Abstract
The deubiquitinating enzyme associated molecule with the SH3 domain of STAM (AMSH) is crucial for the removal of ubiquitin molecules during receptor-mediated endocytosis and lysosomal receptor sorting. AMSH interacts with signal transducing adapter molecule (STAM) 1 or 2, which enhances the activity of AMSH through an unknown mechanism. This stimulation is dependent on the ubiquitin-interacting motif of STAM. Here we investigate the specific mechanism of AMSH stimulation by STAM proteins and the role of the STAM Vps27/Hrs/STAM domain. We show that, in the presence of STAM, the length of the ubiquitin chains affects the apparent cleavage rate. Through measurement of the chain cleavage kinetics, we found that, although the kcat of Lys63-linked ubiquitin chain cleavage was comparable for di- and tri-ubiquitin, the Km value was lower for tri-ubiquitin. This increased affinity for longer chains was dependent on the Vps27/Hrs/STAM domain of STAM and required that the substrate ubiquitin chain contain homogenous Lys63-linkages. In addition, STAM directed AMSH cleavage toward the distal isopeptide bond in tri-ubiquitin chains. Finally, we generated a structural model of AMSH-STAM to show how the complex binds Lys63-linked ubiquitin chains and cleaves at the distal end. These data show how a deubiquitinating enzyme-interacting protein dictates the efficiency and specificity of substrate cleavage.
Keywords: deubiquitylation (deubiquitination), endosomal sorting complex required for transport (ESCRT), metalloprotease, polyubiquitin chain, ubiquitin, AMSH, STAM, VHS, ubiquitin-interacting motif
Introduction
The ubiquitin system plays a role in a wide range of cellular processes, including protein degradation, cell signaling, transcription regulation, and DNA damage response (1). The regulation of numerous cellular processes by ubiquitin is ascribed to the ability of ubiquitin to form a large spectrum of distinct modifications, from monoubiquitination of a target protein to polyubiquitin chains (2). Polyubiquitin chains are formed by connecting one of seven lysines or the N-terminal α-amine within one ubiquitin to Gly76 of another ubiquitin (3, 4). These linkages are described by the lysine within ubiquitin that donates the amine (e.g. Lys63 or Lys48). Similar to other posttranslational modifications, ubiquitination is a reversible modification, and the removal of ubiquitin from substrates or the reduction of the polyubiquitin chain length, called “trimming,” is catalyzed by a group of enzymes called deubiquitinating enzymes (DUBs).3 About 100 DUBs are encoded by the human genome. They are divided into six families on the basis of their structure and catalytic mechanism (5). Five of the families are cysteine proteases, including ubiquitin-specific proteases, ubiquitin C-terminal hydrolases, ovarian tumor domain DUBs, Machado-Joseph disease proteases, and monocyte chemotactic protein-induced protein DUBs, whereas the sixth family, JAB1/MPN/MOV34 domain DUBs, are metalloproteases (5). DUBs that cleave polyubiquitin chains exhibit different cleavage specificities depending on the ubiquitin chain linkage. Some DUBs only cleave chains of a single linkage, whereas others cleave several linkage types and still others exhibit linkage-independent activity, cleaving all types of ubiquitin chains (6–8). Therefore, regulation of DUB activity is required to guarantee on-target ubiquitination in the cell.
Previous work by many research groups has identified several types of regulatory mechanisms applied to DUBs that enable cells to control DUB activity. Posttranslational modifications such as phosphorylation, ubiquitination, and SUMOylation play a pivotal role in the activation of several DUBs (9). For some DUBs, interaction with a specific partner is required for their allosteric activation or inhibition (10). Moreover, the recent finding that several E2 enzymes interact with and regulate the DUB OTUB1 suggests a novel cross-talk between ubiquitin conjugating enzymes and DUBs that does not operate through ubiquitination of the DUB (11).
A subset of the identified DUB binding partners also binds ubiquitin or ubiquitin chains. Although a detailed mechanism of how these partners regulate DUB activity has not yet been determined, it has been proposed that a ubiquitin binding partner facilitates substrate binding for the DUB, thereby affecting DUB activity (12). Several DUBs contain ubiquitin-binding domains or interact with a binding partner that has a ubiquitin binding surface required for proper substrate specificity (13). These additional substrate interaction domains can restrict off-target activity against ubiquitin chains containing different linkages (14, 15). Alternatively, DUBs containing additional ubiquitin binding sites enable efficient cleavage of longer ubiquitin chains because multiple substrate binding sites within the DUB would have an additive effect on substrate affinity. This holds true for the enzyme TRABID, a DUB that contains three N-terminal ubiquitin-binding Npl4-like zinc-finger domains that collectively contribute to the preferential cleavage of longer ubiquitin chains (14). Therefore, it is possible that ubiquitin binding partners of DUBs enforce cleavage specificity not only on the basis of chain topology but also on the basis of the length of the chain.
Lys63-linked ubiquitin chains play a key role in endosomal complexes required for transport (ESCRT) signaling and receptor degradation via the lysosomes (16). Not surprisingly, several proteins that function in ESCRT signaling have roles in ubiquitin metabolism, and some possess ubiquitin-binding domains, including signal transducing adaptor molecule (STAM) 1 and 2 (17). STAM binds monoubiquitin or ubiquitin chains via its Vps27/Hrs/STAM (VHS) domain and its ubiquitin-interacting motif (UIM). However, it shows preferential binding of Lys63-linked ubiquitin chains (18, 19). Besides ubiquitin binding domains, STAM contains an SH3 domain that mediates an interaction with the DUB associated molecule with the SH3 domain of STAM (AMSH), via the SH3 binding sequence (Fig. 1) (20). AMSH is a Lys63-specific DUB belonging to the family of JAB1/MPN/MOV34 metalloproteases (21). AMSH regulates endocytic sorting of membrane proteins such as EGF receptor and CXCR4 (21–23), a process that is mediated by ESCRT. STAM not only recruits AMSH to the endocytic pathway, facilitating deubiquitination of cargo proteins, but also stimulates AMSH DUB activity. In vitro experiments showed that STAM stimulates AMSH DUB activity against Lys63-linked ubiquitin chains. Mutations in the SH3 domain of STAM that disrupt binding to AMSH as well as mutations in the UIM domain have been shown to prevent AMSH stimulation (20, 24, 25). Similarly, mutations in the SH3 binding sequence of AMSH, located just before the catalytic domain, prevent stimulation. The catalytic domain of AMSH contains distal and proximal ubiquitin binding sites that provide the specificity toward Lys63-linked ubiquitin chains (26). Therefore, the additional ubiquitin binding sites provided by STAM are not necessary for ubiquitin chain linkage specificity. To determine the role of the ubiquitin binding domains of STAM on AMSH activity, we determined the kinetics of ubiquitin chain cleavage by the STAM-AMSH complex. We found that STAM directs AMSH specificity to cleave Lys63-linked ubiquitin chains of more than two ubiquitin molecules in length. This stimulation is due in large part to the VHS domain because truncations lacking the VHS domain of STAM show no preference for longer chains. Finally, we modeled the structure of the AMSH-STAM and Lys63-linked tri-Ub complex to show how STAM could direct AMSH to ubiquitin chains.
