Abstract
Synthetic polymers are widely used in daily life. Due to increasing environmental concerns related to global warming and the depletion of oil reserves, the development of microbial-based fermentation processes for the production of polymer building block chemicals from renewable resources is desirable to replace current petroleum-based methods. To this end, strains that efficiently produce the target chemicals at high yields and productivity are needed. Recent advances in metabolic engineering have enabled the biosynthesis of polymer compounds at high yield and productivities by governing the carbon flux towards the target chemicals. Using these methods, microbial strains have been engineered to produce monomer chemicals for replacing traditional petroleum-derived aliphatic polymers. These developments also raise the possibility of microbial production of aromatic chemicals for synthesizing high-performance polymers with desirable properties, such as ultraviolet absorbance, high thermal resistance, and mechanical strength. In the present review, we summarize recent progress in metabolic engineering approaches to optimize microbial strains for producing building blocks to synthesize aliphatic and high-performance aromatic polymers.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-016-0411-0) contains supplementary material, which is available to authorized users.
Keywords: Metabolic engineering, Bio-polymers, Lactic acid, Succinic acid, Adipic acid, Putrescine, Cadaverine, High-performance polymers, Corynebacterium glutamicum, Escherichia coli
Background
Since the discovery and commercialization of synthetic polymers, these materials have become essential for everyday life [1]. Currently, almost all polymer building block chemicals are produced by petroleum-based chemical processes. Although such processes are able to produce a wide variety of materials at relatively low cost, these methods are inherently non-sustainable and have detrimental impacts on the environment. For these reasons, there is increasing global demand to replace petroleum-based production processes with microbial synthetic procedures that utilize renewable resources. The bio-based production of polymer building block chemicals is also advantageous because the synthetic reactions can be performed at near-standard temperatures and pressures, which markedly reduces the amount of required energy.
Despite the clear advantages of bio-based chemicals, poly-l-lactic acid (PLLA) is to date perhaps the only good example of successful industrialization of a 100 % bio-based polymer [2]. The main limitation for shifting to microbial synthetic processes is the high production cost. Specifically, the volumetric and specific productivities, and yields of target compounds by microbial fermentation, are often much lower than those obtained by chemical synthetic processes. For these reasons, the engineering of microbial strains that rapidly reach high-cell densities and have productivities and yields of target compounds at close to the theoretical maxima is needed for the commercialization of bio-based products. Genomic sequencing has opened the door for systems metabolic engineering for many industrially important microorganisms, such as Escherichia coli, Corynebacterium glutamicum, and Saccharomyces cerevisiae. In combination with genetic engineering tools and knowledge of metabolism and pathway regulation, sequence information has facilitated the rational design of strains with high productivity and yields of target compounds [3–8]. In addition, recent development of -omics techniques and computational tools have drastically accelerated the process of strain optimization [9].
In this review, we summarize recent knowledge of gene targets for metabolic engineering that efficiently convert glucose to building block chemicals (such as d-lactic acid, succinic acid, adipic acid, putrescine, and cadaverine) performed primarily in C. glutamicum and E. coli, that permit the synthesis of aliphatic polymer. We then expand the scope of our discussion to the production of other building block chemicals (such as d-phenyllactic acid, 3-amino-4-hydroxybenzoic acid, and cinnamic acid) for the synthesis of aromatic polymers.
Building block chemicals for aliphatic polymer synthesis
d-Lactic acid
Lactic acid (2-hydroxypropanoic acid) is synthesized in one step from pyruvate, the end product of the glycolytic pathway, by lactate dehydrogenase (LDH), which is encoded by the ldhA gene (Fig. 1). Lactic acid has two optical isomers, l- and d-lactic acid, whose synthesis is dependent on the chiral-specific L- or D-LDH enzyme expressed by the microorganism. The optical purity of lactic acid is critical for its polymeric characteristics, as small amounts of optical impurities drastically change properties such as crystallinity, which directly affects thermal resistance [10]. PLLA is the most common bio-based and biodegradable polymer, and is frequently utilized as a film due to its high transparency [11]. However, because this polymer has low melting and glass transition temperatures, PLLA’s use in practical applications is limited [12]. Stereocomplex PLA (scPLA) composed of both PLLA and poly-d-lactic acid can circumvent this defect [12]. To produce high-quality scPLA, microbial strains that produce l- and d-lactic acid with high optical purity are required. As the microbial production of l-lactic acid is well established [13, 14], this section focuses on recent advances in metabolic engineering approaches for producing optically pure d-lactic acid.
Fig. 1.
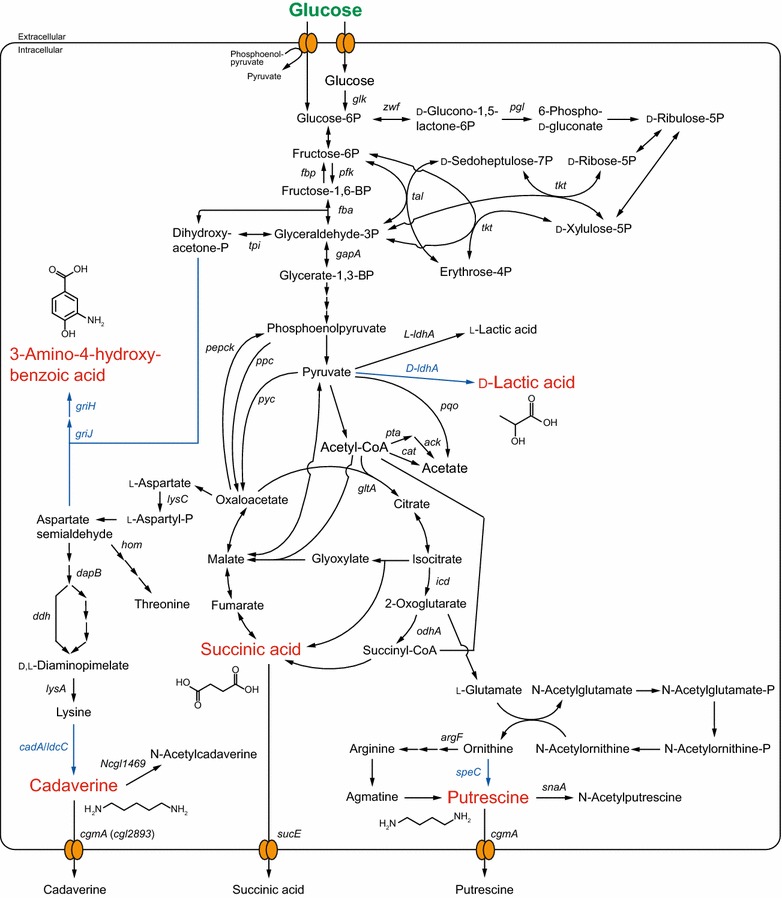
Schematic representation of the metabolic pathway in C. glutamicum for the production of building block chemicals (d-lactic acid, succinic acid, putrescine, cadaverine, and 3, 4-AHBA) for polymer synthesis. Substrate and target chemicals are presented in green and red, respectively. Heterologous genes and lines indicating the corresponding reactions are shown in blue. The deletion, overexpression, or nucleotide substitution of the genes indicated in the metabolic pathways leads to improved production of the target chemicals. Corresponding enzymes and functions are listed in Additional file 1: Table S1
Corynebacterium glutamicum, which is well known as a producer of amino acids such as glutamate and lysine [15, 16], displays arrested cell growth under oxygen-deprived conditions and also produces the organic acids l-lactate, succinate, and acetate [17]. The culturing in mineral salts medium of C. glutamicum at high cell-density under oxygen-deprived conditions led to the high volumetric productivity of organic acids [18]. Introduction of the D-LDH-encoding Lactobacillus delbrueckiildhA gene into a C. glutamicum mutant lacking the endogenous L-LDH-encoding gene yielded a strain that produced 120 g/L d-lactic acid of greater than 99.9 % optical purity (Table 1) [19]. Further disruption in this strain of the endogenous ppc gene (encoding phosphoenolpyruvate carboxylase, the primary source of succinic acid production) decreased succinic acid yield, but also reduced the glucose consumption rate [32]. The simultaneous overexpression of five glycolytic genes, namely glk (encoding glucokinase), gapA (encoding glyceraldehyde phosphate dehydrogenase), pfk (encoding phosphofructokinase), tpi (encoding triosephosphate isomerase), and fba (encoding bisphosphate aldolase), compensated for this impairment of glucose consumption and enabled the engineered C. glutamicum strain to produce 195 g/L d-lactic acid, corresponding to a yield of 1.80 mol/mol glucose (Fig. 1; Table 1) [20].
