Abstract
The genetic regulatory network controlling the innate immune system is well understood in many species. However, the role of the epigenetic mechanisms underlying the expression of immunoregulatory genes is less clear, especially in livestock species. Histone H3 lysine 9 dimethylation (H3K9me2) is an epigenetic modification associated with transcriptional silencing within the euchromatin regions. Euchromatic histone-lysine N-methyltransferase 2 (EHMT2; also known as G9a) is a crucial enzyme responsible for regulating the dynamics of this epigenetic modification. It has been shown that histone modifications play a role in regulating type I interferon (IFN) response. In the present study, we investigated the role of EHMT2 in the epigenetic regulation of bovine antiviral innate immunity and explored its therapeutic potential against viral infections. We evaluated the effects of pharmacological and RNAi-mediated inhibition of EHMT2 on the transcription of IFN-β and other IFN-inducible antiviral genes, as well as its effect on foot-and-mouth disease virus (FMDV) and vesicular stomatitis virus (VSV) replication in bovine cells. We show that treatment of primary bovine cells with the synthetic EHMT2 inhibitor (UNC0638) either before or shortly after virus infection resulted in a significant increase in transcript levels of bovine IFN-β (boIFN-β; 300-fold) and other IFN-inducible genes, including IFN-stimulated gene 15 (ISG-15), myxovirus resistance 1 (Mx-1), Mx-2, RIG-I, 2′,5′-oligoadenylate synthetase 1 (OAS-1), and protein kinase R (PKR). Expression of these factors correlated with a significant decrease in VSV and FMDV viral titers. Our data confirm the involvement of EHMT2 in the epigenetic regulation of boIFN-β and demonstrate the activation of a general antiviral state after EHMT2 inhibition.
Introduction
Host–pathogen interaction initiates a concerted signal transduction pathway that leads to the assembly of the transcriptional machinery on the appropriate response genes to induce the expression of several molecules involved in innate immunity (Doly and others 1998). The regulation of gene expression is governed by epigenetic programs mediated by several enzymes (Fernandez-Morera and others 2010). Modulating the activity of these epigenetic enzymes can therefore influence the innate immune response to pathogens.
Epigenetic control of gene expression is a multifaceted and complex process, including DNA methylation and histone modifications and increasing evidence suggests that epigenetic mechanisms are involved in health and disease stages of the host. Posttranslational histone modification (methylation/acetylation) plays an important role in the regulation of gene transcription and chromatin remodeling. Histone methylation can activate or repress transcription depending on the specific residues and the incorporated modifications (Tachibana and others 2002). Histone H3 lysine 9 dimethylation (H3K9me2) is an epigenetic modification mainly associated with transcriptional silencing within the euchromatin regions (Rice and others 2003).
Three histone methyltransferases, euchromatic histone-lysine N-methyltransferase 2 (EHMT2), G9a-related protein (GLP, also known as EuHMTase1 in human), and histone-lysine N-methyltransferase SETDB1 (ESET/SetDB1), are known to catalyze the formation of H3K9me2 (Schultz and others 2002; Tachibana and others 2002; Wang and others 2003). EHMT2 has been proposed as the key enzyme responsible for establishing and maintaining H3K9me2-mediated gene silencing and thus contributing to specific epigenetic modifications (Rice and others 2003). A number of recent scientific reports have suggested a close association of EHMT2 with the development of several human diseases like leukemia, prostate carcinoma, hepatocellular carcinoma, malaria, cocaine addiction, and mental retardation (Kondo and others 2008; Schaefer and others 2009; Maze and others 2010; Malmquist and others 2012). EHMT2 is also involved in the maintenance of HIV-I latency (Imai and others 2010). This wide range of disease-related activity of EHMT2 warrants further studies that can unravel the underlying mechanisms and therapeutic potential of this protein.
The discovery of small-molecule histone methyltransferase inhibitors like BIX01294 and UNC0638 (EHMT2 and GLP inhibitors) represent a critical tool for investigating these proteins and assessing their potential as therapeutic targets (Kubicek and others 2007; Vedadi and others 2011). The EHMT2 inhibitor UNC0638 is very well characterized in terms of specificity and safety. It has been used to inhibit the EHMT2 methyltransferase function in several mouse and human cell types (Vedadi and others 2011). The crystal structure of the EHMT2 methyltransferase in complex with this inhibitor has also been resolved (PDB code: 3RJW), validating the specificity of the drug (UNC0638) in human cells (Vedadi and others 2011).
Virus infection in cells activates several transcription factors to induce the expression of several molecules involved in innate immunity, including type I interferons (IFNs), which are considered the first line of defense against virus invasion. These cytokines have numerous biological activities and immunomodulatory effects (Doly and others 1998). Once expressed, IFNs bind to the IFN-α/β receptor in an autocrine and paracrine manner and activate transcription of several IFN-stimulated genes (ISGs) such as 2′,5′-oligoadenylate synthetase 1 (OAS-1), protein kinase R (PKR), IFN-stimulated gene 15 (ISG-15), and myxovirus resistance 1 (Mx-1) among many others, creating an antiviral state in the host. Therefore, induction of type I IFN is a very powerful strategy of the host to fight virus infections.
The IFN-β promoter is under negative control and the gene remains silent in differentiated cells unless activated by some exogenous signals like virus infection (Doly and others 1998). Interestingly, histone modification has been shown to play a major role in the negative regulation of the IFN-β promoter (Shestakova and others 2001). In fact, the lack of H3K9me2 marks on IFN-β loci in murine plasmacytoid dendritic cells (pDC), preprograms them to produce more type I IFN (Fang and others 2012). To date, very few studies have revealed the underlying epigenetic mechanisms during the immune response to viral infection (Fernandez-Morera and others 2010) and, as such, it is largely unknown.
Vesicular stomatitis virus (VSV) and foot-and-mouth disease virus (FMDV) cause clinically indistinguishable acute vesicular disease in cloven-hoofed animals causing a huge economic impact on the livestock industry. VSV is a direct threat in the south and western regions of the United States, due to their proximity to endemic areas and the complexity of natural cycles that involve livestock, wild animals, and insect vectors (Rieder and Conzelmann 2009). FMDV is considered a major global animal health problem and the World Organization for Animal Health and the United Nations' Food Agriculture Organization have called for its global control and eradication. Vaccination against FMDV remains the most important control measure. However, it takes several days for the vaccine to take effect, and the resulting immunity tends to be relatively short-lived unless the animal is repeatedly vaccinated (Rodriguez and Gay 2011).
