Abstract
A number of molecular typing methods have been developed for characterization of Staphylococcus aureus isolates. The utility of these systems depends on the nature of the investigation for which they are used. We compared two commonly used methods of molecular typing, multilocus sequence typing (MLST) (and its clustering algorithm, Based Upon Related Sequence Type [BURST]) with the staphylococcal protein A (spa) typing (and its clustering algorithm, Based Upon Repeat Pattern [BURP]), to assess the utility of these methods for macroepidemiology and evolutionary studies of S. aureus in the United States. We typed a total of 366 clinical isolates of S. aureus by these methods and evaluated indices of diversity and concordance values. Our results show that, when combined with the BURP clustering algorithm to delineate clonal lineages, spa typing produces results that are highly comparable with those produced by MLST/BURST. Therefore, spa typing is appropriate for use in macroepidemiology and evolutionary studies and, given its lower implementation cost, this method appears to be more efficient. The findings are robust and are consistent across different settings, patient ages, and specimen sources. Our results also support a model in which the methicillin-resistant S. aureus (MRSA) population in the United States comprises two major lineages (USA300 and USA100), which each consist of closely related variants.
Introduction
Avariety of molecular typing methods have been developed for the characterization of Staphylococcus aureus, including pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and staphylococcal protein A (spa) typing.4,9,12 The suitability of a typing system depends on the nature of the investigation in which it is used. Local outbreak investigations require very high discriminatory power to differentiate between closely related strains. For macroepidemiology and evolutionary studies, in addition to discriminatory power, it is also important to produce unambiguous results that are readily comparable among different laboratories, and it is necessary to have a system for classifying the relationships among closely related strains to monitor changes and patterns in clonal lineages over time or space.
PFGE's high discriminatory power made it a popular choice for outbreak investigations before the adoption of more recently developed sequence-based typing methods, including MLST and spa typing. Despite its popularity, the results of PFGE are difficult to compare across laboratories and over time, so its use in long-term macroepidemiology studies has been questioned.1 Furthermore, PFGE is a technically difficult and laborious method that requires interlaboratory standardization.2
MLST, which differentiates isolates on the basis of nucleotide variation at seven housekeeping loci (slowly evolving genes involved in basic cellular functions), has proven very useful for macroepidemiology and evolutionary studies.5,18 spa typing, which relies only on the assessment of repeats at the x region of spa, exhibits excellent discriminatory power and shares with MLST the advantages of unambiguous typing results that can be compared between laboratories and over time. spa typing is also both easier and less costly to perform than MLST or PFGE.
spa typing has been shown to be highly concordant with MLST and some studies suggest that it is suitable for macroepidemiology and evolutionary investigations based on studies of European and international isolates.8,11,19 However, other studies question the use of a single locus method such as spa typing for macroepidemiologic investigations since recombination events might distort the underlying clonal relationships.14
The primary output of MLST is a sequence type (ST), such as MLST ST8, assigned to each bacterial isolate. The primary output of spa typing is a spa type for each isolate, such as spa type t008. MLST and spa typing can each be used in conjunction with software-based clustering algorithms that group related isolates into clonal complexes (CCs). Therefore, MLST CCs are clusters of MLST STs and spa CCs are clusters of spa types. CCs are named after the ST presumed to be the founder (the ST with the most single locus variants), for example, MLST CC8 is named after its presumed founder, MLST ST8.8 The algorithms used with MLST and spa typing to cluster MLST STs into MLST CCs and spa types into spa CCs are Based Upon Related Sequence Types (BURSTs) and Based Upon Repeat Pattern (BURP), respectively.
Using MLST, the S. aureus population in the United States has been classified into three major groups: the USA300 lineage of community methicillin-resistant S. aureus (MRSA), the USA100 lineage of hospital MRSA, and polyclonal methicillin-susceptible S. aureus (MSSA).15 The USA300 MRSA lineage is MLST ST8, spa type t008, MLST CC8, and spa CC008 and harbors the genes for Panton-Valentine Leukocidin (PVL).12,15 USA300 is a common cause of S. aureus infection in children, skin and soft tissue, and in outpatient settings.12,15
The USA100 MRSA lineage is MLST ST5, spa type t002, MLST CC5, and spa CC002 and is PVL-negative.12,15 USA100 is less commonly associated with infections in children, skin and soft tissue, and in outpatient settings and is a common cause of S. aureus infection in inpatient settings and the elderly.12,15 The third major group, polyclonal MSSA, consists of several different STs and CCs and is seen in a diversity of settings, sources, and age groups.15
This investigation builds on previous studies and further evaluates the comparability of results obtained from MLST/BURST and spa typing/BURP, as well as their ability to classify the major lineages of S. aureus in the United States for epidemiology and evolutionary studies. Both MLST and spa typing possess desirable characteristics in this regard as they provide unambiguous typing results, they are highly discriminatory, and their results can easily be compared between laboratories and over time.
