studies.
Most type II ACPs have a flexible long α1α2 loop between helix I and helix II, resulting in low hNOEs and in the deuterium exchange experiment (4, 35, 59, 60). In addition, most ACPs, such as Ec-ACP, oxytetracycline-ACP, and frenolicin-ACP, have large amplitude motions and show lower NOE values in the α1α2 loop. The average hNOE value of the α1α2 loop in frenolicin-ACP is 0.49 ± 0.038 s−1, whereas that in Ef-ACP is 0.78 ± 0.007 s−1 (59, 63, 64). This loop rigidity results from the hydrogen bonds between the side chain oxygen of Ser15 in helix I and the backbone amide of Ile20 in the loop and a hydrophobic interaction between Phe18 (α1α2 loop) and Phe45 (helix II). As shown in Table 3, the Tm of the S15G mutant was 18 °C lower compared with that of wild type Ef-ACP, implying that this hydrogen bonding interaction is essential for the thermal stability of Ef-ACP.
Previous molecular dynamics simulations on Ec-ACP showed that there were two different acyl chain binding pockets, resulting in the splitting of the hydrophobic binding cavity (14). Subpocket I is located between helices II, III, and IV, and subpocket II is located more toward helix I, between helices I, II, and IV. The entrance of these pockets is close to the N-terminal end of helix II, which is near the prosthetic group binding site, at Ser36 in Ec-ACP. In Ec-ACP, a side chain reorientation of Leu42 and Leu46 is required to act as a switch to determine the direction of acyl chain binding between the two pockets (14). In contrast, for Ef-ACP, a hydrophobic interaction between Phe45 at helix II and Phe18 at the beginning of the α1α2 loop may block the entrance into subpocket II, which otherwise would accommodate the growing acyl chain between helices I, II, and IV as shown in Fig. 8.
FIGURE 8.
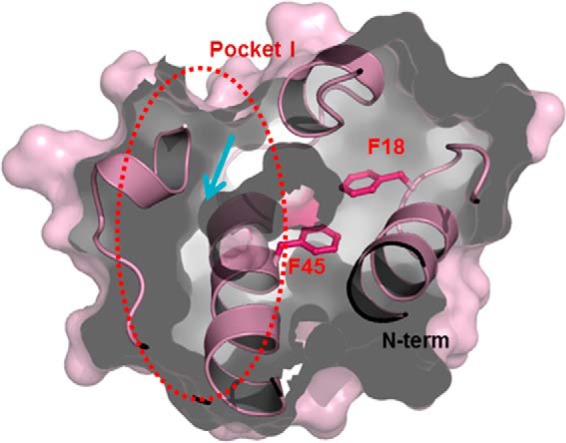
Surface representation of subpockets in the hydrophobic cavity. Phe45 in Ef-ACP has hydrophobic interactions with Phe18, resulting in blockage of the entrance into the alternate binding cavity of Ef-ACP between helices I, II, and IV. The arrows represent the direction of the path of the growing acyl chain.