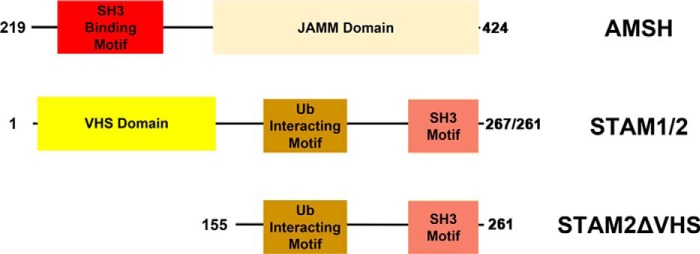
FIGURE 1.
Domain structure of AMSH and STAM 1/2 for the constructs used in this work. AMSH denotes residues 219–424, STAM 1/2 denotes residues 1–267/261, and STAM 2 ΔVHS denotes residues 155–261. JAMM, JAB1/MPN/MOV34.
Materials and Methods
Cloning, Expression, and Purification
The human AMSH, STAM1, and STAM2 open reading frames were amplified from a human spleen complementary DNA library (BioChain) and cloned into a pET32a vector containing an N-terminal tobacco etch virus-cleavable thioredoxin-His6 tag using infusion ligase-free cloning (Clontech). All AMSH, STAM1, and STAM2 fragments were generated in the above vector. Mutants of ubiquitin were generated by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene) following the protocol of the manufacturer.
All proteins were expressed in Escherichia coli T7 Express (New England Biolabs) and grown in Luria-Bertani medium. Cultures were inoculated using 1% (v/v) of a saturated overnight culture and were grown at 37 °C to an A600 of 0.4–0.6. Proteins were induced at 16 °C overnight by addition of 150 μm isopropyl-β-d-thiogalactoside. Cells were harvested by centrifugation (6200 × g, 15 min) and stored at −80 °C for later use.
Ubiquitin WT and mutants were expressed and purified as described previously (27). AMSH WT and deletions were purified by resuspending cell pellets in lysis buffer (50 mm sodium phosphate (pH 8.0), 300 mm NaCl, 15 mm imidazole, and 5% glycerol) after adding 1 mm PMSF. Cells were disrupted using a microfluidizer (Microfluidics), and the lysate was centrifuged at 68,900 × g for 1 h to remove cell debris. The lysate was subjected to immobilized metal affinity chromatography using 5 ml His-Trap columns (GE Healthcare), and protein was eluted with a linear imidazole gradient of 15–300 mm in 30 column volumes. Fractions containing purified protein were pooled, and tobacco etch virus protease was added in a ratio of 1:100 before dialysis overnight at 4 °C into lysis buffer without imidazole. Cleaved protein was then purified by a second round of metal affinity chromatography, and the cleaved protein was collected from the flow-through. Protein was then dialyzed overnight at 4 °C into dialysis buffer (20 mm Tris (pH 7.8), 100 mm NaCl, and 5 mm β-mercaptoethanol) and subjected to ion exchange chromatography using a 5-ml Q-Sepharose column (GE Healthcare). Protein was eluted with a 30-column volume linear gradient of 100–600 mm NaCl in dialysis buffer. Fractions containing purified protein were pooled, concentrated, and flash-frozen in liquid N2 before being stored at −80 °C.
STAM (1 and 2) WT and deletions were purified according to the AMSH purification protocol until the second round of metal affinity chromatography. Proteins were then purified by a Superdex 75 gel filtration column (GE Healthcare) equilibrated in 20 mm Tris (pH 7.5), 150 mm NaCl, and 5 mm β-mercaptoethanol. The elution peak was concentrated and flash-frozen in liquid N2.
Ubiquitin Chain Formation
Lys63-linked di-Ub and tri-Ub and mixed tri-Ub (possessing both Lys48 and Lys63 linkages) were synthesized following the Pickart and Raasi method for controlled synthesis of ubiquitin chains, with slight modifications (27). All enzymes required for these syntheses, viz. E1 (human), Ubc13/MMS2 (yeast), CDC34 (human), RAD5 (846–1169, yeast), and Yuh1 (yeast), were also expressed and purified as recombinant proteins in E. coli as described previously (28, 29). Lys63 linked di-Ub was synthesized using two ubiquitin mutants (UbK63R and Ub74, which is missing the last two glycine residues). These mutants (200 μm each) were incubated overnight at 37 °C in the presence of 1 μm Uba1 (E1), 2 μm Rad5 (E3), 2 μm Ubc13, and 2 μm Mms2 in buffer containing 50 mm Tris (pH 7.6), 10 mm MgCl2, 0.6 mm DTT, and 2 mm ATP. Then the reaction was diluted 20-fold in 50 mm ammonium acetate (pH 4.5), 100 mm NaCl, 5 mm β-mercaptoethanol, and loaded on a Mono-S column. Lys63-linked di-Ub was eluted using a linear NaCl gradient of 100–600 mm in 30 column volumes. Fractions containing Lys63-linked di-Ub were pooled, dialyzed against 10 mm Tris (pH 7.5) and 100 mm NaCl, concentrated, and stored at −80 °C. Lys63 linked tri-Ub was synthesized in two steps. The first step was similar to that of Lys63-linked di-Ub formation, except that UbD77, which has an additional Asp after Gly76, was used instead of Ub74. Then Lys63 di-Ub was incubated with 2.7 μm YUH1, which removes Asp77, for 1 h at 37 °C and used in a second chain extension reaction step. In this step (200 μm), Lys63 di-Ub was incubated overnight at 37 °C with 400 μm Ub74 in the presence of the conjugating enzymes listed previously. Then Lys63-linked tri-Ub was purified using a Mono-S column as described before. Mixed Lys63-Lys48 tri-ubiquitin chains were synthesized in two steps, as described above, with the following modifications. For the first step reaction, UbD77 K63R or K48R was incubated with Ub K63R,K48C in the presence of Ubc13/Mms2 or CDC34 to form Lys63-linked di-UB or Lys48-linked di-Ub, respectively. After removal of Asp77 by YUH1, Ub74 was added to the di-Ub chain to form a Lys48 linkage (using CDC34) or Lys63 linkage (using Ubc13/Mms2). Both mixed tri-Ub (Lys63-Lys48 linkages or Lys48-Lys63 linkages) were concentrated and stored at −80 °C. Lys63-linked ubiquitin chains of non-uniform lengths were synthesized in 50 mm HEPES (pH 8.0) buffer containing 0.1 mm DTT, 150 mm NaCl, 1 mm ATP, and 2.5 mm MgCl2. Reaction mix containing 1 μm hUBC13, 1 μm UEV1a, 5 μm ubiquitin, and 0.1 μm E1 enzyme was incubated at 37 °C for 30 min. Then Apyrase at 0.12 milliunits (New England Biolabs) was added for 10 min at 37 °C to stop the synthesis reaction.