It has been established that the BURST method of grouping MLST STs into CCs provides an accurate way of representing clonal lineages with a common ancestry.4 Furthermore, it has been established in previous studies that MLST and spa typing can yield highly concordant results, as can the MLST ST and spa type clonal clustering methods, BURST and BURP, respectively.8,19,20 The MLST with BURST approach to classify MLST CCs has already proven valuable in understanding the macroepidemiology and evolutionary history of S. aureus, particularly in the United States.15
To better assess the utility of spa typing for these purposes, we performed an in-depth comparison of MLST STs/MLST CCs (BURST) with spa types/spa CCs (BURP) in terms of concordance and index of diversity (DIs) in light of different isolate characteristics in a sample of S. aureus clinical isolates from an integrated healthcare system across the United States. The sample consisted of different patient age groups, sites of infection, settings for sampling, and methicillin susceptibility. Prior comparisons of MLST and spa typing neither analyzed demographic information about the patients from whom the isolates were collected nor focused on isolates from the United States.19,20 We also compared the effects of methicillin resistance status and PVL testing on typing results and analyzed how the major clonal lineages of MRSA in the United States can be characterized using different typing approaches.
Methods
Setting
Kaiser Permanente of Northern California (KPNC) is a nonprofit, integrated healthcare delivery system providing care to ∼3.3 million members. The member population reflects the general population in the northern California region, although as an insured population, it under-represents persons with very low levels of income. KPNC provides services in more than 15 counties and operates more than 50 outpatient clinics and 20 hospitals throughout northern California.
Microbiology tests ordered for KPNC members are performed by KPNC's regional laboratory in Berkeley, CA. Test orders and results are recorded in KPNC's Laboratory Utilization and Reporting System (LURS). Laboratory results from hospital discharges and ambulatory settings, including emergency departments, are archived in LURS. LURS uniquely identifies each specimen by an accession number and each patient by a medical record number, which identifies members of KPNC and is used to track patient interactions with the health plan.
Clinical isolates
The clinical isolates included in this study were from microbiology tests ordered between March 6, 2010, and March 25, 2011, as part of usual care by KPNC providers. Suspect Staphylococcus colonies (catalase positive, Gram-positive cocci in clusters) were identified by clumping factor and/or protein A or by coagulation of coagulase plasma. Using an 18–24-hour culture from non-selective medium, isolated S. aureus colonies were selected for antimicrobial susceptibility testing using commercial systems (MicroScan, Siemens Healthcare Diagnostics, Inc., Sacramento, CA, or Vitek 2, bioMerieux, Hazelwood, MO, for urine isolates).
Isolates displaying an oxacillin minimum inhibitory concentration (MIC) of >4 μg/ml were subjected to additional confirmatory testing for MRSA. A suspension of the suspect isolate was prepared and used to inoculate a BBL Oxacillin Screen Agar Plate (Becton, Dickinson and Company, Sparks, MD), which was incubated in an ambient air incubator at 33–35°C for 24 hours. Isolates were characterized as MRSA if the isolates both grew on the Oxacillin Screen Agar Plate and displayed oxacillin MIC values of >4 μg/ml. This method was used throughout the study period.
On an ongoing basis, a subset (depending on staff availability) of isolates positive for S. aureus was set aside for molecular testing and their accession numbers were recorded. Periodically, these samples were sent to the study sponsor's laboratory where molecular testing was performed. The accession numbers and medical record numbers were used by the KPNC investigators to link the isolates and the patients to clinical and demographic characteristics. Other than the fact that the isolate was S. aureus, the laboratory performing the molecular typing was blinded to the clinical characteristics of the isolate and the demographics of the patients.
A total of 875 unique isolates for 875 unique patients were available for molecular testing. Because our focus was on the old and the young, we excluded 30 isolates for persons between 18 and 50 years of age. Of the remaining 845 isolates, both spa typing and MLST typing were performed on the first 366 isolates, which is the sample used for all analyses in this report.
Molecular typing
We performed molecular characterization (MLST, spa typing, and PVL testing) on all 366 isolates. MLST was performed as previously described.7 Briefly, internal fragments of seven housekeeping genes were amplified by PCR and sequenced using the ABI Big Dye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems, Foster City, CA). MLST STs were assigned using the online S. aureus MLST database (www.mlst.net). MLST STs were clustered into MLST CCs using eBURST, with the minimum number of common alleles set at six of seven.6
spa typing was performed on isolates by amplifying and sequencing the spa repeat region using primers as previously described.13 spa sequence analysis was performed using Ridom StaphType (VERSION 2.0; Ridom GmbH, Würzburg, Germany), and the resulting spa types were then clustered into related spa CCs using the BURP algorithm with parameters set to exclude spa types that are shorter than five repeats and if costs were less than or equal to four.
We tested for the presence of the PVL genes using PCR amplification and gel electrophoresis using primers and PCR conditions as described previously.7
DIs and concordance
The DI is a metric that measures the probability that a typing system will assign a different type to two strains randomly sampled from a microbial population.10 A DI of 1 indicates a completely diverse community, while a DI of 0 indicates a community composed entirely of a single clone. DIs were calculated for MLST STs, MLST CCs, spa types, and spa CCs.
Concordance measures the probability that two different typing systems will agree on whether a pair of randomly selected isolates is the same type or different types. A concordance value of 1 indicates that the two typing systems always agree on how to classify a pair of isolates; a concordance value of 0 means that the two typing systems always disagree. Concordance estimates between MLST STs and spa types, and between MLST CCs and spa CCs were assessed.
