Abstract
Background
Studies in Southeast Asia reported a strong relationship between polymorphisms at the propeller domain of the Kelch 13 (K13) protein encoded by the Plasmodiumfalciparumk13(pfk13) gene and delayed parasite clearance after artemisinin treatment. In Africa, P. falciparum remains susceptible and combination therapy regimens which include an artemisinin component display good efficacy. Using quantitative real-time PCR (qPCR), sub-microscopic persistence of P. falciparum has previously been reported in one-third of children treated with artemisinin combination therapy (ACT) in western Kenya. In this study, further investigation was made to evaluate whether these sub-microscopic residual parasites also harbour mutations at the propeller region of pfk13 and whether the mutations, if any, affect treatment outcome.
Methods
The pfk13 propeller domain was genotyped in DNA samples obtained in 2009 from Kenyan children treated with artemether–lumefantrine (AL) and dihydroartemisinin–piperaquine (DP). Paired samples at pre-treatment (day 0) and day of treatment failure (day 28 or 42) for 32 patients with documented recurrent parasitaemia were available for genotyping. Additional day 3 DNA samples were available for 10 patients.
Results
No mutation associated with artemisinin resistance in Southeast Asia was observed. Only one DP-treated patient harboured a non-synonymous mutation at codon 578 (A578S) of pfk13-propeller gene in the day 0 sample, but this allele was replaced by the wild-type (A578) form on day 3 and on the day of recurrent parasitaemia. The mutation at amino acid codon 578 showed no association with any phenotype. Polymorphisms in pfk13 were not responsible for parasite persistence and gametocyte carriage in the children treated with ACT.
Conclusion
This study contributes to the ongoing surveillance of suspected artemisinin resistance parasites in Africa by providing baseline prevalence of k13-propeller mutations in western Kenya with samples collected from a longitudinal study.
Clinical Trials Registration NCT00868465.
Keywords: k13-propeller, Artemisinin resistance, Western Kenya, Africa, Slow clearance, Sub-microscopic, qPCR
Background
Artemisinin combination therapy (ACT) is widely used to treat uncomplicated Plasmodium falciparum malaria in endemic areas. Significantly reduced susceptibility to artemisinins, characterized by delayed parasite clearance in vivo and enhanced survival of early stage parasites in vitro, has been reported in Southeast Asia [1, 2]. Most recently, these parasite phenotypes have been associated with mutations in the pfk13 gene of P. falciparum, encoding the kelch-domain protein K13 [3]. Further studies reported direct evidence of resistance to artemisinin using genetic modification of different pfk13 loci [4]. This work has led to an operational definition of partial artemisinin resistance: ≥5 % of patients carrying K13 resistance-associated mutations, all of whom have been found to have either persistent parasitaemia by microscopy on day 3, or a parasite clearance half-life of ≥5 h after treatment with ACT or artesunate monotherapy [5].
In Africa, ACT is the first-line anti-malarial treatment and remains efficacious and safe [3, 6]. However, resistance to previous generations of anti-malarial drugs such as chloroquine, sulfadoxine-pyrimethamine (SP) emerged in the 1970s in Southeast Asia and eventually spread to the Indian sub-continent and then to Africa [7]. It is, therefore, critical that continuous monitoring of the therapeutic response of ACT is carried out in endemic areas in order to detect early warning signs and effectively track and contain the development and spread of artemisinin resistance. Currently, delayed clearance of microscopically detectable parasites has not been observed in Africa. In addition, there are limited data about the in vitro susceptibility of ring stage of parasite development against artemisinins as well as the presence and prevalence of pfk13 mutations [8]. Recently, a cross-sectional survey was carried out to determine the prevalence of pfk13 mutations in 14 sub-Saharan African countries. The study reported the absence of mutations that were associated with artemisinin resistance in Southeast Asia, with the exception of codon 543 [9]. In previous study, the presence of residual sub-microscopic P. falciparum parasites in western Kenyan children on day 3 after ACT treatment was reported and this was associated with subsequent recrudescence and transmission [10]. In addition, the authors have shown in the same Kenyan patient that P. falciparum parasites carrying certain genotypes at pfmdr1, pfcrt, pfubp1, and pfap2mu genes were found to survive more often after ACT treatment at sub-microscopic level [11]. The aim of this study was to investigate whether these sub-microscopic residual parasites also harboured mutations at the propeller region of pfk13 and whether the mutations, if any, affect treatment outcome.
