Abstract
Purpose of review
The goal of this review is to summarize recent advances into the pathogenesis and treatment of systemic sclerosis (SSc) from genomic and proteomic studies.
Recent findings
Intrinsic gene expression-driven molecular subtypes of SSc are reproducible across three independent datasets. These subsets are a consistent feature of SSc and are found in multiple end-target tissues, such as skin and esophagus. Intrinsic subsets as well as baseline levels of molecular target pathways are potentially predictive of clinical response to specific therapeutics, based on three recent clinical trials. A gene expression-based biomarker of modified Rodnan skin score (MRSS), a measure of SSc skin severity, can be used as a surrogate outcome metric and has been validated in a recent trial. Proteome analyses have identified novel biomarkers of SSc that correlate with SSc clinical phenotypes.
Summary
Integrating intrinsic gene expression subset data, baseline molecular pathway information, and serum biomarkers along with surrogate measures of MRSS provides molecular context in SSc clinical trials. With validation, these approaches could be used to match patients with the therapies from which they are most likely to benefit and thus increase the likelihood of clinical improvement.
Keywords: Systemic sclerosis, intrinsic subset, pathway signatures, personalized medicine
INTRODUCTION
Systemic sclerosis (SSc; scleroderma) is a rare systemic autoimmune disease with no known cause and no FDA-approved treatments. Clinical trials for SSc have typically selected patients based upon early active disease, patients that are in diffuse subset, or based on internal organ involvement such as pulmonary fibrosis (PF) or pulmonary arterial hypertension (PAH). Trials have typically not quantified molecular heterogeneity. Recently published studies incorporate molecular data into clinical trials for SSc.
In this review, we will discuss insights into pathogenesis and heterogeneity of SSc using data from the recently published genome-wide gene expression studies of SSc. We will also briefly cover advances in proteomic studies in SSc.
Analyzing SSc heterogeneity in skin: intrinsic subsets and clinical phenotypes
The intrinsic subsets of SSc are gene expression-driven molecular subtypes of disease that partially reproduce and further expand upon the clinical phenotypes of diffuse and limited cutaneous SSc (dcSSc, lcSSc) [1,2]. SSc patients can be assigned to one of the four intrinsic subsets based upon the increased expression of genes associated with distinct biological processes and specific signaling pathways. The fibroproliferative subset is associated with cell cycle and cell growth; the inflammatory subset is enriched in immune response and defense response; and the normal-like subset is characterized by increased gene expression associated with fatty acid metabolism and a lack of inflammatory or proliferative signatures. The limited intrinsic subset tends to be composed of patients with limited skin involvement, but has a variable set of functional annotations suggesting that this group of patients displays significant heterogeneity that has not been comprehensively characterized.
Recently, several studies have attempted to gain additional insights into the relationship between different SSc subsets by using novel computational approaches and capitalizing on publicly available gene expression datasets.