DIs and their confidence intervals were calculated using the Ridom StaphType software (VERSION 2.0; Ridom GmbH, Würzburg, Germany). Concordance rates (Wallace Statistic) and their confidence intervals were calculated using an online tool developed by Pinto et al.16
Graphical comparison
Venn Diagrams were constructed using web-based software called Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Results
Summary of molecular typing results
Using the MLST method, we identified 65 different MLST STs among the 366 isolates tested. One isolate could not be assigned an MLST ST because we could only amplify six loci (that isolate still provided enough information to be characterized into an MLST CC by BURST). We identified 35 novel MLST STs not present in the online S. aureus MLST database. The most commonly observed MLST STs were ST8 (38%, n = 138) and ST5 (17%, n = 62). The BURST algorithm assigned the MLST STs into 27 groups, including 12 MLST CCs and 15 MLST singletons. The two most commonly observed MLST CCs were CC8 (40%, n = 145) and CC5 (20%, n = 74).
By the spa typing method, we identified 118 different spa types among the 366 isolates tested. Five isolates could not be typed. We identified 16 novel spa types that were not in the Ridom StaphType database. The most commonly observed spa types were t008 (31%, n = 112) and t002 (12%, n = 43). The BURP algorithm assigned the spa types into 47 different spa CCs, including 14 spa CCs and 33 spa singletons. The two most commonly observed spa CCs were spa CC008 (40%, n = 146) and spa CC002 (16%, n = 58).
Indices of diversity
DI assessment of S. aureus isolates by molecular sequence-based typing with and without clonal grouping methods is presented in Table 1. The DIs among all isolates ranged from a low of 0.787 for MLST CCs to a high of 0.890 for spa types. For each typing method, the DIs varied little across patient age groups, inpatient/outpatient settings, or isolate source.
Table 1.
Diversity Index Assessment of Staphylococcus aureus Isolates by Molecular Sequence-Based Typing Method With or Without Clonal Grouping Methods
| Molecular sequence-based typing | |||||
|---|---|---|---|---|---|
| MLST | spa typing | ||||
| Group | Sample size | MLST ST | MLST CC | spa type | spa CC |
| All | |||||
| All isolates | 366 | 0.820 | 0.787 | 0.890 | 0.797 |
| 95% CI | 0.786–0.853 | 0.753–0.820 | 0.862–0.917 | 0.760–0.834 | |
| Age group | |||||
| Children ≤18 years | 116 | 0.761 | 0.733 | 0.848 | 0.704 |
| 95% CI | 0.680–0.842 | 0.648–0.817 | 0.780–0.915 | 0.609–0.799 | |
| Adults ≥50 years | 250 | 0.837 | 0.795 | 0.899 | 0.818 |
| 95% CI | 0.802–0.872 | 0.761–0.829 | 0.870–0.927 | 0.782–0.855 | |
| Patient setting | |||||
| Inpatient | 68 | 0.875 | 0.786 | 0.91 | 0.812 |
| 95% CI | 0.822–0.928 | 0.719–0.852 | 0.860–0.959 | 0.746–0.878 | |
| Outpatient | 298 | 0.803 | 0.777 | 0.877 | 0.782 |
| 95% CI | 0.762–0.844 | 0.735–0.818 | 0.842–0.911 | 0.735–0.828 | |
| Isolate source | |||||
| Skin | 287 | 0.786 | 0.759 | 0.873 | 0.76 |
| 95% CI | 0.741–0.831 | 0.712–0.805 | 0.836–0.909 | 0.709–0.812 | |
| Nonskin | 79 | 0.876 | 0.797 | 0.918 | 0.835 |
| 95% CI | 0.822–0.930 | 0.734–0.861 | 0.876–0.960 | 0.776–0.895 | |
| Methicillin susceptibility | |||||
| MRSA | 170 | 0.575 | 0.513 | 0.716 | 0.521 |
| 95% CI | 0.497–0.653 | 0.442–0.584 | 0.649–0.784 | 0.445–0.597 | |
| MSSAa | 196 | 0.925 | 0.902 | 0.969 | 0.929 |
| 95% CI | 0.908–0.943 | 0.885–0.919 | 0.957–0.982 | 0.911–0.947 | |
Of note, despite overlapping 95% CI, MLST STs versus spa types are statistically significantly different at alpha = 0.05.
CC, clonal complex; CI, confidence interval; MLST, multilocus sequence typing; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible S. aureus; spa, staphylococcal protein A; ST, sequence type.
However, methicillin resistance status did affect DIs for all typing methods. DIs for MRSA were significantly lower than DIs for MSSA, indicating that MRSA is more clonal. The DI for spa types within MRSA was significantly higher than the DIs observed using other methods (p < 0.05, despite slightly overlapping confidence intervals, based on manual computation of confidence intervals).
The DI for spa types within MSSA was also significantly higher than the DIs using other methods, but the magnitude of the difference was relatively smaller. This indicates that much of the enhanced discriminatory power of spa types relative to other methods is derived from its ability to discriminate within MRSA.
Concordance between typing methods
Concordance between molecular sequence-based typing and clonal grouping methods for S. aureus clinical isolates is presented in Table 2. Across all isolates, MLST STs and spa types were highly concordant (0.906). Concordance between MLST CCs and spa CCs was even higher (0.966). Concordance results were very consistent across isolates from different patient age groups, inpatient/outpatient settings, and isolate sources.
Table 2.
