Abstract
Systemic sclerosis (SSc) is characterized by inflammation, vascular dysfunction, and ultimately fibrosis. Progress in understanding disease pathogenesis and developing effective disease treatments has been hampered by an incomplete understanding of SSc heterogeneity. To clarify this, we have used genomic approaches to identify distinct patient subsets based on gene expression patterns in SSc skin and other end-target organs. Here, we review what is known about the gene expression-based subsets in SSc, currently defined as the inflammatory, fibroproliferative, limited, and normal-like subsets. The inflammatory subset of patients is characterized by infiltrating immune cells that include T cells, macrophages, and possibly dendritic cells, although little is known about the mediators these cells secrete and the pathways that govern cell activation. Prior studies have suggested a role for pathogens as a trigger of immune responses in SSc, and recent data have identified viral and mycobiome components as potential environmental triggers. We present a model based on analyses of gene expression data and a review of the literature, which suggests that the gene expression subsets observed in patients possibly represent distinct, interconnected molecular states of disease, to which an innate immune response is central that results in the generation of clinical disease.
Introduction
Systemic sclerosis (SSc) is a clinically heterogeneous autoimmune disease characterized by fibrosis of the skin and internal organs, vascular abnormalities, and persistent immune activation. While the etiology of SSc remains poorly understood, the earliest clinical symptoms are primarily associated with the vascular system, characterized by vasospastic episodes, referred to as Raynaud’s phenomenon. However, despite similarities in early symptoms, substantial heterogeneity exists between patients with respect to disease progression and the organs affected, hindering our understanding of pathophysiology and complicating the interpretation of clinical trials.
To overcome issues of clinical heterogeneity inherent in SSc, high-throughput gene expression has been used to better understand the pathways and processes that drive the disease. These analyses can be used to identify the major cell types driving pathogenesis in a complex tissue sample as well as to define reproducible gene expression profiles indicative of different forms of disease. Three independent skin biopsy datasets have been generated that identify reproducible gene expression subtypes characteristic of different forms or states of SSc [1–4]. Recent gene expression analyses of SSc lung biopsies provide an important addition to these efforts, expanding our understanding of disease-associated gene expression to a second end-target tissue [5, 6]. Here, we provide an update regarding insights into SSc pathogenesis using gene expression profiling, with an emphasis on the role of the innate immune system as a potential initiator and driver of disease pathology.
Identification of gene expression-based intrinsic subsets of disease
The initial gene expression studies in SSc skin biopsies focused on small cohorts and the identification of differences between SSc and healthy controls [7, 8]. These studies revealed both inflammatory and fibrotic gene expression signatures that characterized diseased tissue. One of these studies [7] showed a surprising result, which was the nearly identical disease-specific patterns of gene expression in biopsies taken from lesional forearm and non-lesional back skin of an SSc patient. This suggested that aberrant gene expression could be found even in unaffected tissues, highlighting the truly systemic nature of the disease [7]. A second result from these studies was that fibroblasts grown in culture do not accurately recapitulate the aberrant gene expression observed in SSc skin, with similar results seen using paired skin biopsies and fibroblast cultures in Gardner et al. [8], as well as non-paired samples in Whitfield et al. [7].
Clinical heterogeneity is a major factor confounding our understanding of SSc. Early studies examining heterogeneity in tumors were able to demonstrate the existence of reproducible gene expression subsets within a given tumor type [9–13]. Using the same approach to understand the variability in end-target tissues affected by SSc was therefore logical; however, this type of analysis had not been previously used for autoimmune diseases. The first reported study of gene expression heterogeneity in SSc by Milano et al. [1] identified four “intrinsic” gene expression subsets among patients with SSc. These included a fibroproliferative subset, which exhibited strong induction of proliferation genes, an inflammatory subset characterized by robust upregulation of genes associated with both innate and adaptive immune responses, a limited subset centered on a cluster of clinically limited patients, and a normal-like subset, which consisted of both healthy controls and a subset of patients with both limited and diffuse SSc. This study provided a proof-of-concept that heterogeneity within SSc clinical groups could be measured using genome-wide molecular profiling.
An analysis of two subsequent cohorts of patients confirmed and expanded these observations. Pendergrass et al. [2] reproduced the fibroproliferative, inflammatory, and normal-like subsets, driven by similar proliferation and immune-related signals; the limited SSc subset was not found due to the absence of lSSc patients in this cohort. Pendergrass et al. included serial biopsies from an investigator-initiated trial of rituximab, which showed no clinical efficacy [14] but demonstrated stable gene expression within patients over the time period analyzed (6–12 months).
In a third independent study, Hinchcliff et al. [3] examined the effects of mycophenylate mofetil (MMF), a potent immunosuppressant commonly used to treat SSc. Intrinsic subset assignments were again reproducible in patients for as long as 24 months, indicative of long-term stability in terms of gene expression. Furthermore, four patients classified in the inflammatory gene expression subset demonstrated clear clinical improvement with MMF treatment. In contrast, three patients classified as either fibroproliferative or normal-like failed to improve, consistent with the use of intrinsic subset assignment as a predictor of treatment outcomes.