Fluorescein-labeled Ub Chains
For labeling Lys63-linked di-Ub, synthesis was done in the presence of UB74/K48C, thereby introducing Cys on the proximal ubiquitin. For labeling Lys63-linked tri-Ub, synthesis was done either in the presence of Ub74/K48C (for introducing Cys on the proximal ubiquitin) or in the presence of UbK63R/K48C (for introducing Cys on the distal ubiquitin). Then 50–100 μm of ubiquitin chains was incubated with a 4-fold excess of fluorescein-5-maleimide in buffer containing 150 mm NaCl, 20 mm Tris (pH 7.5), and 1 mm tris(2-carboxyethyl)phosphine for 2 h at room temperature and quenched by adding a 10-fold excess β-mercaptoethanol for 5 min. The labeled chains were purified using a PD miniTrap G25 gravity column pre-equilibrated in 150 mm NaCl, 20 mm Tris (pH 7.5), and 1 mm tris(2-carboxyethyl)phosphine following the protocol of the manufacturer. The labeled ubiquitin chains were visualized on SDS-PAGE gel and scanned in a Typhoon laser at 488 nm. The final concentrations were measured spectrophotometrically at 280 and 495 nm and stored at −80 °C until use.
In Vitro AMSH DUB Assay
Gel-based assays for AMSH stimulation by STAM were performed in reaction buffer containing 50 mm HEPES (pH 7.5), 150 mm NaCl, and 2 mm DTT. AMSH (residues 219–424) 50 nm was mixed with 5 μm of STAM 1 (1–267), STAM2 (1–261), or STAM2 missing the VHS domain (155–261) with 4 μm ubiquitin chain substrate. Reactions were performed at 37 °C and initiated by addition of the AMSH enzyme. Aliquots were removed at the specified time points, and the reactions were quenched by addition of denaturing SDS-PAGE loading dye containing β-mercaptoethanol. Samples were analyzed by gel electrophoresis. Gels were stained with Coomassie Brilliant Blue. In the absence of STAM, the AMSH concentration was 200 nm in cleavage reactions.
Assay to Detect the Position of Cleavage
Lys63-linked tri-Ub labeled on the distal/proximal ubiquitin (0.12 μm) were incubated with AMSH alone or with 5 μm STAM2 or STAM2 ΔVHS. Reactions were done at room temperature and 4 °C in the absence and presence of STAM2 (or STAM2 missing the VHS), respectively. Also, the AMSH concentration was 200 nm, except in the presence of STAM2, where it was reduced to 50 nm. Reactions were stopped at different time points by adding denaturing loading dye containing β-mercaptoethanol. Then samples were loaded on SDS-PAGE, and labeled ubiquitin chains were visualized using a Typhoon laser scanner at 488 nm.
Immunoblotting
Lys63-linked ubiquitin chains of non-uniform lengths were incubated with 50 nm AMSH alone or with 5 μm STAM2 or STAM2 ΔVHS at 37 °C. The cleavage of chains was stopped at different time points by adding denaturing SDS-PAGE loading dye containing β-mercaptoethanol. Samples were separated on a SDS-PAGE gel and transferred to a PVDF membrane. The membrane was denatured in a solution containing 6 m guanidine HCl, 20 mm Tris-HCl (pH 7.5), 1 mm PMSF, and 5 mm β-mercaptoethanol for 30 min at 4 °C and then washed extensively in Tris-buffered saline and Tween 20 (TBST). Membranes were blocked overnight at 4 °C with 5% BSA in TBST and incubated for 1 h with ubiquitin antibody (1:1000, catalog no. P4D1, Santa Cruz Biotechnology) at room temperature, followed by anti-mouse HRP-conjugated secondary antibody.
Steady-state Kinetic Assays of AMSH-STAM Deubiquitinating Activity against Lys63-linked Di-Ub or Tri-Ub
Steady-state enzyme kinetic assays were performed at 17 °C in a reaction buffer containing 50 mm HEPES (pH 7.5), 150 mm NaCl, and 2 mm DTT. AMSH (50 nm) and STAM2 (1–261, 5 μm) were mixed with specified amounts of Lys63-linked di-Ub or tri-Ub. After 1 min, reactions were stopped by the addition of denaturing SDS-PAGE loading dye containing β-mercaptoethanol and analyzed by SDS-PAGE followed by staining with Coomassie G-250 (30). The product bands corresponding to ubiquitin, di-Ub, or tri-Ub substrates were quantified by densitometry with ImageJ software (31). Ubiquitin and di-Ub standards were used to generate calibration plots and to calculate the product concentration. Reaction velocities were then calculated for each di-Ub or tri-Ub concentration and fitted to the Michaelis-Menten equation with GraphPad Prism software.
Competition Assays
For competition experiments, we followed Cooper et al. (32). Briefly, all experiments were performed at room temperature in buffer containing 50 mm HEPES (pH 7.5), 150 mm NaCl, and 2 mm DTT. The assays were performed by incubating 50 nm AMSH (219–424) and 5 μm of STAM2 with and without the VHS domain (1–261 or 155–261) with 0.7 μm labeled di-Ub in the presence of increasing concentrations of unlabeled di-Ub or tri-Ub for 10 min. The reactions were quenched by the addition of denaturing SDS-PAGE loading dye containing β-mercaptoethanol. Samples were analyzed by gel electrophoresis on 12% polyacrylamide and scanned in a Typhoon laser scanner. Bands were integrated using ImageJ, and the IC50 was calculated with GraphPad Prism software.
Model of the AMSH-STAM-Ubiquitin Complex
The AMSH-STAM-ubiquitin complex was homology-modeled for unknown parts in Phyre2 and YASARA (Yet Another Scientific Artificial Reality Application) in intensive/slow search mode (33, 34). Each domain was built and refined separately in YASARA, followed by addition of domain-connecting sequences (35). Ubiquitin was docked to the STAM UIM using the HADDOCK (High Ambiguity Driven protein-protein DOCKing) web server, and the RAP80 UIM-Lys63-linked di-ubiquitin structure was used to suggest interaction residues for docking (36, 37). The STAM-AMSH interaction site was modeled using 1UJ0 as a template and aligned with the AMSH homology model. The structures of the Lys48-linked ubiquitin chains were aligned to the distal AMSH-bound ubiquitin (38, 39).
Results
The AMSH-STAM Complex Shows Preferential Cleavage of Tri-Lys63 Ub over Di-ubiquitin
Although STAM has been shown to stimulate the DUB activity of AMSH against Lys63 di-Ub, whether STAM directs AMSH cleavage toward longer ubiquitin chains has not been tested. To that end, we compared the kinetics of Lys63 di-Ub cleavage to that of Lys63-linked tri-Ub cleavage. As shown in Fig. 2A, although Lys63-linked tri-Ub disappeared after 15 min, di-Ub survived for more than half an hour, suggesting that the former is a preferred substrate for cleavage by the AMSH-STAM complex. To further understand this observation, we measured the steady-state kinetics of Lys63 linked di-Ub or tri-Ub cleavage. As shown in Fig. 2, B and C, although both tri-Ub and di-Ub have a similar kcat value (44.1 ± 1.6 min−1 and 46.3 ± 2.6 min−1 for tri- and di-Ub, respectively), the Km value for tri-Ub is 4-fold lower (2.1 ± 0.3 μm and 8.8 ± 1.3 μm for tri- and di-ubiquitin, respectively).
FIGURE 2.