Concordance Between Sequence-Based Typing Clonal Grouping Methods for S. aureus Clinical Isolates
| Subgroup | Sample size | MLST STs with spa types with spa | MLST CCs with spa CCs |
|---|---|---|---|
| All | |||
| All isolates | 366 | 0.906 | 0.966 |
| 95% CI | 0.886–0.927 | 0.954–0.979 | |
| Age group | |||
| Children ≤18 years | 116 | 0.899 | 0.97 |
| 95% CI | 0.850–0.947 | 0.946–0.996 | |
| Adults ≥50 years | 250 | 0.906 | 0.964 |
| 95% CI | 0.884–0.928 | 0.948–0.979 | |
| Patient setting | |||
| Inpatient | 68 | 0.89 | 0.962 |
| 95% CI | 0.846–0.934 | 0.926–0.998 | |
| Outpatient | 298 | 0.908 | 0.967 |
| 95% CI | 0.884–0.932 | 0.953–0.981 | |
| Isolate source | |||
| Skin | 287 | 0.893 | 0.967 |
| 95% CI | 0.865–0.921 | 0.952–0.982 | |
| Nonskin | 79 | 0.91 | 0.953 |
| 95% CI | 0.866–0.955 | 0.916–0.991 | |
| Methicillin-susceptibility | |||
| MRSA | 170 | 0.801 | 0.971 |
| 95% CI | 0.747–0.855 | 0.947–0.995 | |
| MSSA | 196 | 0.947 | 0.959 |
| 95% CI | 0.934–0.960 | 0.945–0.972 | |
The concordance between MLST STs and spa types differed depending on methicillin resistance status, with lower concordance (0.801) in MRSA isolates and higher (0.947) in MSSA isolates. In contrast, the concordance between MLST CCs and spa CCs was higher in MRSA isolates (0.971) than in MSSA isolates (0.959), reflecting the more clonal nature of MRSA. A likely cause of the lower concordance between MLST STs and spa types in MRSA is that spa types are more discriminating within MRSA clones.
Characteristics of major MRSA clonal lineages
Overall, 46% of isolates in our sample were MRSA (n = 170). The majority (91%, n = 154) of MRSA isolates were found within two clonal lineages regardless of the typing method. The first includes MLST ST8, MLST CC8, spa type t008, and spa CC008, which are the molecular types of the USA300 lineage. The second includes MLST ST5, MLST CC5, spa type t002, and spa CC002, which are the molecular types of the USA100 lineage (Table 3).
Table 3.
Characteristics of the Two Major Methicillin-Resistant Staphylococcus aureus Lineages by Sequence-Based Typing and Clonal Grouping Methods
| USA300 lineage | USA100 lineage | |||||||
|---|---|---|---|---|---|---|---|---|
| Molecular sequence-based typing method | Clonal grouping method | Molecular sequence-based typing method | Clonal grouping method | |||||
| Subgroup | MLST ST | spa type | MLST CC | spa CC | MLST ST | spa type | MLST CC | spa CC |
| Type/clonal group | ST8 | t008 | CC8 | CC008 | ST5 | t002 | CC5 | CC002 |
| Number of isolates | 105 | 85 | 111 | 111 | 34 | 30 | 42 | 37 |
| % of all MRSA isolates | 61.8% | 50.0% | 65.3% | 65.3% | 20.0% | 17.6% | 24.7% | 21.8% |
| 95% CI | 54.5–69.1% | 42.5–57.5% | 58.1–72.5% | 58.14–72.46% | 14.0–26.0% | 11.9–23.3% | 18.2–31.2% | 15.59–28.01% |
| PVL | ||||||||
| PVL+ | 95.2% | 96.5% | 94.7% | 95.5% | 0.0% | 0.0% | 0.0% | 0.0% |
| 95% CI | 91.1–99.3% | 92.6–100% | 90.5–98.9% | 91.64–99.36% | 0.0–0.4% | 0.0–0.4% | 0.0–0.3% | 0.0–0.3% |
| PVL− | 4.8% | 3.5% | 5.3% | 4.5% | 100.0% | 100.0% | 100.0% | 100.0% |
| 95% CI | 0.7–8.9% | −0.4–7.4% | 1.1–9.5% | 0.64–8.36% | 99.6–100.0% | 99.6–100.0% | 99.7–100.0% | 99.7–100.0% |
| Age group | ||||||||
| Children ≤18 years | 36.2% | 37.6% | 36.3% | 36.9% | 0.0% | 0.0% | 0.0% | 0.0% |
| 95% CI | 27.0–45.4% | 27.3–47.9% | 27.4–45.3% | 27.9–45.9% | 0.0–0.4% | 0.0–0.4% | 0.0–0.3% | 0.0–0.3% |
| Adults ≥50 years | 64.8% | 62.4% | 63.7% | 63.1% | 100.0% | 100.0% | 100.0% | 100.0% |
| 95% CI | 54.8–73.2% | 51.7–72.3% | 54.75–72.65% | 54.1–72.1% | 99.6–100.0% | 99.6–100.0% | 99.7–100.0% | 99.7–100.0% |
| Patient setting | ||||||||
| Inpatient | 13.3% | 9.4% | 12.4% | 12.6% | 41.2% | 50.0% | 47.6% | 45.9% |
| 95% CI | 6.8–19.8% | 3.2–15.6% | 6.27–18.53% | 6.4–18.8% | 24.7–57.8% | 32.1–67.9% | 32.5–62.7% | 29.8–62.0% |
| Outpatient | 86.7% | 90.6% | 87.6% | 87.4% | 58.8% | 50.0% | 52.4% | 54.1% |
| 95% CI | 80.2–93.2% | 84.4–96.8% | 81.47–93.73% | 81.2–93.6% | 42.3–75.3% | 32.1–67.9% | 37.3–67.5% | 38.0–70.2% |
| Isolate source | ||||||||
| Skin | 87.6% | 85.9% | 88.5% | 88.3% | 50.0% | 53.3% | 47.6% | 48.6% |
| 95% CI | 81.3–93.9% | 78.5–93.3% | 82.57–94.43% | 82.3–94.3% | 33.2–66.8% | 35.5–71.2% | 32.5–62.7% | 32.5–64.7% |
| Nonskin | 12.4% | 14.1% | 11.5% | 11.7% | 50.0% | 46.7% | 52.4% | 51.4% |
| 95% CI | 6.1–18.7% | 6.7–21.5% | 5.57–17.43% | 5.7–17.7% | 33.2–66.8% | 28.9–64.6% | 37.3–67.5% | 35.3–67.5% |
A total of 170 MRSA isolates were analyzed.