Two other recent investigator-initiated clinical trials for abatacept [15] and nilotinib [16] have also shown subset-specific responses to therapy. Chakravarty et al. [15] found that patients who improve while on abatacept therapy map to the inflammatory gene expression subset. Comparison of pre- and post-treatment biopsies revealed a decrease in CD28 co-stimulatory signaling, which is the molecular target of abatacept. This pathway change was only observed in patients who improved during treatment, with no changes seen in non-responders or placebo controls. These data suggest that patients in the inflammatory subset are the most likely to improve with abatacept therapy. Another study by Gordon et al. [16] showed that patients who improve with nilotinib, a tyrosine kinase inhibitor (TKI), have high expression of genes associated with increased transforming growth factor β (TGF-β) and platelet-derived growth factor (PDGF) signaling at baseline, suggesting a mechanistic link between pathway activation and clinical improvement. Those who failed to improve did not show high-level expression of these genes. This study argues for targeting patients with TGF-β/PDGF pathway activation with TKIs.
Most recently, the inflammatory and normal-like gene expression subsets have been reproduced in skin biopsies derived from an independent cohort and analyzed by an independent group of investigators; however, they were unable to reproduce the fibroproliferative subset identified in prior studies. An important result from this study that provides insight into the etiology of the subsets was the observation that normal-like patients have the longest disease duration and likely represent end-stage, inactive disease [17].
Meta-analyses of gene expression in skin and genetic polymorphisms
To better identify the genes and processes common across three published SSc datasets [1–3], Mahoney et al. [4] developed a data mining procedure termed mutual information consensus clustering (MICC) to identify co-expression modules conserved across datasets. As co-expressed genes tend to be functionally related, this method permits identification of processes central to the pathology of SSc. Two groups of genes (consensus gene clusters) were consistently identified. A direct analysis of gene-gene interactions within and between the two consensus gene clusters was performed using the IMP gene-gene interaction Bayesian network in association [18] with 41 SSc-associated polymorphic genes identified by genome-wide association studies (GWAS) [19]. Genes were shown to cluster into five distinct communities, each associated with a distinct biological process important to SSc pathogenesis. Three groups of genes that were strongly associated with the inflammatory subset showed enrichment for processes associated with response to interferon signaling, B cell receptor signaling, adaptive immune processes, monocyte chemotaxis, and M2 macrophage activation. Another group, consisting of genes from both the inflammatory and fibroproliferative clusters, exhibited enrichment for genes associated with TGF-β and PDGF signaling as well as extracellular matrix (ECM) remodeling. These findings suggest that these processes, in part, connect the inflammatory response to ECM deposition. Finally, a set of genes with increased expression in fibroproliferative patients was strongly associated with cell proliferation. An important result from this study was that 30 of 41 SSc-associated polymorphic genes were associated with inflammatory genes and processes or formed bridges between inflammatory processes and the ECM. These results implicate a mechanistic link between genetic risk factors and important disease processes in the network. A second implication from this study is that the intrinsic gene expression subsets are likely mechanistically interconnected with the inflammatory signature, leading to activation of ECM deposition and TGF-β and PDGF signaling, which ultimately results in the proliferative response. These data suggest that the intrinsic subsets are long-lived but interconnected, although capturing the transition from one subset to another experimentally has been difficult.
Expansion of gene expression profiling beyond skin
Gene expression profiles have been shown to be broadly consistent between lesional and non-lesional skin within a given patient [7]; however, these phenotypes are quickly lost in culture, limiting the use of in vitro techniques for the study of SSc [8]. While this phenomenon suggests the disease is systemic, it begs the question of whether or not the subsets observed in the skin are found in other end-target tissues. In this regard, both fibrotic and immune processes have been observed in two separate studies of SSc lung disease. Gene expression has been analyzed in late-stage lung samples from SSc patients that included individuals with both pulmonary arterial hypertension (PAH) and interstitial lung disease (ILD) [5]. These studies revealed increased expression of genes involved in fibrosis in SSc-associated ILD, including type I and type III collagen, IGFBPs, MMP-7, CTGF, osteopontin, and tissue inhibitors of metalloproteases 1 (TIMP-1). SSc-PAH lungs shared functional groups with idiopathic PAH lung samples, showing enrichment for interferon, IL-4, IL-17, and antigen presentation signaling. SSc-associated PAH also showed increased expression of inflammatory genes, including chemokines CCL2, CXCL10, and CX3CL1. A recent study by Christmann et al. used open lung biopsies of patients with SSc-related ILD and healthy controls to study early lung pathogenesis in SSc [6]. A total of 21 patients with either dSSc (n= 11) or lSSc (n=10) was included in this study, representing both early- and late-stage disease. Significantly, upregulated processes included collagen expression, TGF-β signaling, IFN signaling, and M2 macrophage activation, broadly consistent with gene activation clusters in Mahoney et al. Such widespread concordance between the skin and lung implicates major immune-related processes as drivers of pathology in SSc.
Moreover, gene expression analysis of esophageal biopsies of patients with SSc has shown that the intrinsic subsets can be found in a tissue other than the skin [20]. Both the inflammatory and fibroproliferative subsets were identified in SSc esophageal biopsies, suggesting that intrinsic subset gene expression may be a consistent feature of SSc tissues and may represent pathophysiological states of disease. Efforts are currently underway to perform a multi-tissue network analysis for common gene expression found in all SSc end-target tissues, which should provide insight into common mechanisms of pathogenesis (Taroni, Mahoney, Pioli, and Whitfield, In preparation).