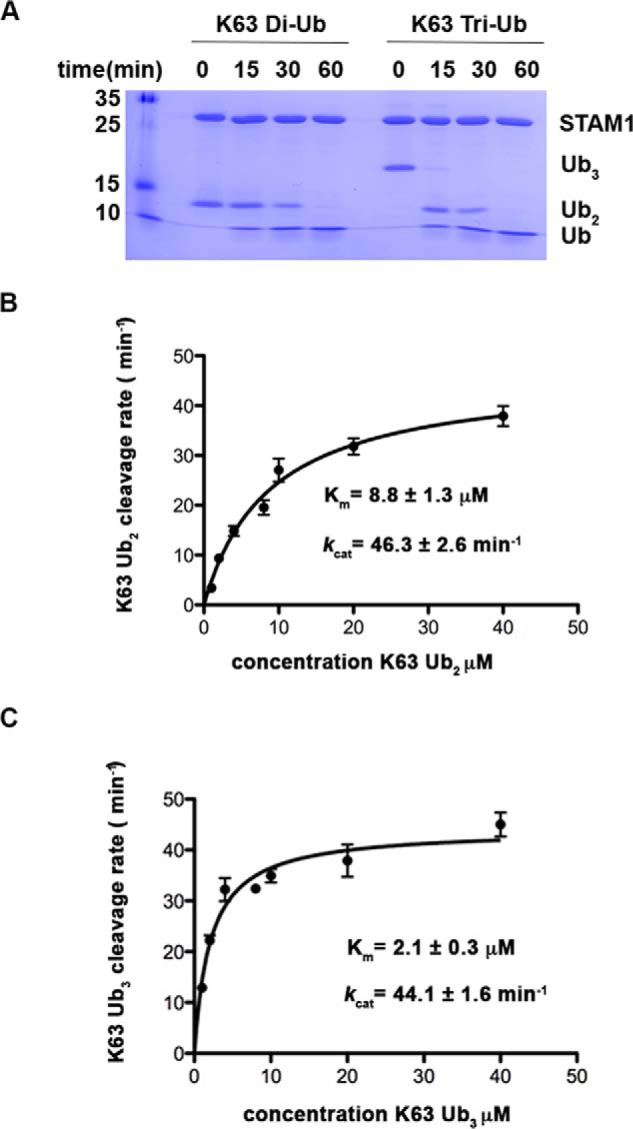
Preferential cleavage of Lys63-linked tri-Ub by the AMSH-STAM complex. A, Coomassie-stained gel showing Lys63 di/tri-Ub (4 μm) cleavage by AMSH (50 nm) and STAM1 (5 μm). B and C, steady-state kinetic saturation curves for cleavage of Lys63-linked di-Ub (B) and Lys63-linked tri-Ub (C) by AMSH (50 nm) and STAM2 (5 μm). Each rate was measured in two sets of experiments with quadruplicate measurements in each experiment, and error bars represent the mean ± S.E. for each measurement.
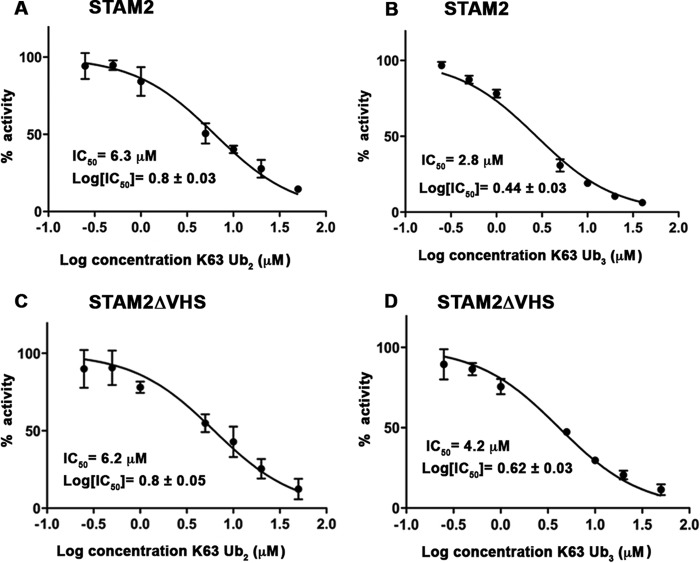
Reduction in the kinetic parameter Km suggests an increase in the affinity for substrate. Therefore, we were motivated to determine whether the preferential cleavage of tri-Ub was due to better binding of tri-Ub over di-Ub to the AMSH-STAM complex. To that end, we performed in vitro deubiquitination assays of fluorescently labeled di-Ub in the presence of increasing amounts of unlabeled di- or tri-Ub chains (Fig. 3). In these experiments, we measured the IC50 of di- or tri-Ub chains, which reflects the relative affinities of these substrates to the complex of AMSH-STAM. As shown in Fig. 3, both di- and tri-Ub chains inhibited AMSH-STAM-mediated cleavage of the fluorescent labeled Lys63-linked di-Ub. The IC50 value for tri-Ub was 2-fold lower than that of di-Ub (2.8 μm and 6.3 μm for tri- and di-Ub, respectively), indicating a preferential binding of tri-Ub. Taken together, our results suggest that the higher binding affinity of tri-ubiquitin to the AMSH-STAM complex contributes to its preferred cleavage by the AMSH-STAM complex.
FIGURE 3.
The VHS domain contributes to the preferential binding of Lys63-linked tri-Ub to the AMSH-STAM2 complex. A and B, competition experiments in which the rate of fluorescein-labeled, Lys63-linked di-Ub cleavage was measured at increasing concentrations of unlabeled Lys63-linked di-Ub (A) or Lys63-linked tri-Ub (B). C and D, as A and B, respectively, but in the presence of STAM2 missing the VHS domain. Each rate was measured in triplicate, and error bars represent the mean ± S.E.