PVL, Panton-Valentine Leukocidin genes.
One hundred and ten isolates were both MLST CC08 and spa CC008, and there was one MLST CC8 isolate that could not be spa typed and one spa CC008 isolate that was a singleton clone consisting of ST80006 (a novel ST). Thirty-seven isolates were both MLST CC5 and spa CC002, and there were four MLST CC5 isolates that could not be spa typed and one MLST CC5 isolate that was spa CC688. The distribution of the remaining 16 MRSA isolates is as follows: eight isolates belonged to the MLST CC30/spa CC021 group, two belonged to the MLST CC59/spa CC216 group, two belonged to the MLST CC22/spa type t005 group, and four were singletons.
As expected, the number of isolates that were assigned to a particular lineage depended on the typing method used. Using the relatively inclusive MLST method, 62% of MRSA isolates were classified as MLST ST8 and thus within the USA300 lineage (Table 3). In contrast, the more discriminatory spa typing method classified only 50% of MRSA isolates as spa type t008 and within the USA300 lineage.
However, if the clonal grouping methods were applied to cluster types closely related to MLST ST8 into MLST CCs or cluster types closely related to spa type t008 into spa CCs, then the results were highly concordant with 65% of MRSA isolates classified as MLST CC8 and 65% of MRSA isolates classified as spa CC008. Similar results were seen within the USA100 clonal lineage, demonstrating the concordance of typing methods as well as the robustness of using MLST CCs or spa CCs combined with methicillin resistance status to classify MRSA in the United States into major clonal lineages.
The two major clonal lineages differed in the frequency that they were isolated from outpatients, skin infections, and children and this was stable regardless of the typing method (Table 3). Based on either MLST or spa typing, approximately one third of isolates from the USA300 clonal lineage were from children and ∼90% were from outpatient settings and skin infections.
In contrast, none of the isolates from the USA100 clonal lineage were from children and only approximately half were from outpatients and skin infections. Regardless of the typing method, the USA300 lineage (ST8/CC8/t008/CC008) was ∼95% PVL-positive, while the USA100 lineage (ST5/CC5/t002/CC002) did not contain any PVL-positive isolates. The 5% PVL-negative MRSA isolates of the USA300 lineage were highly concordant for typing results, that is, they were MLST ST8, MLST CC8, and spa CC008.
Characterization of the USA300 lineage
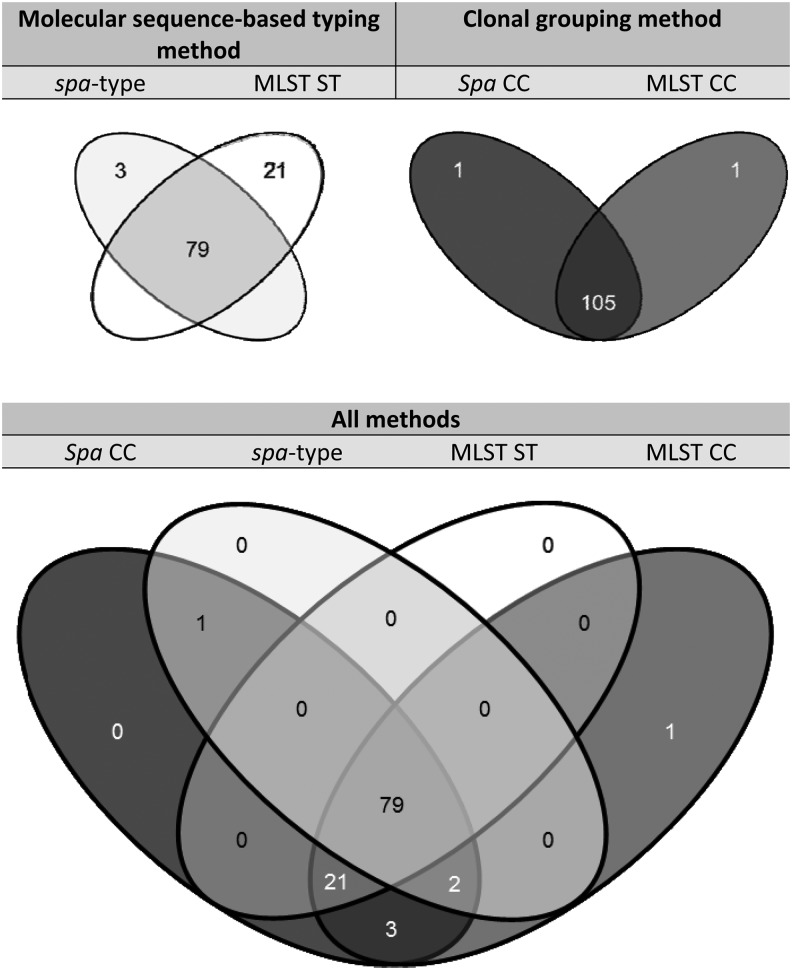
The analysis of USA300 by typing method is represented in a Venn diagram (Fig. 1). It depicts the PVL-positive MRSA isolates that had any of the following typing results indicating evidence for inclusion in the USA300 lineage: MLST ST8, MLST CC8, spa type t008, or spa CC008, a total of 107 isolates. Because the presence of the genes for PVL is a reliable marker for USA300, we excluded the five isolates that are classified as members of the USA300 lineage based on MLST or spa typing (MLST ST8, MLST CC8, and spa CC008), but are PVL negative.12,15
FIG. 1.