Pathway activation events underlying the intrinsic gene expression subsets in skin
Given the success of MMF and abatacept as treatments for patients in the inflammatory subset [3, 15] and the suggestion that TKIs may benefit patients with TGF-β and PDGF activation [16, 21], a greater emphasis has been placed on understanding the pathways that underlie each of the four intrinsic subsets with the ultimate goal of identifying therapeutic targets. To this end, two studies comparing in vitro-derived fibroblast gene expression signatures for TGF-β and IL-4/IL-13 against the Milano et al. skin biopsy dataset revealed strong correlations with the fibroproliferative and inflammatory subsets, respectively, indicating a potential role for each of these pathways in disease pathology [22, 23].
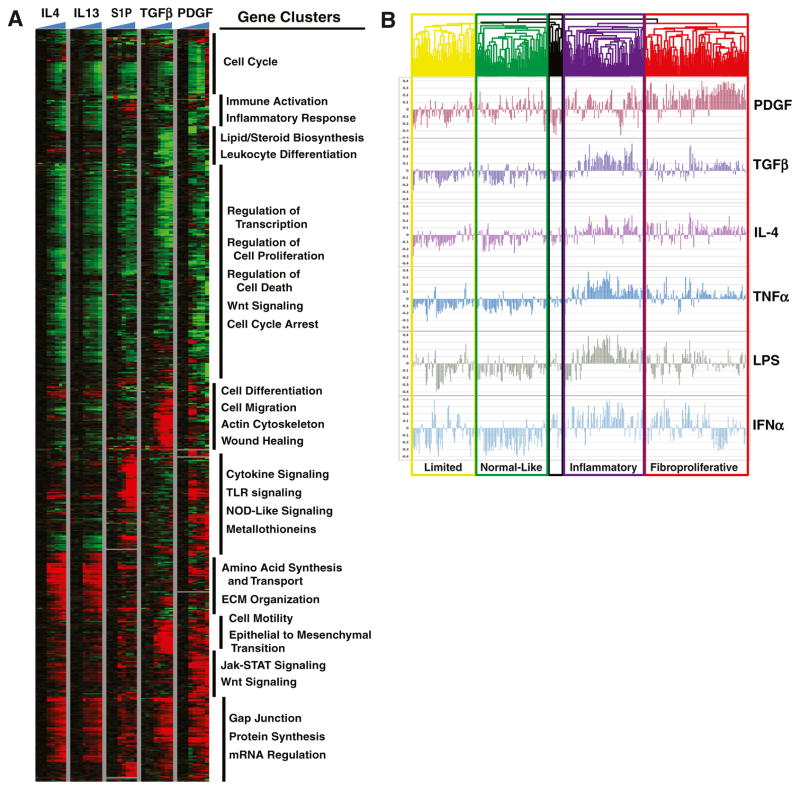
A more expansive follow-up study compared 13 gene signatures experimentally derived in dermal fibroblasts against the three skin biopsy datasets described above, along with additional 82 arrays associated with the existing Hinchcliff et al. dataset [24]. Hierarchical clustering of this expanded Milano-Pendergrass-Hinchcliff (MPH) dataset recreated all four intrinsic subsets and provided a sharper distinction between the limited and normal-like subsets, based on a strong expression of genes associated with lipid signaling and oxidative reduction found in the normal-like group that is absent in limited patients. Alignment of pathway-specific gene signatures showed enrichment in specific patient subsets. The strongest association was seen between genes activated by PDGF in fibroblasts and the fibroproliferative subset (Fig. 1). TGF-β showed a correlation with a subset of fibroproliferative patients and, interestingly, also showed a strong correlation with patients in the inflammatory subset (Fig. 1b). Consistent with the results of Sargent et al. [22], TGF-β was strongly correlated with disease severity [24].
Fig. 1.
Pathway activation signatures show differential expression across the intrinsic gene expression subsets. a Normal and SSc dermal fibroblasts were treated with different pro-fibrotic and immune mediators that have been implicated in SSc. A subset of pathways and fibroblast time courses is shown. b Hierarchical clustering was performed on 329 microarray hybridizations from 287 unique biopsies representing 111 patients: 70 dSSc, 10 lSSc, 26 healthy controls, 4 morphea, and 1 eosinophilic fasciitis from three independent data sets [1–3], as published in [24]. The array tree is color coded to indicate intrinsic subset designations (yellow = limited, green = normal-like, purple = inflammatory, red = fibroproliferative, and black = unassigned). Pearson correlation coefficients were calculated between each pathway and a sample and plotted. Adapted from Johnson et al. PLoS One (2015) with permission [24]
Several different pathways showed enrichment in the inflammatory patient subset, including genes induced by sphingosine-1-phosphate (S1P), IL-4, tumor necrosis factor α (TNF-α), lipopolysaccharide (LPS), and polyinosinic/ polycytidylic acid (poly(I-C); Fig. 1). Interferon α (IFN-α) signaling was also elevated in this subset, although correlation with this pathway failed to reach statistical significance in the analysis. A common theme linking each of the immune activation pathways associated with the inflammatory subset is their convergence on NF-κB. Engagement of Toll-like receptors (TLRs) by their respective ligands initiates a cascade of signal transduction that culminates in activation of NF-κB, resulting in the production of acute phase cytokines, including TNF-α and IFN-α. TGF-β is also known to activate NF-κB via induction of TGF-β-associated kinase 1 (TAK1), suggesting a common theme linking these signals. Indeed, TLR signaling has been implicated directly in the persistent activation of fibroblasts in SSc. In this model, tissue injury leads to the generation and release of damage-associated molecular patterns (DAMPs), which include matrix components (such as hyaluronic acid), nucleic acids, and immune complexes. Binding of DAMPs to TLRs signals fibroblasts to activate tissue repair processes and collagen production. Prolonged damage results in sustained fibroblast activation and unchecked fibrogenesis [25]. From these data and the literature, we have put forth hypotheses regarding the pathogenesis of SSc based on the assumption that the intrinsic gene expression subsets may be longitudinally connected (Fig. 2).