Despite aggressive vaccination campaigns in the endemic areas, sporadic outbreaks of the disease are common and result in huge economic losses thus impacting millions of people dependent on livestock production (Rodriguez and Gay 2011). Therefore, rapid control strategies such as the use of novel antivirals either prophylactically and/or therapeutically assume importance. In the present study, we demonstrate the role of histone methyltransferase EHMT2 in regulation of bovine IFN-β (boIFN-β) gene expression and in the establishment of an antiviral state in primary bovine fibroblast cells. In addition, we have investigated the prophylactic and therapeutic use of EHMT2 inhibitors to control replication of 2 viruses of veterinary importance, FMDV and VSV.
Materials and Methods
Cell lines
Fetal bovine fibroblasts (FBF) were isolated from bovine fetuses and grown in Dulbecco's Modified Eagle's Medium (DMEM-F12) (Life Technologies/Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals), 0.1% fibroblast growth factor, 100 U/mL penicillin, and 10 μg/mL streptomycin (Life Technologies/Invitrogen). All experiments with primary cells were performed with culture passages 3 to 5 to prevent further differentiation. Madin-Darby bovine kidney (MDBK, ATCC No. CCL-22) cells were grown and maintained in DMEM-F12 supplemented with 10% FBS, 100 U/mL penicillin, and 10 μg/mL streptomycin and they were used for the IFN bioassay. MDBK-t2 cells [transfected with plasmid expressing the human MxA promoter linked to a chloramphenicol acetyltransferase (CAT) reporter and with resistance to blasticidin] were kindly provided by B. Charleston (Institute for Animal Health, Pirbright, UK) and used for the CAT-ELISA.
IBRS-2 cells were obtained from the Foreign Animal Disease Diagnostic Laboratory (APHIS) at Plum Island Animal Disease Center (PIADC), Greenport, NY and used for viral titrations of FMDV. These cells were maintained in a minimal essential medium (Gibco-BRL/Invitrogen) containing 10% FBS and supplemented with 100 U/mL penicillin, 10 μg/mL streptomycin (Life Technologies/Invitrogen), and nonessential amino acids (Life Technologies/Invitrogen).
Viral infections
A VSV strain serotype Indiana, kindly provided by Dr. Judith Ball (Texas A&M), was used for challenge assays. VSV serotype New Jersey (VSV NJ) was used for IFN bioassay. FMDV serotype A12 was generated from a full-length virus infectious clone as previously described (Rieder and others 1993). Cells were infected with VSV at a multiplicity of infection (MOI) of 0.01. Cells and supernatants were harvested 36 hours postinfection (hpi). FMDV viral infections were carried out at an MOI of 0.01. Virus was adsorbed in FBS-free media for 1 h followed by acid neutralization and addition of fresh media. Cells and viral supernatants were collected 41 hpi. Viral supernatants were analyzed by microtitration in MDBK or IBRS2 cells using standard methods for TCID50. The titer was calculated using the method of Reed and Muench (1938). All experimental infections using VSV (NJ) or FMDV were conducted at the USDA-ARS Plum Island Animal Disease Center under biosafety level (BSL)-3Ag conditions. Infections with VSV serotype Indiana were conducted under BSL-2 as approved by the Institutional Biosafety Committee, Texas A&M University.
EHMT2 inhibitor treatment and poly (I:C) stimulation
For the prophylactic series of experiments, FBF were seeded in 12-well tissue culture plates. The next day, 2 set of cells were exposed to 5 μM of EHMT2 inhibitor (UNC0638) obtained from Sigma-Aldrich or DMSO (vehicle) control. After 4 days of treatment, cells were transfected with or without 0.5 μg/mL of poly (I:C) (Invivogen) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cell lysates and supernatants from a subset of cells were harvested 6 h poststimulation with poly (I:C) for RNA analysis, while the other set of cells was utilized for viral challenge.
For experiments testing the therapeutic application of EHMT2 modulation, FBF cells previously seeded in 12-well plates were infected with 0.01 MOI of VSV as described above. The infected cells were treated with vehicle or 5 μM EHMT2 inhibitor at 4, 8, or 12 hpi. Cells and supernatants were harvested 36 hpi for RNA analysis by quantitative real-time polymerase chain reaction (qPCR) and viral titers (TCID50), respectively. Three independent replicates were performed with each virus and each treatment was performed in duplicate.
siRNA targeting bovine EHMT2
FBF were transfected with 50 nM of siRNA (Invitrogen) targeting bovine EHMT2, SetDB1, or a Cy3-labeled negative control (Invitrogen) using calcium phosphate technique as described previously (Donze and Picard 2002). Twenty-four hours later, the transfection efficiency was monitored by fluorescent microscopy. To confirm the depletion of target mRNA (bovine EHMT2 and SetDB1), cell lysates were harvested from half the plates 48 h posttransfection for RNA analysis by qPCR. The remaining cells were stimulated with poly (I:C) to evaluate the IFN response.
IFN bioassay
The antiviral activity induced by type I IFN secretion was tested in the supernatants of vehicle or EHMT2 inhibitor-treated primary bovine cells by a VSV infection inhibition assay on MDBK cells as previously described (Rodriguez-Pulido and others 2011). Briefly, supernatants to be tested were serially diluted and applied to MDBK cells previously seeded in 96-well plates. Twenty-four hours later, treated cells were infected with VSV NJ at a MOI of 2. Twenty-four to 48 h after infection, cells were evaluated for cytopathic effect (CPE). A recombinant boIFN-α standard (Kingfisher Biotech) was used as a positive control in the assay. The antiviral activity is expressed as the reciprocal of the highest 2-fold dilution of supernatant able to suppress VSV-induced CPE on MDBK cells in 50% of the wells assayed.
Quantitative real-time reverse transcription-qPCR
Total RNA was isolated from cells using the RNAeasy kit (Qiagen) as per manufacturer's protocol. DNAseI-treated total RNA was quantified and used to produce cDNA with the qScript kit (Quanta Biosciences) according to the manufacturer's instructions. Relative mRNA levels were determined by comparative threshold cycle (CT) analysis (Livak and Schmittgen 2001) for boIFN-β, ISG-15, RIG-I, Mx-1, Mx-2, OAS-1, PKR, EHMT2, SetDB1, and GAPDH (reference gene) by reverse transcription-qPCR using the PerfeCTa® SYBR® Green FastMix, ROX (Quanta Biosciences) on an ABI Prism 7500 thermocycler (Applied Biosystems). Relative mRNA levels were expressed as fold change over transfection control. The primers used in these studies are given in Table 1.