Table 1.
Summary of microbial production of polymer building block chemicals from glucose with notable productivities
| Monomer | Organism | Titer (g/L) | Yield (molproduct/molsubstrate) | Cultivation time (h) | References |
|---|---|---|---|---|---|
| d-Lactic acid | C. glutamicum R ΔldhA/pCRB204 | 120 | 1.73 | 30a | [19] |
| C. glutamicum R LPglc279/pCRB215 | 195 | 1.80 | 80a | [20] | |
| E. coli B0013-070B | 122 | 1.69 | 28 | [21] | |
| Succinic acid | C. glutamicum R ΔldhA/pCRA717 | 146 | 1.40 | 46a | [22] |
| C. glutamicum ATCC13032 BOL-3/pAN6-gap | 134 | 1.67 | 54a | [23] | |
| C. glutamicum ATCC13032 SA5 | 109 | 1.32 | 98a | [24] | |
| E. coli AFP111/pTrc99A-pyc | 99 | 1.10 | 76 | [25] | |
| E. coli KJ134 | 72 | 1.53 | 95 | [26] | |
| E. aerogenes ES08 ΔptsG | 55 | 1.32 | 60 | [27] | |
| Putrescine | E. coli XQ52/p15SpeC | 24 | – | 32 | [28] |
| C. glutamicum b | 19 | 0.33 | 34 | [29] | |
| Cadaverine | C. glutamicum DAP-16 | 88 | 0.50 | 40 | [30] |
| E. coli XQ56/p15CadA | 9.6 | 0.07 | 30 | [31] |
aOnly production phase
bStrain name unknown
Escherichia coli naturally produces optically pure d-lactic acid and has many advantages as a host for microbial production, such as simple nutritional requirements and well-established systems for genetic manipulation [33]. However, E. coli performs mixed-acid fermentation, in which the principal products are d-lactate, succinate, acetate, formate, and ethanol [33]. For this reason, attempts to increase d-lactic acid production by E. coli have mainly focused on minimizing the production of by-products without decreasing the growth or sugar consumption rates [34–37]. For example, Zhou and colleagues metabolically engineered E. coli for d-lactic acid production by the deletion of ackA (encoding acetate kinase), pta (encoding phosphotransacetylase), and poxB (encoding pyruvate oxidase) to minimize acetate production, in addition to deleting adhE (encoding alcohol dehydrogenase) to prevent ethanol fermentation, ppsA (encoding phosphoenolpyruvate synthase) and pflB (encoding pyruvate formate lyase) to promote pyruvate accumulation, and frdA (encoding fumarate reductase) to prevent succinic acid accumulation (Fig. 2) [38]. The resultant strain produced highly optically pure (>99.9 %) d-lactic acid at concentrations reaching 125 g/L in 39 h, corresponding to a yield of 0.87 g/g glucose. This group subsequently demonstrated that replacing the ldhA gene promoter with the λ PR and PL promoters and shifting the temperature from 33 to 42 °C (thereby permitting the strict separation of the growth and d-lactic acid production phases) improved d-lactate productivity by minimizing the inhibitory effect of the produced d-lactate on cell growth and increasing the activity of LDH [21]. Using this approach, the engineered strain produced 122 g/L d-lactic acid in 28 h at a yield of 0.84 g/g glucose (Table 1).
Fig. 2.
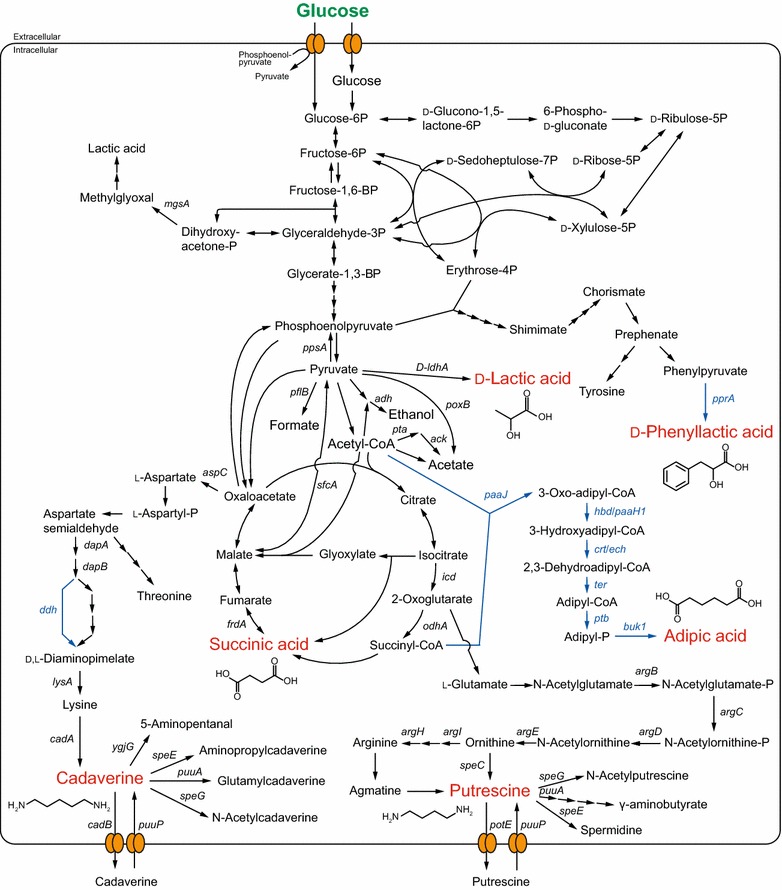
Schematic representation of the metabolic pathway in E. coli for the production of building block chemicals (d-lactic acid, succinic acid, adipic acid, putrescine, cadaverine, and phenyllactic acid) for polymer synthesis. Substrate and target chemicals are presented in green and red, respectively. Heterologous genes and lines indicating the corresponding reactions are shown in blue. The deletion, overexpression, or nucleotide substitution of the genes indicated in the metabolic pathways leads to improved production of the target chemicals. Corresponding enzymes and functions are listed in Additional file 1: Table S1
Succinic acid
Succinic acid (butanedioic acid) is a dicarboxylic acid of the tricarboxylic acid (TCA) cycle (Fig. 1) and is utilized as a building block for several commercially important polymers, such as polybutylene succinate adipate [39, 40]. Moreover, in combination with diamines, putrescine, and cadaverine, succinic acid can also be used to produce 100 % bio-based nylon materials [41, 42]. Therefore, the potential of bio-based processes to replace chemical-based succinic acid production has been extensively studied [6, 25, 42–44].