STAM Introduces Specificity on the Basis of the Position of the Isopeptide Bond
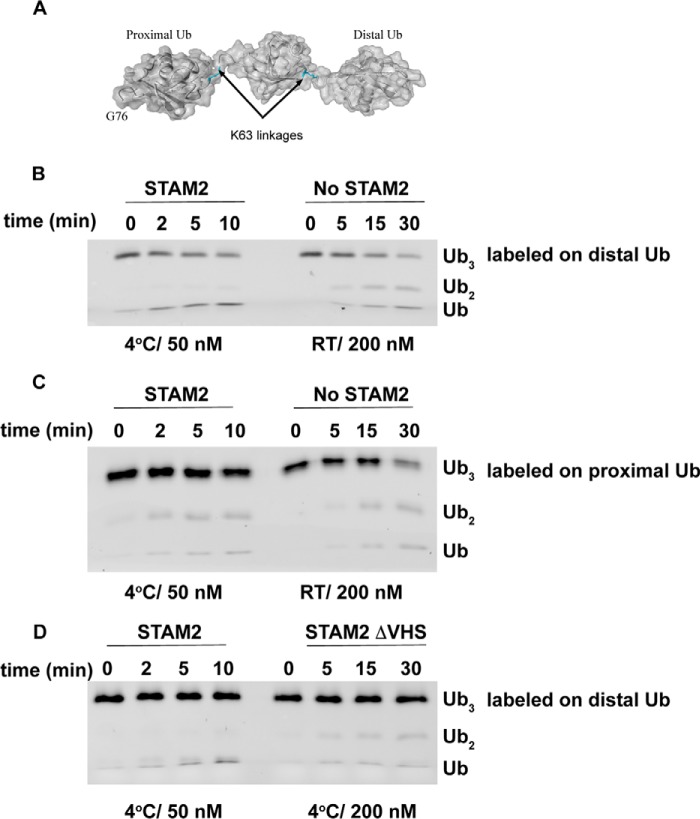
Lys63-linked tri-Ub possesses two isopeptide bonds, raising the question of whether one of the bonds is preferred over the other for cleavage by AMSH. To that end, we first fluorescently labeled Lys63-linked tri-Ub chains on the distal ubiquitin to determine whether STAM confers endo- or exodeubiquitinase activity (Fig. 4). As shown in figure 4B, tri-Ub chains labeled on the distal Ub showed only mono-Ub as the cleavage product in the presence of STAM, suggesting that the distal bond is preferred for cleavage. However, in the absence of STAM, we observed both di-Ub and mono-Ub as cleavage products, suggesting that the distal bond is no more preferred than the proximal bond. Notably, to discount the possibility that the mono-Ub band observed in the presence of STAM is due to fast cleavage at both distal and proximal linkages, we slowed down the reaction by decreasing the temperature to 4 °C and using only 50 nm AMSH. However, in the absence of STAM, we worked at room temperature and with 200 nm AMSH to discount the possibility that di-Ub band is due to the slow activity of AMSH in the absence of STAM. To further support this observation, we labeled Lys63-linked tri-Ub chains on the proximal Ub. As expected, in the presence of STAM, we observed di-Ub, which is the product of tri-Ub cleavage at the distal bond (Fig. 4C). Moreover, because this di-Ub can serve as a substrate for cleavage by itself, we also observed the mono-Ub product when Lys63-linked tri-Ub chains were labeled on the proximal ubiquitin.
FIGURE 4.
STAM2 directs the cleavage of Lys63 tri-Ub by AMSH to the distal bond. A, schematic of the distal and proximal ubiquitin positions and bonds in Lys63-linked tri-Ub. B, cleavage of Lys63-linked tri-Ub labeled on the distal ubiquitin (0.12 μm) in the presence or absence of 5 μm STAM2. RT, room temperature. C, similar to B but with Lys63-linked tri-Ub labeled on the proximal ubiquitin. D, cleavage of Lys63-linked tri-Ub labeled on the distal ubiquitin (0.12 μm) in the presence STAM2 possessing or missing the VHS domain (5 μm). The temperature and concentration of AMSH are indicated for each reaction.
To better understand this observation, we asked whether the VHS domain plays a role in this process. To that end, we tested the cleavage of labeled Ub chains in the presence of STAM, which was missing the VHS domain. As expected, the lack of the VHS domain abrogated the ability of STAM to direct cleavage to the distal bond. The cleavage pattern in the absence of the VHS domain resembled that of AMSH alone (Fig. 4D). Taken together, our results suggest that STAM not only increases substrate-binding affinity, as described previously, but also directs the cleavage to a specific isopeptide bond in the chain.
The VHS Domain Contributes to the Cleavage of Lys63-linked Ub Chains Possessing Various Lengths
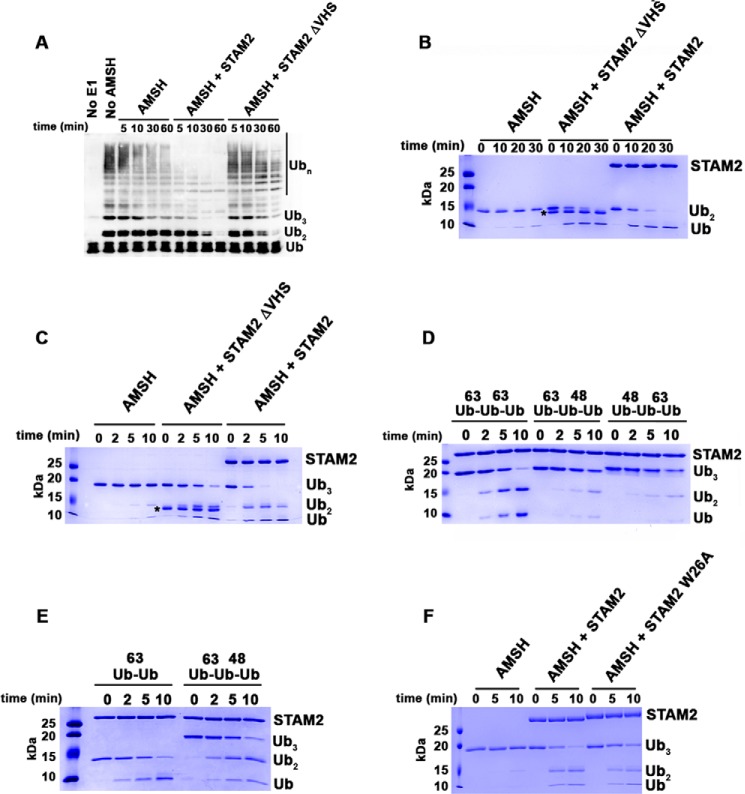
So far, our model substrates for studying the role of the VHS domain were Lys63-linked di- and tri-ubiquitin chains comprised of two or three ubiquitin molecules. Therefore, to examine whether our proposed mechanism for the VHS domain holds true for ubiquitin chains longer than tri-Ub, we synthesized Lys63-linked ubiquitin chains of non-uniform lengths using the E2 UBC13 and Uev1A. Then we tested the cleavage of these chains by the AMSH-STAM complex and a complex lacking the STAM VHS domain. As shown in Fig. 5A, the lack of the VHS domain significantly reduced the disappearance of longer ubiquitin chains. However, STAM missing the VHS domain still enhances the overall rate of ubiquitin chain cleavage. These data support our finding that the STAM VHS domain is required for cleavage of chains longer than two ubiquitin molecules.
FIGURE 5.
The VHS domain of STAM2 directs AMSH to cleave longer Lys63-linked ubiquitin chains. A, cleavage of Lys63-linked ubiquitin chains by AMSH (50 nm) at 37 °C in the absence of STAM2 or in the presence of 5 μm STAM2 possessing and missing the VHS domain. Detection of ubiquitin chains was done by anti-Ub Western blot analysis. B, cleavage of Lys63-linked di-Ub (4 μm) by AMSH (50 nm) at 37 °C in the absence of STAM2 or in the presence of STAM2 (5 μm) possessing and missing the VHS domain. The asterisk denotes the position of bands corresponding to STAM2ΔVHS. C, similar to B, but tri-Ub is the substrate. D, cleavage of Lys63-linked tri-Ub chains or mixed tri-Ub chains (4 μm) by AMSH (50 nm) in the presence of STAM2 (5 μm) at room temperature. E, cleavage of mixed tri-Ub compared with Lys63-linked di-Ub. The experiment was performed as described in D. F, cleavage of Lys63-linked tri-Ub by AMSH in the absence of STAM2 or in the presence of STAM2 WT or mutant (W26A). The experiment was performed as described in B.