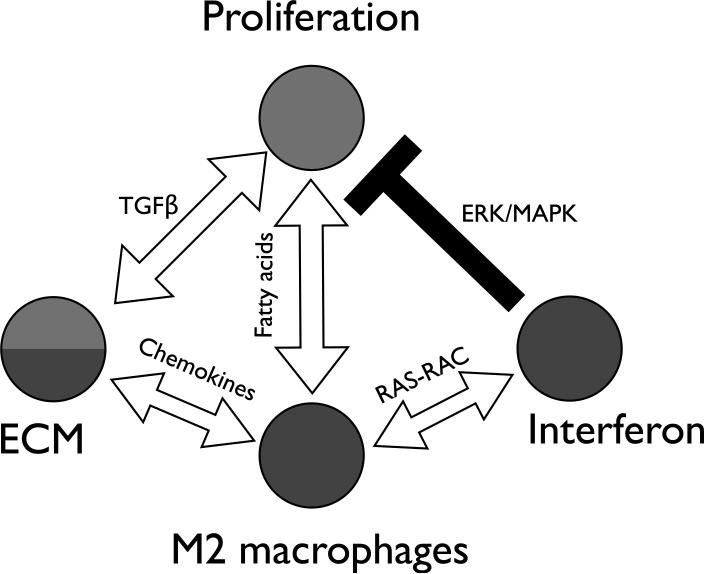
Mahoney et al. defined the genes and molecular processes that are reproducible across different SSc datasets analyzing gene expression in skin [3*]. The authors developed a novel procedure based on the concept of mutual information of partitions and applied it to the analysis of three independent SSc gene expression datasets [1,2,4]. This approach identified several gene expression modules, or ‘consensus clusters’, that are conserved and associated with the intrinsic subsets. Network analysis of the consensus clusters corresponding to the fibroproliferative and inflammatory intrinsic subsets with Integrative Multi-species Prediction (IMP) [5] showed five distinct and interacting subnetworks: adaptive immunity, interferon, cell proliferation, M2 macrophage and extracellular matrix (ECM). Genes linked to known SSc polymorphisms were found to interact primarily within the inflammatory subnetwork (e.g. IRF5, IRF7, IRF8 and NOTCH4) suggesting that an aberrant inflammatory response is an initiating point of the disease. Genes known to be important in SSc pathogenesis were either subnetwork hubs (e.g. IFI44 for the interferon subnetwork) or connected subnetworks (e.g. PLAUR connecting ECM, M2 macrophage and adaptive immunity subnetworks). These data suggested that the initiating inflammatory response is linked to the fibroproliferative response via the ECM subnetwork and transforming growth factor beta (TGF-β) signaling (Figure 1).
Figure 1. Relationships between the SSc molecular networks in skin.
Shown is a summary of core, conserved molecular processes in SSc skin. This includes an inflammatory subnetwork that contains both interferon and M2 macrophage modules that interact via Ras-Rac signaling. ECM module appears to represent a transition state between inflammatory and fibroproliferative subnetworks and is connected to M2 macrophages via chemokine signaling and to proliferation module via TGF-β signaling. The proliferation module is connected to the M2 macrophage module via fatty acid signaling and to the interferon module via negative regulation by ERK/MAPK pathway. Adapted with permission from Figure 7 in [3*]. ERK – extracellular-signal-regulated kinases, MAPK – mitogen-activated protein kinases.
Johnson et al. [6*] analyzed SSc-specific signaling pathways in dermal fibroblasts and their enrichment in each intrinsic subset. The fibroproliferative subset showed the strongest correlation with the platelet-derived growth factor (PDGF) pathway, consistent with its association with cell cycle and proliferation. The inflammatory subset showed significant correlations with several pathway signatures including IL4, tumor necrosis factor alpha, and nuclear factor kappa B signaling. TGF-β signaling showed enrichment in both inflammatory and fibroproliferative patients, paralleling the results of [3] and suggesting that TGF-β signaling may span fibroproliferative and inflammatory subsets. Activation of TGF-β signaling was associated with more severe disease confirming prior results of Sargent et al. [7]. Normal-like and limited intrinsic subsets were primarily characterized by lack of specific pathway signatures. A weak association between the inflammatory subset and interferon alpha (IFNα) signaling was observed and IFNα signaling was associated with early disease suggesting it is an early event in pathogenesis. These data suggest that an early inflammatory response characterized by IFNα signaling and innate immune cells, leads to TGF-β signaling and transition from inflammatory to fibroproliferative subsets, which ultimately results in activation of PDGF signaling (recently reviewed in [8]).
Assassi et al. [9**] recently analyzed SSc gene expression heterogeneity in an independent cohort using a separate technology platform. Analysis of the differentially expressed genes between SSc patients and healthy controls showed two groups of patients with signatures of increased keratin expression and separate inflammatory signature (termed fibro-inflammatory in this study) enriched in both inflammatory and fibrotic pathways. The inflammatory signature showed significant overlap with inflammatory genes from the intrinsic subset of Milano et al. [1]. A subset of patients showed the normal-like gene expression signature [1]; these patients had the longest disease duration suggesting that patients transition to normal-like when their disease reaches an inactive state. Analysis of cell type-specific gene signatures showed macrophage and fibroblast cell types had the highest correlation with the fibro-inflammatory signature identified in the study. In general, the inflammatory profile of SSc patients was very heterogeneous as the majority of patients showed increased dendritic and macrophage signatures and a few SSc patients had the increased expression of natural killer cell, CD4 and CD8 T-cell signatures.
Apart from the question of inter-dataset reproducibility and functional relationship of intrinsic subsets, it was not known whether subsets were a skin-specific feature of SSc or if they could be found in other organs and tissues. Recent data have suggested that intrinsic gene expression subsets are, in fact, a common feature of end-target tissues. Taroni et al. [10*] analyzed the gene expression data from esophageal biopsies of SSc patients. The authors identified three stable groups of esophageal samples, two of which showed significant enrichment in functional terms associated with immune response and proliferation. These esophageal molecular subsets were functionally equivalent to the inflammatory and fibroproliferative intrinsic subsets from skin. These results suggest that the intrinsic subsets likely represent common processes of SSc pathogenesis across multiple organs and tissues. Of course, a broader and more formal test of this hypothesis, involving the analysis of many different tissues, is required to support this hypothesis.
Patient stratification in drug trials and clinical response biomarkers
Several recent studies have examined the relationship between the intrinsic subset of a patient at baseline and a clinical response to a specific therapeutic (Table 1). Clinical response in these cases was defined as improvement in modified Rodnan skin score (MRSS) between pre- and post-treatment time points.
Table 1.
Summary of SSc clinical trials discussed in this review.
| Trials analyzed | Improvers | Non-improvers | Reference |
|---|---|---|---|
| Imatinib mesylate | ND, but response signature overlapped with fibroproliferative subset | NA | Chung et al. 2009 |
| Mycophenolate mofetil (MMF) | Inflammatory | Fibroproliferative | Hinchcliff et al. 2013 |
| Abatacept | Inflammatory; patients with high baseline levels of CD28 signaling | Normal-like; patient with low baseline levels of CD28 signaling | Chakravarty et al. 2015 |
| Nilotinib | Intrinsic subset was variable; patients with high baseline levels of TGFBR/PDGFRB signaling | Fibroproliferative; patients with low baseline levels of TGFBR/PDGFRB signaling | Gordon et al. 2015 |
For each trial, information about improvers and non-improvers in terms of intrinsic subsets and relevant targeted pathways is provided (where applicable). NA – not available, ND – not determined.
Chung et al. [11] reported two patients with dcSSc that improved in response to treatment with imatinib mesylate (Gleevec™; Novartis International AG, Basel, Switzerland), a tyrosine kinase inhibitor (TKI) that specifically targets Abelson tyrosine-protein kinase 1, KIT and PDGF receptor (PDGFR). The genes that changed in response to imatinib were those typically found with high expression in the fibroproliferative SSc samples, although the intrinsic subset of the patients who improved was not determined in this study.
More recently, Hinchcliff et al. [4] analyzed data from an investigator-initiated trial of mycophenolate mofetil (MMF, CellCept™; Hoffmann-La Roche, Basel, Switzerland) in dcSSc. MMF is an immunosuppressant that inhibits B and T lymphocyte proliferation. The patients that improved while taking MMF were primarily classified as inflammatory whereas none of the fibroproliferative patients improved.
Several recent studies have confirmed that the intrinsic gene expression subset assignment of a patient, or the activation state of the targeted molecular pathway at baseline, may predict improvement to a given therapy. Chakravarty et al. [12*] analyzed clinical outcomes and gene expression changes in a placebo-controlled study of abatacept (Orencia™; Bristol-Myers Squibb Company, New York City, New York, United States), an immunomodulator that inhibits T cell CD28 costimulation by preferentially binding to CD80/86 receptors. Most patients treated with abatacept showed significant improvement in MRSS after 6 months of therapy. Four out of five improvers were assigned to the inflammatory intrinsic subset whereas the single non-improver was classified as normal-like. Abatacept improvers showed a significant decrease in immune response and cytokine signaling pathways post-treatment. Specifically, the expression levels of genes associated with CD28 signaling were significantly higher in abatacept improvers than in the non-improver at baseline, consistent with the abatacept molecular mechanism of action, suggesting that only patients with baseline activation of the pathway improve. Improvers showed significant downregulation of CD28-dependent costimulatory signaling, while this pathway was unchanged in non-improver and placebo-treated patients.
Nilotinib (Tasigna™; Novartis International AG, Basel, Switzerland) is a TKI with a kinase inhibition profile similar to imatinib. Studies in bleomycin-induced skin fibrosis and Tsk1/+ mice have suggested TKIs can inhibit fibrosis if the relevant targets display high baseline activation levels [13]. Gordon et al. [14*] evaluated nilotinib safety and efficacy in an open-label investigator-initiated trial in SSc. Based on improvement in MRSS at 12 months compared to baseline, four out of six patients were classified as improvers. Improvers were characterized by high expression of TGF-β receptor (TGFBR) and PDGFR-β (PDGFRB) signaling pathways at baseline that significantly decreased post-treatment but were unchanged in non-improvers. Improvers had variable intrinsic gene expression subset assignments but trended toward inflammatory, whereas both non-improvers were from the fibroproliferative intrinsic subset. These data are consistent with the TGF-β pathway spanning fibroproliferative and inflammatory intrinsic gene expression subsets.
Incorporating intrinsic gene subset or molecular pathway gene expression information entails generation of signatures comprising hundreds or dozens of genes, respectively. An argument can be made that a more tractable gene signature, or biomarker, consisting of a small number of genes might be particularly useful in the clinical trial setting.
Pharmacodynamic biomarkers
MRSS is the most commonly used outcome measure in SSc clinical trials, but is potentially confounded by inter-observer variability [15]. There has been significant interest in developing quantitative surrogate markers for MRSS that would be objective and possibly report changes across shorter time scales. Prior work identified a skin score biomarker comprising four genes (COMP, IFI44, SIGLEC1 and THBS1) changes in which highly correlated with changes in MRSS [16]. Given that two genes from this biomarker (COMP and THBS1) are highly regulated by TGF-β, the change in their levels was used as the primary efficacy measure in the study of fresolimumab (GC1008; Genzyme Corporation, Cambridge, Massachusetts, United States), immunomodulator and a monoclonal antibody inhibiting all isoforms of TGF-β [17**]. Both COMP and THBS1 decreased after fresolimumab treatment. This decrease in the biomarker genes paralleled a significant decrease in MRSS and rapid improvement of skin disease, suggesting that targeting TGF-β may result in significant improvement in SSc patients.
In a follow-up paper [18*] the authors refined their approach towards defining a skin score biomarker using NanoString assays and created two different biomarkers. The 2-gene SSc skin biomarker was defined mathematically using a generalized estimating equation and comprised THBS1 and MS4A4A. The weighted model or weighted SSc skin biomarker included all genes that had a significant longitudinal correlation with MRSS and consisted of THBS1, CTGF, CCL2, CD163, MS4A4A and WIF. Both biomarkers strongly correlated with MRSS and moderately correlated with fibrosis and inflammation.
Serum biomarkers
Serum biomarkers have been difficult to identify. van Bon et al. [19**] isolated plasmacytoid dendritic cells (pDCs) which are implicated in SSc pathogenesis from patients with SSc and healthy controls and performed proteome-wide analysis of the proteins secreted by these cells using mass spectrometry. They validated their observations in five SSc patient cohorts and compared these data to patients with three other autoimmune conditions. Chemokine (C-X-C motif) ligand 4 (CXCL4) was the main protein secreted by SSc pDCs, with its levels significantly higher in SSc compared to healthy controls and patients with other autoimmune diseases. The levels of CXCL4 correlated with skin and lung fibrosis as well as PAH. According to the authors, CXCL4 was the only cytokine that predicted risk and progression of SSc.
In the next study, van Bon et al. [20*] performed a proteomic analysis of SSc patients, with dSSc and lcSSc, healthy controls as well as an additional SSc replication cohort. One peptide with differential expression between patients with SSc and healthy controls corresponded to S100A8/A9, a Toll-like receptor agonist. This result was reproduced in a replication cohort. The highest levels of S100A8/A9 were found in patients with lcSSc that had lung fibrosis.
Lambrecht et al. [21] used an enzyme-linked immunosorbent assay to analyze a cohort of SSc patients for serum levels of growth differentiation factor 15 (GDF15), a distant member of TGF-β family. Increased levels of GDF15 were observed in groups of patients with both lcSSc and dcSSc and its baseline levels correlated strongly with SSc disease activity and symptoms of lung fibrosis. Similar picture was observed in the bleomycin-induced fibrosis mouse model that showed an increase in GDF15 levels in lung tissue.
Another potential biomarker of SSc fibrosis that has been recently investigated is lysyl oxidase (LOX), the enzyme that is involved in crosslinking ECM proteins. Rimar et al. [22] evaluated serum LOX levels in SSc patients, healthy controls and disease control patients. Patients with SSc had significantly higher LOX levels than healthy controls and they were significantly higher in dcSSc compared to lcSSc group. LOX levels showed correlation with MRSS in SSc patients without lung fibrosis.
Brkic et al. [23] investigated IFN type I gene signature in whole blood samples from several groups of patients with SSc and healthy controls. All SSc groups had significantly higher IFN signature scores compared with healthy controls. The presence of IFN type I signature in monocytes highly correlated with the expression levels of B cell activating factor (BAFF) and high collagen synthesis, suggesting a connection with SSc pathogenesis.
CONCLUSION
Recent studies have resulted in important insights into the pathogenesis of SSc and identification of many potential biomarker signatures. Patients can be classified based on their intrinsic gene expression subsets that may indicate specific treatment options. Baseline expression levels of pathways that drive SSc or are expected to be modulated by therapies provide complementary data and can be predictive of the treatment response. TGF-β signaling remains central to SSc pathogenesis, and in a recent study its therapeutic targeting resulted in marked patient improvement. Surrogate biomarkers of clinical endpoints provide objective assessment of response in clinical trials. These approaches can complement standard clinical measures and patient-reported outcomes. This new information could be used to rationally recruit, target and treat SSc patients in clinical trials.
KEY POINTS.
Insights into the SSc pathogenesis can be gained by analyzing gene expression and proteomic data from patients with SSc and other autoimmune diseases.
We have gained a better understanding of the SSc intrinsic subsets, the importance of baseline levels of druggable pathways and validation of a surrogate biomarker for MRSS.
Discovery of serum biomarkers that correlate with SSc clinical phenotypes provides promise for identifying circulating factors for disease severity.
Incorporating these data into SSc clinical trials could result in personalized treatments for SSc and other rare diseases.
Acknowledgements
The authors would like to thank Jaclyn N. Taroni for critical reading of the manuscript.
Financial support and sponsorship
VM and MLW have been supported by the Scleroderma Research Foundation, Scleroderma Foundation, by the NIH (P50AR060780, P30AR061271, R21AR068035, R44AR061920) and by the Dr. Ralph and Marian Falk Medical Research Trust.
Footnotes
Conflicts of interest
MLW has filed patents for gene expression biomarkers in systemic sclerosis and is a Scientific Founder of Celdara Medical LLC.
The drug products mentioned in this manuscript are not labeled for use in systemic sclerosis and are still under investigation.
References
- 1.Milano A, Pendergrass SA, Sargent JL, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One. 2008;3:e2696. doi: 10.1371/journal.pone.0002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pendergrass SA, Lemaire R, Francis IP, et al. Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol. 2012;132:1363–1373. doi: 10.1038/jid.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Mahoney JM, Taroni J, Martyanov V, et al. Systems level analysis of systemic sclerosis shows a network of immune and profibrotic pathways connected with genetic polymorphisms. PLoS Comput Biol. 2015;11:e1004005. doi: 10.1371/journal.pcbi.1004005. [Reports a novel computational approach to identify genes and molecular processes that are conserved across the intrinsic subsets from three independent datasets and patient cohorts. It shows the subsets are likely to be mechanistically connected and the inflammatory modules are enriched for SSc susceptibility genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinchcliff M, Huang CC, Wood TA, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol. 2013;133:1979–1989. doi: 10.1038/jid.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong AK, Park CY, Greene CS, et al. IMP: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 2012;40:W484–490. doi: 10.1093/nar/gks458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Johnson ME, Mahoney JM, Taroni J, et al. Experimentally-derived fibroblast gene signatures identify molecular pathways associated with distinct subsets of systemic sclerosis patients in three independent cohorts. PLoS One. 2015;10:e0114017. doi: 10.1371/journal.pone.0114017. [Characterizes experimentally derived pathway signatures generated in dermal fibroblasts across three independent gene expression datasets probing genome-wide gene expression in SSc skin biopsies. Results suggest that IFNα signaling is an early event and confirm that activation of TGF-β signaling is associated with increased disease severity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sargent JL, Milano A, Bhattacharyya S, et al. A TGFbeta-responsive gene signature is associated with a subset of diffuse scleroderma with increased disease severity. J Invest Dermatol. 2010;130:694–705. doi: 10.1038/jid.2009.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson ME, Pioli PA, Whitfield ML. Gene expression profiling offers insights into the role of innate immune signaling in SSc. Semin Immunopathol. 2015;37:501–509. doi: 10.1007/s00281-015-0512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **9.Assassi S, Swindell WR, Wu M, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheumatol. 2015 doi: 10.1002/art.39289. ‘Accepted Article’, doi: 10.1002/art.39289. [The authors reproduce the inflammatory and normal-like subgroups observed in other studies using an independent patient cohort, different platform and a different analysis procedure. This study provides a clear association of the normal-like subset with the longest disease duration. The inflammatory group was associated with higher MRSS.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Taroni JN, Martyanov V, Huang CC, et al. Molecular characterization of systemic sclerosis esophageal pathology identifies inflammatory and proliferative signatures. Arthritis Res Ther. 2015;17:194. doi: 10.1186/s13075-015-0695-1. [First report of gene expression analyses of esophageal biopsies in SSc patients. Shows that the intrinsic subsets can be found in an end target-tissue other than skin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung L, Fiorentino DF, Benbarak MJ, et al. Molecular framework for response to imatinib mesylate in systemic sclerosis. Arthritis Rheumatol. 2009;60:584–591. doi: 10.1002/art.24221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Chakravarty EF, Martyanov V, Fiorentino D, et al. Gene expression changes reflect clinical response in a placebo-controlled randomized trial of abatacept in patients with diffuse cutaneous systemic sclerosis. Arthritis Res Ther. 2015;17:159. doi: 10.1186/s13075-015-0669-3. [Shows that patients with SSc who improved in response to abatacept were inflammatory at baseline and had high baseline expression levels of CD28 signaling pathway, targeted by this therapy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurer B, Distler A, Dees C, et al. Levels of target activation predict antifibrotic responses to tyrosine kinase inhibitors. Ann Rheum Dis. 2013;72:2039–2046. doi: 10.1136/annrheumdis-2013-203729. [DOI] [PubMed] [Google Scholar]