Several microorganisms, including Anaerobiospirillum succiniciproducens and Actinobacillus succinogenes, naturally produce high amounts of succinic acid as an end-product of anaerobic fermentation [45–48]. Under anaerobic conditions, succinic acid is produced mainly from phosphoenolpyruvate and pyruvate through anapleotic pathways and the reductive branch of the TCA cycle via the intermediates oxaloacetate, malate, and fumarate (Fig. 1) [19, 49]. Corynebacterium glutamicum produces small amounts of succinic acid under anaerobic conditions. However, by deleting the L-ldhA gene and overexpressing the pyc gene (encoding pyruvate carboxylase), Okino et al. engineered C. glutamicum to produce 146 g/L succinic acid with a yield of 1.40 mol/mol glucose in a two-stage (aerobic growth and anaerobic fed-batch production) system [22]. Despite this marked increase in succinic acid production, a large amount of acetate was still produced as a by-product. Additional deletions of genes in acetate-producing pathways, including pta (encoding phosphotransacetylase), ackA (encoding acetate kinase), cat (encoding acetyl-CoA:CoA transferase), and pqo (encoding pyruvate oxidoreductase) [50], in combination with the overexpression of a mutant pycP458S gene, fdh gene (encoding formate dehydrogenase) from Mycobacterium vaccae, and gapA gene, further increased the yield of succinic acid to 1.67 mol/mol glucose, corresponding to a titer of 133.8 g/L (Table 1) [23]. Overexpression of gltA (encoding citrate synthase) helped to channel more carbon towards the glyoxylate pathway, and overexpression of the previously identified sucE gene (encoding succinate exporter) in C. glutamicum [51, 52] was also employed. In combination, overexpression of these two genes provided 9 and 19 % increases in succinate yield and productivity, respectively [24].
Escherichia coli uses mixed-acid fermentation under anaerobic conditions to generate various products, including succinate, d-lactate, acetate, formate, and ethanol, as described above. Most studies aimed at increasing succinic acid production by E. coli have focused on eliminating the production of by-products and balancing the cellular redox state [43]. Using this approach, an E. coli strain was engineered for producing succinic acid in a single-step fermentation strategy. Specifically, deletion of the ldhA, adhE (encoding alcohol dehydrogenase), pflB, focA (encoding formate transporter), pta-ackA, mgsA (encoding methylglyoxal synthase), poxB (encoding pyruvate oxidase), and combined deletion of aspC (encoding aspartate aminotransferase) and sfcA (encoding malic enzyme) genes markedly reduced by-product formation and stimulated the reductive pathway, resulting in the production of 71.5 g/L succinic acid with a yield of 1.53 mol/mol glucose (Fig. 2; Table 1) [26].
Although the above-described C. glutamicum and E. coli strains were metabolically engineered to efficiently produce succinic acid, these strains are limited to growth at neutral pH conditions due to their sensitivity to acid stress [7, 53]. The production of organic acids is ideally performed at low pH to avoid the need for alkali solutions for pH neutralization during fermentation, and more importantly, to reduce the costs of downstream purification, which typically requires large quantities of acid [54, 55]. Saccharomyces cerevisiae is a promising candidate to overcome this limitation because of its high tolerance to acid stress, as demonstrated by its ability to grow at pH 3.0 [56, 57]. However, even after extensive metabolic engineering, the maximum succinic acid titer generated by S. cerevisiae remained too low for viable commercial production [58, 59]. Recently, Tajima and colleagues showed that the metabolic engineering of a newly isolated Gram-negative bacterium, Enterobacter aerogenes AJ110637, led to a producer of succinic acid under low pH conditions. This bacterium rapidly assimilated glucose at pH 5.0 [60]. Because the strain produced succinate, lactate, formate, and acetate (in addition to ethanol and 2, 3-butanediol) by mixed-acid fermentation, four genes [ldhA, adhE, pta, and budA (encoding α-acetolactate decarboxylase)] involved in by-product formation were deleted to minimize by-product accumulation. The gene-deleted strain was further engineered by overexpression of the pck gene (encoding phosphoenolpyruvate carboxykinase) from A. succinogenes and the pyc gene (encoding pyruvate carboxylase) from C. glutamicum, providing production of 11.2 g/L succinic acid at pH 5.7 [61]. However, this titer was 50 % lower than that obtained at pH 7.0, demonstrating that lowering the culture pH negatively impacts succinic acid production. To increase the acid tolerance of this strain, this group attempted to maximize adenosine-5′-triphosphate (ATP) yield, as used in E. coli [62, 63]. To accomplish this, the ptsG gene (encoding glucose-phosphotransferase system permease) was deleted, together with individual overexpression of the pck gene from A. succinogenes instead of the two anapleotic pathway genes. Further deletion of poxB and pflB, along with overexpression of frdABCD (encoding fumarate reductase), resulted in the production of 55.4 g/L succinic acid at pH 5.7 (Table 1) [27].
Adipic acid
Polyamide, commonly known as nylon, has recurring amide groups linking the monomers to chains, and shows high durability and strength. More than 6 million tons of nylon are produced annually, and this polymer is considered indispensable for modern life [64]. The most common commercial polyamides are nylon-6 and nylon-6, 6, which account for more than 90 % of the global market. Adipic acid (1, 4-butanedicarboxylic acid) is a building block dicarboxylic acid that permits (in combination with 1, 6-hexamethylenediamine) the synthesis of nylon-6, 6 polyamide [65]. Currently, nearly all adipic acid is commercially produced petro-chemically from benzene via cyclohexane [66], and approximately 65 % of adipic acid is used for synthesizing nylon-6, 6 polyamide [67]. Therefore, the development of bio-based methods for adipic acid production in place of petroleum-based processes is expected to allow the synthesis of “green” polymers. Although a cellular metabolic degradation pathway for adipic acid has been described in Pseudomonas and Acinetobacter sp. [68, 69], the biosynthetic route towards adipic acid from the carbon source, such as glucose, through central metabolic pathways has not been reported. Until recently, bio-based adipic acid was obtained through the chemical conversion of the precursors glucaric acid and cis, cis-muconic acid, which can be biologically synthesized in metabolically engineered E. coli via myo-inositol or through the shikimate pathway from glucose [67]. Yu and colleagues described direct production of adipic acid from glucose by reversal of the adipate degradation pathway [70]. Specifically, adipic acid was produced in six enzymatic steps from acetyl-CoA and succinyl-CoA through 3-oxoadipyl-CoA, 3-hydroxyadipyl-CoA, 2, 3-dehydroadipyl-CoA, adipyl-CoA, and adipyl-phosphate (Fig. 1). To construct the complete pathway from acetyl-CoA and succinyl-CoA to adipic acid in E. coli, this group selected six enzyme genes for overexpression from E. coli, Clostridium acetobutylicum, and Euglena gracilis, and performed multiple gene deletions to minimize the accumulation of by-products and direct carbon flux towards the two precursors, acetyl-CoA and succinyl-CoA (Fig. 2). When engineered using this approach, the recombinant E. coli strain produced 639 µg/L adipic acid [70]. Deng and Mao later reported that the moderately thermophilic soil bacterium Thermobifida fusca naturally possesses the genes responsible for converting acetyl-CoA and succinyl-CoA to adipic acid; this bacterium produces 2.23 g/L adipic acid after 72 h cultivation at 55 °C [71].
Putrescine
Diamine is a building block chemical for synthesizing polyamide with dicarboxylic acid. To achieve the production of 100 % bio-based polyamide, an efficient microbial production of diamines that replaces traditional petroleum-based synthesis is required. 1, 6-Hexamethylenediamine, a building block for synthesizing nylon-6, 6, has not been produced by microbial fermentation. However, diamines with different carbon atom numbers also can be used for synthesizing bio-based polyamide. For example, a four-carbon diamine, putrescine (1, 4-diaminobutane), is a promising target for microbial fermentation; this compound is industrially produced by chemical synthesis via the addition of hydrogen cyanide to acrylonitrile through succinonitrile [72]. Nylon-4, 6 (distributed by DSM as Stanyl®, which is synthesized from putrescine and adipic acid) has been demonstrated to possess mechanical and physical properties comparable, or even superior, to those of nylon-6, 6 in terms of melting point, glass transition temperature, tensile strength, solvent resistance, and rate of crystallization [73]. In addition, polymerization with sebacic acid, a ten-carbon dicarboxylic acid derived from castor plant oil, yields a 100 % bio-based nylon-4, 10; this polymer, which is distributed as by DSM as EcoPaXX®, has a high melting point and high crystallization rate and has been used as an engineering plastic [74].