Superimposing only the Cα atoms in Ef-ACP onto other type II ACPs gave root mean square deviation values of 2.62 Å for Ec-ACP, 2.38 Å for M. tuberculosis ACP, 3.24 Å for P. falciparum ACP, and 2.87 Å for B. subtilis ACP (32–34, 37). Sequence alignments shown in Fig. 1 indicated that the similarity between Ef-ACP and other ACPs was high (Gram-positive: B. subtilis ACP, 72.2%; M. tuberculosis ACP, 63.0%; and S. aureus ACP, 74.7%; Gram-negative: Ec-ACP, 69.6%; Acinetobacter baumanni ACP, 69.6%; P. falciparum ACP, 67.1%; and V. harveyi ACP, 68.4%). Comparisons of the structures of other ACPs with that of Ef-ACP indicate that they all have similar global folds with similar secondary structural elements, with the four-helix bundle fold being well conserved among ACPs (Fig. 9, D–G). In comparison with the B. subtilis ACP structure, the distinctive difference between that of Ef-ACP appears at the α2α3 loop, which protrudes outward in the latter. The distance between the Cα of phosphopantethylated Ser39 and the Cα of Ser58 at the α2α3 loop was 16.6 Å, whereas the corresponding distances in M. tuberculosis ACP, Ec-ACP, B. subtilis ACP, and P. falciparum ACP are 12.9, 12.9, 13.5, and 14.4 Å, respectively. Therefore, this results in the outward movement of helix III to expand the hydrophobic cavity for stable binding of the growing acyl chains in Ef-ACP compared with other ACPs. Because Phe45-mediated hydrophobic interaction may block the entrance into subpocket II in Ef-ACP, to accommodate the long growing acyl chain, Ser58 in the α2α3 loop of Ef-ACP might facilitate the movement of this flexible loop and of helix III further outside the main structure compared with other ACPs. The relaxation rates confirmed the flexibility of Ser58 in the α2α3 loop as well. The R2 rates were uniform in all regions of Ef-ACP with the exception of the R2 rates of Ile57 and Ser58, implying the flexibility of the α2α3 loop in Ef-ACPs. Within the α2α3 loop of Ec-ACP, Ile54 and Pro55, showed uniform R2 value because proline had a tendency to form a rigid bent structure in the loop in our previous study (59). These results imply that the Ile57 and Ser58 loop region might have a conformational exchange to release and accept the acyl chain and generate enough room to accommodate the longer acyl chains.
It has been reported that the α2α3 loop is important in interactions between ACP and FabA in fatty acid synthesis because perturbing the α2α3 loop is required for these protein-protein interactions (60). In previous studies, Ile54 in Ec-ACP, which corresponds to Ile57 in Ef-ACP, shows a large chemical shift perturbation in the ACP spectra by the addition of FabA (65). Here, conformational exchange in Ile57 and Ser58 in the α2α3 loop was observed for the first time, and the flexibility of the α2α3 loop might play an important role in its interaction with other FAS enzymes that could promote release of the growing acyl moieties from the hydrophobic pocket of ACP.
It has been reported that the Glu and Asp residues in helix II of Ec-ACP are important to recognize FabA, FabB, FabD, FabG, MCAT, AcpS, and FabH (61, 65–68). For example, Arg249 in the basic surface of FabH is critical for interaction with Glu41 of Ec-ACP (69). NMR and mutagenesis studies have shown that Glu41 and Glu48 in E. coli are important for FabG binding (67). Also, negatively charged residues in helix II of Ec-ACP have electrostatic interactions with basic residues of AcpS (70). Because Ef-ACP also has many negatively charged residues in helix II and suitable for binding with FAS proteins similar to Ec-ACP, molecules that mimic helix II and the α2α3 loop of Ef-ACP may inhibit these interactions with various FAS proteins and can be attractive candidates as therapeutic agents against drug-resistant strains of E. faecalis.
In conclusion, Ef-ACP has unique features, including hydrophobic interactions between Phe45 and Phe18 and hydrogen bonds between Ser15 and Ile20 in the α1α2 loop, resulting in the high thermal stability of Ef-ACP. Furthermore, Ser58 in the α2α3 loop of Ef-ACP can provide loop flexibility and allow the provision of room to accommodate a longer acyl chain. Further studies on the structures and dynamics of acylated Ef-ACP will provide insight to understand the optimum length of the acyl chain bound in the hydrophobic pocket of Ef-ACP and to clarify the specific recognition mechanisms between acyl-ACP and functionally different enzyme partners in the FAS pathway. This study may provide insights to develop useful inhibitors for fatty acid synthesis as potent antibiotics against pathogenic drug-resistant strains.
Author Contributions
Y.-G. P. and Y. K. designed the study and wrote the paper. Y.-G. P., K.-W. J., E. B., G.-S. H., and M.-C. J. performed CD and NMR experiments and analyzed the data. Y. P. and H. S. cloned and purified the proteins. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank the High Field NMR Research Program of the Korea Basic Science Institute.