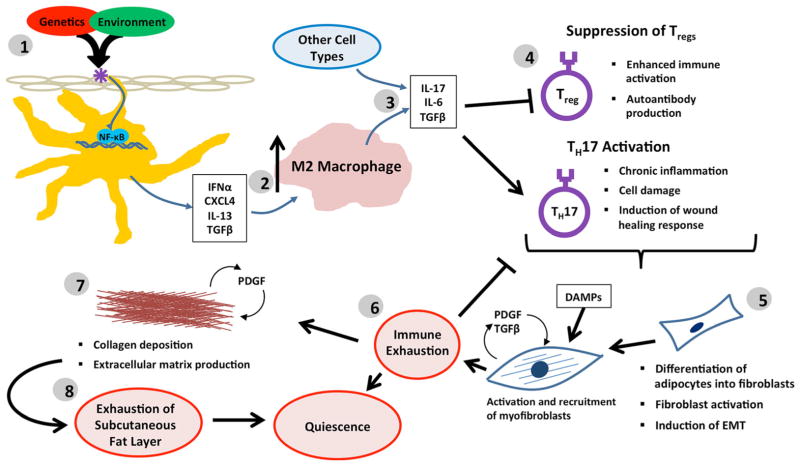
Fig. 2.
Progressive model of SSc pathogenesis. 1 In this model, SSc pathogenesis is initiated by a disease trigger in a permissive genetic background, resulting in an innate immune response signaling through NF-κB. These early responses may be mediated in part through pDCs and macrophages, which induce the expression of TH2-like cytokines, along with fibrotic mediators, such as TGF-β. 2 This early TH2 bias results in persistent M2 macrophage activation, which further exacerbates the fibrotic phenotype. 3 Over time, the gradual decrease in pDC involvement results in a loss of IFN-α signaling, 4 resulting in transition to a more TH17-like disease and the suppression of Treg function. 5 This persistent inflammation further perpetuates the chronic fibrosis phenotype driven by TGF-β, which then stimulates production of pro-fibrotic PDGF. 6 Eventual resolution of inflammation allows for downregulation of innate immune responses. 7 Continued proliferation is supported through differentiation of resident adipocytes into fibroblasts, resulting in a persistent replicative phenotype, in combination with a decrease in lipid signaling. 8 Exhaustion of the adipocyte layer results in the loss of proliferating cells, ultimately resulting in transition to a more quiescent form of the disease
Potential innate immune triggers in SSc
In addition to mounting gene expression data, several additional lines of evidence point to a role for innate immune activation in the pathogenesis of SSc. Notably, aberrant responses to TLR stimulation have been noted in SSc patients [26, 27], and inflammasome activation has been implicated as a mediator of fibrosis in murine models of SSc [28]. Because these mechanisms also mediate immune defense against microbial invasion, it is possible that SSc is triggered by pathogenic encounter. In this regard, metagenomic analysis of skin biopsies from patients with early dSSc demonstrates significant overrepresentation of transcripts from the environmental fungus Rhodotorula glutinis compared with normal controls [29]. Notably, each of the patients identified in this study mapped to the inflammatory intrinsic gene expression subset, suggesting a TLR-mediated event may be responsible for the subsequent induction of NF-κB activation observed in this patient cohort. In further support of a potential role for a microbial trigger in SSc, Epstein-Barr viral nucleic acids and proteins have been detected in both lesional and non-lesional skin of dSSc patients, and EBV infection of SSc fibroblasts induced TLR activation [26].
Antigen-presenting cells as a driver of pathology in SSc
Evidence of macrophage activation in SSc skin [4, 23] and lung [6, 30], coupled with increased expression of genes associated with monocyte/macrophage recruitment and differentiation in SSc [4, 31], suggests a role for antigen-presenting cells (APCs) in disease pathogenesis. Because macrophages have significant plasticity and are subject to modulation by local microenvironmental factors, in vivo macrophage activation spans a broad spectrum of polarization states; classically activated (M1) and alternatively activated (M2) macrophages form the extreme ends of this activation spectrum [32]. Gene expression and molecular profiling studies suggest macrophages are skewed toward an M2 activation state in SSc, as demonstrated by enhanced expression of CCL18, CD163, and IL-10R [4, 6, 33]. As recent studies have shown, M2 polarization may be elicited with a variety of stimuli, including immune complexes, IL-4, IL-13, IL-10, and TGF-β [34]. Significantly, immune complexes have been detected in SSc patients [35, 36]; elevated expression of IL-4, IL-13, and IL-10 has been identified in SSc patient sera [37]; and numerous studies have implicated TGF-β in the pathogenesis of SSc [22, 38, 39]. Therefore, it is possible that these stimuli contribute to the phenotypic and functional activation state characteristic of SSc macrophages. M2 macrophages produce a number of pro-fibrotic factors including TGF-β, CCL2, and CCL18, which are known to play important roles in SSc pathogenesis [34]. These cells are also potent sources of TGF-β [40], which mediates T cell activation [41], and IL-10 [40], which may further enhance M2 activation. As such, it is likely that these cells play a key role in the initiation of fibrosis in SSc and are responsible for the activation of inflammation and adaptive immune responses observed in SSc.