Putrescine can be synthesized from two alkaline amino acids, l-ornithine or its downstream product l-arginine, through a single decarboxylation reaction catalyzed by ornithine decarboxylase or arginine decarboxylase, respectively (Figs. 1, 2) [29]. To date, the highest titer of microbial-produced putrescine was achieved using an engineered strain of E. coli. In this strain, designated XQ52/p15SpeC, potE (encoding putrescine/ornithine antiporter) was overexpressed in combination with the deletion of puuP (encoding putrescine importer) and of genes encoding enzymes of competitive and degradation routes for putrescine (including puuA (encoding glutamate-putrescine ligase), speE (encoding spermidine synthase), speG (encoding spermidine acetyltransferase), and argI (encoding a component of ornithine transcarbamylase) (Fig. 2). In addition, the native promoters of key biosynthetic genes [argECBH operon, argD (encoding N-acetyl-ornithine aminotransferase) and speC (encoding ornithine decarboxylase)] were replaced with stronger promoters, and argR (encoding a transcriptional repressor) and rpoS (encoding a stress-responsive RNA polymerase sigma factor) also were deleted (Fig. 1). The resultant strain was capable of producing 24.2 g/L putrescine (Table 1) [28].
Corynebacterium glutamicum also is a promising host for putrescine production because of this species’ capability of large-scale production of l-glutamic acid [75], as well as higher tolerance to putrescine compared to E. coli and S. cerevisiae [76]. Although the putrescine metabolic pathway has not been identified in C. glutamicum, introduction of the speC gene from E. coli enabled C. glutamicum to synthesize putrescine [76]. Recently, the Wendisch group has energetically identified engineering targets for increasing putrescine production in C. glutamicum (Fig. 1). This group demonstrated that deletion of argF (encoding ornithine transcarbamylase) and argR was effective for increasing putrescine production due to an increase in the ornithine supply; however, argF deletion resulted in arginine auxotrophy. This problem was circumvented by the fine-tuning of argF expression through modifications of the promoter, translational start codon, and ribosome-binding site, resulting in a 60 % increase in putrescine production [77]. Furthermore, this group also identified a gene responsible for putrescine acetylation, snaA, and showed deletion of snaA minimized the generation of acetylputrescine as a by-product, resulting in a further 41 % increase in putrescine production [78]. The Wendisch group also identified a putative putrescine transporter, CgmA, which was first identified as a cadaverine transporter (Cg2893; see cadaverine section), and demonstrated that overexpression of the cgmA gene increased putrescine production by 24 %, although cgmA overexpression in a snaA-deletion strain did not result in further increases in putrescine production [77]. The decreased activity of 2-oxoglutarate dehydrogenase (ODH) in C. glutamicum is associated with glutamate overproduction [79, 80]. To examine the effect of excess glutamate on putrescine production by C. glutamicum, ODH activity was decreased five-fold. This effect required replacement of the start codon of a gene (odhA) that encodes a subunit of the ODH complex, as well as mutating the gene (odhI) encoding an inhibitory protein for ODH complex (creating a Thr15-to-Ala substitution in OdhI to remove a phosphorylation site, because phosphorylated OdhI inhibits the function of ODH) [81, 82]. This genetic engineering strategy improved putrescine production by 28 %, corresponding to a yield of 0.26 g/g glucose, a value that is higher than that achievable with E. coli [83].
Very recently, park and colleagues reported the metabolic engineering of a strain of C. glutamicum capable of producing 92.5 g/L l-arginine in fed-batch fermentation [84]. Construction of this strain involved removing regulatory repressors of the arg operon, optimizing nicotinamide adenosine dinucleotide phosphate levels, disrupting the l-glutamate exporter gene (cgl1270) to increase production of the l-arginine precursor, and flux optimizing the rate-limiting l-arginine biosynthetic reactions. This engineered strain would be suitable for overproducing ornithine; thus, the strain might be rendered useful for the efficient production of putrescine by introducing the decarboxylase-encoding gene and metabolic engineering of targets as described above.
Cadaverine
Cadaverine (1, 5-diaminopentane), a five-carbon diamine, is another candidate for the synthesis of “green” nylon [41]. Cadaverine is synthesized by the one-step decarboxylation of l-lysine, which is produced from oxaloacetate of the TCA cycle (Figs. 1, 2). The microbial production of cadaverine was first demonstrated in a metabolically engineered strain of C. glutamicum. Although C. glutamicum lacks the decarboxylase gene for converting l-lysine to cadaverine, the introduction of cadA (encoding lysine decarboxylase) from E. coli, in combination with the deletion of the endogenous hom gene (which encodes a homoserine dehydrogenase), enabled the production of 2.6 g/L cadaverine [85]. E. coli also has been engineered to produce 9.6 g/L cadaverine by deleting genes of the cadaverine degradation pathway and overexpressing lysine pathway genes (Fig. 2) [31].
Corynebacterium glutamicum is so far a superior host for large-scale, bio-based cadaverine production because of its ability to produce large quantity of l-lysine [86]. Several genetic mutations (lysCT311I encoding aspartokinase, homV59A, and pycP458S) have been identified that improve lysine production through the deregulation of feedback resistance [87]. Recently, the Wittmann group extensively examined cadaverine production by C. glutamicum. In addition to the mutations of lysCT311I, homV59A, and pycP458S, chromosomal overexpression of the lysine pathway genes dapB (encoding dihydrodipicolinate reductase) and pyc by replacing the promoters, integration of a second copy of ddh (encoding diaminopimelate dehydrogenase) and lysA (encoding diaminopimelate decarboxylase), and deletion of pepck (encoding phosphoenolpyruvate carboxykinase) markedly increased cadaverine production (Fig. 1) [88]. In that study, another lysine carboxylase-encoding gene from E. coli, ldcC, was employed instead of cadA because the LdcC protein prefers neutral pH [89]. However, approximately 20 % of the intracellular cadaverine produced by the resultant strain was acetylated [88]. The Wittmann group therefore identified a gene responsible for cadaverine acetylation (Ncgl1469 encoding diaminopentane acetyltransferase) in C. glutamicum by targeted, single-gene deletion of 17 potential N-acetyltransferases [90]. Notably, the identified gene shared low homology with the snaA gene, responsible for putrescine acetylation. Deletion of the Ncgl1469 gene increased the yield of cadaverine by 11 %. Genome-wide transcriptional analysis led to the further identification of an exporter gene (cg2893), which was later identified as a putrescine transporter (CgmA; see putrescine section). Cadaverine secretion was improved by 20 % when cg2893 was overexpressed [91]. Further metabolic engineering of C. glutamicum was conducted to replace the common ATG start codon of the icd gene (encoding isocitrate dehydrogenase) with the rare GTG (generating a variant designated icdGTG) to increase the flux through the anapleotic pathway, and to overexpress the tkt operon genes zwf (encoding glucose-6-phosphate dehydrogenase), tal (encoding transaldolase), tkt (encoding transketolase), opcA (encoding a putative subunit of glucose-6-phosphate dehydrogenase), and pgl (encoding 6-phosphogluconolactonase) by promoter exchange (Fig. 1). The resultant strain produced 88 g/L cadaverine, corresponding to a molar yield of 50 % (Table 1) [30]. The cadaverine produced by this strain was polymerized with sebacic acid to synthesize 100 % bio-polyamide (nylon-5, 10), which displayed a comparable melting point (215 °C) and glass transition temperature (50 °C), and even higher transparency, to that of the petrochemical polymers nylon-6, and nylon-6, 6 [30].