This work was supported by Basic Science Research Program Grant 2013R1A1A2058021 and Priority Research Centers Program Grant 2009-0093824 through the National Research Foundation of Korea, funded by the Ministry of Education, Science, and Technology. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 2N50) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- FAS
- fatty acid synthase
- ACP
- acyl carrier protein
- Ef-ACP
- Enterococcus faecalis ACP
- Ec-ACP
- Escherichia coli ACP
- AcpS
- holo-ACP synthase
- hNOE
- heteronuclear Overhauser effect
- HSQC
- heteronuclear single quantum coherence spectroscopy
- PDB
- Protein Data Bank.
References
- 1. Uauy R., Mena P., and Rojas C. (2000) Essential fatty acids in early life: structural and functional role. Proc. Nutr. Soc. 59, 3–15 [DOI] [PubMed] [Google Scholar]
- 2. Cronan J. E., Jr. (2004) The structure of mammalian fatty acid synthase turned back to front. Chem. Biol. 11, 1601–1602 [DOI] [PubMed] [Google Scholar]
- 3. Cronan J. E. (2006) Remarkable structural variation within fatty acid megasynthases. Nat. Chem. Biol. 2, 232–234 [DOI] [PubMed] [Google Scholar]
- 4. Chan D. I., and Vogel H. J. (2010) Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem. J. 430, 1–19 [DOI] [PubMed] [Google Scholar]
- 5. White S. W., Zheng J., Zhang Y. M., and Rock (2005) The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831 [DOI] [PubMed] [Google Scholar]
- 6. Byers D. M., and Gong H. (2007) Acyl carrier protein: structure-function relationships in a conserved multifunctional protein family. Biochem. Cell Biol. 85, 649–662 [DOI] [PubMed] [Google Scholar]
- 7. Cronan J. E. (2014) The chain-flipping mechanism of ACP (acyl carrier protein)-dependent enzymes appears universal. Biochem. J. 460, 157–163 [DOI] [PubMed] [Google Scholar]
- 8. Shen B., Summers R. G., Gramajo H., Bibb M. J., and Hutchinson C. R. (1992) Purification and characterization of the acyl carrier protein of the Streptomyces glaucescens tetracenomycin C polyketide synthase. J. Bacteriol. 174, 3818–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geiger O., Spaink H. P., and Kennedy E. P. (1991) Isolation of the Rhizobium leguminosarum NodF nodulation protein: NodF carries a 4′-phosphopantetheine prosthetic group. J. Bacteriol. 173, 2872–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heaton M. P., and Neuhaus F. C. (1994) Role of the d-alanyl carrier protein in the biosynthesis of d-alanyl-lipoteichoic acid. J. Bacteriol. 176, 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Issartel J. P., Koronakis V., and Hughes C. (1991) Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature 351, 759–761 [DOI] [PubMed] [Google Scholar]
- 12. Gong H., Murphy A., McMaster C. R., and Byers D. M. (2007) Neutralization of acidic residues in helix II stabilizes the folded conformation of acyl carrier protein and variably alters its function with different enzymes. J. Biol. Chem. 282, 4494–4503 [DOI] [PubMed] [Google Scholar]
- 13. Crosby J., and Crump M. P. (2012) The structural role of the carrier protein–active controller or passive carrier. Nat. Prod. Rep. 29, 1111–1137 [DOI] [PubMed] [Google Scholar]
- 14. Chan D. I., Stockner T., Tieleman D. P., and Vogel H. J. (2008) Molecular dynamics simulations of the apo-, holo-, and acyl-forms of Escherichia coli acyl carrier protein. J. Biol. Chem. 283, 33620–33629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuch A., Willems R. J. L., Werner G., Coque T. M., Hammerum A. M., Sundsfjord A., Klare I., Ruiz-Garbajosa P., Simonsen G. S., van Luit-Asbroek M., Hryniewicz W., and Sadowy E. (2012) Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. J. Antimicrob. Chemother. 67, 551–558 [DOI] [PubMed] [Google Scholar]
- 16. Orr K. E., Holliday M. G., Jones A. L., Robson I., and Perry J. D. (2002) Survival of enterococci during hospital laundry processing. J. Hosp. Infect. 50, 133–139 [DOI] [PubMed] [Google Scholar]
- 17. Wade J. J. (1997) Enterococcus faecium in hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 16, 113–119 [DOI] [PubMed] [Google Scholar]
- 18. Silva Laport M., da Silva M. R., Costa Silva C., do Carmo de Freire Bastos M., and Giambiagi-deMarval M. (2003) Heat-resistance and heat-shock response in the nosocomial pathogen Enterococcus faecium. Curr. Microbiol. 46, 313–317 [DOI] [PubMed] [Google Scholar]
- 19. Paulsen I. T., Banerjei L., Myers G. S., Nelson K. E., Seshadri R., Read T. D., Fouts D. E., Eisen J. A., Gill S. R., Heidelberg J. F., Tettelin H., Dodson R. J., Umayam L., Brinkac L., Beanan M., Daugherty S., DeBoy R. T., Durkin S., Kolonay J., Madupu R., Nelson W., Vamathevan J., Tran B., Upton J., Hansen T., Shetty J., Khouri H., Utterback T., Radune D., Ketchum K. A., Dougherty B. A., and Fraser C. M. (2003) Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299, 2071–2074 [DOI] [PubMed] [Google Scholar]
- 20. Huycke M. M., Sahm D. F., and Gilmore M. S. (1998) Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4, 239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neely A. N., and Maley M. P. (2000) Survival of enterococci and staphylococci on hospital fabrics and plastic. J. Clin. Microbiol. 38, 724–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seedat J., Zick G., Klare I., Konstabel C., Weiler N., and Sahly H. (2006) Rapid emergence of resistance to linezolid during linezolid therapy of an Enterococcus faecium infection. Antimicrob. Agents Chemother. 50, 4217–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kristich C. J., Li Y. H., Cvitkovitch D. G., and Dunny G. M. (2004) Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186, 154–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brusic V., and Petrovsky N. (2005) Immunoinformatics and its relevance to understanding human immune disease. Expert Rev. Clin. Immunol. 1, 145–157 [DOI] [PubMed] [Google Scholar]
- 25. Nallapareddy S. R., Singh K. V., Sillanpää J., Zhao M., and Murray B. E. (2011) Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect. Immun. 79, 2901–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arias C. A., and Murray B. E. (2008) Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti Infect. Ther. 6, 637–655 [DOI] [PubMed] [Google Scholar]
- 27. Murray B. E. (1990) The life and times of the Enterococcus. Clin. Microbiol. Rev. 3, 46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vergis E. N., Hayden M. K., Chow J. W., Snydman D. R., Zervos M. J., Linden P. K., Wagener M. M., Schmitt B., and Muder R. R. (2001) Determinants of vancomycin resistance and mortality rates in enterococcal bacteremia. a prospective multicenter study. Ann. Intern. Med. 135, 484–492 [DOI] [PubMed] [Google Scholar]
- 29. Lee J. Y., Jeong K. W., Shin S., Lee J. U., and Kim Y. (2012) Discovery of novel selective inhibitors of Staphylococcus aureus β-ketoacyl acyl carrier protein synthase III. Eur. J. Med. Chem. 47, 261–269 [DOI] [PubMed] [Google Scholar]
- 30. Jeong K. W., Lee J. Y., Kang D. I., Lee J. U., Shin S. Y., and Kim Y. (2009) Screening of flavonoids as candidate antibiotics against Enterococcus faecalis. J. Nat. Prod. 72, 719–724 [DOI] [PubMed] [Google Scholar]
- 31. Holak T. A., Kearsley S. K., Kim Y., and Prestegard J. H. (1988) Three-dimensional structure of acyl carrier protein determined by NMR pseudoenergy and distance geometry calculations. Biochemistry 27, 6135–6142 [DOI] [PubMed] [Google Scholar]
- 32. Wu B. N., Zhang Y. M., Rock C. O., and Zheng J. J. (2009) Structural modification of acyl carrier protein by butyryl group. Protein Sci. 18, 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu G. Y., Tam A., Lin L., Hixon J., Fritz C. C., and Powers R. (2001) Solution structure of B. subtilis acyl carrier protein. Structure 9, 277–287 [DOI] [PubMed] [Google Scholar]
- 34. Wong H. C., Liu G., Zhang Y. M., Rock C. O., and Zheng J. (2002) The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J. Biol. Chem. 277, 15874–15880 [DOI] [PubMed] [Google Scholar]
- 35. Park S. J., Kim J. S., Son W. S., and Lee B. J. (2004) pH-induced conformational transition of H. pylori acyl carrier protein: insight into the unfolding of local structure. J. Biochem. 135, 337–346 [DOI] [PubMed] [Google Scholar]
- 36. Barnwal R. P., Van Voorhis W. C., and Varani G. (2011) NMR structure of an acyl-carrier protein from Borrelia burgdorferi. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma A. K., Sharma S. K., Surolia A., Surolia N., and Sarma S. P. (2006) Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs. Biochemistry 45, 6904–6916 [DOI] [PubMed] [Google Scholar]
- 38. Lim J., Kong R., Murugan E., Ho C. L., Liang Z. X., and Yang D. (2011) Solution structures of the acyl carrier protein domain from the highly reducing type I iterative polyketide synthase CalE8. PLoS One 6, e20549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perez D. R., Leibundgut M., and Wider G. (2015) Interactions of the acyl chain with the Saccharomyces cerevisiae acyl carrier protein. Biochemistry 54, 2205–2213 [DOI] [PubMed] [Google Scholar]
- 40. Płoskoń E., Arthur C. J., Evans S. E., Williams C., Crosby J., Simpson T. J., and Crump M. P. (2008) A mammalian type I fatty acid synthase acyl carrier protein domain does not sequester acyl chains. J. Biol. Chem. 283, 518–528 [DOI] [PubMed] [Google Scholar]
- 41. Zhang Y. M., Marrakchi H., White S. W., and Rock C. O. (2003) The application of computational methods to explore the diversity and structure of bacterial fatty acid synthase. J. Lipid Res. 44, 1–10 [DOI] [PubMed] [Google Scholar]
- 42. Lambalot R. H., and Walsh C. T. (1995) Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J. Biol. Chem. 270, 24658–24661 [DOI] [PubMed] [Google Scholar]
- 43. Rock C. O., Cronan J. E. Jr., and Armitage I. M. (1981) Molecular properties of acyl carrier protein derivatives. J. Biol. Chem. 256, 2669–2674 [PubMed] [Google Scholar]
- 44. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 45. Goddard T. D., and Kneller D. G. (2004) SPARKY 3, University of California, San Francisco [Google Scholar]
- 46. Cordier F., Dingley A. J., and Grzesiek S. (1999) A doublet-separated sensitivity-enhanced HSQC for the determination of scalar and dipolar one-bond J-couplings. J. Biomol. NMR 13, 175–180 [DOI] [PubMed] [Google Scholar]
- 47. Shen Y., Delaglio F., Cornilescu G., and Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Güntert P., Mumenthaler C., and Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 49. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 50. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., and Thornton J. M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 51. Koradi R., Billeter M., and Wüthrich K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 52. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., and Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 53. Jeong K. W., Kang D. I., Lee E., Shin A., Jin B., Park Y. G., Lee C. K., Kim E. H., Jeon Y. H., Kim E. E., and Kim Y. (2014) Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3. Biochemistry 53, 4814–4825 [DOI] [PubMed] [Google Scholar]
- 54. Lee J., Jeong K. W., Jin B., Ryu K. S., Kim E. H., Ahn J. H., and Kim Y. (2013) Structural and dynamic features of cold-shock proteins of Listeria monocytogenes, a psychrophilic bacterium. Biochemistry 52, 2492–2504 [DOI] [PubMed] [Google Scholar]