The importance of macrophages in the pathogenesis of SSc was demonstrated by a comparative gene expression profiling study that used the sclGVHD mouse as a model of human inflammatory intrinsic patient subset [23]. This work showed IL-13 pathway activation in both sclGVHD and the inflammatory intrinsic subset and identified CCL2 as a downstream target of the IL-13 signaling pathway. In addition to its pro-fibrotic properties, NF-κB-regulated CCL2 is a potent M2 macrophage activator and chemotactic factor. Significantly, blockade of CCL2 protected sclGVHD mice from clinical and pathological disease, suggesting that inhibition of macrophage recruitment and activation may be useful therapeutically.
In addition to macrophages, plasmacytoid dendritic cells (pDCs) have also been implicated as important mediators of SSc disease progression. As key regulators of innate and adaptive immunity, pDCs circulate in the peripheral blood, present antigen, and secrete copious type I interferons (mainly IFN-α and IFN-β) in response to stimulation [42]. Type I interferons modulate immune cell differentiation and proliferation as well as inflammatory cytokine production. Increased expression of type I interferon genes has been detected in the peripheral blood and sera of SSc patients [43, 44], and pDCs have been identified as the major source of type I interferons in these patients [44, 45]. Distinct from their role in interferon production, proteomic analysis of pDCs isolated from patients with SSc demonstrated that these cells secreted elevated levels of CXCL4 and other chemokines, which correlated directly with disease severity [46]. Furthermore, expression of CXCL4 was highest coincident with early disease and diminished over time, suggesting a role in the initiation of disease. Given the role of pDCs as the primary source of IFN-α, coupled with the strong IFN signaling responses seen in the skin and lung, these data implicate pDCs as early drivers of SSc pathogenesis.
Effects of APCs on adaptive immunity
Gene expression profiles of the skin and lung from SSc patients have shown increased expression of chemokines and chemokine receptor genes that are associated with recruitment of T helper type 2 (TH2) cells [4, 47]. Consistent with the potent induction of M2 macrophages and pDCs early in disease, the earliest adaptive immune responses in SSc appear dominantly TH2-skewed, driven by a combination of IL-4, IL-13, and TGF-β [34]. Indeed, these cytokines are present in the skin, sera, and bronchoalveolar fluid of SSc patients [22, 23, 37, 48, 49], as are both M2 macrophages [50, 51] and TH2 cells [52], highlighting the importance of these factors in SSc pathogenesis. Intriguingly, the spontaneous rate of CD8 T cell apoptosis is elevated in SSc compared with controls [53], which may account for the enhanced CD4:CD8 ratio in SSc.
However, despite the strong TH2-like immune response observed early in the disease, these signals diminish over time. The graft-versus-host disease model used to evaluate the effects of IL-13 on fibrosis examined mice at 2 and 5 weeks post-splenocyte transfer and revealed a strong IL-13 signature at 2 weeks, which was attenuated at 5 weeks [23]. This suggests that initial activation of CCL2 and other fibrotic genes occurs through IL-13 signaling but is likely sustained through the activity of other signaling pathways and cell types. In this regard, mounting evidence suggests a role for TH17 cells in the pathogenesis of SSc, with clear differences between diffuse and limited disease [54–58]. This progression is observed in many other autoimmune diseases, including multiple sclerosis, systemic lupus erythematosus, psoriasis, neuromyelitis optica, Crohn’s disease, inflammatory bowel disease, and rheumatoid arthritis, all of which exhibit a strong TH17-like bias [59–62].
Under normal conditions, type I IFNs are potent inhibitors of TH17 activity [57]. However, the documented decrease in IFN-α-producing pDCs [46] and subsequent time-dependent attenuation in IFN-associated signaling [24] suggests a mechanism by which T cells may shift from a TH2- to TH17-dominant phenotype with increased disease progression. Such a shift is likely mediated by cytokines (especially IL-6) that are produced by myeloid and/or TH2 cells. In this model, myeloid-derived TGF-β, in combination with IL-6, suppresses Treg production and promotes differentiation of TH17 cells [63]. The strong TGF-β and TNF-α gene expression signatures observed in the inflammatory subset, in conjunction with pervasive inflammatory infiltrates, are consistent with a TH17-like immune response [64]. In support of this hypothesis, immunohistochemical analysis of SSc skin and gene expression profiling of PBMCs from SSc patients indicated expansion of circulating TH17 cells and increased infiltration of IL-17+ cells in SSc skin. Furthermore, increased frequencies of activated TH17 cells have been noted in all SSc subtypes, and intracellular expression of TGF-β, which induces TH17 differentiation, is specifically elevated in patients with late disease.
Cumulative data suggest a progressive model of SSc disease pathogenesis
Collectively, the analyses presented here suggest a progressive model of SSc pathogenesis (Fig. 2). Our experimental data have shown that a patient’s intrinsic subset assignment is relatively stable in serial biopsies taken over the course of 6, 12, and sometimes even 24 months [2, 3], but our computational network analyses and meta-analyses of multiple patient cohorts have suggested that the intrinsic subsets are long-lived but likely mechanistically interconnected [4, 24]. Capturing patients transitioning between the inflammatory, fibroproliferative, or normal-like gene expression subsets in longitudinal studies is required to truly prove they are interconnected, and this has been hampered by the slow and variable disease progression, combined with the limited resolution one can obtain with serial biopsies from patients. A recent study by Assassi et al. [17] extends preliminary results shown by Pendergrass et al. [2] and confirms that patients in the normal-like subset have the longest disease duration, indicating it is likely the late, inactive form of the disease.
In the progressive model (Fig. 2), we propose that a disease trigger, in the presence of a permissive genetic background, initiates an innate immune response through the activation of TLRs, which culminates in NF-κB activation, marking the inflammatory subset as the likely initiation point for most SSc patients. We believe that the intensity and duration of patients’ active immune responses likely have some bearing on their prognosis and overall outcomes. These early responses are likely mediated in part through pDCs and macrophages, which induce the expression of TH2-like cytokines, along with fibrotic mediators, such as TGF-β. This early TH2 bias results in persistent M2 macrophage activation, which further exacerbates the fibrotic phenotype. Over time, the gradual decrease in pDC involvement results in a loss of IFN-α signaling, causing a transition to a more TH17-like disease and the suppression of Treg function. This persistent inflammation further perpetuates the chronic fibrosis phenotype driven by TGF-β, which then stimulates the production of pro-fibrotic PDGF. Eventual resolution of inflammation allows for downregulation of innate immune responses and a transition into the fibroproliferative subset. Continued proliferation is supported through differentiation of resident adipocytes into fibroblasts, resulting in a persistent replicative phenotype, in combination with a decrease in lipid signaling [65]. Finally, exhaustion of the adipocyte layer may result in the loss of the proliferative signature, ultimately resulting in transition to a more quiescent form of the disease, consistent with the normal-like subset of patients.
Contributor Information
Patricia A. Pioli, Email: patricia.a.pioli@dartmouth.edu.
Michael L. Whitfield, Email: michael.whitfield@dartmouth.edu.
References
- 1.Milano A, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One. 2008;3:e2696. doi: 10.1371/journal.pone.0002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pendergrass SA, Lemaire R, Francis IP, Mahoney JM, Lafyatis R, Whitfield ML. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol. 2012;132:1363–1373. doi: 10.1038/jid.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinchcliff M, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol. 2013;133:1979–1989. doi: 10.1038/jid.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahoney JM, et al. Systems level analysis of systemic sclerosis shows a network of immune and profibrotic pathways connected with genetic polymorphisms. PLoS Comput Biol. 2015;11:e1004005. doi: 10.1371/journal.pcbi.1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu E, Shi H, Jordan RM, Lyons-Weiler J, Pilewski JM, Feghali-Bostwick CA. Lung tissues in patients with systemic sclerosis have gene expression patterns unique to pulmonary fibrosis and pulmonary hypertension. Arthritis Rheum. 2011;63:783–794. doi: 10.1002/art.30159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christmann RB, et al. Association of interferon- and transforming growth factor beta-regulated genes and macrophage activation with systemic sclerosis-related progressive lung fibrosis. Arthritis Rheumatol. 2014;66:714–725. doi: 10.1002/art.38288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitfield ML, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci U S A. 2003;100:12319–12324. doi: 10.1073/pnas.1635114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner H, et al. Gene profiling of scleroderma skin reveals robust signatures of disease that are imperfectly reflected in the transcript profiles of explanted fibroblasts. Arthritis Rheum. 2006;54:1961–1973. doi: 10.1002/art.21894. [DOI] [PubMed] [Google Scholar]
- 9.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 10.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 11.Garber ME, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafyatis R, et al. B cell depletion with rituximab in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum. 2009;60:578–583. doi: 10.1002/art.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarty EF, et al. A pilot randomized placebo-controlled study of abatacept for the treatment of diffuse cutaneous systemic sclerosis. Arthritis Res Ther. 2015 doi: 10.1186/s13075-015-0669-3. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon JK, et al. Nilotinib in the treatment of early diffuse systemic sclerosis: an open-label, pilot clinical trial. Arthritis Res Ther. 2015 doi: 10.1186/s13075-015-0721-3. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Assassi S, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheum. 2015 doi: 10.1002/art.39289. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong AK, Park CY, Greene CS, Bongo LA, Guan Y, Troyanskaya OG. IMP: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 2012;40:W484–W490. doi: 10.1093/nar/gks458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assassi S, Radstake TR, Mayes MD, Martin J. Genetics of scleroderma: implications for personalized medicine? BMC Med. 2013;11:9. doi: 10.1186/1741-7015-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taroni J, et al. Genome-wide gene expression analysis of systemic sclerosis esophageal biopsies identifies disease-specific molecular subsets. Arthritis Res Ther. 2015 In press. [Google Scholar]
- 21.Chung L, et al. Molecular framework for response to imatinib mesylate in systemic sclerosis. Arthritis Rheum. 2009;60:584–591. doi: 10.1002/art.24221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sargent JL, et al. A TGFbeta-responsive gene signature is associated with a subset of diffuse scleroderma with increased disease severity. J Invest Dermatol. 2009;130:694–705. doi: 10.1038/jid.2009.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenblatt MB, et al. Interspecies comparison of human and murine scleroderma reveals IL-13 and CCL2 as disease subset-specific targets. Am J Pathol. 2012;180:1080–1094. doi: 10.1016/j.ajpath.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson ME, et al. Experimentally-derived fibroblast gene signatures identify molecular pathways associated with distinct subsets of systemic sclerosis patients in three independent cohorts. PLoS One. 2015;10:e0114017. doi: 10.1371/journal.pone.0114017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharyya S, Wei J, Tourtellotte WG, Hinchcliff M, Gottardi CG, Varga J. Fibrosis in systemic sclerosis: common and unique pathobiology. Fibrogenesis Tissue Repair. 2012;5:S18. doi: 10.1186/1755-1536-5-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farina A, et al. Epstein-Barr virus infection induces aberrant TLR activation pathway and fibroblast-myofibroblast conversion in scleroderma. J Invest Dermatol. 2014;134:954–964. doi: 10.1038/jid.2013.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Bon L, et al. Distinct evolution of TLR-mediated dendritic cell cytokine secretion in patients with limited and diffuse cutaneous systemic sclerosis. Ann Rheum Dis. 2010;69:1539–1547. doi: 10.1136/ard.2009.128207. [DOI] [PubMed] [Google Scholar]
- 28.Artlett CM, Sassi-Gaha S, Rieger JL, Boesteanu AC, Feghali-Bostwick CA, Katsikis PD. The inflammasome activating caspase 1 mediates fibrosis and myofibroblast differentiation in systemic sclerosis. Arthritis Rheum. 2011;63:3563–3574. doi: 10.1002/art.30568. [DOI] [PubMed] [Google Scholar]
- 29.Arron ST, et al. High Rhodotorula sequences in skin transcriptome of patients with diffuse systemic sclerosis. J Invest Dermatol. 2014;134:2138–2145. doi: 10.1038/jid.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christmann RB, et al. Interferon and alternative activation of monocyte/macrophages in systemic sclerosis-associated pulmonary arterial hypertension. Arthritis Rheum. 2011;63:1718–1728. doi: 10.1002/art.30318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathes AL, et al. Global chemokine expression in systemic sclerosis (SSc): CCL19 expression correlates with vascular inflammation in SSc skin. Ann Rheum Dis. 2014;73:1864–1872. doi: 10.1136/annrheumdis-2012-202814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higashi-Kuwata N, et al. Characterization of monocyte/ macrophage subsets in the skin and peripheral blood derived from patients with systemic sclerosis. Arthritis Res Ther. 2010;12:R128. doi: 10.1186/ar3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atamas SP, White B. Cytokine regulation of pulmonary fibrosis in scleroderma. Cytokine Growth Factor Rev. 2003;14:537–550. doi: 10.1016/s1359-6101(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 35.Chen ZY, et al. Immune complexes and antinuclear, antinucleolar, and anticentromere antibodies in scleroderma. J Am Acad Dermatol. 1984;11:461–467. doi: 10.1016/s0190-9622(84)70191-5. [DOI] [PubMed] [Google Scholar]
- 36.Seibold JR, Medsger TA, Jr, Winkelstein A, Kelly RH, Rodnan GP. Immune complexes in progressive systemic sclerosis (scleroderma) Arthritis Rheum. 1982;25:1167–1173. doi: 10.1002/art.1780251004. [DOI] [PubMed] [Google Scholar]
- 37.Hasegawa M, Fujimoto M, Kikuchi K, Takehara K. Elevated serum levels of interleukin 4 (IL-4), IL-10, and IL-13 in patients with systemic sclerosis. J Rheumatol. 1997;24:328–332. [PubMed] [Google Scholar]
- 38.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. Faseb J. 2004;18:816–827. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
- 39.Varga J, Pasche B. Transforming growth factor beta as a therapeutic target in systemic sclerosis. Nat Rev Rheumatol. 2009;5:200–206. doi: 10.1038/nrrheum.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 41.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol. 2011;29:163–183. doi: 10.1146/annurev-immunol-031210-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Assassi S, et al. Systemic sclerosis and lupus: points in an interferon-mediated continuum. Arthritis Rheum. 2010;62:589–598. doi: 10.1002/art.27224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eloranta ML, et al. Type I interferon system activation and association with disease manifestations in systemic sclerosis. Ann Rheum Dis. 2010;69:1396–1402. doi: 10.1136/ard.2009.121400. [DOI] [PubMed] [Google Scholar]
- 45.Kim D, et al. Induction of interferon-alpha by scleroderma sera containing autoantibodies to topoisomerase I: association of higher interferon-alpha activity with lung fibrosis. Arthritis Rheum. 2008;58:2163–2173. doi: 10.1002/art.23486. [DOI] [PubMed] [Google Scholar]
- 46.van Bon L, et al. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. NEJM. 2014;370:433–443. doi: 10.1056/NEJMoa1114576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luzina IG, Atamas SP, Wise R, Wigley FM, Xiao HQ, White B. Gene expression in bronchoalveolar lavage cells from scleroderma patients. Am J Respir Cell Mol Biol. 2002;26:549–557. doi: 10.1165/ajrcmb.26.5.4683. [DOI] [PubMed] [Google Scholar]
- 48.Falanga V, Gerhardt CO, Dasch JR, Takehara K, Ksander GA. Skin distribution and differential expression of transforming growth factor β1 and β2. J Dermatol Sci. 1992;3:131–136. doi: 10.1016/0923-1811(92)90026-8. [DOI] [PubMed] [Google Scholar]
- 49.Sfikakis PP, McCune BK, Tsokos M, Aroni K, Vayiopoulos G, Tsokos GC. Immunohistological demonstration of transforming growth factor-β isoforms in the skin of patients with systemic sclerosis. Clin Immunol Immunopathol. 1993;69:199–204. doi: 10.1006/clin.1993.1170. [DOI] [PubMed] [Google Scholar]
- 50.Higashi-Kuwata N, Makino T, Inoue Y, Takeya M, Ihn H. Alternatively activated macrophages (M2 macrophages) in the skin of patient with localized scleroderma. Exp Dermatol. 2009;18:727–729. doi: 10.1111/j.1600-0625.2008.00828.x. [DOI] [PubMed] [Google Scholar]
- 51.Pechkovsky DV, et al. Alternatively activated alveolar macrophages in pulmonary fibrosis—mediator production and intracellular signal transduction. Clin Immunol. 2010;137:89–101. doi: 10.1016/j.clim.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 52.Truchetet M-E, Brembilla NC, Montanari E, Allanore Y, Chizzolini C. Increased frequency of circulating Th22 in addition to Th17 and Th2 lymphocytes in systemic sclerosis: association with interstitial lung disease. Arth Res Ther. 2011;13:R166. doi: 10.1186/ar3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kessel A, et al. Increased CD8+ T cell apoptosis in scleroderma is associated with low levels of NF-kappa B. J Clin Immunol. 2004;24:30–36. doi: 10.1023/B:JOCI.0000018060.36183.bb. [DOI] [PubMed] [Google Scholar]
- 54.Mathian A, et al. Activated and resting regulatory T cell exhaustion concurs with high levels of interleukin-22 expression in systemic sclerosis lesions. Ann Rheum Dis. 2012;71:1227–1234. doi: 10.1136/annrheumdis-2011-200709. [DOI] [PubMed] [Google Scholar]
- 55.Murata M, et al. Clinical association of serum interleukin-17 levels in systemic sclerosis: is systemic sclerosis a Th17 disease? J Dermatol Sci. 2008;50:240–242. doi: 10.1016/j.jdermsci.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Radstake TR, et al. The pronounced Th17 profile in systemic sclerosis (SSc) together with intracellular expression of TGFβ and IFNγ distinguishes SSc phenotypes. PLoS One. 2009;4:e5903. doi: 10.1371/journal.pone.0005903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodríguez-Reyna TS, et al. Th17 peripheral cells are increased in diffuse cutaneous systemic sclerosis compared with limited illness: a cross-sectional study. Rheumatol Int. 2012;32:2653–2660. doi: 10.1007/s00296-011-2056-y. [DOI] [PubMed] [Google Scholar]
- 58.Deleuran B, Abraham DJ. Possible implication of the effector CD4+ T-cell subpopulation TH17 in the pathogenesis of systemic scleroderma. Nat Clin Pract Rheum. 2007;3:682–683. doi: 10.1038/ncprheum0618. [DOI] [PubMed] [Google Scholar]
- 59.Axtell RC, Raman C. Janus-like effects of type I interferon in autoimmune diseases. Immunol Rev. 2012;248:23–35. doi: 10.1111/j.1600-065X.2012.01131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brand S. Crohn’s disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn’s disease. Gut. 2009;58:1152–1167. doi: 10.1136/gut.2008.163667. [DOI] [PubMed] [Google Scholar]
- 61.Camporeale A. IL-6, IL-17, and STAT3: a holy trinity in auto-immunity? Front Biosci. 2012;17:2306–2326. doi: 10.2741/4054. [DOI] [PubMed] [Google Scholar]
- 62.Kimura A, Kishimoto T. Th17 cells in inflammation. Int Immunopharmacol. 2011;11:319–322. doi: 10.1016/j.intimp.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 63.Zheng SG. Regulatory T cells versus Th17: differentiation of Th17 versus Treg, are they mutually exclusive? IL-17, IL-22 and their producing cells: role in inflammation and autoimmunity: springer. 2013:91–107. [Google Scholar]
- 64.Steinman L. A brief history of TH17, the first major revision in the TH1/TH2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- 65.Marangoni RG, et al. Myofibroblasts in murine cutaneous fibrosis originate from adiponectin-positive intradermal progenitors. Arthritis Rheumatol. 2015;67:1062–1073. doi: 10.1002/art.38990. [DOI] [PMC free article] [PubMed] [Google Scholar]