Building block chemicals for aromatic polymer synthesis
The above sections focused on building block chemicals for synthesizing aliphatic polymers. In this final section, we describe the production of aromatic chemicals that potentially can be used to synthesize high-performance plastics that possess desirable properties such as ultraviolet (UV) absorbance, higher thermal resistance, and mechanical strength compared to aliphatic polymers. These next-generation bio-polymers may be applicable for the production of performance fabrics and electronics, and for use in the automobile and air industries. To be used for applications in these fields, the materials must have a glass transition temperature close to 200 °C, in addition to high mechanical strength and Young’s modulus [92]. To address this issue, the production of aromatic “bio-monomers” by microbial fermentation or bioconversion has been the subject of considerable research in the past decade, although the productivity of most chemicals remains limited.
d-phenyllactic acid (d-PhLA), one candidate precursor, is synthesized through the shikimate pathway via erythrose-4-phosphate, itself a product of the pentose phosphate pathway (Fig. 2). Optically pure d-PhLA was produced from glucose at a titer of 29 g/L by a recombinant E. coli strain expressing the pprA gene (encoding phenylpyruvate reductase) from Wickerhamia fluorescens [93]. More recently, d-PhLA was produced from the lignocellulosic biomass of kraft pulp [94] and pretreated bagasse [95] in a single-pod reaction of simultaneous saccharification and fermentation.
Cinnamic acid is a phenylalanine derivative that also has been produced from glucose by recombinant Pseudomonas putida [96] and Streptomyces lividans [97] overexpressing the pal genes (encoding phenylalanine ammonia lyase) from Rhodosporidium toruloides and Streptomyces maritimus, respectively (Fig. 1). The hydroxycinnamate derivatives of 4-hydroxycinnamic acid (p-coumaric acid) [98] and 3, 4-dihydroxycinnamic acid (caffeic acid) [99] were used as building blocks for the synthesis of aromatic bio-based polyesters with a glass transition temperature of 169 °C. It has also been demonstrated that the chemocatalytic processing of bio-monomers confers multiple properties to the resulting biopolymers. For instance, a bio-based copolymer formed from caffeic acid and p-coumaric acid showed strong adhesive characteristics [99], and caffeic acid was recently produced from glucose by recombinant E. coli [100].
3-Amino-4-hydroxybenzoic acid (3, 4-AHBA) serves as a subunit of poly-benzoxazole [101], which is a commercially available textile with extremely high thermal and mechanical properties. In contrast to most aromatic compounds, which are formed in multistep reactions via the shikimate pathway [102], 3, 4-AHBA is biosynthesized via a unique pathway. In Streptomyces griseus cells, 3, 4-AHBA is formed from the glycolytic intermediate dihydroxyacetone phosphate and aspartate metabolite aspartate-semialdehyde in two-step aldol condensation reactions catalyzed by the gene products of griI and griH, respectively (Fig. 1) [103]. Thus, the 3, 4-AHBA synthetic pathway can be engineered in other microorganisms by introducing the corresponding heterologous genes, thereby potentially permitting high 3, 4-AHBA productivity from renewable feedstocks. As an example, C. glutamicum heterologously expressing the griI and griH genes produced 1.0 g/L 3, 4-AHBA from sweet sorghum juice [104].
Aromatic polyimides are alternative building blocks for high-performance bio-based polymers due to their excellent thermo-mechanical performance, high chemical stability, and low coefficient of thermal expansion. A phenylpropanoid derivative of 4-aminocinnamic acid was produced by the bioconversion of the non-standard amino acid 4-aminophenylalanine using a recombinant E. coli strain [92]. Bio-based polyimide was subsequently produced from a photodimer of 4-aminocinnamic acid through a chemocatalytic reaction. The resulting polyimide films exhibited ultra-high thermal resistance with a glass transition temperature of over 250 °C (the highest value for all bio-based plastics reported to date); these films also had high tensile strength and Young’s modulus [92]. The 4-aminocinnamic acid precursor 4-aminophenylalanine can be produced from glucose by microbial fermentation [105], suggesting that the fermentation and subsequent bioconversion of 4-aminophenylalanine can be produced using 4-aminocinnamic acid as a building-block material for the synthesis of bio-based polyimides from renewable sugars.
Compared to current aliphatic polymers, emerging bio-based aromatic polymers are value-added molecules with high thermal and mechanical properties; these polymers therefore might serve as engineering plastics. Further developments to increase the compatibility of aromatic compounds for bioprocessing will be needed to achieve high productivity of aromatic bio-monomers from renewable feedstocks.
Conclusions
The present review aimed to provide a broad view of metabolic engineering strategies for producing building block chemicals for use in generating aliphatic polymers. We further described the current state of knowledge for the production of building block chemicals of next-generation, high-performance aromatic polymers. As described above, advances in metabolic engineering have markedly improved the productivities and yields of microbially produced polymer building blocks. Following the success of industrial l-lactic acid production through microbial fermentation, several bio-based approaches for succinic acid synthesis recently have been commercialized [55]. However, further improvements related to productivity and yield are required for many chemicals, particularly those that are synthesized via peripheral metabolic pathways. To realize this goal, novel methods for the rational design and optimization of enzymes and transporters to improve substrate specificity and reaction rates likely will be necessary. These developments are expected to enable efficient redirection and acceleration of carbon flux towards the target chemicals and extracellular secretion, respectively.
Authors’ contributions
YT wrote the “d-Lactic acid”, “Succinic acid”, “Putrescine”, and “Cadaverine” sections. HK wrote the “Building block chemicals for aromatic polymer synthesis” section. KS wrote the “Adipic acid” section. AK reviewed and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Owing to space constraints, we were unable to describe and reference many manuscripts that we feel have contributed valuable data to the field. We express our sincerest apologies to the authors of those manuscripts. The writing of this review was supported by Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), MEXT, Japan.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- LDH
lactate dehydrogenase
- NADPH
nicotinamide adenosine dinucleotide phosphate
- ODH
2-oxoglutarate dehydrogenase
- PLLA
poly-L-lactic acid
- TCA
tricarboxylic acid
Additional file
10.1186/s12934-016-0411-0 Engineering target genes covered in this review.
Contributor Information
Yota Tsuge, Email: ytsuge@people.kobe-u.ac.jp.
Hideo Kawaguchi, Email: kawaguchi_h@port.kobe-u.ac.jp.
Kengo Sasaki, Email: sikengo@people.kobe-u.ac.jp.
Akihiko Kondo, Email: akondo@kobe-u.ac.jp.