- 14*.Gordon JK, Martyanov V, Magro C, et al. Nilotinib (Tasigna™) in the treatment of early diffuse systemic sclerosis: an open-label, pilot clinical trial. Arthritis Res Ther. 2015;17:213. doi: 10.1186/s13075-015-0721-3. [Demonstrates that patients with SSc who responded to nilotinib had high baseline levels of TGFBR and PDGFRB signaling pathways, targeted by this drug, whereas patients who did not improve had low baseline levels of these pathways.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pope JE, Baron M, Bellamy N, et al. Variability of skin scores and clinical measurements in scleroderma. J Rheumatol. 1995;22:1271–1276. [PubMed] [Google Scholar]
- 16.Farina G, Lafyatis D, Lemaire R, Lafyatis R. A four-gene biomarker predicts skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol. 2010;62:580–588. doi: 10.1002/art.27220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Rice LM, Padilla CM, McLaughlin SR, et al. Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients. J Clin Invest. 2015;125:2795–2807. doi: 10.1172/JCI77958. [Clinical trial of an anti-TGF-β inhibitor that shows a rapid improvement in patients’ MRSS. Reports use of a surrogate skin score biomarker in the SSc clinical trial. The expression levels of genes comprising skin score biomarker decreased paralleling improvement in MRSS post-treatment.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Rice LM, Ziemek J, Stratton EA, et al. A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol. 2015 doi: 10.1002/art.39287. ‘Accepted Article’, doi: 10.1002/art.39287. [The authors used two different approaches to generate longitudinal gene biomarkers of skin disease in SSc. Both biomarkers showed high correlation with MRSS and moderate correlation with skin fibrosis and inflammation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.van Bon L, Affandi AJ, Broen J, et al. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. N Engl J Med. 2014;370:433–443. doi: 10.1056/NEJMoa1114576. [Proteomic analysis of serum proteins in SSc. Showed that levels of cytokine CXCL4 differentiated between patients with SSc, other autoimmune conditions and healthy controls. CXCL4 correlated with skin and lung fibrosis, PAH and predicted SSc risk and progression.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.van Bon L, Cossu M, Loof A, et al. Proteomic analysis of plasma identifies the Toll- like receptor agonists S100A8/A9 as a novel possible marker for systemic sclerosis phenotype. Ann Rheum Dis. 2014;73:1585–1589. doi: 10.1136/annrheumdis-2013-205013. [S100A8/A9, a TLR agonist, was differentially expressed between patients with SSc and healthy controls and was enriched in limited SSc patients with lung fibrosis.] [DOI] [PubMed] [Google Scholar]
- 21.Lambrecht S, Smith V, De Wilde K, et al. Growth differentiation factor 15, a marker of lung involvement in systemic sclerosis, is involved in fibrosis development but is not indispensable for fibrosis development. Arthritis Rheumatol. 2014;66:418–427. doi: 10.1002/art.38241. [DOI] [PubMed] [Google Scholar]
- 22.Rimar D, Rosner I, Nov Y, Slobodin G, Rozenbaum M, Halasz K, Haj T, Jiries N, Kaly L, Boulman N, Daood R, Vadasz Z. Lysyl oxidase is a potential biomarker of fibrosis in systemic sclerosis. Arthritis Rheumatol. 2014;66:726–730. doi: 10.1002/art.38277. [DOI] [PubMed] [Google Scholar]
- 23.Brkic Z, van Bon L, Cossu M, et al. The interferon type I signature is present in systemic sclerosis before overt fibrosis and might contribute to its pathogenesis through high BAFF gene expression and high collagen synthesis. Ann Rheum Dis. 2015 doi: 10.1136/annrheumdis-2015-207392. doi:10.1136/annrheumdis-2015-207392. [DOI] [PubMed] [Google Scholar]