Methods
DNA samples were previously obtained from filter-paper blood spots from children (age 8 months–10 years) treated with either artemether–lumefantrine (AL) or dihydroartemisinin–piperaquine (DP) in a clinical trial carried out in 2009 in Mbita, western Kenya and fully described elsewhere [6, 10]. The study was carried out 5 years after Kenya had officially changed the first line of treatment from SP to AL [12]. The protocol was approved by the Kenya Medical Research Institute Ethical Review Committee and the London School of Hygiene and Tropical Medicine Ethics Committee (reference 5455). Written informed consent was obtained from a parent or guardian of each participating child. This is an ancillary study of pfk13 propeller domain polymorphism in DNA samples available from the 2009 study at the following time points: for 32 patients with documented recurrent parasitaemia, paired samples at day 0 (pretreatment) and day of treatment failure (28 or 42, PCR-uncorrected); for 10 patients, additional day 3 DNA samples (Table 1).
Table 1.
Sample availability for k13 genotyping
| Sample ID | D0 | D1 | D2 | D3 | D28 | D42 |
|---|---|---|---|---|---|---|
| K0025 | X | X | ||||
| K0344 | X | X | ||||
| K0385 | X | X | ||||
| K0544 | X | X | ||||
| K0598 | X | X | ||||
| K0701 | X | X | X | |||
| K0719 | X | X | X | |||
| K0774 | X | X | ||||
| K0804 | X | X | ||||
| K0840 | X | X | ||||
| K0875 | X | X | ||||
| K0881 | X | X | ||||
| K1010 | X | X | ||||
| K1149 | X | X | X | |||
| K1152 | X | X | ||||
| K1307 | X | X | ||||
| K1348 | X | X | X | |||
| K1368 | X | X | X | X | X | |
| K1438 | X | X | X | |||
| K1478 | X | X | ||||
| K1521 | X | X | ||||
| K1861 | X | X | X | |||
| K1917 | X | X | ||||
| K1943 | X | X | ||||
| K2023 | X | X | ||||
| K2024 | X | X | ||||
| K3077 | X | X | ||||
| K3212 | X | X | X | |||
| K3215 | X | X | X | |||
| K3404 | X | X | X | |||
| K3762 | X | X | ||||
| K3801 | X | X |
Table shows sample which were available for k13 genotyping on different time points. Days 0 and day of fail (days 28 or 42) samples were available for k13 genotyping for 32 patients. Of the 32, ten patients had additional samples on day 3 while for patient K3168, additional samples on days 1 and 2 were included retrospectively for further k13 genotype analysis
The K13-propeller gene fragment (coordinates 1726169–1726997 on chromosome 13 of 3D7 isolate (PF3D7_1343700) was amplified by nested PCR using previously published primers [13]. For the first round of PCR, 250 nM primer (Eurofins, Germany), 1X hot fire pol® blend master mix (Solis Biodyne, Estonia) and 5 μl of DNA in a total volume of 20 µl was run under the following cycling conditions: 95 °C for 15 min then 30 cycles at 95 °C for 30 s, 58 °C for 2 min and extension at 72 °C for 2 min and final extension at 72 °C for 10 min. For the second round of PCR, a total volume of 25 μl made of 18.75 μl nuclease free water, 250 nM primer, 1X hot fire pol® blend master mix (Solis Biodyne, Estonia) final concentration and 5 μl of DNA was run under the following cycling conditions: 95 °C for 15 min then 40 cycles at 95 °C for 30 s, 60 °C for 1 min and extension at 72 °C for 1 min and final extension at 72 °C for 10 min. A positive control (K13_5) with one of the mutations identified in Cambodia (provided by D Ménard) was included in every experiment.