References
- 1.Feldman D. Polymer history. Des Monomers Polym. 2008;11:1–15. doi: 10.1163/156855508X292383. [DOI] [Google Scholar]
- 2.Pang X, Zhuang X, Tang Z, Chen X. Polylactic acid (PLA): research, development and industrialization. Biotechnol J. 2010;5:1125–1136. doi: 10.1002/biot.201000135. [DOI] [PubMed] [Google Scholar]
- 3.Becker J, Lange A, Fabarius J, Wittmann C. Top value platform chemicals: bio-based production of organic acids. Curr Opin Biotechnol. 2015;36:168–175. doi: 10.1016/j.copbio.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Zhou L, Tian K, Kumar A, Singh S, Prior BA, Wang Z. Metabolic engineering of Escherichia coli: a sustainable industrial platform for bio-based chemical production. Biotechnol Adv. 2013;31:1200–1203. doi: 10.1016/j.biotechadv.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Redden H, Alper HS. Frontiers of yeast metabolic engineering: diversifying beyond ethanol and Saccharomyces. Curr Opin Biotechnol. 2013;24:1023–1030. doi: 10.1016/j.copbio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Tsuge Y, Hasunuma T, Kondo A. Recent advances in the metabolic engineering of Corynebacterium glutamicum for the production of lactate and succinate from renewable resources. J Ind Microbiol Biotechnol. 2015;42:375–389. doi: 10.1007/s10295-014-1538-9. [DOI] [PubMed] [Google Scholar]
- 7.Wendisch VF, Bott M, Eikmanns BJ. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol. 2006;9:268–274. doi: 10.1016/j.mib.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Zhang F, Rodriguez S, Keasling JD. Metabolic engineering of microbial pathways for advanced biofuels production. Curr Opin Biotechnol. 2011;22:775–783. doi: 10.1016/j.copbio.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Nielsen J. Advances in metabolic pathway and strain engineering paving the way for sustainable production of chemical building blocks. Curr Opin Biotechnol. 2013;24:965–972. doi: 10.1016/j.copbio.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto K, Taguchi S. Enzyme and metabolic engineering for the production of novel biopolymers: crossover of biological and chemical processes. Curr Opin Biotechnol. 2013;24:1054–1060. doi: 10.1016/j.copbio.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 11.Svagan AJ, Åkesson A, Cárdenas M, Bulut S, Knudsen JC, Risbo J, Plackett D. Transparent films based on PLA and montmorillonite with tunable oxygen barrier properties. Biomacromolecules. 2012;13:397–405. doi: 10.1021/bm201438m. [DOI] [PubMed] [Google Scholar]
- 12.Tsuji H, Ikada Y. Stereocomplex formation between enantiomeric poly(lactic acid)s. XI. Mechanical properties and morphology of solution-cast films. Polymer. 1999;40:6699–6708. doi: 10.1016/S0032-3861(99)00004-X. [DOI] [Google Scholar]
- 13.Abdel-Rahman MA, Tashiro Y, Sonomoto K. Recent advances in lactic acid production by microbial fermentation processes. Biotechnol Adv. 2013;31:877–902. doi: 10.1016/j.biotechadv.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Tashiro Y, Sonomoto K. Fermentative production of lactic acid from renewable materials: recent achievements, prospects and limits. J Biosci Bioeng. 2015;119:10–18. doi: 10.1016/j.jbiosc.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Hermann T. Industrial production of amino acids by coryneform bacteria. J Biotechnol. 2003;104:155–172. doi: 10.1016/S0168-1656(03)00149-4. [DOI] [PubMed] [Google Scholar]
- 16.Wendisch VF. Microbial production of amino acids and derived chemicals: synthetic biology approaches to strain development. Curr Opin Biotechnol. 2014;30:51–58. doi: 10.1016/j.copbio.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol. 2004;7:182–196. doi: 10.1159/000079827. [DOI] [PubMed] [Google Scholar]
- 18.Okino S, Inui M, Yukawa H. Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol. 2005;68:475–480. doi: 10.1007/s00253-005-1900-y. [DOI] [PubMed] [Google Scholar]
- 19.Okino S, Suda M, Fujikura K, Inui M, Yukawa H. Production of D-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol. 2008;78:449–454. doi: 10.1007/s00253-007-1336-7. [DOI] [PubMed] [Google Scholar]
- 20.Tsuge Y, Yamamoto S, Kato N, Suda M, Vertès AA, Yukawa H, Inui M. Overexpression of the phosphofructokinase encoding gene is crucial for achieving high production of D-lactate in Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol. 2015;99:4679–4689. doi: 10.1007/s00253-015-6546-9. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Niu DD, Tian KM, Chen XZ, Prior BA, Shen W, Shi GY, Singh S, Wang ZX. Genetically switched D-lactate production in Escherichia coli. Metab Eng. 2012;14:560–568. doi: 10.1016/j.ymben.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol. 2008;81:459–464. doi: 10.1007/s00253-008-1668-y. [DOI] [PubMed] [Google Scholar]
- 23.Litsanov B, Brocker M, Bott M. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol. 2012;78:3325–3337. doi: 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu N, Xia H, Yang J, Zhao X, Chen T. Improved succinate production in Corynebacterium glutamicum by engineering glyoxylate pathway and succinate export system. Biotechnol Lett. 2014;36:553–560. doi: 10.1007/s10529-013-1376-2. [DOI] [PubMed] [Google Scholar]
- 25.Vemuri GN, Eiteman MA, Altman E. Succinate production in dual-phase Escherichia coli fermentations depends on the time of transition from aerobic to anaerobic conditions. J Ind Microbiol Biotechnol. 2002;28:325–332. doi: 10.1038/sj.jim.7000250. [DOI] [PubMed] [Google Scholar]
- 26.Jantama K, Zhang X, Moore JC, Shanmugam KT, Svoronos SA, Ingram LO. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng. 2008;101:881–893. doi: 10.1002/bit.22005. [DOI] [PubMed] [Google Scholar]
- 27.Tajima Y, Yamamoto Y, Fukui K, Nishio Y, Hashiguchi K, Usuda Y, Sode K. Impact of an energy-conserving strategy on succinate production under weak acidic and anaerobic conditions in Enterobacter aerogenes. Microb Cell Fact. 2015;14:80. doi: 10.1186/s12934-015-0269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian ZG, Xia XX, Lee SY. Metabolic engineering of Escherichia coli for the production of putrescine: a four carbon diamine. Biotechnol Bioeng. 2009;104:651–662. doi: 10.1002/bit.22502. [DOI] [PubMed] [Google Scholar]
- 29.Schneider J, Wendisch VF. Biotechnological production of polyamines by bacteria: recent achievements and future perspectives. Appl Microbiol Biotechnol. 2011;91:17–30. doi: 10.1007/s00253-011-3252-0. [DOI] [PubMed] [Google Scholar]
- 30.Kind S, Neubauer S, Becker J, Yamamoto M, Völkert M, Abendroth Gv, Zelder O, Wittmann C. From zero to hero—production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum. Metab Eng. 2014;25:113–123. doi: 10.1016/j.ymben.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Qian ZG, Xia XX, Lee SY. Metabolic engineering of Escherichia coli for the production of cadaverine: a five carbon diamine. Biotechnol Bioeng. 2011;108:93–103. doi: 10.1002/bit.22918. [DOI] [PubMed] [Google Scholar]
- 32.Tsuge Y, Yamamoto S, Suda M, Inui M, Yukawa H. Reactions upstream of glycerate-1, 3-bisphosphate drive Corynebacterium glutamicum (D)-lactate productivity under oxygen deprivation. Appl Microbiol Biotechnol. 2013;97:6693–6703. doi: 10.1007/s00253-013-4986-7. [DOI] [PubMed] [Google Scholar]
- 33.Chang DE, Jung HC, Rhee JS, Pan JG. Homofermentative production of D- or L-lactate in metabolically engineered Escherichia coli RR1. Appl Environ Microbiol. 1999;65:1384–1389. doi: 10.1128/aem.65.4.1384-1389.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou S, Causey TB, Hasona A, Shanmugam KT, Ingram LO. Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl Environ Microbiol. 2003;69:399–407. doi: 10.1128/AEM.69.1.399-407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou S, Shanmugam KT, Yomano LP, Grabar TB, Ingram LO. Fermentation of 12 % (w/v) glucose to 1.2 M lactate by Escherichia coli strain SZ194 using mineral salts medium. Biotechnol Lett. 2006;28:663–670. doi: 10.1007/s10529-006-0032-5. [DOI] [PubMed] [Google Scholar]