- 55. Bai Y., Milne J. S., Mayne L., and Englander S. W. (1993) Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Frederick A. F., Kay L. E., and Prestegard J. H. (1988) Location of divalent ion sites in acyl carrier protein using relaxation perturbed 2D NMR. FEBS Lett. 238, 43–48 [DOI] [PubMed] [Google Scholar]
- 57. Horvath L. A., Sturtevant J. M., and Prestegard J. H. (1994) Kinetics and thermodynamics of thermal denaturation in acyl carrier protein. Protein Sci. 3, 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chan D. I., Chu B. C., Lau C. K., Hunter H. N., Byers D. M., and Vogel H. J. (2010) NMR solution structure and biophysical characterization of Vibrio harveyi acyl carrier protein A75H: effects of divalent metal ions. J. Biol. Chem. 285, 30558–30566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim Y., Kovrigin E. L., and Eletr Z. (2006) NMR studies of Escherichia coli acyl carrier protein: dynamic and structural differences of the apo- and holo-forms. Biochem. Biophys. Res. Commun. 341, 776–783 [DOI] [PubMed] [Google Scholar]
- 60. Andrec M., Hill R. B., and Prestegard J. H. (1995) Amide exchange rates in Escherichia coli acyl carrier protein: correlation with protein structure and dynamics. Protein Sci. 4, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Flaman A. S., Chen J. M., Van Iderstine S. C., and Byers D. M. (2001) Site-directed mutagenesis of acyl carrier protein (ACP) reveals amino acid residues involved in ACP structure and acyl-ACP synthetase activity. J. Biol. Chem. 276, 35934–35939 [DOI] [PubMed] [Google Scholar]
- 62. Keating M. M., Gong H., and Byers D. M. (2002) Identification of a key residue in the conformational stability of acyl carrier protein. Biochim. Biophys. Acta 1601, 208–214 [DOI] [PubMed] [Google Scholar]
- 63. Li Q., Khosla C., Puglisi J. D., and Liu C. W. (2003) Solution structure and backbone dynamics of the holo form of the frenolicin acyl carrier protein. Biochemistry 42, 4648–4657 [DOI] [PubMed] [Google Scholar]
- 64. Findlow S. C., Winsor C., Simpson T. J., Crosby J., and Crump M. P. (2003) Solution structure and dynamics of oxytetracycline polyketide synthase acyl carrier protein from Streptomyces rimosus. Biochemistry 42, 8423–8433 [DOI] [PubMed] [Google Scholar]
- 65. Nguyen C., Haushalter R. W., Lee D. J., Markwick P. R., Bruegger J., Caldara-Festin G., Finzel K., Jackson D. R., Ishikawa F., O'Dowd B., McCammon J. A., Opella S. J., Tsai S. C., and Burkart M. D. (2014) Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature 505, 427–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Worsham L. M., Earls L., Jolly C., Langston K. G., Trent M. S., and Ernst-Fonberg M. L. (2003) Amino acid residues of Escherichia coli acyl carrier protein involved in heterologous protein interactions. Biochemistry 42, 167–176 [DOI] [PubMed] [Google Scholar]
- 67. Zhang Y. M., Wu B., Zheng J., and Rock C. O. (2003) Key residues responsible for acyl carrier protein and β-ketoacyl-acyl carrier protein reductase (FabG) interaction. J. Biol. Chem. 278, 52935–52943 [DOI] [PubMed] [Google Scholar]
- 68. Arthur C. J., Williams C., Pottage K., Płoskoń E., Findlow S. C., Burston S. G., Simpson T. J., Crump M. P., and Crosby J. (2009) Structure and malonyl CoA-ACP transacylase binding of Streptomyces coelicolor fatty acid synthase acyl carrier protein. ACS Chem. Biol. 4, 625–636 [DOI] [PubMed] [Google Scholar]
- 69. Zhang Y. M., Rao M. S., Heath R. J., Price A. C., Olson A. J., Rock C. O., and White S. W. (2001) Identification and analysis of the acyl carrier protein (ACP) docking site on β-ketoacyl-ACP synthase III. J. Biol. Chem. 276, 8231–8238 [DOI] [PubMed] [Google Scholar]
- 70. Parris K. D., Lin L., Tam A., Mathew R., Hixon J., Stahl M., Fritz C. C., Seehra J., and Somers W. S. (2000) Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites. Structure 8, 883–895 [DOI] [PubMed] [Google Scholar]