Polymorphisms at pfk13 were determined by direct sequencing of amplicons using ABI BigDye Terminator v3.1 cycling sequencing kit and analysis on an ABI 3730 sequencer (Applied Biosystems, USA) as described previously [11] and chromatogram sequences were analysed using Geneious v6.1.5 (Biomatters, USA) in comparison to pfk13-propeller sequence region of 3D7 isolate (PF3D7_134700).
Results
Pfk13 sequences for 32 samples on day 0, seven samples on day 3 and 30 samples on day of failure were successfully obtained and analysed for sequence variation. Of these 69 sequenced isolates, 68 harboured pfk13 loci encoding propeller domains identical to the reference. A single DP-treated patient (K1368) harboured a non-synonymous mutation at codon 578 of pfk13-propeller gene in the day 0 sample, but this allele was replaced by the wild-type form on day 3 and on the day of recurrent parasitaemia (day of failure) (Table 2). The K13-propeller sequence was deposited in Gene Bank (accession number KT261646). To understand the clearance dynamics of the mutant parasites in the patient, pfk13 was retrospectively sequenced in day 1 and day 2 parasites from this individual. Sequence analysis of day 0, day 1 and day 2 samples revealed the presence of mutation at codon A578S, suggesting that the mutant parasite was present 2 days after DP treatment (Figure 1). The treatment outcome and other phenotype and genotype data for the patient are found in Table 2. The patient cleared parasites as measured by microscopy on day 1 after DP treatment and no gametocytes were detected on day 7. Recurrent parasitaemia was observed on day 42 but was classified as new infection after PCR-correction. One additional patient carried a synonymous mutation at pfk13 codon P553P on day of failure but that was not observed on day 0 or day 3.
Table 2.
Phenotype and genotype outcomes of patient K1368
| Patient ID | Phenotype | ||||
|---|---|---|---|---|---|
| D3 Parasite positive | Gametocyte D7 positivity | Mosquito infection | Recurrent parasitaemia | PCR correction | |
| K1368 | No | No | No | Day 42 | New infection |
| Patient ID | Genotype | ||||
|---|---|---|---|---|---|
| D0 Pfk13 at 578 | D3 Pfk13 at 578 | D0 Pfmdr1 at 86/184/1246 | Day 0 Pfcrt at 72–76 | D0 PfAP2mu S160 N | |
| K1368 | S | A | NFD | CVMNK | S |
The patient phenotypic data show typical normal clearing parasites and all samples before treatment show the wild type alleles with the exception of pfk13, where the mutant is present at codon 578. However, 3 days after treatment only the wild type of codon 578 was observed. When the pfk13 genotyping on day 3 was repeated no amplification was observed
Fig. 1.
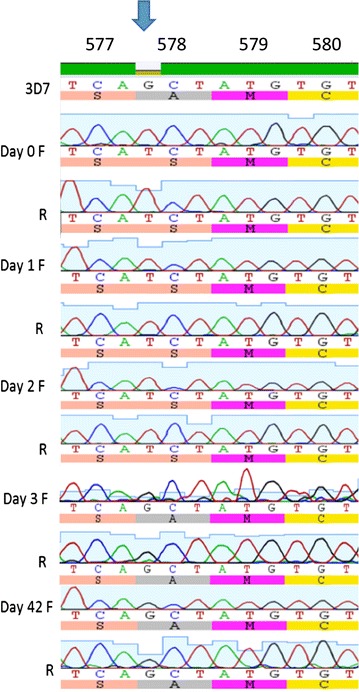
DNA and amino acid sequence of a propeller region of PfK13 showing mutation at codon 578. Sequence of samples from patient K1368 at day 0, 1 and 2 show mutation at amino acid position 578 while the samples on day 3 and day 42 have no mutations. DNA amplification of sample on day 3 using pfK13 [13] and pgmet primers [18] repeatedly failed. DNA and amino acid sequence of 3D7 isolate was used as a reference. Each sample has a sequence using forward (F) and reverse (R) primers. Arrow indicates mutation and top numbers indicate amino acid position
Discussion
No evidence was found that artemisinin resistance-associated mutations in the pfk13-encoded propeller domain, including the major resistance-associated mutation C580Y, contribute to ACT treatment failure in Mbita, western Kenya. Only one of the 32 patients from this longitudinal study had mutation in the propeller region of pfk13 gene, despite evidence of parasite persistence in over 30 % of children [10]. One patient transiently carried parasites harbouring mutation at codon 578 (A578S) on enrolment, and on days 1 and 2 after DP treatment. However, these parasites were apparently cleared by DP in this individual, and persistent parasites detected at day 3 and day 42 harboured the wild type A578 allele only. However, it is not clear whether the wild type was already present on days 0, 1 and 2 but below the detection limit of the assay. The importance of multiplicity of infection was discussed in detail in previous work and the findings broadly agree with Farnert et al., who also showed that there were genetic changes in samples collected in the same patient as short as 6 h apart [10, 14]. In fact, the msp1 and msp2 genotyping of the day 0 sample showed that the patient was infected with at least three different parasite clones. Blood from this patient was presented to Anopheles gambiae mosquitoes at day 7 by membrane feeding, as reported in previous study, but no infected mosquitoes were generated [6].