- 36.Zhou L, Tian KM, Niu DD, Shen W, Shi GY, Singh S, Wang ZX. Improvement of D-lactate productivity in recombinant Escherichia coli by coupling production with growth. Biotechnol Lett. 2012;34:1123–1130. doi: 10.1007/s10529-012-0883-x. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Y, Eiteman MA, DeWitt K, Altman E. Homolactate fermentation by metabolically engineered Escherichia coli strains. Appl Environ Microbiol. 2007;73:456–464. doi: 10.1128/AEM.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou L, Zuo ZR, Chen XZ, Niu DD, Tian KM, Prior BA, Shen W, Shi GY, Singh S, Wang ZX. Evaluation of genetic manipulation strategies on D-lactate production by Escherichia coli. Curr Microbiol. 2011;62:981–989. doi: 10.1007/s00284-010-9817-9. [DOI] [PubMed] [Google Scholar]
- 39.Sauer M, Porro D, Mattanovich D, Branduardi P. Microbial production of organic acids: expanding the markets. Trends Biotechnol. 2008;26:100–108. doi: 10.1016/j.tibtech.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Zeikus JG, Jain MK, Elankovan P. Biotechnology of succinic acid production and markets for derived industrial products. Appl Microbiol Biotechnol. 1999;51:545–552. doi: 10.1007/s002530051431. [DOI] [Google Scholar]
- 41.Kind S, Wittmann C. Bio-based production of the platform chemical 1, 5-diaminopentane. Appl Microbiol Biotechnol. 2011;91:1287–1296. doi: 10.1007/s00253-011-3457-2. [DOI] [PubMed] [Google Scholar]
- 42.Choi S, Song CW, Shin JH, Lee SY. Biorefineries for the production of top building block chemicals and their derivatives. Metab Eng. 2015;28:223–239. doi: 10.1016/j.ymben.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 43.Thakker C, Martínez I, San KY, Bennett GN. Succinate production in Escherichia coli. Biotechnol J. 2012;7:213–224. doi: 10.1002/biot.201100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wieschalka S, Blombach B, Bott M, Eikmanns BJ. Bio-based production of organic acids with Corynebacterium glutamicum. Microb Biotechnol. 2013;6:87–102. doi: 10.1111/1751-7915.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carvalho M, Matos M, Roca C, Reis MA. Succinic acid production from glycerol by Actinobacillus succinogenes using dimethylsulfoxide as electron acceptor. N Biotechnol. 2014;31:133–139. doi: 10.1016/j.nbt.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Guettler MV, Rumler D, Jain MK. Actinobacillus succinogenes sp. nov., a novel succinic-acid-producing strain from the bovine rumen. Int J Syst Bacteriol. 1999;49:207–216. doi: 10.1099/00207713-49-1-207. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Zheng XY, Fang XJ, Liu SW, Chen KQ, Jiang M, Wei P, Ouyang PK. A complete industrial system for economical succinic acid production by Actinobacillus succinogenes. Bioresour Technol. 2011;102:6147–6152. doi: 10.1016/j.biortech.2011.02.093. [DOI] [PubMed] [Google Scholar]
- 48.Nghiem NP, Davison BH, Suttle BE, Richardson GR. Production of succinic acid by Anaerobiospirillum succiniciproducens. Appl Biochem Biotechnol. 1997;63:565–576. doi: 10.1007/BF02920454. [DOI] [PubMed] [Google Scholar]
- 49.Song H, Lee SY. Production of succinic acid by bacterial fermentation. Enzyme Microb Technol. 2006;39:352–361. doi: 10.1016/j.enzmictec.2005.11.043. [DOI] [Google Scholar]
- 50.Yasuda K, Jojima T, Suda M, Okino S, Inui M, Yukawa H. Analyses of the acetate-producing pathways in Corynebacterium glutamicum under oxygen-deprived conditions. Appl Microbiol Biotechnol. 2007;77:853–860. doi: 10.1007/s00253-007-1199-y. [DOI] [PubMed] [Google Scholar]
- 51.Huhn S, Jolkver E, Krämer R, Marin K. Identification of the membrane protein SucE and its role in succinate transport in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2011;89:327–335. doi: 10.1007/s00253-010-2855-1. [DOI] [PubMed] [Google Scholar]
- 52.Fukui K, Koseki C, Yamamoto Y, Nakamura J, Sasahara A, Yuji R, Hashiguchi K, Usuda Y, Matsui K, Kojima H, Abe K. Identification of succinate exporter in Corynebacterium glutamicum and its physiological roles under anaerobic conditions. J Biotechnol. 2011;154:25–34. doi: 10.1016/j.jbiotec.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Warnecke T, Gill RT. Organic acid toxicity, tolerance, and production in Escherichia coli biorefining applications. Microb Cell Fact. 2005;4:25. doi: 10.1186/1475-2859-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng KK, Zhao XB, Zeng J, Wu RC, Xu YZ, Liu DH, Zhang JA. Downstream processing of biotechnological produced succinic acid. Appl Microbiol Biotechnol. 2012;95:841–850. doi: 10.1007/s00253-012-4214-x. [DOI] [PubMed] [Google Scholar]
- 55.Jansen ML, van Gulik WM. Towards large scale fermentative production of succinic acid. Curr Opin Biotechnol. 2014;30:190–197. doi: 10.1016/j.copbio.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Liu X, Jia B, Sun X, Ai J, Wang L, Wang C, Zhao F, Zhan J, Huang W. Effect of initial pH on growth characteristics and fermentation properties of Saccharomyces cerevisiae. J Food Sci. 2015;80:M800–M808. doi: 10.1111/1750-3841.12813. [DOI] [PubMed] [Google Scholar]
- 57.Valli M, Sauer M, Branduardi P, Borth N, Porro D, Mattanovich D. Intracellular pH distribution in Saccharomyces cerevisiae cell populations, analyzed by flow cytometry. Appl Environ Microbiol. 2005;71:1515–1521. doi: 10.1128/AEM.71.3.1515-1521.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raab AM, Gebhardt G, Bolotina N, Weuster-Botz D, Lang C. Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metab Eng. 2010;12:518–525. doi: 10.1016/j.ymben.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 59.Yan D, Wang C, Zhou J, Liu Y, Yang M, Xing J. Construction of reductive pathway in Saccharomyces cerevisiae for effective succinic acid fermentation at low pH value. Bioresour Technol. 2014;156:232–239. doi: 10.1016/j.biortech.2014.01.053. [DOI] [PubMed] [Google Scholar]
- 60.Tajima Y, Kaida K, Hayakawa A, Fukui K, Nishio Y, Hashiguchi K, Fudou R, Matsui K, Usuda Y, Sode K. Study of the role of anaerobic metabolism in succinate production by Enterobacter aerogenes. Appl Microbiol Biotechnol. 2014;98:7803–7813. doi: 10.1007/s00253-014-5884-3. [DOI] [PubMed] [Google Scholar]
- 61.Tajima Y, Yamamoto Y, Fukui K, Nishio Y, Hashiguchi K, Usuda Y, Sode K. Effects of eliminating pyruvate node pathways and of coexpression of heterogeneous carboxylation enzymes on succinate production by Enterobacter aerogenes. Appl Environ Microbiol. 2015;81:929–937. doi: 10.1128/AEM.03213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X, Jantama K, Moore JC, Jarboe LR, Shanmugam KT, Ingram LO. Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc Natl Acad Sci USA. 2009;106:20180–20185. doi: 10.1073/pnas.0905396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh A, Cher Soh K, Hatzimanikatis V, Gill RT. Manipulating redox and ATP balancing for improved production of succinate in E. coli. Metab Eng. 2011;13:76–81. doi: 10.1016/j.ymben.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 64.Aizenshtein EM. Chemical fibres in global and Russian markets in 2012. Fibre Chem. 2014;45:329–335. doi: 10.1007/s10692-014-9538-0. [DOI] [Google Scholar]
- 65.Luedeke VD. Adipic acid: encyclopedia of chemical processing and design vol. 2. In: McKetta JJ, Cunningham WA, editors. New York: Marcel Dekker Inc.; 1977. p. 128–46.