The VHS Domain of STAM Is Essential for the Preferential Cleavage of Tri-Lys63-linked Ubiquitin Chains
The basis for STAM VHS domain-stimulated chain cleavage by AMSH was not clear from our experiment with the heterogeneous chain mixture. Therefore, we tested the stimulation of cleavage of di-Ub and tri-Ub by AMSH. A STAM construct lacking the VHS domain did not show a detectable defect in its ability to stimulate cleavage of di-Ub by AMSH (Fig. 5B). However, removal of the VHS domain diminished the ability of STAM to stimulate tri-Ub cleavage by AMSH, supporting our previous findings shown in Fig. 5A (Fig. 5C). These data suggest that the VHS domain plays a role in the stimulation of ubiquitin chains longer than di-Ub. To further understand the role of the VHS domain, we tested whether the VHS domain enhances the affinity of the AMSH-STAM complex for tri-Ub over di-Ub. As expected, the IC50 of di-Ub was not affected by deletion of the VHS domain and was comparable with that measured for the full AMSH-STAM complex (compare Fig. 3, A and C). However, the IC50 for Tri-Ub was increased in the absence of the VHS domain compared with the value determined for STAM with the VHS domain, suggesting that the VHS domain plays a role in the binding of tri-Ub chains (compare Fig. 3, B and D).
Our results suggest that the VHS domain within STAM contributes to stimulation when the ubiquitin chains are composed of more than two ubiquitin molecules (i.e. at least two isopeptide bonds). However, it was not clear whether stimulation by the VHS domain requires two adjacent Lys63-linked bonds or whether it is insensitive to the linkage of the non-scissile bond(s). To that end, we made two types of mixed chains: one group with Lys48-linked isopeptide bond at the distal end and another group with this bond at the proximal end. First we tested the disappearance of the mixed tri-ubiquitin chains by the AMSH-STAM complex. As shown in Fig. 5D, both substrates disappeared more slowly compared with tri-Ub chains composed of two Lys63-linked isopeptide bonds. However, when we compared the cleavage of the mixed chains with that of Lys63-linked di-Ub, we found that their disappearance rate was similar, supporting the conclusion that the contribution of the VHS domain to cleavage requires two adjacent Lys63-linked isopeptide bonds (Fig. 5E).
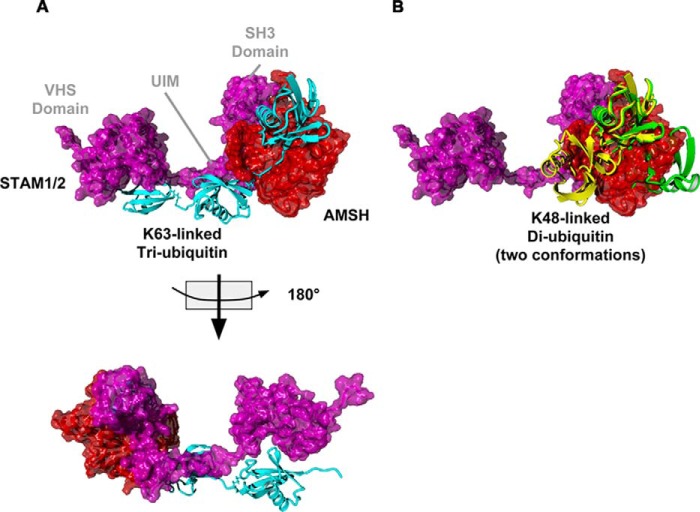
Structural Model for the AMSH·STAM·Tri-Ub Complex
The structural basis for the preference of AMSH-STAM for longer ubiquitin chains was not clear given the current structures of AMSH, STAM, and associated complexes (26, 40, 41). We modeled the structure of STAM bound to AMSH using the existing structures of AMSH-like protease bound to di-ubiquitin and the known structures of domains of STAM and homology modeling of the UIM of STAM to provide structural support for our biochemical findings. Secondary structure prediction supported unstructured regions between the VHS, UIM, and SH3 domain (data not shown). To show the interaction between the UIM and the ubiquitin hydrophobic patch, we used HADDOCK to identify a favorable complex followed by energy minimization to reduce structural errors (see “Materials and Methods” for details). We also modeled the proximal ubiquitin onto the chain of AMSH using existing structures of Lys63-linked ubiquitin chains (42). On the basis of the biochemical data showing that the VHS domain is required for specificity toward longer chains (Fig. 5), we modeled the VHS as interacting with the proximal end of the chain and the UIM interacting with the middle ubiquitin (Fig. 6). Supporting the structural model is a mutation at the VHS domain of STAM2 (W26A) that diminished the ability of STAM to stimulate cleavage (Fig. 5F). The final model shows that the sequence of STAM is long enough to bridge AMSH to two ubiquitin molecules proximal to the DUB-bound ubiquitin. Moreover, modeling a Lys48-linked chain showed that, although the UIM may be able to bind to a ubiquitin molecule, the proximal ubiquitin molecule would be out of position to interact with the VHS domain. These data support the cleavage of mixed chains, shown in Fig. 5, D and E, where mixed chains were cleaved no better than Lys63-linked di-ubiquitin.
FIGURE 6.
Model of the AMSH-STAM complex bound to Lys63-linked tri-Ub and Lys48-linked di-Ub. A, AMSH (red) bound to STAM1/2 (magenta) and Lys63-linked tri-Ub (cyan). B, the AMSH-STAM complex bound to two conformation of Lys48-linked di-Ub (yellow and green).
Discussion
DUBs are key players in regulating ubiquitination in the cell because of their activity to remove or trim ubiquitin chains (43). However, in many cases, the proper function of a DUB depends on associating with other proteins that modulate the activity or specificity of the DUB. These additional proteins, thereby, provide additional mechanisms for regulating DUB function (10). Here we focused on the Lys63-linked specific DUB AMSH, and its partner, STAM, which possesses two ubiquitin-binding domains. It has been shown previously that the UIM of STAM enhances AMSH DUB activity by facilitating the binding of Lys63 ubiquitin chains (20, 24, 25). The UIM of STAM, together with the AMSH proximal and distal ubiquitin binding sites, can easily bind di-ubiquitin (24). However, whether the another ubiquitin-binding domain of STAM, the VHS domain, contributes to the DUB activity and/or specificity of AMSH was not clear. Here we show that the VHS domain does not contribute to the rate of catalysis of Lys63-linked di-ubiquitin cleavage by AMSH. However, the VHS domain enhances AMSH cleavage of ubiquitin chains composed of more than two ubiquitin molecules. This specificity toward tri-ubiquitin disappears in the absence of the VHS domain. Our results suggest that the ubiquitin-binding domains in DUB binding partners can increase the number of ubiquitin molecules that are recognized by the DUB, leading to enhanced cleavage of chains where all the binding sites are occupied. The rate-limiting step of proteases is thought to be chemistry. Therefore, our competition values are likely a reflection of differences in affinity (44). Supporting this is the IC50 value of AMSH and Lys63 di-Ub, which is similar to the value of the KD between them (data not shown). At ubiquitin concentrations below the IC50 value, the 2- to 5-fold differences in IC50 value observed here would be maximized, resulting in a noticeable preference toward cleavage of longer chains because of better binding. Moreover, these additional ubiquitin binding domains can direct cleavage toward a specific isopeptide bond. In the presence of the VHS domain, the AMSH-STAM complex is directed to cleave the distal isopeptide bond in tri-ubiquitin chains. Our proposed model of the STAM-AMSH complex bound to tri-ubiquitin shows how STAM directs the specificity of AMSH. The VHS and UIM domains of STAM bind the proximal and inner ubiquitin molecules, respectively, directing AMSH toward the distal ubiquitin. Therefore, not only does STAM enhance the affinity of AMSH for substrate, but STAM directs AMSH to function as an exodeubiquitinase. Having additional ubiquitin binding domains that are separate from the AMSH active site would enhance the affinity of the complex for the ubiquitin chain and, as a result, the preference for chains longer than two ubiquitin proteins. Following cleavage of a tri-ubiquitin or a longer ubiquitin chain, the new distal ubiquitin is bound to the UIM and could be transferred to the AMSH active site, permitting processive breakdown of a Lys63-linked ubiquitin chain.