The A578S polymorphism has previously been reported in a cross-sectional study in western Kenya, Kisumu, and in four other African countries [9], but the current study is the first study to investigate the role of this variant in gametocyte carriage and infectiousness to mosquito. These findings are consistent with recent published multisite studies in Southeast Asia and Africa that showed mutation at codon 578 in pfk13 and its lack of association with parasite clearance half-life in patients treated with ACT [3]. In Bangladesh, the mutation at codon 578 has also been observed in clinical isolates and using computational modelling, the authors suggested that it has an effect on tertiary structure of the protein [15]. Although the parasite harbouring the mutant A578S allele was successfully cleared by DP, the contribution of piperaquine in clearing these parasites is not clear. This polymorphism lies adjacent to C580Y, and further in vivo and in vitro studies to clarify the significance of the A578S mutation are warranted, particularly in different genetic backgrounds. Mutations in pfcrt, pfmdr1, pfap2mu, and pfubp1 also contribute to treatment outcomes after either artemisinin monotherapy or ACT treatment [11, 16]. Studies introducing transgenic variants of pfk13 and other candidate loci into parasite isolates with an African genetic background could clarify whether pfk13 mutations or polymorphisms in other genes are most relevant in determining artemisinin sensitivity in clinical isolates in Africa [4, 17].
The rarity of pfk13 variants in the current small study precludes from making meaningful assessment of the role of A578S and other propeller domain mutations in treatment outcomes in Africa. However, it can be confirmed that the sub-microscopic clearance phenotype observed in western Kenya is not directly related to the parasite clearance half-life observed in Southeast Asia. The sub-microscopic residual parasites have been shown previously to be important phenotypes as children harbouring those parasites were significantly more likely to be infectious to mosquitoes and were more likely to have recurrent asexual parasitaemia on day 28 or 42 [10]. Factors associated with the host, drug and parasite should be investigated to determine the exact cause of the sub-microscopic residual parasites.
This study contributes to the ongoing surveillance of suspected artemisinin resistance parasites in Africa by providing baseline prevalence of k13-propeller mutations in western Kenya with samples collected from a longitudinal study, and thus one of the first to relate genotype and phenotype. This information from samples collected in 2009 does not reflect the current status of ACT, nor the prevalence of k13-propeller mutations now. Therefore, continuous monitoring of ACT in western Kenya and the collation of phenotypic and genotypic data to track the emergence and spread of parasites with reduced susceptibility to artemisinins is recommend.
Authors’ contributions
PS, TB, CJS, and KBB contributed to study design. PS collected clinical data. JM, GH, CJS and KBB did laboratory work. JM, CJS and KBB analysed the data. JM, CJS and KBB wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the European Community’s Seventh Framework Programme [grant 201889 to Project MALACTRES (Multi-drug resistance in malaria under combination therapy: assessment of specific markers and development of innovative, rapid and simple diagnostics)], the Bill and Melinda Gates Foundation (grant OPP1024438 to TB), Public Health England (to CJS) and the EDCTP WANECAM Consortium (to KBB).