- 66.Niu W, Draths KM, Frost JW. Benzene-free synthesis of adipic acid. Biotechnol Prog. 2002;18:201–211. doi: 10.1021/bp010179x. [DOI] [PubMed] [Google Scholar]
- 67.Polen T, Spelberg M, Bott M. Toward biotechnological production of adipic acid and precursors from biorenewables. J Biotechnol. 2013;167:75–84. doi: 10.1016/j.jbiotec.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 68.Ahvazi B, Coulombe R, Delarge M, Vedadi M, Zhang L, Meighen E, Vrielink A. Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity. Biochemical J. 2000;349:853–861. doi: 10.1042/bj3490853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng Q, Thomas SM, Rouviere P. Biological conversion of cyclic alkanes and cyclic alcohols into dicarboxylic acids: biochemical and molecular basis. Appl Microbiol Biotech. 2002;58:704–711. doi: 10.1007/s00253-002-0958-z. [DOI] [PubMed] [Google Scholar]
- 70.Yu JL, Xia XX, Zhong JJ, Qian ZG. Direct biosynthesis of adipic acid from a synthetic pathway in recombinant Escherichia coli. Biotechnol Bioeng. 2014;111:2580–2586. doi: 10.1002/bit.25293. [DOI] [PubMed] [Google Scholar]
- 71.Deng Y, Mao Y. Production of adipic acid by the native-occurring pathway in Thermobifida fusca B6. J Appl Microbiol. 2015;119:1057–1063. doi: 10.1111/jam.12905. [DOI] [PubMed] [Google Scholar]
- 72.Sanders J, Scott E, Weusthuis R, Mooibroek H. Bio-refinery as the bio-inspired process to bulk chemicals. Macromol Biosci. 2007;7:105–117. doi: 10.1002/mabi.200600223. [DOI] [PubMed] [Google Scholar]
- 73.Roerdink E, Warnier JMM. Preparation and properties of high molar mass nylon-4, 6: a new development in nylon polymers. Polymer. 1985;26:1582–1588. doi: 10.1016/0032-3861(85)90098-9. [DOI] [Google Scholar]
- 74.Harmsen PFH, Hackmann MM, Bos HL. Green building blocks for bio-based plastics. Biofuel Bioprod Biorefin. 2014;8:306–324. doi: 10.1002/bbb.1468. [DOI] [Google Scholar]
- 75.Kimura E. l-Glutamate production: handbook of Corynebacterium glutamicum. In: Eggeling R, Bott M, editors. Boca Raton: CRC Press; 2005. p. 439–64.
- 76.Schneider J, Wendisch VF. Putrescine production by engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2010;88:859–868. doi: 10.1007/s00253-010-2778-x. [DOI] [PubMed] [Google Scholar]
- 77.Schneider J, Eberhardt D, Wendisch VF. Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system. Appl Microbiol Biotechnol. 2012;95:169–178. doi: 10.1007/s00253-012-3956-9. [DOI] [PubMed] [Google Scholar]
- 78.Nguyen AQ, Schneider J, Wendisch VF. Elimination of polyamine N-acetylation and regulatory engineering improved putrescine production by Corynebacterium glutamicum. J Biotechnol. 2015;201:75–85. doi: 10.1016/j.jbiotec.2014.10.035. [DOI] [PubMed] [Google Scholar]
- 79.Asakura Y, Kimura E, Usuda Y, Kawahara Y, Matsui K, Osumi T, Nakamatsu T. Altered metabolic flux due to deletion of odhA causes L-glutamate overproduction in Corynebacterium glutamicum. Appl Environ Microbiol. 2007;73:1308–1319. doi: 10.1128/AEM.01867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawahara Y, Takahashi-Fuke K, Shimizu E, Nakamatsu T, Nakamori S. Relationship between the glutamate production and the activity of 2-oxoglutarate dehydrogenase in Brevibacterium lactofermentum. Biosci Biotechnol Biochem. 1997;61:1109–1112. doi: 10.1271/bbb.61.1109. [DOI] [PubMed] [Google Scholar]
- 81.Niebisch A, Kabus A, Schultz C, Weil B, Bott M. Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J Biol Chem. 2006;281:12300–12307. doi: 10.1074/jbc.M512515200. [DOI] [PubMed] [Google Scholar]
- 82.Schultz C, Niebisch A, Gebel L, Bott M. Glutamate production by Corynebacterium glutamicum: dependence on the oxoglutarate dehydrogenase inhibitor protein OdhI and protein kinase PknG. Appl Microbiol Biotechnol. 2007;76:691–700. doi: 10.1007/s00253-007-0933-9. [DOI] [PubMed] [Google Scholar]
- 83.Nguyen AQ, Schneider J, Reddy GK, Wendisch VF. Fermentative production of the diamine putrescine: system metabolic engineering of Corynebacterium glutamicum. Metabolites. 2015;5:211–231. doi: 10.3390/metabo5020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park SH, Kim HU, Kim TY, Park JS, Kim SS, Lee SY. Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat Commun. 2014;5:4618. doi: 10.1038/ncomms5618. [DOI] [PubMed] [Google Scholar]
- 85.Mimitsuka T, Sawai H, Hatsu M, Yamada K. Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci Biotechnol Biochem. 2007;71:2130–2135. doi: 10.1271/bbb.60699. [DOI] [PubMed] [Google Scholar]
- 86.Eggeling L, Bott M. A giant market and a powerful metabolism: l-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2015;99:3387–3394. doi: 10.1007/s00253-015-6508-2. [DOI] [PubMed] [Google Scholar]
- 87.Ikeda M, Ohnishi J, Hayashi M, Mitsuhashi S. A genome-based approach to create a minimally mutated Corynebacterium glutamicum strain for efficient l-lysine production. J Ind Microbiol Biotechnol. 2006;33:610–615. doi: 10.1007/s10295-006-0104-5. [DOI] [PubMed] [Google Scholar]
- 88.Kind S, Jeong WK, Schröder H, Wittmann C. Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab Eng. 2010;12:341–351. doi: 10.1016/j.ymben.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 89.Yamamoto Y, Miwa Y, Miyoshi K, Furuyama J, Ohmori H. The Escherichia coli ldcC gene encodes another lysine decarboxylase, probably a constitutive enzyme. Genes Genet Syst. 1997;72:167–172. doi: 10.1266/ggs.72.167. [DOI] [PubMed] [Google Scholar]
- 90.Kind S, Jeong WK, Schröder H, Zelder O, Wittmann C. Identification and elimination of the competing N-acetyldiaminopentane pathway for improved production of diaminopentane by Corynebacterium glutamicum. Appl Environ Microbiol. 2010;76:5175–5180. doi: 10.1128/AEM.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kind S, Kreye S, Wittmann C. Metabolic engineering of cellular transport for overproduction of the platform chemical 1, 5-diaminopentane in Corynebacterium glutamicum. Metab Eng. 2011;13:617–627. doi: 10.1016/j.ymben.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 92.Suvannasara P, Tateyama S, Miyasato A, Matsumura K, Shimoda T, Ito T, Yamagata Y, Fujita T, Takaya N, Kaneko T. Biobased polyimides from 4-aminocinnamic acid photodimer. Macromolecules. 2014;47:1586–1593. doi: 10.1021/ma402499m. [DOI] [Google Scholar]
- 93.Fujita T, Nguyen HD, Ito T, Zhou S, Osada L, Tateyama S, Kaneko T, Takaya N. Microbial monomers custom-synthesized to build true bio-derived aromatic polymers. Appl Microbiol Biotechnol. 2013;97:8887–8894. doi: 10.1007/s00253-013-5078-4. [DOI] [PubMed] [Google Scholar]
- 94.Kawaguchi H, Uematsu K, Ogino C, Teramura H, Niimi-Nakamura S, Tsuge Y, Hasunuma T, