Abstract
The Broad-Complex gene (BR-C) encodes transcription factors that dictate larval-pupal metamorphosis in insects. The expression of BR-C is induced by molting hormone (20-hydroxyecdysone (20E)), and this induction is repressed by juvenile hormone (JH), which exists during the premature larval stage. Krüppel homolog 1 gene (Kr-h1) has been known as a JH-early inducible gene responsible for repression of metamorphosis; however, the functional relationship between Kr-h1 and repression of BR-C has remained unclear. To elucidate this relationship, we analyzed cis- and trans elements involved in the repression of BR-C using a Bombyx mori cell line. In the cells, as observed in larvae, JH induced the expression of Kr-h1 and concurrently suppressed 20E-induced expression of BR-C. Forced expression of Kr-h1 repressed the 20E-dependent activation of the BR-C promoter in the absence of JH, and Kr-h1 RNAi inhibited the JH-mediated repression, suggesting that Kr-h1 controlled the repression of BR-C. A survey of the upstream sequence of BR-C gene revealed a Kr-h1 binding site (KBS) in the BR-C promoter. When KBS was deleted from the promoter, the repression of BR-C was abolished. Electrophoresis mobility shift demonstrated that two Kr-h1 molecules bound to KBS in the BR-C promoter. Based on these results, we conclude that Kr-h1 protein molecules directly bind to the KBS sequence in the BR-C promoter and thereby repress 20E-dependent activation of the pupal specifier, BR-C. This study has revealed a considerable portion of the picture of JH signaling pathways from the reception of JH to the repression of metamorphosis.
Keywords: development, endocrinology, hormone, insect, signaling
Introduction
The molting and metamorphosis of insects are intricately regulated by the actions and interactions of ecdysteroids and juvenile hormone (JH).2 In holometabolous insects, 20-hydroxyecdysone (20E, the primary active ecdysteroid) induces the larval-larval molt in the presence of JH. When the JH titer declines to a trace level in the final larval instar, 20E induces larval-pupal and pupal-adult molts (metamorphosis). Thus, JH plays a key role in preventing larvae from undergoing precocious metamorphosis (1).
Our understanding of the molecular mechanism of JH-mediated repression of insect metamorphosis has significantly advanced (2): JH carried to a target cell is received by a JH receptor, methoprene tolerant (Met) (3–6); JH-liganded Met interacts with steroid receptor coactivator (7–12); the JH/Met/steroid receptor coactivator complex activates Krüppel homolog 1 (Kr-h1), a repressor of metamorphosis, by interacting with a JH response element (kJHRE) in the Kr-h1 gene (9, 10, 12–15); Kr-h1 represses the larval-pupal metamorphosis (16–20). To date, however, the mechanism of the repression of metamorphosis by Kr-h1, including target gene(s), binding site(s), and partner(s), has remained unknown.
The Broad-Complex (BR-C) protein is a transcription factor that is composed of a Bric-a-brac/Tramtrack/Broad-Complex (BTB) domain and an alternatively spliced zinc finger domain (Z1-Z6) (21–25). BR-C expression is induced by 20E, and BR-C works as a pupal specifier in the larval-pupal transition of holometabolous insects (26–33). In Manduca sexta and Bombyx mori, removal of the corpora allata (allatectomy), the primary organs for JH synthesis, induced BR-C expression and subsequent precocious pupation, and application of exogenous JH to allatectomized larvae inhibited BR-C expression and pupation in these larvae (33, 34). JH-mediated repression of the activation of BR-C by 20E has been also reported in cultured insect epidermis, but the inhibitory effect of JH declined after the transition to the pupal commitment phase (30, 35). A study using the Kr-h1 mutant of Drosophila melanogaster showed that Kr-h1 was required for the repression of BR-C expression in the fat body of young D. melanogaster larvae (36); however, the molecular mechanism underlying the JH-mediated repression of BR-C, particularly whether the BR-C expression is directly repressed by Kr-h1 or repressed by Kr-h1-inducible gene(s), has remained unclear.
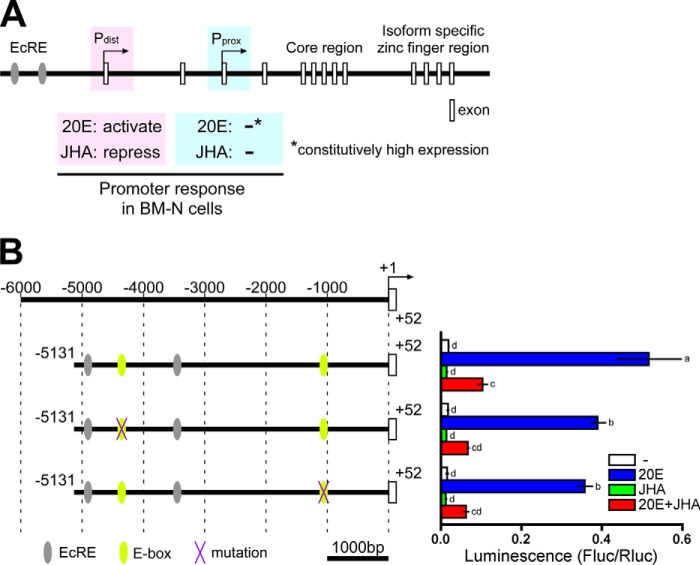
The BR-C gene of B. mori (BmBR-C) has two transcriptional start sites: the distal promoter (Pdist) and the proximal promoter (Pprox) (37–39). In the B. mori cell line BM-N, Pdist was activated by 20E, and this activation was repressed by JH, whereas Pprox was constitutively activated regardless of the presence/absence of 20E and JH (Fig. 1A) (39, 40). The activation of Pdist by 20E was mediated by two ecdysone response elements, one of which interacted with ecdysone receptor/ultraspiracle (Fig. 1A) (39, 40).
FIGURE 1.
Genomic structure of BmBR-C, hormonal responses of its promoter with reference to its E-box sequences. A, schematic representation of the genomic structure of BmBR-C. Predicted exons are shown as boxes. The BmBR-C gene has two transcriptional start sites (the distal promoter (Pdist) and the proximal promoter (Pprox)). In BM-N cells, Pdist was activated by 20E via two ecdysone response elements (EcRE; gray ellipses), and the activation was repressed by JHA, whereas the Pprox transcript was constitutively highly expressed regardless of 20E and methoprene (JHA) (40). B, functional characterization of the E-box sequences in BmBR-C_Pdist. Yellow ellipses indicate two E-box sequences (−4362 to −4357 and −1066 to −1061) located in the upstream region of BmBR-C_Pdist (−5131 to +52). Purple X marks indicate mutations of E-box sequence. BM-N cells were cotransfected with reporter plasmids that express firefly luciferase (Fluc) under the regulation of the regions indicated in the figure (−5131 to +52) and a reference reporter plasmid carrying Renilla luciferase (Rluc). Cells were treated with 1 μm 20E and/or 10 μm methoprene (JHA) for 2 days. Reporter activities were measured using a dual-luciferase reporter assay system. Data represent the means ± S.D. (n = 3). Bars with the same letter are not significantly different (Tukey-Kramer test, α = 0.05).
In the present study we aimed to clarify the molecular mechanism of JH-mediated repression of larval-pupal metamorphosis using a Bombyx cell line. Based on the results of the present study, we present a substantive portion of the picture of JH signaling pathways from the reception of JH to the repression of metamorphosis.
Experimental Procedures
Cell Lines
The BM-N cell line, derived from the ovary of B. mori (RIKEN BRC), was maintained at 25 °C in IPL-41 medium (Gibco, Invitrogen) containing 10% FBS (HyClone, Logan, UT).
Chemicals
Methoprene (juvenile hormone analog (JHA), SDS Biotech, Tokyo, Japan) was a gift from Dr. Sho Sakurai. 20E was purchased from Sigma.
Construction of Reporter and Expression Plasmids
A reporter plasmid (pGV_Pdist_−5131/+52) carrying 5′-flanking regions of the distal promoter of BmBR-C was prepared as described previously (39, 40). Deleted and mutated reporter plasmids of pGV_Pdist_−5131/+52 were constructed by inverse PCR. The primers and templates used for construction of the reporter plasmids are listed in Tables 1 and 2.
TABLE 1.