Table 1.
Primers for Amplifying Bovine Target Genes for Quantitative Real-Time Polymerase Chain Reaction Analysis
| Gene | Primer | Primer sequences |
|---|---|---|
| Bovine | For | 5′-CTACAGCTTGCTTCGATTCCAA-3′ |
| IFN-β | Rev | 5′-CTGCCCCAGGAGTTTCTGAC-3′ |
| Bovine | For | 5′-GACTCATTGCCCCAGGTCATT-3′ |
| RIG-I | Rev | 5′-TCGGCTGTGTTTTTGGCAT-3′ |
| Bovine | For | 5′-GTACGAGCCGAGTTCTCCAA-3′ |
| Mx-1 | Rev | 5′-ATGTCCACAGCAGGCTCTTC-3′ |
| Bovine | For | 5′-CTTCAGAGACGCCTCAGTCG-3′ |
| Mx-2 | Rev | 5′-TGAAGCAGCCAGGAATAGTG-3′ |
| Bovine | For | 5′-GCGTGTACAAGCGGACCAGT-3′ |
| ISG-15 | Rev | 5′AGCGGGTGCTCATCATCC-3′ |
| Bovine | For | 5′-CCAAAGTTGTGAAGGGTGGC-3′ |
| OAS-1 | Rev | 5′-TGATCGTCCCCTGAGGGTC-3′ |
| Bovine | For | 5′-TGCCAAACTGGCTTATGAAAAG-3′ |
| PKR | Rev | 5′-TCACCACACGCAGCACTGA-3′ |
| Bovine | For | 5′-ACCAACTGGGACGACATGGAGAAA-3′ |
| GAPDH | Rev | 5′-TGTCCTGGA AGAGAAGGCAATGGT-3′ |
| Bovine | For | 5′-TGGGAAAAATGAAAGCGAGA-3′ |
| IFN-β promoter | Rev | 5′-CAGGAGAACCATCTGGAGGA-3′ |
| Bovine | For | 5′-TCATCATCATCGGCATTGTT-3′ |
| ISG-15 promoter | Rev | 5′-TCTCTGCAGACACTGGTTGG-3′ |
Chloramphenicol acetyltransferase-ELISA
Relative quantitation of IFN levels in the supernatants of primary bovine cells was determined by a Mx/CAT reporter gene assay previously described (Fray and others 2001). Briefly, supernatants of primary bovine cells to be tested were incubated for 24 h at 37°C and 5% CO2 on MDBK-t2 cells previously seeded into 24-well tissue culture plates. Next, MDBK-t2 cells were lysed and a commercially available enzyme-linked immunosorbent assay (ELISA) kit (Roche Applied Sciences) was used for CAT detection according to the manufacturer's protocol. Units of IFN per milliliter of the samples were calculated from a standard curve using known amounts of recombinant human IFN-α2A (PBL Interferon Source).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (Golding and others 2010). Briefly, FBF cells were grown to maximum confluence in T-75 tissue culture flasks. Cells were treated with EHMT2 inhibitor or vehicle for 4 days. On day 4, cells were washed twice with phosphate-buffered saline, trypsinized, and resuspended in a growth medium containing 0.1 volume of crosslinking solution (Kondo and others 2008). The cell pellets were sonicated to generate sheared chromatin ranging from 100 to 500 bp. The sheared chromatin was immunoprecipitated with antibodies recognizing dimethylated histone 3 lysine 9 (H3K9me2), acetylated histone 3 lysine 9 (H3K9Ac), or acetylated histone 3 lysine 14 (H3K14Ac).
All antibodies were purchased from Millipore and Anti-Rabbit IgG from Santa Cruz. The retrieved DNA fragments were purified using a Qiagen PCR purification kit and subjected to qPCR to measure the relative enrichment of candidate gene promoter regions by methods previously utilized in our laboratory (Golding and others 2010). The sequences of the primers are given in Table 1. ChIP samples were normalized to 1% input and data were analyzed using a formula described previously (Mukhopadhyay and others 2008) for quantitative analysis of enrichment of histone marks on candidate gene promoter regions.
Statistical methods
Statistical analysis was performed using Student's t-test and 1-way analysis of variance followed by Tukey's multiple comparison test (GraphPad Prism 5.0; Graph Pad Software). The data are presented as mean ± SE. The differences were considered significantly different when P < 0.05.
Results
Depletion of EHMT2 enhances IFN response to poly (I:C)
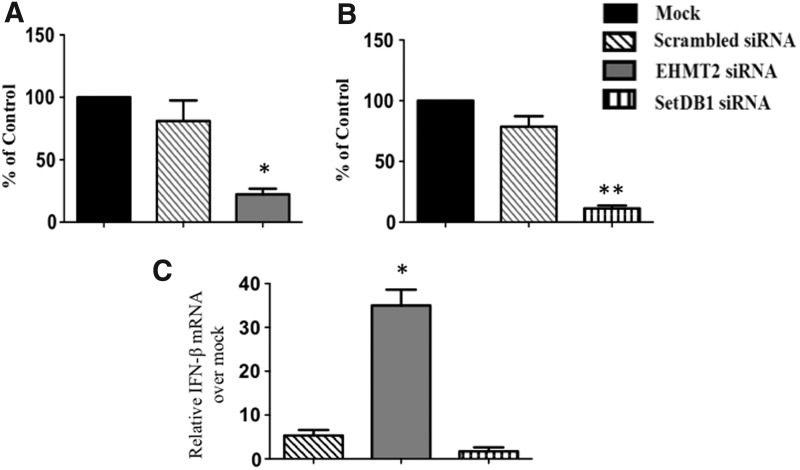
DNA methylation plays a minor, if any, role in the regulation of the IFN-β promoter (Shestakova and others 2001), however, the role of histone modifications in IFN regulation has not been fully characterized. Recently, it has been shown that the lack of H3K9me2 mark on IFN-β loci in murine pDCs preprograms them to produce type I IFN. Therefore, we hypothesized that transient inhibition of EHMT2 will enhance type I IFN responses in bovine cells. To ablate the repressive effect of EHMT2 on transcription of boIFN-β gene, we utilized RNA interference (RNAi). FBF cells were mock transfected or transfected with siRNAs (50 μM) targeting bovine EHMT2 or SetDB1 and thereafter stimulated with poly (I:C). Cells were harvested 48 h posttransfection for transcript analysis. We observed a 70%–80% reduction (P < 0.05) in bovine EHMT2 mRNA in cells treated with EHMT2 siRNA compared to mock or scrambled siRNA-transfected controls (Fig. 1A). Depletion of bovine EHMT2 mRNA resulted in a significant induction (P < 0.02) of boIFN-β transcripts (>30-fold increase) compared to a less than 10-fold increase in cells transfected with scrambled siRNA control (Fig. 1C). SiRNA-mediated knockdown of bovine SetDB1 (Fig. 1B), another histone-lysine methyltransferase that plays a central role in the silencing of euchromatic genes, failed to enhance boIFN-β upon stimulation with poly (I:C) (Figs. 1C). These results suggested that the enhancement of IFN-β expression following exogenous stimulus is a specific response to EHMT2 inhibition.
FIG. 1.
Specific depletion of euchromatic histone-lysine N-methyltransferase 2 (EHMT2) in fetal bovine fibroblast (FBF) cells potently induce type I interferon (IFN) transcripts. FBF cells were mock transfected or transfected with siRNAs targeting bovine EHMT2 or SetDB1. Cells were stimulated with 0.5 μg/mL of poly (I:C) and harvested 6 h later for quantitative real-time polymerase chain reaction (qPCR) analysis. (A) Targeting EHMT2 (B) Targeting SetDB1. (C) Bovine IFN-β (boIFN-β) levels in FBF cells stimulated with poly (I:C). mRNA levels were normalized to endogenous GAPDH and represented as fold increase compared to the control. The data represent mean ± SE from 3 independent experiments performed in duplicates. *P < 0.05, **P < 0.001 determined by 2-tailed Student's t-test.
Pharmacological inhibition of EHMT2 in bovine cells enhanced IFN production in response to poly (I:C) stimulus
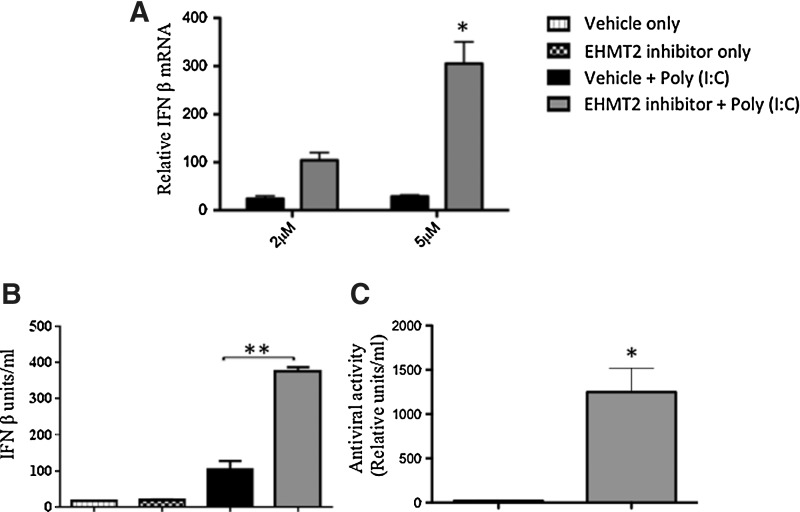
We also utilized a pharmacological approach for EHMT2 inhibition using a small molecule inhibitor of EHMT2 (UNC0638) that competes with EHMT2 for substrate binding (Vedadi and others 2011), thereby inhibiting its enzymatic activity. We first established that an optimum dose of 5 μM of EHMT2 inhibitor induced high levels of boIFN-β (>300-fold) (Fig. 2A), without causing any adverse or toxic effect in the treated cells as determined by microscopic examination of cell monolayer. FBF cells were treated with DMSO (vehicle control) or EHMT2 inhibitor for 4 days followed by poly (I:C) stimulation for 6 h.
FIG. 2.
Pharmacological inhibition of EHMT2 led to potent induction of boIFN-β in FBF cells. FBF cells were treated with the vehicle or EHMT2 inhibitor at 2 and 5 μM for 4 days. Four days after treatment, cells were stimulated with 0.5 μg/mL poly (I:C) and 6 h later, cells and supernatants were harvested for qPCR and chloramphenicol acetyltransferase (CAT)-ELISA (A) EHMT2 inhibitor dose–response on boIFN-β mRNA levels. (B) Quantitation of IFN levels in the supernatants by an Mx/CAT reporter gene assay. Units of boIFN-β per milliliter of the samples were calculated from a standard curve using recombinant human IFN-α2A (PBL Interferon Source). (C) Relative comparison in the antiviral activity in the supernatants as determined by IFN bioassay. The data represent mean ± SE from at least 2 independent experiments. *P < 0.05, **P < 0.001 determined by 2-tailed Student's t-test.
Poly (I:C) treatment of cells exposed to EHMT2 inhibitor led to an increase (P < 0.03) in the transcript levels of boIFN-β (>300-fold), relative to the vehicle control, which displayed an expected poly (I:C)-induced increase of ∼50 fold (Fig. 2A). Relative quantification of IFN levels in the supernatants of FBF cells was conducted by an Mx/CAT reporter gene assay. Poly (I:C) enhanced the boIFN-β levels (∼400 U/mL) in EHMT2 inhibitor-treated cells compared to the vehicle control (∼100 U/mL; Fig. 2B; P < 0.001). To assess the functionality of boIFN-β secreted in the supernatants of stimulated FBF cells, we utilized an IFN bioassay on MDBK cells infected with VSV. Pharmacological inhibition of EHMT2 in the presence of poly (I:C) induced ∼1,200 antiviral activity units compared to the vehicle control (18 antiviral activity units) (Fig. 2C)
Inhibition of EHMT2 establishes an antiviral state in bovine cells
IFNs bind to IFN-α/β receptor on infected and noninfected neighboring cells to activate the JAK–STAT signal transduction pathway leading to the transcription of several other IFN-stimulated antiviral genes (ISGs), and the combinatorial action of numerous ISGs is a hallmark of an effective IFN-induced antiviral response (Doly and others 1998). We investigated whether the increase in IFN production in bovine cells, seen as a result of EHMT2 inhibition along with external stimulus [poly (I:C)], can establish an antiviral state in these cells.
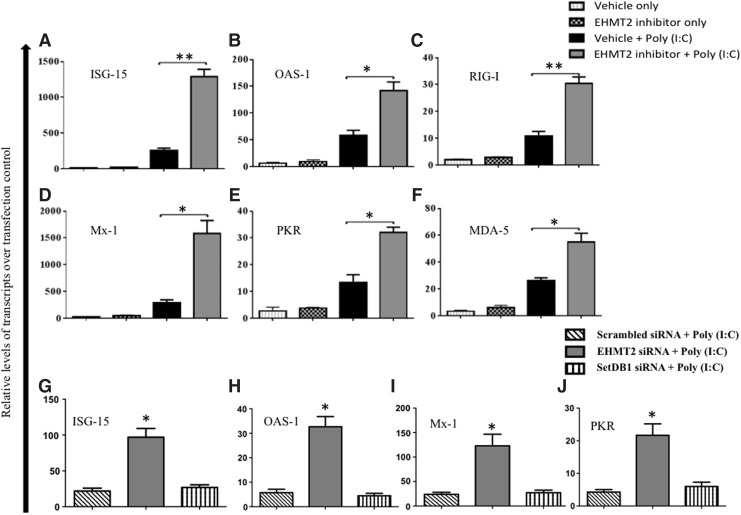
Pharmacological or RNAi-induced inhibition of EHMT2 in FBF cells followed by poly (I:C) stimulation resulted in a significant induction of ISG-15, OAS-1, PKR, Mx-1, RIG-I, and MDA-5 compared to the vehicle only or mock-transfected cells (Fig. 3A–J). ISG-15 is an IFN-inducible ubiquitin-like protein (UBL), which has been recently recognized as a key component in mammalian antiviral immunity (Skaug and Chen 2010). An increase in ISG-15 transcripts was observed in cells treated with EHMT2 inhibitor (P < 0.05) or EHMT2 siRNA (P < 0.01) compared to cells treated with the vehicle control or scrambled siRNA (Fig. 3A, G). A similar trend was seen with other ISGs. Inhibition of EHMT2 led to a significant increase (P < 0.05) in Mx-1 (Fig. 3D, I), OAS-1 (Fig. 3B, H), and PKR (Fig. 3E, J) transcripts than those obtained in control groups.
FIG. 3.
Pharmacological inhibition or depletion of EHMT2 in FBF cells resulted in an induction of IFN-stimulated genes (ISGs). FBF cells were treated with the EHMT2 inhibitor (5 μM) or vehicle for 4 days followed by 0.5 μg/mL poly (I:C) stimulation. The cells were harvested for RNA analysis by qPCR. Relative mRNA levels of (A) ISG-15, (B) 2′,5′-oligoadenylate synthetase 1 (OAS-1), (C) RIG-I, (D) myxovirus resistance 1 (Mx-1), (E) protein kinase R (PKR), and (F) MDA-5 in FBF cells stimulated with poly (I:C). For depletion of EHMT2, FBF cells were mock transfected or transfected with siRNAs targeting bovine EHMT2 or SetDB1. Forty-eight hours later, cells were stimulated with poly (I:C) (0.5 μg/mL) and 6 h later, cells were harvested for RNA analysis by qPCR. Relative mRNA levels of (G) ISG-15, (H) OAS-1, (I) Mx-1, and (J) PKR. The mRNA levels of these genes are expressed as fold increase over the transfection control. Independent gene expression was normalized with GAPDH as control. The data represent mean ± SE from 3 independent experiments. *P < 0.05, **P < 0.001, determined by 2-tailed Student's t-test.
Inhibition of EHMT2 either before or after infection with FMDV or VSV impedes virus replication
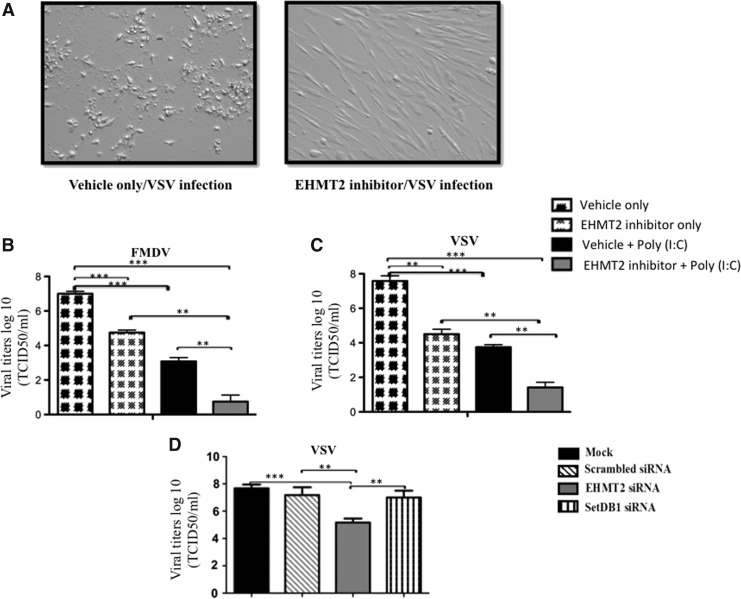
We investigated the impact of EHMT2 inhibition in bovine cells against viral diseases of veterinary importance such as foot-and-mouth disease (FMD) and vesicular stomatitis (VS). Viral titers were measured in FBFs where EHMT2 was depleted with a specific siRNA or by treating with an EHMT2 inhibitor (UNC0638) before viral infection.
EHMT2 inhibitor treatment before infection inhibited viral replication as evident by a healthy monolayer, as opposed to vehicle-treated cells (Fig. 4A), which exhibited widespread CPE. Pharmacological inhibition of EHMT2 resulted in the reduction (P < 0.001) of FMDV and VSV viral titers compared to the vehicle control (Fig. 4B, C). As expected, reduction in viral titers (FMDV and VSV) was more pronounced in cells treated with EHMT2 inhibitor followed by poly (I:C) stimulation compared to treatment with EHMT2 inhibitor only (Fig. 4B, C). Similarly, depletion of EHMT2 in FBF resulted in reduced (P < 0.001) VSV titers compared to the scrambled siRNA control (Fig. 4D).
FIG. 4.
Pharmacological inhibition or depletion of EHMT2 confers protection in bovine cells against foot-and-mouth disease virus (FMDV) and vesicular stomatitis virus (VSV) infections. FBF cells were treated with the EHMT2 inhibitor (5 μM) or vehicle for 4 days. Treated cells were unstimulated or stimulated with poly (I:C) (0.5 μg/mL) for 6 h, followed by infection with VSV or FMDV [multiplicity of infection (MOI) = 0.01]. Supernatants were collected at 36 or 41 hours postinfection (hpi) for determining viral titers (TCID50). For depletion of EHMT2, FBF cells were mock transfected or transfected with siRNAs targeting bovine EHMT2 or SetDB1 for 48 h followed by infection with VSV. Thirty-six hours later, supernatants were collected for determining viral titers (TCID50) in MDBK cells. (A) Left panel represents control FBF, exhibiting cytopathic effects (CPE). Right panel represents EHMT2 inhibitor-treated FBF monolayer. (B–D) Represents VSV and FMDV titers. Viral titers (log10) expressed as TCID50 per milliliter. The data (mean ± SE) are from 3 independent experiments done in duplicates. **P < 0.001, ***P < 0.0001 determined by 1-way ANOVA followed by Tukey's multiple comparison test.