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- ACT
artemisinin combination therapy
- AL
artemether–lumefantrine
- DP
dihydroartemisinin–piperaquine
- EDCTP
European and developing countries clinical trials partnership
- pfk13
Plasmodium falciparum kelch 13 gene
- qPCR
quantitative (real-time) PCR
Contributor Information
Julian Muwanguzi, Email: julian.muwanguzi@lshtm.ac.uk.
Gisela Henriques, Email: gi_pt@yahoo.com.
Patrick Sawa, Email: psawa@icipe.org.
Teun Bousema, Email: teun.bousema@lshtm.ac.uk.
Colin J. Sutherland, Email: colin.sutherland@lshtm.ac.uk
Khalid B. Beshir, Email: khalid.beshir@lshtm.ac.uk
References
- 1.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in vitro and ex vivo drug-response studies. Lancet Infect Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, et al. Drug resistance K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. Status report on artemisinin resistance. 2014. WHO/HTM/GMP/2014.9. http://www.who.int/malaria/publications/atoz/status_rep_artemisinin_resistance_sep2014.pdf?ua=1. Accessed 10 March 2015.
- 6.Sawa P, Shekalaghe SA, Drakeley CJ, Sutherland CJ, Mweresa CK, Baidjoe AY, et al. Malaria transmission after artemether–lumefantrine and dihydroartemisinin–piperaquine: a randomized trial. J Infect Dis. 2013;207:1637–1645. doi: 10.1093/infdis/jit077. [DOI] [PubMed] [Google Scholar]
- 7.Roper C, Alifrangis M, Ariey F, Talisuna A, Menard D, Mercereau-Puijalon O, et al. Molecular surveillance for artemisinin resistance in Africa. Lancet Infect Dis. 2014;14:668–670. doi: 10.1016/S1473-3099(14)70826-6. [DOI] [PubMed] [Google Scholar]
- 8.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, et al. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis. 2015;211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beshir KB, Sutherland CJ, Sawa P, Drakeley CJ, Okell L, Mweresa CK, et al. Residual Plasmodium falciparum parasitemia in Kenyan children after artemisinin-combination therapy is associated with increased transmission to mosquitoes and parasite recurrence. J Infect Dis. 2013;208:2017–2024. doi: 10.1093/infdis/jit431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henriques G, Hallett RL, Beshir KB, Gadalla NB, Johnson RE, Burrow R, et al. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis. 2014;210:2001–2008. doi: 10.1093/infdis/jiu358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amin AA, Walley T, Kokwaro GO, Winstanley PA, Snow RW. Reconciling national treatment policies and drug regulation in Kenya. Health Policy Plan. 2007;22:111–112. doi: 10.1093/heapol/czl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnert A, Lebbad M, Faraja L, Rooth I. Extensive dynamics of Plasmodium falciparum densities, stages and genotyping profiles. Malar J. 2008;7:241. doi: 10.1186/1475-2875-7-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohon AN, Alam MS, Bayih AG, Folefoc A, Shahinas D, Haque R, et al. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009–2013) Malar J. 2014;13:431. doi: 10.1186/1475-2875-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriques G, van Schalkwyk DA, Burrow R, Warhurst DC, Thompson E, Baker DA, et al. The Mu subunit of Plasmodium falciparum clathrin-associated adaptor protein 2 modulates in vitro parasite response to artemisinin and quinine. Antimicrob Agents Chemother. 2015;59:2540–2547. doi: 10.1128/AAC.04067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beshir KB, Hallett RL, Eziefula AC, Bailey R, Watson J, Wright SG, et al. Measuring the efficacy of anti-malarial drugs in vivo: quantitative PCR measurement of parasite clearance. Malar J. 2010;9:312. doi: 10.1186/1475-2875-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]