Venn diagram showing the overlapping representations of the USA300 lineage (107 isolates that are methicillin-resistant Staphylococcus aureus, Panton-Valentine Leukocidin positive, and multilocus sequence typing [MLST] ST8, MLST CC8, staphylococcal protein A (spa) type 008, and/or spa CC008) as identified using four different typing methods: MLST CC8, MLST ST8, spa type t008, and spa CC008.
For the 107 PVL-positive isolates, the majority (74%, n = 79) shared identical typing results: ST8, CC8, t008, and CC008. Importantly, the overlap between MLST CC8 and spa CC008 accounted for 105 of the 107 isolates. There was one PVL-positive MRSA isolate in MLST CC8, but not in spa CC008, and there was one PVL-positive MRSA isolate in spa CC008, but not MLST CC8. Within spa CC008, 81 of 106 PVL-positive MRSA isolates were spa type t008, whereas within MLST CC8, 100 out of 106 isolates were MLST ST8.
Compared with the MLST and spa clonal groupings, the overlap between MLST ST8 and spa type t008 was less substantial. Only 79 isolates were MLST ST8 and spa t008 (center of diagram). Notably, there were 21 isolates that were MLST ST8, but not spa t008 (lower right region of diagram). In addition, there were 2 isolates that were spa t008, but not MLST ST8 (lower left region of diagram).
Discussion
A primary purpose of this study was to determine the usefulness of MLST relative to spa typing for epidemiology and evolutionary studies of S. aureus. We observed a high degree of concordance between MLST STs and spa types (0.906), and the concordance was even higher between MLST CCs and spa CCs (0.966). The MLST CC versus spa CC concordance reported here is similar to previously published values of 0.937, 0.968, and 0.986,7,19,16 while the MLST ST versus spa type concordance reported here is slightly lower than previously reported values, 0.963 and 0.954.7,19 Minor differences in the estimates can possibly be attributed to geographic differences between the S. aureus populations, particularly for MRSA, as the Hallin and Strommenger studies were performed in Europe, while our isolates were from one region in the United States.
In keeping with previous studies, the calculated DIs in the current study showed that overall spa types had slightly higher discriminatory power than MLST STs (0.890 and 0.820, respectively). However, with BURP and BURST algorithms to cluster spa types into spa CCs and MLST STs into MLST CCs, the DIs were seen to be equivalent (0.797 and 0.787, respectively).
Within typing methods, the DIs were generally consistent regardless of patient age, location, or specimen site. Likewise, the concordance rates were remarkably consistent when we analyzed the data by patient age, location, or specimen site. This indicates that use of either MLST CCs or spa CCs is appropriate for macroepidemiology and evolutionary studies that track clonal lineages of S. aureus through time and/or geography. Nevertheless, spa typing may be preferred over MLST in that when the spa types are looked at before clonal grouping by BURP, there is the added advantage of additional discriminatory power between closely related strains.
In contrast to the consistency of results for patient age, location, or specimen site, methicillin resistance status had a profound effect on typing results. Within MRSA, the DIs were all relatively lower, reflecting the clonal nature of MRSA in the United States, but the DI for spa types was substantially higher than the DIs for MLST STs, MLST CCs, or spa CCs. Within MRSA, the concordance between spa types and MLST STs was 0.801, which is significantly lower than the overall rate of 0.906, while the concordance between spa CCs and MLST CCs was 0.973, similar to the overall rate of 0.966.
This leads us to conclude that the principle difference between spa typing and MLST is that spa types have higher discriminatory power within clonal lineages of MRSA versus MLST STs. spa typing is more likely to assign two closely related MRSA isolates to different types than does MLST. This increased discriminatory power is intraclonal and thus probably of limited value in understanding broad macroepidemiological and evolutionary trends. Nevertheless, when spa types are clustered into clonal lineages via the BURP algorithm, the resulting spa CCs are highly comparable with MLST CCs. On the other hand, where discriminatory power is highly valued for studies of MRSA, such as outbreak investigations within a single institution, spa typing is preferred over MLST.
Both MLST and spa typing identified two major clonal lineages of MRSA: MLST ST8, MLST CC8, spa type t008, and/or spa CC008 (representing the USA300 community MRSA lineage) and MLST ST5, MLST CC5, spa type t002, and/or spa CC002 (representing the USA100 traditional hospital MRSA lineage). That both MLST and spa typing-based methods identify the same major lineages provides evidence that both typing systems accurately identify true clonal lineages based on common descent.
We further explored the relationship between spa typing and MLST by analyzing the characteristics of the two major MRSA clones as identified by each of the methods. We assessed the proportion of isolates within each MRSA clone that was isolated from children under age 18, from outpatients, and from skin infections because the two major clones differ in the proportions from these categories.15 We also determined the proportion of isolates carrying the genes for PVL as this has been reported as a good marker of community MRSA in the United States.3 We observed that regardless of typing method, the USA300 lineage MRSA were all very similar in terms of the proportions of isolates from children, outpatients, and skin, and all were around 95% PVL positive.
Across typing methods, the USA100 lineage had no isolates from children, fewer isolates from outpatients and skin, and all isolates were PVL negative. These observations lead to two conclusions. First, spa typing and MLST are similarly effective in distinguishing between the two major clones of MRSA in the United States. Second, the more inclusive spa CC and MLST CC classifications of the MRSA clones should be preferred because using spa types or MLST STs alone without the clonal grouping algorithms could potentially exclude rare or novel variants that are actually representatives of one of the major clonal lineages.
This is especially true for spa typing due to the tendency of spa types to be more discriminatory within clonal lineages of MRSA. Studies assessing USA300, for example, might miss members of that lineage using spa types alone, as evidenced here where 24 isolates were not spa type t008, but were spa CC008.
That the major U.S. MRSA clonal lineages, as defined by MLST CCs or spa CCs, have similar profiles (in terms of patient demographics, presence of PVL, etc.) to the lineages as defined by spa types or MLST STs supports a model in which there are two major clonal lineages of MRSA in the United States, which combined represented ∼90% of the MRSA in our study, and that rare and novel variants represent the accumulation of molecular variation within the lineages. In contrast, these data are inconsistent with a model in which there exist narrowly defined “true” versions of USA300 and USA100, and rare and novel variants represent distinct evolutionary branches.
Note that this analysis applies to MRSA lineages in the United States; analysis of isolates from several European countries indicates the presence of a variety of MRSA clones and variants, possibly due to the dissemination of varied clonal types among the different European countries.17
The Venn diagram approach was useful for assessing the overlap among spa typing and MLST in how they describe the much studied USA300 clone. Out of 107 PVL-positive MRSA isolates that were within the more inclusive clonal groups (MLST CC8 and/or spa CC008), 79 (74%) were identical across the four possible categories, MLST ST8, MLST CC8, spa type t008, and spa CC008. Thus, there was substantial overlap among methods, and we can conclude that they are all describing the same clone.
It is worth noting that the overlap between MLST CC8 and spa CC008 was nearly perfect, with 105 of 107 isolates (98%) overlapping. This provides strong evidence that both the BURST (MLST CC) and BURP (spa CC) clustering methods are independently identifying the same clonal lineage and use of either of these methods will capture the majority of isolates in the USA300 lineage.
Notably, the overlap between spa types and MLST STs was not as great as the overlap between MLST CCs and spa CCs. This supports the model in which USA300 is best represented as a clonal lineage within which minor variants have arisen over time. That spa types identified only 82 out of a potential 107 isolates (77%) as USA300 versus 100 of 107 for MLST STs (93%) is consistent with spa types overdiscriminating within MRSA clones and excluding minor variants of the lineage. Nevertheless, the BURP algorithm successfully linked related spa types into a single spa CC representing the USA300 lineage.
Conclusions
Two main conclusions are derived from this study. First, spa typing may be preferred over MLST. In agreement with previous studies,8,11,19 we show high concordance between MLST and spa typing and further demonstrate that DI and concordance values are consistent for age groups, settings, and infection source. Both MLST STs and spa types provide good discriminatory power; however, spa types provide the strongest discriminatory power. Furthermore, when spa types are combined with BURP analysis, the resulting spa CCs are adequate for describing the clonal structure of the S. aureus population in the United States.
It is important to note that the use of spa typing or MLST without the appropriate clustering algorithm may result in clinically relevant isolates being overlooked. The greater discriminatory power of spa types combined with the ability to cluster closely related spa types into spa CCs via the BURP algorithm, demonstrated here to be highly concordant with BURST-derived MLST CC classification, demonstrate that spa typing is a method that is suited both to macroepidemiological and evolutionary studies (provided the BURP algorithm is used to cluster spa types), as well as studies involving closely related strains such as MRSA outbreaks in a single institution. Given that spa typing requires fewer PCR amplification and sequencing reactions than MLST, its relative ease of use and lower cost make it a good choice for typing S. aureus isolates in the United States.
Second, spa CCs or MLST CCs combined with methicillin resistance status are sufficient to accurately classify the majority of S. aureus isolates in the United States into clonal lineages. We find that for U.S. S. aureus isolates, methicillin resistance (without the need for the more detailed SCC mec typing to determine the subtype of methicillin resistance) combined with the spa type/spa CC or MLST ST and CC designation is sufficient to classify an isolate into USA100, USA300, a minor MRSA clone, or the diverse class of polyclonal MSSA.
Of note, we found that PVL testing was largely unnecessary for assigning an isolate to one of the major MRSA lineages as long as the CC and methicillin resistance status were known. This is because we observed the PVL genes in over 95% of MRSA isolates from spa CC8/MLST CC8, and we never observed them in spa CC002/MLST CC5. Thus, determination of spa CCs or MLST CCs combined with methicillin resistance status (microbiologically or through testing for the presence of the mecA gene) is sufficient to identify at least 95% of isolates from the USA300 or USA100 lineages.
Acknowledgments
The authors thank James LaPan from the Kaiser Permanente Northern California Regional Lab for providing the specimens used in these analyses. The authors gratefully acknowledge the GSK U.S. Sequencing Facility, with particular thanks to Ganesh Sathe. The authors also thank Heather Santiago (GSK Vaccines) and Jennifer Dorts and Marie Cloes (Business and Decision Life Sciences, on behalf of GSK Vaccines) for managing the publication. This study has been funded by GlaxoSmithKline Biologicals SA. This original research is a collaboration among researchers from Kaiser Permanente Vaccine Study Center and Research Division, and GSK Vaccines. Decisions on study design, data collection and analysis, decision where to publish, and preparation of the manuscript were done solely and collectively by the authors.
Authors' Contributions
Conception and design: F.P.O., J.A.S., G.T.R., R.B., R.M.M., and H.A.M.; Acquisition of data/isolates: G.T.R., R.B., and E.T.; Molecular typing and analysis of isolates: F.P.O., M.L.B., N.M.C., and H.A.M.; Interpretation of data: F.P.O., J.A.S., G.T.R., R.B., M.L.B., R.M.M., and H.A.M.; Drafting of the manuscript: F.P.O., J.A.S., G.T.R., and H.A.M.; Critical revision of the manuscript for important intellectual content: F.P.O., J.A.S., G.T.R., R.B., M.L.B., R.M.M., N.M.C., E.T., and H.A.M.; Statistical analysis: F.P.O., J.A.S., R.M.M., and H.A.M.; Obtaining funding: J.A.S.; Supervision: F.P.O., J.A.S., and H.A.M. All authors read and approved the final manuscript.
Disclosure Statement
F.P.O., J.A.S., and R.M.M. were employees of the GSK group of companies at the time of this study. J.A.S. held stock options of the GSK group of companies at that time. M.L.B., E.T., N.M.C., and H.A.M. are employees of the GSK group of companies, and H.A.M. holds stock options and restricted shares of the GSK group of companies. The institution of G.T.R. has received a grant and support for travelling from the GSK group of companies and grants from Pfizer, Merck, and Purdue Pharma L.P. The institution of R.B. has received research grants from the GSK group of companies (including funding for this study) and from Merck and Pfizer. J.A.S. currently works at Pfizer.
References
- 1.Blanc D.S., Francioli P., and Hauser P.M. 2002. Poor value of pulsed-field gel electrophoresis to investigate long-term scale epidemiology of methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 2:145–148 [DOI] [PubMed] [Google Scholar]
- 2.Cookson B.D., Aparicio P., Deplano A., Struelens M., Goering R., and Marples R. 1996. Inter-centre comparison of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 44:179–184 [DOI] [PubMed] [Google Scholar]
- 3.David M.Z., and Daum R.S. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23:616–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright M.C., Day N.P., Davies C.E., Peacock S.J., and Spratt B.G. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright M.C., Robinson D.A., Randle G., Feil E.J., Grundmann H., and Spratt B.G. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. U. S. A. 99:7687–7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feil E.J., Li B.C., Aanensen D.M., Hanage W.P., and Spratt B.G. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goering R.V., Shawar R.M., Scangarella N.E., O'Hara F.P., Amrine-Madsen H., West J.M., Dalessandro M., Becker J.A., Walsh S.L., Miller L.A., van Horn S.F., Thomas E.S., and Twynholm M.E. 2008. Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J. Clin. Microbiol. 46:2842–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallin M., Deplano A., Denis O., De Mendonca R., De Ryck R., and Struelens M.J. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45:127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmsen D., Claus H., Witte W., Rothgänger J., Claus H., Turnwald D., and Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter P.R., and Gaston M.A. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koreen L., Ramaswamy S.V., Graviss E.A., Naidich S., Musser J.M., and Kreiswirth B.N. 2004. Spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDougal L.K., Steward C.D., Killgore G.E., Chaitram J.M., McAllister S.K., and Tenover F.C. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellmann A., Friedrich A.W., Rosenkotter N., Rothgänger J., Karch H., Reintjes R., and Harmsen D. 2006. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med. 3:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nübel U., Roumagnac P., Feldkamp M., Song J.H., Ko K.S., Huang Y.C., Coombs G., Ip M., Westh H., Skov R., Struelens M.J., Goering R.V., Strommenger B., Weller A., Witte W., and Achtman M. 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 105:14130–14135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Hara F.P., Amrine-Madsen H., Mera R.M., Brown M.L., Close N.M., Suaya J.A., and Acosta C.J. 2012. Molecular characterization of Staphylococcus aureus in the United States 2004–2008 reveals the rapid expansion of USA300 among inpatients and outpatients. Microb. Drug Resist. 18:555–561 [DOI] [PubMed] [Google Scholar]
- 16.Pinto F.R., Melo-Cristino J., and Ramirez M. 2008. A confidence interval for the wallace coefficient of concordance and its application to microbial typing methods. PLoS One 3:e3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rolo J., Miragaia M., Turlej-Rogacka A., Empel J., Bouchami O., Faria N.A., et al. 2012. High genetic diversity among community-associated Staphylococcus aureus in Europe: results from a multicenter study. PLoS One 7:e34768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefani S., Chung D.R., Lindsay J.A., Friedrich A.W., Kearns A.M., Westh H., and Mackenzie F.M. 2012. Methicillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents 39:273–282 [DOI] [PubMed] [Google Scholar]
- 19.Strommenger B., Braulke C., Heuck D., Schmidt C., Pasemann B., Nübel U., and Witte W. 2008. spa Typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J. Clin. Microbiol. 46:574–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strommenger B., Kettlitz C., Weniger T., Harmsen D., Friedrich A.W., and Witte W. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 44:2533–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]