List of primers and templates used for construction of reporter plasmids
| Name | Figure | PCR template | Nucleotide sequence (5′ to 3′) |
|---|---|---|---|
| pGV_Pdist_−5131/+52a | Figs. 1B; 2, B and D; 3, A, B, and D; 5, A and B; 6A | ||
| pGV_Pdist_−5131/+52_ΔE-box(−4362/−4357) | Fig. 1B | pGV_Pdist_−5131/+52 | AAAATCGTTTTTTTCTGTTATTTATTTGTTTGTTTGTTATTTAGTTTTTTGTG |
| TTTTGAACGATTCAATTCTAAATTCTAAGCGCTACG | |||
| pGV_Pdist_−5131/+52_ΔE-box(-1066/-1061) | Fig. 1B | pGV_Pdist_−5131/+52 | AAAAAATGAAAATTAGGTTATATTTTTATGACTCTTTTTTCAAAATTCAAC |
| TTTATTCGTTCTAAAGCTATTTCATTTCATTGAATTGAGACATG | |||
| pGV_Pdist_−5131/−3008 and −511/+52 | Fig. 3, B and C | pGV_Pdist_−5131/+52 | GAAAACACTTTTCGGAACTTTCACTTCATTGG |
| CTAACAACAAAATGGATTGCTCTACCCATATAC | |||
| pGV_Pdist_−4609/−3008 and −511/+52 | Fig. 3, B, C, F, and G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GCTCTCTGTTTATCATGGGTCAATAAAGAAGTC |
| GAGCTCGGTACCTCCCGGGTTATG | |||
| pGV_Pdist_−5131/−4714 and −4086/−3008 and −511/+52 | Fig. 3B | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GAATGGTGGTAAATATCTTCTCTCTCAATGTAGTTAAG |
| CAGATTGCGGATATACACGTTTCGTCATTGAG | |||
| pGV_Pdist_−5131/-4199 and −3575/−3008 and −511/+52 | Fig. 3B | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | CCATTAAACATTTCCATAAAAACCTACGAATGTGAATTAAC |
| GGCCTTGGAATTAATTCTGTACTTCAATCATG | |||
| pGV_Pdist_−4469/−3008 and −511/+52 | Fig. 3C | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | ACGTCTATCCGTGACCTACGCTAACG |
| GAGCTCGGTACCTCCCGGGTTATG | |||
| pGV_Pdist_-4449/−3008 and −511/+52 | Fig. 3C | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | CTAACGCTAAATAGAGTTCCGACCTTGACTC |
| GAGCTCGGTACCTCCCGGGTTATG | |||
| pGV_Pdist_-4429/−3008 and −511/+52 | Fig. 3C | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GACCTTGACTCTAACGGTCGTCGAC |
| GAGCTCGGTACCTCCCGGGTTATG | |||
| pGV_Pdist_−4609/−4410 and −511/+52 | Fig. 3C | pGV_Pdist_ − 4609/ − 3008 and − 511/+52 | GAAAACACTTTTCGGAACTTTCACTTCATTGG |
| CGACCGTTAGAGTCAAGGTCGGAAC | |||
| pGV_Pdist_−4609/−4430 and −511/+52 | Fig. 3C | pGV_Pdist_ − 4609/ − 3008 and − 511/+52 | GAAAACACTTTTCGGAACTTTCACTTCATTGG |
| GGAACTCTATTTAGCGTTAGCGTAGGTCAC | |||
| pGV_Pdist_−4609/−4450 and −511/+52 | Fig. 3C | pGV_Pdist_ − 4609/ − 3008 and − 511/+52 | GAAAACACTTTTCGGAACTTTCACTTCATTGG |
| CGTAGGTCACGGATAGACGTAGAGC | |||
| pGV_Pdist_−5131/+52_ΔKBS | Fig. 2D, 6A | pGV_Pdist_−5131/+52 | TCGACTCTACATTACGTAGCGCTTAGAATTTAG |
| AGAGCGCACAAATAAATTTCATGATCTTTATTAAAAACAC | |||
| pGV_Pdist_ KBS and −511/+52 | Fig. 2D | pGV_Pdist_ − 4469/ − 3008 and − 511/+52 | GAAAACACTTTTCGGAACTTTCACTTCATTGG |
| CGACCGTTAGAGTCAAGGTCGGAAC | |||
| pGV_Pdist_−511/+52 | Fig. 2D | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GAAAACACTTTTCGGAACTTTCACTTCATTGG |
| GAGCTCGGTACCTCCCGGGTTATG | |||
| pGV_Pdist_-1537/+52 | Fig. 1A | pGV_Pdist_−5131/+52 | CATTGCTTTCGTCCGGTGACATCTTATTAG |
| GAGCTCGGTACCTCCCGGGTTATG |
a See Nishita and Takiya (40).
TABLE 2.
List of primers, methods, and templates used for construction of reporter plasmids with transversion and transition
| Name | Figure | PCR template | Nucleotide sequence (5′ to 3′) |
|---|---|---|---|
| pGV_Pdist_−4609/−3008 and −511/+52_TV1 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GAGATCCGTGACCTACGCTAACGCTAAATAGAG |
| ATGAGAGCGCACAAATAAATTTCATGATCTTTATTAAAAACAC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TV2 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | ATGGACCTACGCTAACGCTAAATAGAGTTCCG |
| TCGAGACGTAGAGCGCACAAATAAATTTCATGATC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TV3 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | AGCCGCTAACGCTAAATAGAGTTCCGACCTTG |
| TGAACGGATAGACGTAGAGCGCACAAATAAATTTC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TV4 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GCCCGCTAAATAGAGTTCCGACCTTGACTCTAAC |
| TATTAGGTCACGGATAGACGTAGAGCGC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TV5 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GCCATAGAGTTCCGACCTTGACTCTAACGGTC |
| TATTTAGCGTAGGTCACGGATAGACGTAGAG | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TV6 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | TCTTTCCGACCTTGACTCTAACGGTCGTC |
| GCGTTAGCGTTAGCGTAGGTCACGGATAGAC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TV7 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | ATCCCTTGACTCTAACGGTCGTCGACTC |
| TCCCTCTATTTAGCGTTAGCGTAGGTCACGG | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TV8 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GTCCTCTAACGGTCGTCGACTCTACATTACG |
| CTTTCGGAACTCTATTTAGCGTTAGCGTAGGTC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TV9 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GCCCGGTCGTCGACTCTACATTACGTAGC |
| TCTTCAAGGTCGGAACTCTATTTAGCGTTAGC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TV10 | Fig. 3F | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | GATTCGACTCTACATTACGTAGCGCTTAGAATTTAG |
| AATTTAGAGTCAAGGTCGGAACTCTATTTAGCG | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS1 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | CTCATCCGTGACCTACGCTAACGCTAAATAGAG |
| TACAGAGCGCACAAATAAATTTCATGATCTTTATTAAAAACAC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS2 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | TACGACCTACGCTAACGCTAAATAGAGTTCCG |
| AGCAGACGTAGAGCGCACAAATAAATTTCATGATC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS3 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | TCGCGCTAACGCTAAATAGAGTTCCGACCTTG |
| ACTACGGATAGACGTAGAGCGCACAAATAAATTTC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS4 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | CGGCGCTAAATAGAGTTCCGACCTTGACTCTAAC |
| ATATAGGTCACGGATAGACGTAGAGCGC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS5 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | CGGATAGAGTTCCGACCTTGACTCTAACGGTC |
| ATATTAGCGTAGGTCACGGATAGACGTAGAG | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS6 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | AGATTCCGACCTTGACTCTAACGGTCGTC |
| CGCTTAGCGTTAGCGTAGGTCACGGATAGAC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS7 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | TAGCCTTGACTCTAACGGTCGTCGACTC |
| AGGCTCTATTTAGCGTTAGCGTAGGTCACGG | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS8 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | CAGCTCTAACGGTCGTCGACTCTACATTACG |
| GAATCGGAACTCTATTTAGCGTTAGCGTAGGTC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS9 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | CGGCGGTCGTCGACTCTACATTACGTAGC |
| AGATCAAGGTCGGAACTCTATTTAGCGTTAGC | |||
| pGV_Pdist_−4609/−3008 and −511/+52_TS10 | Fig. 3G | pGV_Pdist_ − 5131/ − 3008 and − 511/+52 | CTATCGACTCTACATTACGTAGCGCTTAGAATTTAG |
| TTATTAGAGTCAAGGTCGGAACTCTATTTAGCG |
An expression plasmid of BmKr-h1 (pIZT_BmKr-h1) was constructed using the Gateway system (Invitrogen). The full ORF of BmKr-h1 (AB360766) was amplified by PCR from full-length cDNA clones (9) using the primer containing attB1 and Kozak sequences and the primer containing an attB2 sequence (Table 3). The amplified fragment was inserted into the pDONR 221 plasmid (Invitrogen) and then into the pIZT/V5-His vector (Invitrogen) modified for the Gateway system. The p65AD (from p65, a subunit of NF-κB) (41) was inserted into the N terminus of BmKr-h1 by inverse PCR using primers shown in Table 3 and a blunt-end ligation kit (Toyobo Co. Ltd., Osaka, Japan). Deleted and mutated pIZT_BmKr-h1 and pIZT_p65AD_BmKr-h1 plasmids were constructed by inverse PCR using primers and templates as shown in Table 3.
TABLE 3.
List of primers used for construction of expression plasmids
| Name | Figure | PCR template | Nucleotide sequence (5′ to 3′) |
|---|---|---|---|
| pIZT_BmKr-h1 | Fig. 2B, 3A, and 6A | Full length cDNA clone | AAAAAGCAGGCTTCGAAGGAGATAGAACCATGATAGGTGACGAGGAGCGAG |
| AGAAAGCTGGGTCCTATGATTCTGTAGCTGGCGGAG | |||
| pIZT_p65AD_BmKr-h1 | Figs. 3, A, B, C, D, F, and G; 5, A and B | pIZT_BmKr-h1 | ATGATAGGTGACGAGGAGCGAGTTCATC |
| GGTTCTATCTCCTTCGAAGCCTGCTTTTTT | |||
| ATGTTCCCTTCTGGGCAAATCTCAAACCAGG | |||
| TTGGCCGCTGGAGCTGATCTGACTC | |||
| pIZT_p65AD | pIZT_p65AD_BmKr-h1 | ACCCAGCTTTCTTGTACAAAGTGGTCCC | |
| CTATTGGCCGCTGGAGCTGATCTGACTC | |||
| pF25A_BmKr-h1 | pIZT_BmKr-h1 | TTTGCGATCGCATGATAGGTGACGAGGAGCGAG | |
| TTTGTTTAAACCTATGATTCTGTAGCTGGCGGAGCAAATTG | |||
| pF25A_BmKr-h1_HA | Fig. 4, A and B | pF25A_BmKr-h1 | TCCCGGACTATGCAGGATCCTATCCATATGACGTTCCAGATTACGCTTGAGTTTAAACGAATTCGGGCTCGGTACCC |
| CGTCATAGGGATAGCCCGCATAGTCAGGAACATCGTATGGGTATGATTCTGTAGCTGGCGGAGCAAATTGTATAAC | |||
| pF25A_BmKr-h1(1–243) | Fig. 4B | pF25A_BmKr-h1 | TGAGTTTAAACGAATTCGGGCTCGGTACCC |
| ACTCTGTAATGGTGCGGTGCGGGG | |||
| pIZT_p65AD_BmKr-h1_ΔZF1 | Fig. 5A | pIZT_p65AD_BmKr-h1 | CGTTCTACTGCTGATGCACATCGCTGC |
| CTGATGAACTCGCTCCTCGTCACCTATC | |||
| pIZT_p65AD_BmKr-h1_ΔZF2 | Fig. 5A | pIZT_p65AD_BmKr-h1 | ACTGGCGAGCGACCATTTGAATGTGAATATTG |
| GCGATGTGCATCAGCAGTAGAACGATG | |||
| pIZT_p65AD_BmKr-h1_ΔZF3 | Fig. 5A | pIZT_p65AD_BmKr-h1 | ACAAAGGAGAGGCCGTATCGGTGCAATG |
| TTCAAATGGTCGCTCGCCAGTATGGGTTC | |||
| pIZT_p65AD_BmKr-h1_ΔZF4 | Fig. 5A | pIZT_p65AD_BmKr-h1 | ACCGGCGAGAGACCGCACGCATG |
| CCGATACGGCCTCTCCTTTGTGTGAATAC | |||
| pIZT_p65AD_BmKr-h1_ΔZF5 | Fig. 5A | pIZT_p65AD_BmKr-h1 | ACAGGTGAGAAACCCTATCGTTGTCCTG |
| TGCGTGCGGTCTCTCGCCGGTAT | |||
| pIZT_p65AD_BmKr-h1_ΔZF6 | Fig. 5A | pIZT_p65AD_BmKr-h1 | ACGGGAGAGCGACCCTATACTTGTG |
| ACGATAGGGTTTCTCACCTGTGTGGG | |||
| pIZT_p65AD_BmKr-h1_ΔZF7 | Fig. 5A | pIZT_p65AD_BmKr-h1 | TTTGGCGAGCGCTGTTATCGTTGCACTG |
| AGTATAGGGTCGCTCTCCCGTGTGAG | |||
| pIZT_p65AD_BmKr-h1_ΔZF8 | Fig. 5A | pIZT_p65AD_BmKr-h1 | GGCGCGGAAGCCCCCCGCA |
| GCGCTCGCCAAAATGTTGAAAACGATGTAAT | |||
| pIZT_p65AD_BmKr-h1_ZF1_amino acid substitution | Fig. 5B | pIZT_p65AD_BmKr-h1 | GCAGTGCGCTTACAGCATTCGCACGATCCTTTCGTTC |
| GGGTGGACAATGTCAAACCAGACTCGCCGGACTGAT | |||
| pIZT_p65AD_BmKr-h1_ZF2_amino acid substitution | Fig. 5B | pIZT_p65AD_BmKr-h1 | CTGCACGACTCGTGCGCTTCTACCGAACCTTTACTGG |
| GCACAGCAAAAGTCTTATGGGACACGTCGGAGCGAT | |||
| pIZT_p65AD_BmKr-h1_ZF3_amino acid substitution | Fig. 5B | pIZT_p65AD_BmKr-h1 | AAGAAAATTTGCAAGTATTCCGTCGTATTTTCACAAA |
| TCACACTAAACATTTTATGGGAATATTCAGATTCAA | |||
| pIZT_p65AD_BmKr-h1_ZF4_amino acid substitution | Fig. 5B | pIZT_p65AD_BmKr-h1 | CCGGAAAACTTCACAGATTTGCTAGAATCTTTACCGG |
| AATGCTCGAATGCCGCATCAGAAACATTGGACCGAT | |||
| pIZT_p65AD_BmKr-h1_ZF5_amino acid substitution | Fig. 5B | pIZT_p65AD_BmKr-h1 | CTGGTCAACTGGTCATATTTTTAAGGACCTTCACAGG |
| ATTGTATGAACGTCTTATGAGAATGTGGAGATGCGT | |||
| pIZT_p65AD_BmKr-h1_ZF6_amino acid substitution | Fig. 5B | pIZT_p65AD_BmKr-h1 | TCCTCTAAACAGCTTAAAGTATTTTCTCGTACTTTCACGGG |
| GGTGAAGCCCTTTCCGGATCCAGGTGCAGGAGAACGAT | |||
| pIZT_p65AD_BmKr-h1_ZF7_amino acid substitution | Fig. 5B | pIZT_p65AD_BmKr-h1 | ATCATGTTCTCAAATTATTTCGTTTTCAATTTTTTGG |
| TATAACCAAAGTCCCTGAGAGAAATTTCAGAAGTAT | |||
| pIZT_p65AD_BmKr-h1_ZF8_amino acid substitution | Fig. 5B | pIZT_p65AD_BmKr-h1 | CAAAAAAACAAATGGAAGCCTTTATTTACAAAGAATTTGGCGC |
For the in vitro transcription and translation system, the BmKr-h1 ORF anchored with SgfI and PmeI sites was amplified by PCR using primers and a template as shown in Table 3. The amplified fragment was digested with Sgf1 and PmeI, and inserted into the pF25A ICE T7 Flexi plasmid (Promega). pF25A plasmids with a C-terminal HA epitope tag (BmKr-h1-HA) and without the C-terminal region downstream of the zinc finger domains (BmKr-h1(1–243)) were constructed by inverse PCR using primers and a template as shown in Table 3.
Double-stranded RNA Synthesis
Template DNA fragments of BmKr-h1 and MalE (control) used for the synthesis of double-stranded RNA were amplified by PCR using primers and templates as shown in Table 4, and PCR products were purified using a Wizard SV Gel and PCR Clean-Up System (Promega). Double-stranded RNAs were synthesized from the amplified DNA using RiboMAX SP6 and T7 Large Scale RNA Production Systems according to the manufacturer's instructions (Promega).
TABLE 4.
List of primers and oligonucleotides used for dsRNA syntheses and EMSA
q-PCR, quantitative real-time PCR.
| Name | Figure | PCR template | Nucleotide sequence (5′ to 3′) |
|---|---|---|---|
| dsRNA | |||
| dsMalE | Fig. 2, C and D | pMAL-c4E (NEB) | GGATCCTAATACGACTCACTATAGGTGATTGCTGCTGACGGGGGT |
| GGATCCTAATACGACTCACTATAGGTTTCTGGGCGTTTTCCATAGTGG | |||
| dsBmKr-h1 | Fig. 2, C and D | pIZT_BmKr-h1 | GGATCCTAATACGACTCACTATAGGATGATAGGTGACGAGGAGCGAG |
| GGATCCTAATACGACTCACTATAGGGCTCTCCCGTGTGAGTACGAG | |||
| qPCR | |||
| BmKr-h1_RNAi_qPCR | Fig. 2C | cDNA | CATCGTTTTCAACATTTTGGCGAG |
| CACATCACTTTACCATCGGCAGC | |||
| EMSA | |||
| Probe (−4480/−4401) | Fig. 4, A and B | Biotin-ON-tgtgcgctctACGTCTATCCGTGACCTACGCTAACGCTAAATAGAGTTCCGACCTTGACTCTAACGGTCGtcgactctac | |
| Biotin-ON-gtagagtcgaCGACCGTTAGAGTCAAGGTCGGAACTCTATTTAGCGTTAGCGTAGGTCACGGATAGACGTagagcgcaca | |||
| Specific competitor (−4480/−4401) | Fig. 4, A and B | tgtgcgctctACGTCTATCCGTGACCTACGCTAACGCTAAATAGAGTTCCGACCTTGACTCTAACGGTCGtcgactctac | |
| gtagagtcgaCGACCGTTAGAGTCAAGGTCGGAACTCTATTTAGCGTTAGCGTAGGTCACGGATAGACGTagagcgcaca | |||
| Non-specific competitor (−4054/−3975) | Fig. 4, A and B | TTAAGAAGTTTCACTTCCGATTTGTTTTATTAAACTAATCAAGTTGTTTTCATTCTGAAATATTATTATTTGAGAGACTA | |
| TAGTCTCTCAAATAATAATATTTCAGAATGAAAACAACTTGATTAGTTTAATAAAACAAATCGGAAGTGAAACTTCTTAA | |||
Transfection and Reporter Assays
BM-N cells were seeded at a density of 1.5 × 105 cells/well in 200 μl medium in a 96-well plate (Sumilon, Sumilomo Bakelite Co., Tokyo, Japan) 1 day before transfection. Transfection of BM-N cells was performed using FuGENE HD (Promega, Madison, WI). The pIZT_RLuc vector containing the Renilla luciferase gene was constructed as the reference for BM-N cells (42). The cells were incubated for 1 day (Fig. 1B) or 2 days (Figs. 2, B, C and D, 3A, and 6A) after transfection and treated with JHA or 20E for 2 days. The treated cells were then processed by using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions and analyzed with a luminometer (ARVO, PerkinElmer Life Sciences).
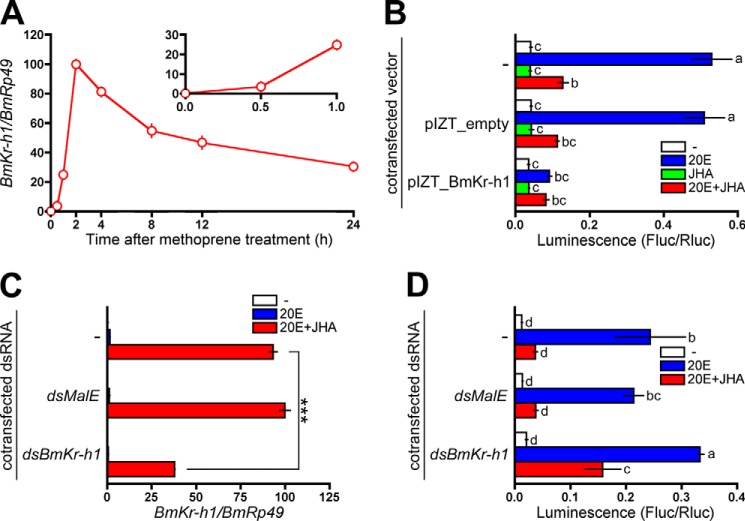
FIGURE 2.
Repression of the BmBR-C promoter by BmKr-h1. A, BM-N cells were treated with 10 μm JHA, and temporal changes in BmKr-h1 expression were monitored by quantitative real-time PCR. B, BM-N cells were cotransfected with a reporter plasmid carrying the −5131 to +52 region (pGL4.14), a reference reporter plasmid, and a BmKr-h1 expression plasmid, and the cells were incubated for 2 days. The transfected cells were treated with 1 μm 20E or 10 μm JHA for 2 days. Reporter activities were measured using a dual-luciferase reporter assay system. C, BM-N cells were transfected with dsBmKr-h1, and the cells were incubated for 2 days. The transfected cells were treated with 1 μm 20E or 10 μm JHA for 2 days, and the RNAi efficiency was monitored by quantitative real-time PCR. D, BM-N cells were cotransfected with a reporter plasmid carrying the −5131 to +52 region (pGL4.14), a reference reporter plasmid, and dsBmKr-h1, and the cells were incubated for 2 days. The transfected cells were treated with 1 μm 20E or 10 μm JHA for 2 days. Reporter activities were measured using a dual-luciferase reporter assay system. Data represent the means ± S.D. (n = 3). B and D, bars with the same letter are not significantly different (Tukey-Kramer test, α = 0.05). C, data were analyzed using Student's t tests (***, p < 0.001).
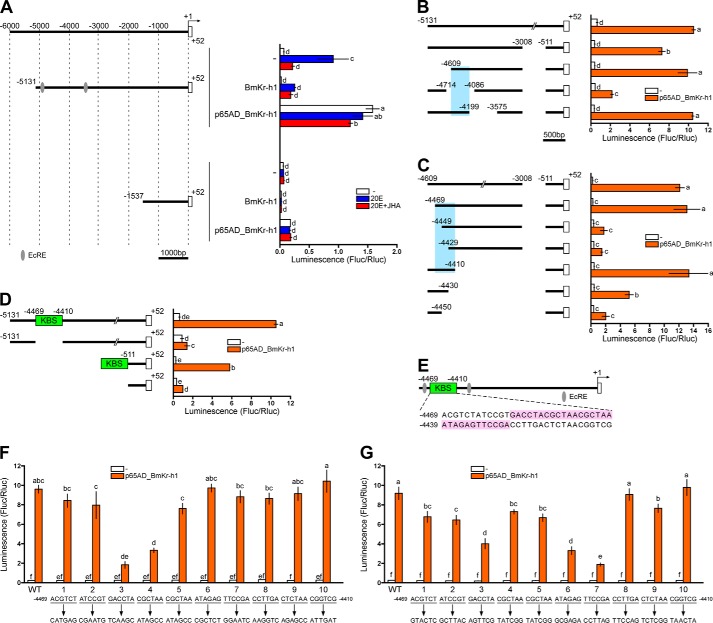
FIGURE 3.
Identification of the BmKr-h1 binding site (KBS) in the BmBR-C promoter. Reporter assays using progressive deletion and mutation constructs were performed to identify the KBS region. BM-N cells were cotransfected with pGL4.14 reporter plasmids carrying the promoter regions indicated in the figure, a reference reporter plasmid, and a p65AD-BmKr-h1 expression plasmid (pIZT). The cells were incubated for 3 days, and reporter activities were measured using a dual-luciferase reporter assay system. Data represent the means ± S.D. (n = 3). Bars with the same letter are not significantly different (Tukey-Kramer test, α = 0.05). A, reporter plasmids containing the 5′-flanking regions of BmBR-C_Pdist (−5131 to +52) and (−1537 to +52) were assayed. Numbers indicate the distance from the transcription start site, and boxes represent the exons. The cells incubated for 2 days after transfection were treated with 1 μm 20E or 10 μm JHA for 2 days. B, reporter activities of progressive deletion constructs are shown below. Sky blue highlighting indicates the KBS region. C, the inset in the plasmid used in B, consisting of the −4609 to −3008 and −511 to +52 region, was progressively reduced from both sides by 20 bp, and the effects were measured by reporter assays. Sky blue highlighting indicates the region containing KBS. D, a reporter carrying the −5131 to +52 region excluding −4469 to −4410 (KBS) and another reporter carrying the KBS region connected to a basal promoter region (−511 to +52) were assayed. E, schematic representation of the location of KBS (-4469 to −4410). The nucleotide sequence is shown under the gene structure. Red ellipses indicate ecdysone response elements (EcRE). The functionality of KBS was assayed with mutations causing a sextuplet transversion (F) or transition (G) in the KBS (−4469 to −4410) region of the reporter (−4609to −3008 and −511 to +52).
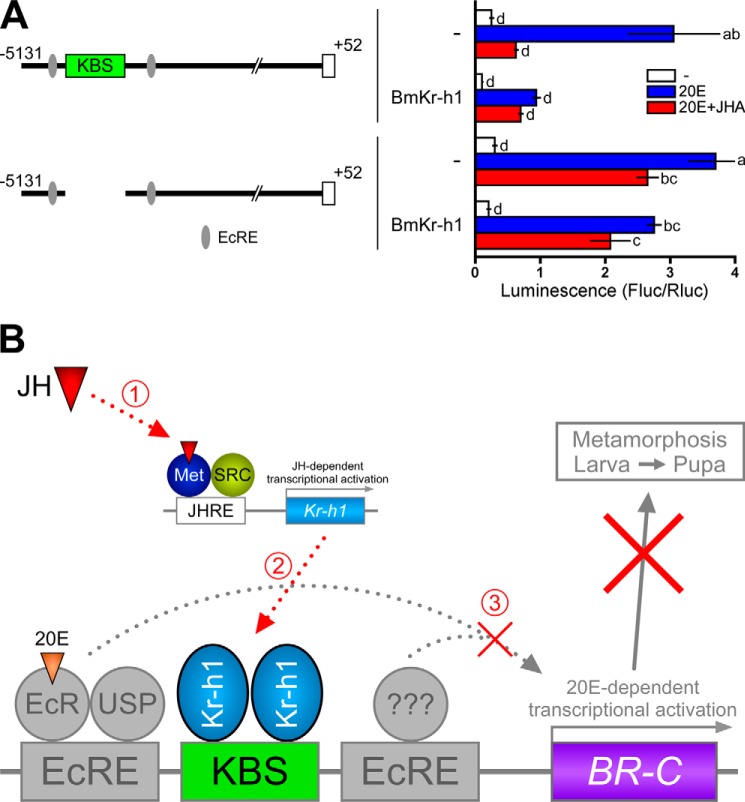
FIGURE 6.
Regulation of the BR-C promoter by hormones, Kr-h1, and KBS. A, BM-N cells were cotransfected with a reporter plasmid carrying the −5131 to +52 region without KBS (pGL4.14), a reference reporter plasmid, and a BmKr-h1 expression plasmid, and the transfected cells were incubated for 2 days. The cells were treated with 1 μm 20E or 10 μm JHA for 2 days, and reporter activities were measured by a dual-luciferase reporter assay system. Data represent the means ± S.D. (n = 3). Bars with the same letter are not significantly different (Tukey-Kramer test, α = 0.05). B, a model explaining the repression of 20E-dependent induction of BmBR-C by JH. SRC, steroid receptor coactivator; EcRe, ecdysone response element.
Quantitative Real-time PCR
Total RNA was extracted from BM-N cells using an RNeasy Plus mini kit (Qiagen, Venlo, The Netherlands) and used to synthesize cDNAs with a PrimeScript RT reagent kit (TaKaRa Bio, Ohtsu, Japan). To examine the time course of BmKr-h1 expression after JHA treatment, the primers designed to amplify both isoforms of BmKr-h1 have been described previously (9). For evaluation of the efficiency of RNAi, the primers designed to amplify both isoforms of BmKr-h1 were shown in Table 4. BmRp49 was used as the internal reference (9). The reaction was carried out in a 10-μl reaction volume containing template cDNA derived from 1 ng of total RNA, SYBR Premix Ex Taq (TaKaRa Bio), and 0.2 μm concentrations of each primer using a LightCycler 480 real-time thermal cycler (Roche Applied Science). The PCR conditions were 95 °C for 5 min and 55 cycles of 95 °C for 5 s and 60 °C for 20 s. The relative amounts of the transcripts were calculated by a crossing point analysis using standard curves generated from a plasmid containing a fragment of each gene. The expression levels of BmKr-h1 were normalized by those of BmRp49, and the levels of BmKr-h1 transcript at 2 h after JHA treatment (Fig. 2A) and of dsMalE after 20E and JHA treatment (Fig. 2C) were set as 100.
Electrophoresis Mobility Shift Assay (EMSA)
EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Pierce). Oligonucleotides labeled with 5′-terminal Biotin-ON were purchased from Eurofins Genomics (Tokyo, Japan); the sequences are shown in Table 4. The oligonucleotide pairs were mixed together, heated at 95 °C for 5 min, and annealed at room temperature for 1.5 h. The resultant double-stranded DNA was used as a probe for EMSA. Specific and nonspecific competitors (Table 4) were also prepared by the method described above. The in vitro transcription and translation of BmKr-h1-HA, BmKr-h1(1–243), and Luciferase (mock) were performed with the TnT T7 Insect Cell Extract Protein Expression System (Promega). Binding reactions were carried out in a 10-μl volume containing 0.02 pmol probe, 1.5 μl TnT reaction, 10 × Binding Buffer (Pierce), 0.5 μg of poly (dI-dC), 2.5% glycerol, 0.05% Nonidet P-40, 10 mm KCl, 1 mm MgCl2, 1 mm ZnSO4, and 2 mm EDTA at 25 °C for 2 or 4 h. In competition and antibody assays, the reaction mixture was incubated with 100-fold (2 pmol) unlabeled competitors and 1 μl of tag antibody (Abcam, Cambridge, UK), respectively. The samples were then electrophoresed at room temperature on a 4% nondenaturing polyacrylamide gel in 0.5 × Tris borate-EDTA. The probes in electrophoresed gels were transferred to positively charged nylon membranes (Roche Applied Science) by electrophoresis. The nylon membranes were then processed using the LightShift Chemiluminescent EMSA kit according to the manufacturer's instructions and detected with a LAS-3000 mini (Fujifilm, Tokyo, Japan).
Results and Discussion
Hormonal Regulation via E-box Sequences of the BR-C Promoter
In BM-N cells, a reporter carrying the upstream region (−5131 to +52; KAIKObase) of BmBR-C_Pdist was activated by 20E, and this activation was repressed by a JH analog methoprene (JHA) (Fig. 1B) in accordance with previous studies (39, 40). In general, bHLH-PAS transcription factors, to which Met belongs, recognize DNA sequences with a hexanucleotide core known as the E-box (CANNTG) (43). The core sequence of kJHRE has been identified as a 12-bp sequence that contains a palindromic canonical E-box (CACGTG), with which two Met paralogs of B. mori (BmMet1 and BmMet2) interact (9, 12). We found two CACGTG E-box sequences in the upstream sequence of BmBR-C_Pdist, i.e. positions −4362 to −4357 and −1066 to −1061 (Fig. 1B). We carried out reporter assays using constructs mutated in the E-box sequences to examine whether these sequences contribute to the JH-mediated repression of the activation of BmBR-C_Pdist. The reporter activities observed in response to 20E and JHA were only minimally affected by the mutation of either of the two E-box sequences (Fig. 1B), indicating that the contribution of the two E-box sequences to the JH-mediated repression was negligible.
Repression of the BR-C Promoter by BmKr-h1
Kr-h1 has been reported to be induced by JH in several insects in vivo and also in several insect cell lines (8–10, 16–20). In BM-N cells, the ordinary expression level of BmKr-h1 transcripts was marginal, but it increased 37-, 254-, and 1025-fold by 0.5, 1, and 2 h after JHA treatment, respectively (Fig. 2A). To examine the involvement of BmKr-h1 in the JH-mediated repression against the activation of BmBR-C_Pdist by 20E, we carried out reporter assays using BM-N cells. Ectopic expression of BmKr-h1 repressed the activation of a reporter carrying the −5131 to +52 region of BmBR-C_Pdist (Fig. 2B). We next performed reporter assays in BM-N cells in combination with RNAi. In cells treated with dsBmKr-h1, the levels of BmKr-h1 transcripts observed following 20E and JHA treatments declined to about one-third that of those in the cells untreated with double-stranded RNA, whereas treatment with dsMalE (control) showed no effects on BmKr-h1 transcript levels (Fig. 2C). RNAi silencing of BmKr-h1 alleviated the repression of the reporter activities by JHA (Fig. 2D), reconfirming that BmKr-h1 repressed the activation of BmBR-C_Pdist.
Identification of KBS in the BR-C Promoter
Insect Kr-h1 proteins commonly have eight C2H2 zinc finger domains, which putatively bind to a specific DNA sequence (9, 17). We hypothesized that BmKr-h1 directly binds to the BmBR-C_Pdist region and represses the activation of BmBR-C_Pdist by 20E. To identify a putative BmKr-h1 binding sequence, we employed a p65AD_BmKr-h1 expression vector in which BmKr-h1 was fused with the activation domain of p65 (p65AD), a subunit of NF-κB (41). Through this modification, if p65AD_BmKr-h1 bound to the BmBR-C_Pdist region, the luciferase reporter downstream of the BmBR-C_Pdist region would be forcibly activated by p65AD. We first confirmed that overexpression of p65AD alone did not affect the reporter activities (data not shown). Then, we tested a construct carrying the upstream region (−5131 to +52) of BmBR-C_Pdist. Although the activation of this reporter by 20E was repressed by the overexpression of native BmKr-h1 (Fig. 2B and Fig. 3A), p65AD-BmKr-h1 consistently activated the reporter regardless of the presence of 20E or 20E/JHA (Fig. 3A). Because the activity of the reporter carrying the region −1537 to +52 was not increased by p65AD-BmKr-h1, the binding site of BmKr-h1 was considered to lie between −5131 and −1537 (Fig. 3A). Next, we tested the reporter activities of several deletion constructs (−5131 to −3008, −4609to −3008, −5131 to −4714/−4086 to −3008, and −5131 to −4199/−3575 to 3008; Fig. 3B). All constructs except the ones carrying the regions −5131 to −4714/−4086 to −3008 showed an increase in luciferase reporter activity in the presence of p65AD-BmKr-h1, suggesting that the binding site of BmKr-h1 lay between −4609 and −4199. Subsequent reporter assays of constructs from which 20-bp fragments were serially deleted from both sides of the sequence −4609to −4199 revealed that the crucial region for the response to JH was −4469 to −4410 (Fig. 3C). This 60-bp region of Kr-h1 binding was referred to as KBS.
To confirm the identification of KBS, we constructed two reporters, one carrying the −5131 to +52 region without KBS and the other carrying the KBS region connected with the basal promoter region (−511 to +52) (Fig. 3D). The deletion of KBS resulted in a disappearance of reporter activity induced by p65AD-BmKr-h1, whereas the reporter carrying KBS and the basal promoter was sufficient for activation by p65AD-BmKr-h1 (Fig. 3D), indicating that KBS included the target sequence for BmKr-h1. Interestingly, KBS was located between the two ecdysone response elements of BmBR-C_Pdist (Fig. 3E).
To pinpoint sequences within KBS that are indispensable for the binding of BmKr-h1, transversion (A ↔ C and T ↔ G) and transition (A ↔ G and T ↔ C) mutations were introduced. Reporter activity was drastically reduced when a mutation was introduced into −4457GACCTA, −4451CGCTAA (Fig. 3F), −4439ATAGAG, or −4433TTCCGA (Fig. 3G). These results indicated that the 30-bp sequence encompassing −4457 to −4428 (GACCTACGCTAACGCTAAATAGAGTTCCGA) is crucial for the binding of BmKr-h1 (Fig. 3E, pink highlight). We referred to the sequence as the KBS core region. Although Krüppel, a gap gene of D. melanogaster involved in the development of segmented embryos, has four zinc finger domains similar to those of Kr-h1, the binding consensus sequence (ACAAAA and AAAAGGGTTAA) of Krüppel (44, 45) shared no homology with that of KBS core region.
BmKr-h1 Directly Binds to KBS
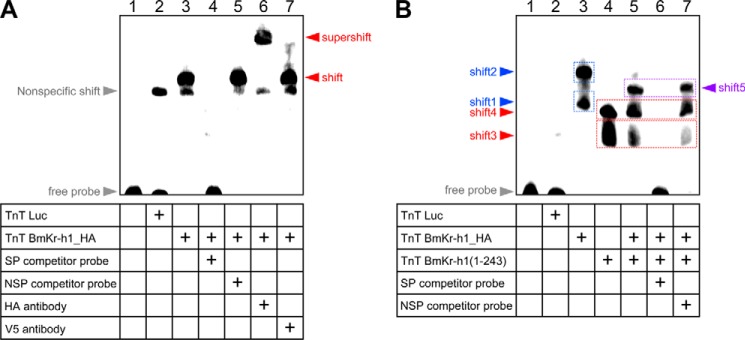
To demonstrate that BmKr-h1 directly binds to KBS, we performed EMSAs using the KBS sequence (60 bp) as a probe. As shown in Fig. 4A, a specific band shift appeared when HA-tagged BmKr-h1 (BmKr-h1_HA) (lane 3) was added to the probe and the mixture was incubated for 4 h, whereas only a nonspecific shift was observed when luciferase was added as a mock binding factor (lane 2). Competition assays showed that the specific band disappeared upon the addition of 100-fold molar excess of an unlabeled KBS probe (lane 4) but not by a nonspecific probe (lane 5). This specific band was supershifted by the addition of an anti-HA tag antibody (lane 6) but not by an anti-V5 tag antibody (negative control, lane 7). These results clearly indicated that BmKr-h1 directly and specifically bound to a DNA sequence in KBS.
FIGURE 4.
BmKr-h1 directly binds to KBS. A and B, EMSA experiments. Luciferase (mock), BmKr-h1-HA, and BmKr-h1(1–243) proteins were synthesized by an in vitro transcription and translation system and incubated with a KBS (60 bp) probe labeled with Biotin-ON for 4 h (A) or 2 h (B). Competition assays were performed using a 100-fold molar excess of unlabeled specific (SP) or nonspecific (NSP) probes. The specificities of shifted bands were verified using polyclonal antibodies against HA and V5 tags (negative control). Note that the disappearance of shift-3, -4, and -5 by the addition of specific probe (lane 6), but not by nonspecific probe (lane 7), showed that these all represented specific BmKr-h1 binding.
Furthermore, we performed EMSAs in which the binding reaction time was shortened from 4 to 2 h to obtain information on the number of BmKr-h1 molecules that interact with the KBS region. Two shifted bands (shift-1 and shift-2) were detected in lane 3, which contained BmKr-h1_HA (Fig. 4B), suggesting that the shift-1 band was composed of one BmKr-h1 molecule, whereas the shift-2 band was composed of two. Likewise, two shifted bands (shift-3 and shift-4) were observed when incubated with modified BmKr-h1 (BmKr-h1(1–243)) in which the C-terminal region downstream of the zinc finger domains was deleted (Fig. 4B, lane 4). When BmKr-h1_HA and BmKr-h1(1–243) were mixed, a new shifted band (shift-5) appeared between shift-2 and shift-4, which was diminished by the addition of the specific probe (Fig. 4B, lane 6) but not by the nonspecific probe (Fig. 4B, lane 7), suggesting that the shift-5 band was composed of both BmKr-h1_HA and BmKr-h1(1–243) molecules.
Functional Characterization of BmKr-h1
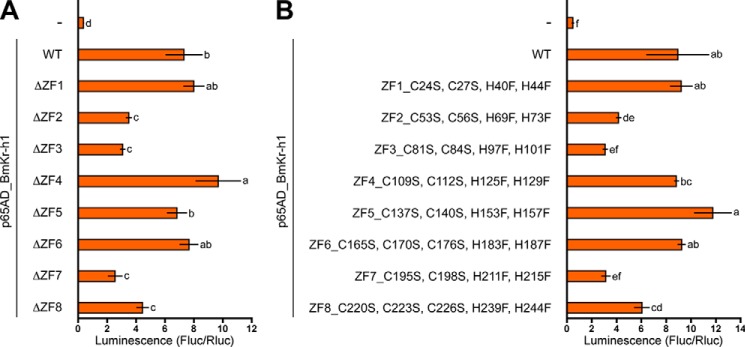
To identify which of the eight zinc finger domains (ZF1–8) in BmKr-h1 is important for binding to the KBS sequence, we constructed eight expression vectors, each lacking one of the eight zinc finger domains. Deletion of ZF2, -3, -7, or -8 resulted in decreases in the reporter activities induced by p65AD-BmKr-h1, whereas no changes in reporter activities were observed when ZF1, -4, -5, or -6 was deleted (Fig. 5A). Because potential denaturation of BmKr-h1 protein conformation after deletion of the entire single zinc finger domain was a concern, we instead constructed expression vectors designed to substitute cysteine and histidine, which coordinate Zn2+ in the zinc finger domain, with serine and phenylalanine, respectively. Decreased reporter activities were observed when ZF2, -3, -7, or -8 were mutated, which was in complete agreement with the results of the deletion constructs (Fig. 5B). C2H2 zinc fingers bind to DNA with a specific affinity conferred by several amino acid residues in the α helix of each finger (46). The amino acid residues of the α helix of each zinc finger domain in BmKr-h1 are diverse (9), suggesting that each zinc finger domain of BmKr-h1 recognizes a different sequence in the KBS core region.
FIGURE 5.
Binding activities of BmKr-h1 mutated in each zinc finger domain. BM-N cells were cotransfected with a reporter plasmid carrying the −5131 to +52 region (pGL4.14), a reference reporter plasmid, and expression plasmids (pIZT) of p65AD_BmKr-h1 carrying deletions of each zinc finger domain (A) or mutated by amino acid substitutions (B). The transfected cells were incubated for 3 days, and reporter activities were measured using a dual-luciferase reporter assay system. Data represent the means ± S.D. (n = 3). Bars with the same letter are not significantly different (Tukey-Kramer test, α = 0.05).
Hormonal Regulation of the BR-C Promoter via Kr-h1 and KBS
Last, we performed a comprehensive analysis of the regulation of BmBR-C by 20E and JH via BmKr-h1 and KBS in BM-N cells. The reporter carrying the −5131 to +52 region containing KBS was activated by 20E, and this activation was repressed by JHA (Fig. 6A). Ectopic expression of BmKr-h1 repressed the activation of the reporter in the absence of JH (Fig. 6A). On the other hand, when KBS was deleted from the reporter carrying −5131 to +52, the repression of reporter activation was not in turn repressed by JH even if BmKr-h1 was ectopically expressed in the cells (Fig. 6A). Taking these results together, we propose the following mechanism of the hormonal regulation of BmBR-C; in the presence of JH, BmKr-h1 gene expression is induced by JH via BmMet/Bm-steroid receptor coactivator (9, 12); subsequently, two molecules of BmKr-h1 bind to the KBS sequence of the BmBR-C promoter and thereby repress 20E-dependent expression of BmBR-C (Fig. 6B).
The response of BmBR-C in BM-N cells to 20E and/or JH is reminiscent of that observed in the larval epidermis and silk gland before pupal commitment (30, 33, 34). At this stage the expression of BmKr-h1 is maintained at high levels by the continuous presence of JH (20). The decline of JH titer at the beginning of the final instar larvae causes the temporal disappearance of BmKr-h1 (20). This might facilitate the subsequent induction of BmBR-C by 20E after the commitment to the larval-pupal transition (30, 34). Thus, the mechanism proposed above is likely to be applicable within the immature larvae until pupal commitment.
In contrast, the function of BmKr-h1 and KBS in the regulation of BmBR-C during metamorphosis seems to be more complex. After pupal commitment, BmKr-h1 re-expresses at a high level (20) along with BmBR-C during the prepupal stage (30). In transgenic silkworms, ectopic expression of BmKr-h1 did not suppress the induction of BmBR-C by 20E in the epidermis of final instar larvae, although the larval-pupal metamorphosis was interrupted (20). Apparently, the BmKr-h1 alone was not sufficient to suppress the expression of BmBR-C in the final instar larval stage. Furthermore, exogenous JH induced re-expression of BR-C in the epidermis of early pupae in M. sexta (28), and Kr-h1 is involved in the induction of BR-C in the pupae of D. melanogaster and Tribolium castaneum (16, 17). These apparent inconsistencies in the action of Kr-h1 on the regulation of BR-C in different developmental stages might result from differences in the cell autonomous factors at each stage, such as the repertoires of transcription factors and co-activator/co-repressors and epigenetic modifications of the promoter as well as the differences in the cell environment including endocrine factors, paracrine factors, and nutrients. Exploration of cofactors involved in the Kr-h1-mediated repression of BRC and TALEN-based genome editing of the KBS region would help to gain a unified understanding of the regulation of BR-C by Kr-h1.
In conclusion, we obtained a comprehensive understanding of the process by which insects avoid precocious entry into metamorphosis. This knowledge would provide important clues to the development of chemicals or treatments that artificially facilitate or delay this process, which may be usable in pest management or enhancement of silk production in sericulture.
Author Contributions
T. K., K. N., and Y. N. designed the research. T. K., K. N., Y. Ito, and Y. N. performed the research. T. K., K. N., Y. N., Y. Ishikawa, and T. S. analyzed the data. T. K., Y. Ishikawa, and T. S. wrote the paper.
Acknowledgment
We thank Dr. Toshimasa Yamazaki for helpful advice on protein analysis.
This work was supported by JSPS KAKENHI Grant 25850230 (to T. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- JH
- juvenile hormone
- JHA
- juvenile hormone analog
- 20E
- 20-hydroxyecdysone
- BR-C
- Broad-Complex
- KBS
- Kr-h1 binding site
- Met
- methoprene tolerant.
References
- 1. Riddiford L. M. (1994) Cellular and molecular actions of juvenile hormone I: general considerations and premetamorphic actions. Adv. Insect Physiol. 24, 213–274 [Google Scholar]
- 2. Jindra M., Palli S. R., and Riddiford L. M. (2013) The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 58, 181–204 [DOI] [PubMed] [Google Scholar]
- 3. Wilson T. G., and Fabian J. (1986) A Drosophila melanogaster mutant resistant to a chemical analog of juvenile hormone. Dev. Biol. 118, 190–201 [DOI] [PubMed] [Google Scholar]
- 4. Ashok M., Turner C., and Wilson T. G. (1998) Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc. Natl. Acad. Sci. U.S.A. 95, 2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miura K., Oda M., Makita S., and Chinzei Y. (2005) Characterization of the Drosophila Methoprene-tolerant gene product. Juvenile hormone binding and ligand-dependent gene regulation. FEBS J. 272, 1169–1178 [DOI] [PubMed] [Google Scholar]
- 6. Charles J. P., Iwema T., Epa V. C., Takaki K., Rynes J., and Jindra M. (2011) Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc. Natl. Acad. Sci. U.S.A. 108, 21128–21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li M., Mead E. A., and Zhu J. (2011) Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc. Natl. Acad. Sci. U.S.A. 108, 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Z., Xu J., Sheng Z., Sui Y., and Palli S. R. (2011) Steroid receptor co-activator is required for juvenile hormone signal transduction through a bHLH-PAS transcription factor, methoprene tolerant. J. Biol. Chem. 286, 8437–8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kayukawa T., Minakuchi C., Namiki T., Togawa T., Yoshiyama M., Kamimura M., Mita K., Imanishi S., Kiuchi M., Ishikawa Y., and Shinoda T. (2012) Transcriptional regulation of juvenile hormone-mediated induction of Krüppel homolog 1, a repressor of insect metamorphosis. Proc. Natl. Acad. Sci. U.S.A. 109, 11729–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kayukawa T., Tateishi K., and Shinoda T. (2013) Establishment of a versatile cell line for juvenile hormone signaling analysis in Tribolium castaneum. Sci. Rep. 3, 1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lozano J., Kayukawa T., Shinoda T., and Belles X. (2014) A role for taiman in insect metamorphosis. PLoS Genet. 10, e1004769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kayukawa T., and Shinoda T. (2015) Functional characterization of two paralogous JH receptors, methoprene-tolerant 1 and 2 in the silkworm, Bombyx mori (Lepidoptera: Bombycidae). Appl. Entomol. Zool. 50, 383–391 [Google Scholar]
- 13. Shin S. W., Zou Z., Saha T. T., and Raikhel A. S. (2012) bHLH-PAS heterodimer of methoprene-tolerant and cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc. Natl. Acad. Sci. U.S.A. 109, 16576–16581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui Y., Sui Y., Xu J., Zhu F., and Palli S. R. (2014) Juvenile hormone regulates Aedes aegypti Krüppel homolog 1 through a conserved E box motif. Insect Biochem. Mol. Biol. 52, 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li M., Liu P., Wiley J. D., Ojani R., Bevan D. R., Li J., and Zhu J. (2014) A steroid receptor coactivator acts as the DNA-binding partner of the methoprene-tolerant protein in regulating juvenile hormone response genes. Mol. Cell. Endocrinol. 394, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Minakuchi C., Zhou X., and Riddiford L. M. (2008) Krüppel homolog 1 (Kr-h1) mediates juvenile hormone action during metamorphosis of Drosophila melanogaster. Mech. Dev. 125, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Minakuchi C., Namiki T., and Shinoda T. (2009) Krüppel homolog 1, an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev. Biol. 325, 341–350 [DOI] [PubMed] [Google Scholar]
- 18. Konopova B., Smykal V., and Jindra M. (2011) Common and distinct roles of juvenile hormone signaling genes in metamorphosis of holometabolous and hemimetabolous insects. PLoS ONE 6, e28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lozano J., and Belles X. (2011) Conserved repressive function of Krüppel homolog 1 on insect metamorphosis in hemimetabolous and holometabolous species. Sci. Rep. 1, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kayukawa T., Murata M., Kobayashi I., Muramatsu D., Okada C., Uchino K., Sezutsu H., Kiuchi M., Tamura T., Hiruma K., Ishikawa Y., and Shinoda T. (2014) Hormonal regulation and developmental role of Krüppel homolog 1, a repressor of metamorphosis, in the silkworm Bombyx mori. Dev. Biol. 388, 48–56 [DOI] [PubMed] [Google Scholar]
- 21. DiBello P. R., Withers D. A., Bayer C. A., Fristrom J. W., and Guild G. M. (1991) The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics 129, 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bayer C. A., Holley B., and Fristrom J. W. (1996) A switch in broad-complex zinc finger isoform expression is regulated posttranscriptionally during the metamorphosis of Drosophila imaginal discs. Dev. Biol. 177, 1–14 [DOI] [PubMed] [Google Scholar]
- 23. Spokony R. F., and Restifo L. L. (2007) Anciently duplicated Broad Complex exons have distinct temporal functions during tissue morphogenesis. Dev. Genes Evol. 217, 499–513 [DOI] [PubMed] [Google Scholar]
- 24. Piulachs M. D., Pagone V., and Bellés X. (2010) Key roles of the Broad-Complex gene in insect embryogenesis. Insect Biochem. Mol. Biol. 40, 468–475 [DOI] [PubMed] [Google Scholar]
- 25. Nagamine K., Kayukawa T., Hoshizaki S., Matsuo T., Shinoda T., and Ishikawa Y. (2014) Cloning, phylogeny, and expression analysis of the Broad-Complex gene in the longicorn beetle Psacothea hilaris. Springerplus 3, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiss I., Bencze G., Fodor G., Szabad J., and Fristrom J. W. (1976) Prepupal larval mosaics in Drosophila melanogaster. Nature 262, 136–138 [DOI] [PubMed] [Google Scholar]
- 27. Kiss I., Beaton A. H., Tardiff J., Fristrom D., and Fristrom J. W. (1988) Interactions and developmental effects of mutations in the Broad-Complex of Drosophila melanogaster. Genetics 118, 247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou X., and Riddiford L. M. (2002) Broad specifies pupal development and mediates the “status quo” action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development 129, 2259–2269 [DOI] [PubMed] [Google Scholar]
- 29. Zhou X., Zhou B., Truman J. W., and Riddiford L. M. (2004) Overexpression of broad: a new insight into its role in the Drosophila prothoracic gland cells. J. Exp. Biol. 207, 1151–1161 [DOI] [PubMed] [Google Scholar]
- 30. Muramatsu D., Kinjoh T., Shinoda T., and Hiruma K. (2008) The role of 20-hydroxyecdysone and juvenile hormone in pupal commitment of the epidermis of the silkworm, Bombyx mori. Mech. Dev. 125, 411–420 [DOI] [PubMed] [Google Scholar]
- 31. Suzuki Y., Truman J. W., and Riddiford L. M. (2008) The role of Broad in the development of Tribolium castaneum: implications for the evolution of the holometabolous insect pupa. Development 135, 569–577 [DOI] [PubMed] [Google Scholar]
- 32. Konopova B., and Jindra M. (2008) Broad-Complex acts downstream of Met in juvenile hormone signaling to coordinate primitive holometabolan metamorphosis. Development 135, 559–568 [DOI] [PubMed] [Google Scholar]
- 33. Zhou B., Hiruma K., Shinoda T., and Riddiford L. M. (1998) Juvenile hormone prevents ecdysteroid-induced expression of broad complex RNAs in the epidermis of the tobacco hornworm, Manduca sexta. Dev. Biol. 203, 233–244 [DOI] [PubMed] [Google Scholar]
- 34. Reza A. M., Kanamori Y., Shinoda T., Shimura S., Mita K., Nakahara Y., Kiuchi M., and Kamimura M. (2004) Hormonal control of a metamorphosis-specific transcriptional factor Broad-Complex in silkworm. Comp. Biochem. Physiol. B 139, 753–761 [DOI] [PubMed] [Google Scholar]
- 35. Zhou B., and Riddiford L. M. (2001) Hormonal regulation and patterning of the Broad-Complex in the epidermis and wing discs of the tobacco hornworm, Manduca sexta. Dev. Biol. 231, 125–137 [DOI] [PubMed] [Google Scholar]
- 36. Huang J., Tian L., Peng C., Abdou M., Wen D., Wang Y., Li S., and Wang J. (2011) DPP-mediated TGF beta signaling regulates juvenile hormone biosynthesis by activating the expression of juvenile hormone acid methyltransferase. Development 138, 2283–2291 [DOI] [PubMed] [Google Scholar]
- 37. Nishita Y., and Takiya S. (2004) Structure and expression of the gene encoding a Broad-Complex homolog in the silkworm, Bombyx mori. Gene 339, 161–172 [DOI] [PubMed] [Google Scholar]
- 38. Nishita Y., and Takiya S. (2006) Differential usage of two promoters of the Broad-Complex gene in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 36, 779–788 [DOI] [PubMed] [Google Scholar]
- 39. Nishita Y. (2014) Ecdysone response elements in the distal promoter of the Bombyx Broad-Complex gene, BmBR-C. Insect Mol. Biol. 23, 341–356 [DOI] [PubMed] [Google Scholar]
- 40. Nishita Y., and Takiya S. (2009) Differential responsiveness of BmBR-C promoters to ecdysone signals. J. Insect Biotechnol. Sericol. 78, 127–138 [Google Scholar]
- 41. Kobayashi I., Kojima K., Uchino K., Sezutsu H., Iizuka T., Tatematsu K., Yonemura N., Tanaka H., Yamakawa M., Ogura E., Kamachi Y., and Tamura T. (2011) An efficient binary system for gene expression in the silkworm, Bombyx mori, using GAL4 variants. Arch. Insect Biochem. Physiol. 76, 195–210 [DOI] [PubMed] [Google Scholar]
- 42. Kanamori Y., Hayakawa Y., Matsumoto H., Yasukochi Y., Shimura S., Nakahara Y., Kiuchi M., and Kamimura M. (2010) A eukaryotic (insect) tricistronic mRNA encodes three proteins selected by context-dependent scanning. J. Biol. Chem. 285, 36933–36944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sailsbery J. K., Atchley W. R., and Dean R. A. (2012) Phylogenetic analysis and classification of the fungal bHLH domain. Mol. Biol. Evol. 29, 1301–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rosenberg U. B., Schröder C., Preiss A., Kienlin A., Côté S., Riede I., and Jäckle H. (1986) Structural homology of the product of the Drosophila Krüppel gene with Xenopus transcription factor IIIA. Nature 319, 336–339 [Google Scholar]
- 45. Pankratz M. J., Hoch M., Seifert E., and Jäckle H. (1989) Krüppel requirement for knirps enhancement reflects overlapping gap gene activities in the Drosophila embryo. Nature 341, 337–340 [DOI] [PubMed] [Google Scholar]
- 46. Iuchi S. (2001) Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci. 58, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]