Preferential cleavage of longer Lys63-linked ubiquitin chains by AMSH raises the possibility that it plays a role in regulating the length of ubiquitin chains. The length of the ubiquitin chain can affect signaling and the rate of protein degradation. However, the persistence of ubiquitin chains of a certain length and how the rate of trimming/cleavage affects cellular function is not entirely clear. This lack of knowledge is mainly due to the difficulty in defining the length of ubiquitin chains attached to substrates because cellular ubiquitin chains represent a balance between two opposing activities: that of ubiquitin-conjugating enzymes and of DUBs (45). It has been shown recently that, in cells treated with EGF, the EGF receptor, whose ubiquitinated form is a substrate of AMSH, is ubiquitinated predominantly with short Lys63-linked di-ubiquitin chains (46). This fits our observation that longer Lys63-linked ubiquitin chains are degraded faster, and, therefore, we expect them to be less abundant. Additionally, in cells lacking AMSH, EGF receptor degradation increases 2-fold, suggesting that small changes in the rate of deubiquitination can have functional effects in cells (21).
In this work, we found a decreased cleavage rate for longer Lys63-linked ubiquitin chains in the absence of the VHS domain. Two studies have found phosphorylation sites in STAM adjacent to the VHS and UIM domains, which suggests a possible regulatory mechanism for the AMSH-STAM complex via regulation of ubiquitin chain binding (47, 48). Moreover, we believe that this modular substrate recognition mechanism and the ability to alter substrate binding without altering the rate of cleavage are useful for further study of AMSH-dependent chain cleavage in vivo.
AMSH possesses two ubiquitin binding sites that hold two Lys63-linked ubiquitin molecules in a productive conformation for cleavage (26). This suggests that each scissile isopeptide bond is recognized independently of the length of the chain and the linkages between ubiquitin molecules not bound in the active site. Here we show that the AMSH-STAM complex recognizes at least the isopeptide bond between ubiquitin molecules adjacent to the scissile isopeptide bond, suggesting a more global mechanism of substrate recognition. Moreover, this bond has to be in the form of Lys63 linkage to be recognized by the AMSH-STAM complex. Our work suggests that introducing ubiquitin binding sites into a DUB via a binding partner enables the DUB to “sense” whether the ubiquitin chain has the right length and type of linkage. Taken together, we propose that not only the types of the linkages involved in mixed chains are important but that also their order within the chain is important for substrate specificity.
In summary, we focused on STAM and studied its effect on AMSH DUB activity and specificity. We showed that the VHS domain of STAM plays a role in AMSH specificity to longer ubiquitin chains. Mutations in the UIM domain that abolish STAM stimulation show that the VHS domain cannot replace the UIM domain (20). These data suggest that the position of each domain relative to the AMSH active site defines activity. This raises the intriguing possibility that other ubiquitin binding domains in the vicinity of AMSH may also contribute to its activity. In the cell, AMSH is recruited to ESCRT-0, which contains the ubiquitin binding protein hepatocyte growth factor-regulated tyrosine kinase substrate in addition to STAM (49). Hepatocyte growth factor-regulated tyrosine kinase substrate interacts with STAM via its GGA and TOM domain near the AMSH-STAM interaction site (50). The VHS and UIM domains of hepatocyte growth factor-regulated tyrosine kinase substrate are possibly in close proximity to STAM. Taken together, this suggests that hepatocyte growth factor-regulated tyrosine kinase substrate ubiquitin binding domains may also affect AMSH activity and suggest a more complicated cross-talk between ubiquitin binding domains and AMSH that requires further research. Finally, DUB activity characterized mainly by using the minimal ubiquitin chains or model substrates that can serve as a substrate for cleavage (i.e. di-ubiquitin or Ub-7-amino-4-methylcoumarin) may not be ideal for mechanistic studies of DUB function (51). Our work exposes the limitations of using di-ubiquitin as a model substrate and emphasizes the need to also study DUBs in the presence of longer ubiquitin chains.
Author Contributions
N. B., P. P., and R. W. designed the research. N. B., P. P., B. M., and E. C. K. performed the research. B. M., C. E. B., and R. W. analyzed the data. C. E. B., E. A. T., and K. E. D. modeled the structure of the AMSH-STAM complex. N. B., C. E. B., and R. W. wrote the manuscript.
This work was supported by United States-Israel Binational Science Foundation Grant 2013261, National Science Foundation Research Experience for Undergraduates Grant CHE-1461175, and Marie Curie Career Integration Grant PCIG13-GA-2013-630755. The authors declare that they have no conflicts of interest with the contents of this article.
- DUB
- deubiquitinating enzyme
- ESCRT
- endosomal sorting complex required for transport
- STAM
- signal-transducing adaptor molecule
- VHS
- Vps27/Hrs/STAM
- UIM
- ubiquitin-interacting motif
- SH
- Src homology
- AMSH
- associated molecule with the SH3 domain of STAM
- Ub
- ubiquitin.
References
- 1. Welchman R. L., Gordon C., and Mayer R. J. (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 2. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 3. Behrends C., and Harper J. W. (2011) Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 18, 520–528 [DOI] [PubMed] [Google Scholar]
- 4. Komander D. (2009) The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37, 937–953 [DOI] [PubMed] [Google Scholar]
- 5. Fraile J. M., Quesada V., Rodríguez D., Freije J. M., and López-Otín C. (2012) Deubiquitinases in cancer: new functions and therapeutic options. Oncogene 31, 2373–2388 [DOI] [PubMed] [Google Scholar]
- 6. Edelmann M. J., Iphöfer A., Akutsu M., Altun M., di Gleria K., Kramer H. B., Fiebiger E., Dhe-Paganon S., and Kessler B. M. (2009) Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 418, 379–390 [DOI] [PubMed] [Google Scholar]
- 7. Faesen A. C., Luna-Vargas M. P., Geurink P. P., Clerici M., Merkx R., van Dijk W. J., Hameed D. S., El Oualid F., Ovaa H., and Sixma T. K. (2011) The Differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 18, 1550–1561 [DOI] [PubMed] [Google Scholar]
- 8. Wang T., Yin L., Cooper E. M., Lai M. Y., Dickey S., Pickart C. M., Fushman D., Wilkinson K. D., Cohen R. E., and Wolberger C. (2009) Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of Otubain 1. J. Mol. Biol. 386, 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kessler B. M., and Edelmann M. J. (2011) PTMs in conversation: activity and function of deubiquitinating enzymes regulated via post-translational modifications. Cell Biochem. Biophys. 60, 21–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ventii K. H., and Wilkinson K. D. (2008) Protein partners of deubiquitinating enzymes. Biochem. J. 414, 161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wiener R., Dibello A. T., Lombardi P. M., Guzzo C. M., Zhang X., Matunis M. J., and Wolberger C. (2013) E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 20, 1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Komander D., Clague M. J., and Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 13. Reyes-Turcu F. E., Ventii K. H., and Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Licchesi J. D., Mieszczanek J., Mevissen T. E., Rutherford T. J., Akutsu M., Virdee S., El Oualid F., Chin J. W., Ovaa H., Bienz M., and Komander D. (2012) An ankyrin-repeat ubiquitin-binding domain determines TRABID's specificity for atypical ubiquitin chains. Nat. Struct. Mol. Biol. 19, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Winborn B. J., Travis S. M., Todi S. V., Scaglione K. M., Xu P., Williams A. J., Cohen R. E., Peng J., and Paulson H. L. (2008) The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 283, 26436–26443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Raiborg C., and Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 17. Bache K. G., Raiborg C., Mehlum A., and Stenmark H. (2003) STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278, 12513–12521 [DOI] [PubMed] [Google Scholar]
- 18. Lange A., Castañeda C., Hoeller D., Lancelin J. M., Fushman D., and Walker O. (2012) Evidence for cooperative and domain-specific binding of the signal transducing adaptor molecule 2 (STAM2) to Lys63-linked diubiquitin. J. Biol. Chem. 287, 18687–18699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ren X., and Hurley J. H. (2010) VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 29, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCullough J., Row P. E., Lorenzo O., Doherty M., Beynon R., Clague M. J., and Urbé S. (2006) Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 16, 160–165 [DOI] [PubMed] [Google Scholar]
- 21. McCullough J., Clague M. J., and Urbé S. (2004) AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma Y. M., Boucrot E., Villén J., Affar el B., Gygi S. P., Göttlinger H. G., and Kirchhausen T. (2007) Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J. Biol. Chem. 282, 9805–9812 [DOI] [PubMed] [Google Scholar]
- 23. Sierra M. I., Wright M. H., and Nash P. D. (2010) AMSH interacts with ESCRT-0 to regulate the stability and trafficking of CXCR4. J. Biol. Chem. 285, 13990–14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davies C. W., Paul L. N., and Das C. (2013) Mechanism of recruitment and activation of the endosome-associated deubiquitinase AMSH. Biochemistry 52, 7818–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim M. S., Kim J. A., Song H. K., and Jeon H. (2006) STAM-AMSH interaction facilitates the deubiquitination activity in the C-terminal AMSH. Biochem. Biophys. Res. Commun. 351, 612–618 [DOI] [PubMed] [Google Scholar]
- 26. Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., and Fukai S. (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 [DOI] [PubMed] [Google Scholar]
- 27. Pickart C. M., and Raasi S. (2005) Controlled synthesis of polyubiquitin chains. Methods. Enzymol. 399, 21–36 [DOI] [PubMed] [Google Scholar]
- 28. Berndsen C. E., Wiener R., Yu I. W., Ringel A. E., and Wolberger C. (2013) A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat. Chem. Biol. 9, 154–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dong K. C., Helgason E., Yu C., Phu L., Arnott D. P., Bosanac I., Compaan D. M., Huang O. W., Fedorova A. V., Kirkpatrick D. S., Hymowitz S. G., and Dueber E. C. (2011) Preparation of distinct ubiquitin chain reagents of high purity and yield. Structure 19, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 30. Lawrence A. M., and Besir H. U. (2009) Staining of proteins in gels with Coomassie G-250 without organic solvent and acetic acid. J. Vis. Exp. 30, 1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cooper E. M., Boeke J. D., and Cohen R. E. (2010) Specificity of the BRISC deubiquitinating enzyme is not due to selective binding to Lys63-linked polyubiquitin. J. Biol. Chem. 285, 10344–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., and Karplus K. (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krieger E., and Vriend G. (2014) YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics 30, 2981–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Vries S. J., van Dijk M., and Bonvin A. M. (2010) The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 [DOI] [PubMed] [Google Scholar]
- 37. Sato Y., Yoshikawa A., Mimura H., Yamashita M., Yamagata A., and Fukai S. (2009) Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lai M. Y., Zhang D., Laronde-Leblanc N., and Fushman D. (2012) Structural and biochemical studies of the open state of Lys48-linked diubiquitin. Biochim. Biophys. Acta 1823, 2046–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trempe J. F., Brown N. R., Noble M. E., and Endicott J. A. (2010) A new crystal form of Lys48-linked diubiquitin. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 994–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaneko T., Kumasaka T., Ganbe T., Sato T., Miyazawa K., Kitamura N., and Tanaka N. (2003) Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain. J. Biol. Chem. 278, 48162–48168 [DOI] [PubMed] [Google Scholar]
- 41. Davies C. W., Paul L. N., Kim M. I., and Das C. (2011) Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability. J. Mol. Biol. 413, 416–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Datta A. B., Hura G. L., and Wolberger C. (2009) The structure and conformation of Lys63-linked tetraubiquitin. J. Mol. Biol. 392, 1117–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Amerik A. Y., and Hochstrasser M. (2004) Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695, 189–207 [DOI] [PubMed] [Google Scholar]
- 44. Frey P. A., and Hegeman A. D. (2007) Enzymatic Reaction Mechanisms, pp. 314–317, Oxford University Press, Oxford [Google Scholar]
- 45. Békés M., Okamoto K., Crist S. B., Jones M. J., Chapman J. R., Brasher B. B., Melandri F. D., Ueberheide B. M., Denchi E. L., and Huang T. T. (2013) DUB-resistant ubiquitin to survey ubiquitination switches in mammalian cells. Cell Rep. 5, 826–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang F., Zeng X., Kim W., Balasubramani M., Fortian A., Gygi S. P., Yates N. A., and Sorkin A. (2013) Lysine 63-linked polyubiquitination is required for EGF receptor degradation. Proc. Natl. Acad. Sci. U.S.A. 110, 15722–15727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., and Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal 3, ra3. [DOI] [PubMed] [Google Scholar]
- 48. Steen H., Kuster B., Fernandez M., Pandey A., and Mann M. (2002) Tyrosine phosphorylation mapping of the epidermal growth factor receptor signaling pathway. J. Biol. Chem. 277, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 49. Hurley J. H., and Emr S. D. (2006) The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35, 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Prag G., Watson H., Kim Y. C., Beach B. M., Ghirlando R., Hummer G., Bonifacino J. S., and Hurley J. H. (2007) The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev. Cell 12, 973–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dang L. C., Melandri F. D., and Stein R. L. (1998) Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry 37, 1868–1879 [DOI] [PubMed] [Google Scholar]