A reduction (P < 0.0001) in FMDV and VSV titers was also observed by poly (I:C) stimulation alone (Fig. 4B, C). This was expected, as poly (I:C) itself is a potent inducer of type I IFN.
After observing the antiviral effects of EHMT2 inhibition against viral challenge, we assessed the therapeutic potential of this strategy during an ongoing viral infection. FBF cells were infected with VSV at an MOI of 0.01, followed by treatment with EHMT2 inhibitor at 4, 8, and 12 hpi. The infected cells and supernatants were harvested 36 hpi. Pharmacological inhibition of EHMT2 induced high (P < 0.05) levels of boIFN-β at all time points after infection with a peak (>150-fold) when the inhibitor was added at 8 hpi (Fig. 5A). A lower boIFN-β mRNA induction (<60-fold) was detected when the inhibitor was added later during infection (12 hpi). A similar trend was observed for other ISGs. Inhibition of EHMT2 resulted in a significant increase in ISG-15 and Mx-1 transcripts at all time points compared to the vehicle control (P < 0.05; Fig. 5B, C). The potent induction of boIFN-β and other ISGs in cells treated with EHMT2 inhibitor at early time points (4 and 8 h) after virus (VSV) infection led to a decrease in viral titers at these time points compared to the vehicle control (P < 0.05; Fig. 5D). In contrast, the addition of EHMT2 inhibitor at 12 hpi did not significantly alter the virus yield (Fig. 5D).
FIG. 5.
Pharmacological inhibition of EHMT2 confers protection in FBF cells against VSV infection therapeutically. FBF cells were infected with VSV MOI = 0.01 and followed by treatment with EHMT2 inhibitor or vehicle at indicated time points (4, 8, and 12 h) postinfection. Cells and supernatants were collected 36 hpi, for RNA analysis and viral titers. mRNA levels of (A) boIFN-β, (B) ISG-15, and (C) Mx-1 normalized to expression of endogenous control GAPDH and are represented as fold increase compared to virus-only control. (D) Viral titers expressed as TCID50 per milliliter. The data are from 3 independent experiments done in duplicates and represent mean ± SE. *P < 0.05, **P < 0.01, ***P < 0.0001 determined by 2-tailed Student's t-test.
Reduction in H3K9me2 marks on boIFN-β and ISG-15 gene promoters due to pharmacological inhibition of EHMT2
EHMT2 is the key enzyme responsible for dimethylation of the lysine 9 residue on histone 3 tail (H3K9me2) (Fang and others 2012). Therefore, we explored the specific histone modifications that are playing a role in EHMT2 inhibitor-mediated antiviral response. We performed ChIP-qPCR with vehicle-treated or EHMT2 inhibitor-treated FBF cells. Antibodies recognizing the specific chromatin modifications H3K9me2, acetylated histone 3 lysine 9 (H3K9Ac), or acetylated histone 3 lysine 14 (H3K14Ac) were used for immunoprecipitation. The primers for ChIP-qPCR were designed to amplify small fragment (∼100 bp) in the promoter region of bovine boIFN-β and ISG-15 genes as shown in Fig. 6A.
FIG. 6.
Reduction in histone H3 lysine 9 dimethylation (H3K9me2) mark on boIFN-β and ISG-15 promoter due to EHMT2 inhibition. (A) Schematic representation of boIFN-β and ISG-15 promoter regions indicating primer locations used in chromatin immunoprecipitation (ChIP)-qPCR. Promoter region of boIFN-β depicting TATA box and positive regulatory domains (PRD) I, II, III, and IV. The promoter region of bovine ISG-15 gene is about 216 bp upstream of transcription start site and depicts TATA box and IFN-stimulated response elements (ISRE). The primer sites to amplify the promoter regions are depicted with arrows. (B) Relative enrichment (1% of the total input) of H3K9me2, H3K9Ac, and H3K14Ac for boIFN-β and (C) ISG-15 gene promoter regions. Background levels are represented by the IgG control. The data represent mean ± SE and is from 3 independent cultures. *P < 0.05, determined by 2-tailed Student's t-test. Color images available online at www.liebertpub.com/jir
Treatment of FBF cells with EHMT2 inhibitor resulted in a decrease of H3K9me2 mark on both boIFN-β and ISG-15 gene promoters, as opposed to vehicle-treated cells (P < 0.05; Fig. 6B, C). As anticipated, the lack of EHMT2 enzymatic activity decreased H3K9me2, and a decrease in this particular mark is observed on the candidate gene promoters. In parallel, we assessed the influence of EHMT2 inhibitor on acetylation of lysine 9/14 residues on H3 (H3K9Ac and H3K14Ac). Several studies have revealed the functional importance of histone H3 acetylation (active transcription) at the IFN-β gene (Fang and others 2012). Treatment of cells with EHMT2 inhibitor did not result in a significant change in the levels of H3K9Ac and H3K14Ac on boIFN-β and ISG-15 gene promoters compared to the vehicle control (Fig. 6B, C). Thus, our results confirmed that boIFN-β and ISG-15 promoters are regulated by EHMT2-mediated H3K9 dimethylation without affecting H3K9 or H3K14 acetylation.
Discussion
The rapid onset and highly contagious nature of viral diseases pose a big challenge to develop control strategies. Conventional vaccines tend to induce protection several days after vaccination (Barnett and Carabin 2002), therefore, the development of antivirals to prevent or limit the rapid spread of the disease assumes importance. FMDV and VSV are highly cytopathic viruses with broad species tropism (Lang and others 2007) and both viruses are very well suited for innate immune studies. The main focus of the present study was to evaluate the effect induced by transient inhibition of EHMT2 on the innate antiviral response in primary bovine cells against FMDV and VSV infection.
The IFN-β transcriptional regulation at the nucleosome level is marked by acetylation of the histone H3 lysine 9 and 14 residues at the IFN-β promoter recruiting CBP-Pol II, TFIID, and other complexes required for transcription (Agalioti and others 2000; Fang and others 2012). The varying ability of cells to produce IFN-β, therefore, depends on posttranslational modifications of histone tails (histone marks) on nucleosomes encompassing the IFN-β promoter region. A recent study has attributed the distribution of H3K9me2 at IFN promoters for cell-specific differences in IFN expression (Fang and others 2012). In the light of this report (Shestakova and others 2001) and the fact that BLAST protein analysis comparing human and bovine EHMT2 revealed 100% homology within the drug interacting SET domain of the protein, we hypothesized that treatment of FBF with EHMT2 inhibitor (UNC0638) in the presence of an IFN triggering stimulus should result in an enhanced IFN response. As expected, inhibition of EHMT2 in nonimmortalized FBF cells resulted in a superior IFN response in the presence of exogenous stimuli compared to control groups (Figs. 1C and 2B, C). We acknowledge that the activation of the IFN response could be attributed to an off-target effect of the drug, however the data from experiments employing RNAi-based suppression of EHMT2 (Fig. 1C) and the correlative decreases in H3K9me2 (Fig. 6B, C) strongly indicate that the response is specifically linked to a decreased enzyme activity.
Consistent with the findings of Vedadi and others, we also established that lower concentrations (2 and 5 μM) of UNC0638 EHMT2 inhibitor did not manifest any toxic effects in FBF with the cell monolayer being healthy. Also, we did not see any remarkable difference in EHMT2 inhibitor effects between 6-day treatment with fresh inhibitor being replenished every 2 days and 4-day treatment without any replenishment, suggesting that the residual activity of UNC0638 was sufficient to exert its activity (Vedadi and others 2011). However, since histone methylation (H3K9me2) occurs at a slow turn-over rate compared to other histone modifications (half-life of H3K9me2 is about 1 day), a maximum effect could be detected only if treatment with the inhibitor is carried out for a longer period (Vedadi and others 2011).
We have shown that the antiviral state was associated with a significant reduction in FMDV and VSV titers in EHMT2 inhibitor-treated cells (Fig. 4). We did not detect an upregulation of ISGs or boIFN-β in EHMT2 inhibitor-treated cells in the absence of poly (I:C) or viral stimulation. This is due to the fact that EHMT2 inhibition increases the likelihood of transcription of IFN and ISGs by keeping the target genes in a transcriptionally primed state, without triggering the antiviral response. However, EHMT2 inhibition resulted in significantly reduced viral titers even in the absence of poly (I:C) stimulus, because the viral components and their replicative intermediates function as pathogen-associated molecular patterns that trigger the innate antiviral response. In cells treated with EHMT2 inhibitor, direct increase in the likelihood of transcription of IFN and ISGs contributes to the rapid development of an antiviral state. Further studies are required to explore the dynamics of the enhancement in the antiviral response mediated by EHMT2 inhibition.
All the ISGs that were upregulated when costimulated with poly (I:C) have an intrinsic antiviral activity and have been shown to be upregulated in bovine cells after type I IFN administration and challenge with FMDV (Diaz-San Segundo and others 2010). In addition, Mx-1 is a potent inhibitor of myxoviruses (Staeheli and Haller 1987), previously shown to effectively inhibit VSV replication (Zhang and others 2013). OAS-1, together with RNaseL, exert its antiviral activity by viral RNA degradation (Silverman 2007) and PKR binds to and phosphorylates eukaryotic initiation factor 2α (eIF2α), thereby inhibiting the host's translation initiation process (Hovanessian 2007).
Several studies have shown that FMDV and VSV are highly sensitive to type I IFN response both in vitro (Chinsangaram and others 2001) and in vivo (Chinsangaram and others 2003). Infection of FMDV in DCs is often abortive owing to the constitutive expression of IFN (Bautista and others 2005). However, viruses have also evolved different mechanisms to antagonize and limit the host IFN response (Weber and others 2004). Therefore, we also evaluated the antiviral potential of our novel epigenetic approach in a therapeutic setting to determine the feasibility of this strategy in controlling VSV or FMDV replication. Under the therapeutic setting, FBF cells were infected with VSV and thereafter treated with EHMT2 inhibitor at different times postinfection. Our results suggested that treatment of infected cells with EHMT2 inhibitor early during the infection can be a useful strategy to limit virus infection and spread of the disease.
Similar to the findings of Fang and others, in MEF, we show that the treatment of FBF with EHMT2 inhibitor for 4 days resulted in a significant reduction in H3K9me2 marks on the promoter regions of boIFN-β and ISG-15 genes. Posttranslational modifications on neighboring residues within the same histone tail can often influence each other and likewise some studies have reported cross-talk between H3K9me2 and H3K14Ac marks (Rice and others 2003; Vedadi and others 2011). However, in the present study, we did not observe any significant change in H3K14Ac or H3K9Ac on the promoter region of boIFN-β and ISG-15 genes with EHMT2 inhibitor treatment compared to the vehicle control.
Depletion of bovine SetDB1 mRNA did not affect the boIFN-β expression. Previous studies have revealed the role of SetDB1 in suppressing a subset of genes encoding developmental regulators in ES cells (Bilodeau and others 2009), however, no reports have suggested its role in IFN and ISG regulation. Similarly, no other reports have highlighted the role of EHMT2 in the regulation of ISGs or cytokines. Others have reported a downregulation by epigenetic silencing of IFN signaling pathway genes during immortalization. For instance, the promoter regions of IRF7, but not IRF5, were epigenetically silenced by methylation of CpG islands in cancer cells (Li and others 2008). In the current report, we have demonstrated, for the first time, the direct involvement of EHMT2 in the regulation of IFN-β and ISG-15 in nonimmortalized primary bovine cells.
In conclusion, our results have highlighted the effect of H3K9me2 on individual gene promoters, including those of boIFN-β and ISG-15, as a limiting factor in what determines the likelihood of transcription. Importantly, we have demonstrated the prophylactic and therapeutic use of EHMT2 inhibitor in modulating antiviral immune responses in bovine cells and highlighted the importance of epigenetic-based therapies for controlling viral diseases of livestock and human importance. Future studies will be directed toward in vivo testing of this strategy in the animal host and also defining the role of other epigenetic marks on the components of innate immunity.
Acknowledgments
This research was supported by the Texas A&M College of Veterinary Medicine and Biomedical Sciences Postdoc Trainee Research Grant, Texas Agrilife Exceptional Research Pilot Programs. N.S. was partially funded through NIH-OD 8R240D011188-02. We thank Luis L. Rodríguez for his supervision and assistance in conducting experiments at Plum Island Animal Disease Center. We would also like to thank Kylee Veazey, Mike Peoples, and Daria Muller for technical assistance.
Author Disclosure Statement
No competing financial interests exist.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667–678 [DOI] [PubMed] [Google Scholar]
- Barnett PV, Carabin H. 2002. A review of emergency foot-and-mouth disease (FMD) vaccines. Vaccine 20:1505–1514 [DOI] [PubMed] [Google Scholar]
- Bautista EM, Ferman GS, Gregg D, Brum MC, Grubman MJ, Golde WT. 2005. Constitutive expression of alpha interferon by skin dendritic cells confers resistance to infection by foot-and-mouth disease virus. J Virol 79:4838–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. 2009. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23:2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinsangaram J, Koster M, Grubman MJ. 2001. Inhibition of L-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J Virol 75:5498–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinsangaram J, Moraes MP, Koster M, Grubman MJ. 2003. Novel viral disease control strategy: adenovirus expressing alpha interferon rapidly protects swine from foot-and-mouth disease. J Virol 77:1621–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-San Segundo F, Moraes MP, de Los Santos T, Dias CC, Grubman MJ. 2010. Interferon-induced protection against foot-and-mouth disease virus infection correlates with enhanced tissue-specific innate immune cell infiltration and interferon-stimulated gene expression. J Virol 84:2063–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doly J, Civas A, Navarro S, Uze G. 1998. Type I interferons: expression and signalization. Cell Mol Life Sci 54:1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze O, Picard D. 2002. RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res 30:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang TC, Schaefer U, Mecklenbrauker I, Stienen A, Dewell S, Chen MS, Rioja I, Parravicini V, Prinjha RK, Chandwani R, MacDonald MR, Lee K, Rice CM, Tarakhovsky A. 2012. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J Exp Med 209:661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Morera JL, Calvanese V, Rodriguez-Rodero S, Menendez-Torre E, Fraga MF. 2010. Epigenetic regulation of the immune system in health and disease. Tissue Antigens 76:431–439 [DOI] [PubMed] [Google Scholar]
- Fray MD, Mann GE, Charleston B. 2001. Validation of an Mx/CAT reporter gene assay for the quantification of bovine type-I interferon. J Immunol Methods 249:235–244 [DOI] [PubMed] [Google Scholar]
- Golding MC, Zhang L, Mann MR. 2010. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell 6:457–467 [DOI] [PubMed] [Google Scholar]
- Hovanessian AG. 2007. On the discovery of interferon-inducible, double-stranded RNA activated enzymes: the 2'-5'oligoadenylate synthetases and the protein kinase PKR. Cytokine Growth Factor Rev 18:351–361 [DOI] [PubMed] [Google Scholar]
- Imai K, Togami H, Okamoto T. 2010. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J Biol Chem 285:16538–16545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR, Issa JP. 2008. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One 3:e2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T. 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25:473–481 [DOI] [PubMed] [Google Scholar]
- Lang KS, Navarini AA, Recher M, Lang PA, Heikenwalder M, Stecher B, Bergthaler A, Odermatt B, Akira S, Honda K, Hengartner H, Zinkernagel RM. 2007. Myd88 protects from lethal encephalitis during infection with vesicular stomatitis virus. Eur J Immunol 37:2434–2440 [DOI] [PubMed] [Google Scholar]
- Li Q, Tang L, Roberts PC, Kraniak JM, Fridman AL, Kulaeva OI, Tehrani OS, Tainsky MA. 2008. Interferon regulatory factors IRF5 and IRF7 inhibit growth and induce senescence in immortal Li-Fraumeni fibroblasts. Mol Cancer Res MCR 6:770–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Malmquist NA, Moss TA, Mecheri S, Scherf A, Fuchter MJ. 2012. Small-molecule histone methyltransferase inhibitors display rapid antimalarial activity against all blood stage forms in Plasmodium falciparum. Proc Natl Acad Sci U S A 109:16708–16713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. 2010. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327:213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. 2008. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc 3:698–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ. and Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497 [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12:1591–1598 [DOI] [PubMed] [Google Scholar]
- Rieder E, Bunch T, Brown F, Mason PW. 1993. Genetically engineered foot-and-mouth disease viruses with poly(c) tracts of two nucleotides are virulent in mice. J Virol 67:5139–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder M, Conzelmann KK. 2009. Rhabdovirus evasion of the interferon system. J Interferon Cytokine Res 29:499–509 [DOI] [PubMed] [Google Scholar]
- Rodriguez LL, Gay CG. 2011. Development of vaccines toward the global control and eradication of foot-and-mouth disease. Expert Rev Vaccines 10:377–387 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Pulido M, Borrego B, Sobrino F, Saiz M. 2011. RNA structural domains in noncoding regions of the foot-and-mouth disease virus genome trigger innate immunity in porcine cells and mice. J Virol 85:6492–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, Tarakhovsky A, Greengard P. 2009. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64:678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16:919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova E, Bandu MT, Doly J, Bonnefoy E. 2001. Inhibition of histone deacetylation induces constitutive derepression of the beta interferon promoter and confers antiviral activity. J Virol 75:3444–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH. 2007. Viral encounters with 2',5'-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81:12720–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Chen ZJ. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143:187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeheli P, Haller O. 1987. Interferon-induced Mx protein: a mediator of cellular resistance to influenza virus. Interferon 8:1–23 [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16:1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, Dimaggio PA, Wasney GA, Siarheyeva A, Dong A, Tempel W, Wang SC, Chen X, Chau I, Mangano TJ, Huang XP, Simpson CD, Pattenden SG, Norris JL, Kireev DB, Tripathy A, Edwards A, Roth BL, Janzen WP, Garcia BA, Petronis A, Ellis J, Brown PJ, Frye SV, Arrowsmith CH, Jin J. 2011. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol 7:566–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG, Zhang Y. 2003. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell 12:475–487 [DOI] [PubMed] [Google Scholar]
- Weber F, Kochs G, Haller O. 2004. Inverse interference: how viruses fight the interferon system. Viral Immunol 17:498–515 [DOI] [PubMed] [Google Scholar]
- Zhang XM, He DN, Zhou B, Pang R, Liu K, Zhao J, Chen PY. 2013. In vitro inhibition of vesicular stomatitis virus replication by purified porcine Mx1 protein fused to HIV-1 tat protein transduction domain (PTD). Antiviral Res 99:149–157 [DOI] [PubMed] [Google Scholar]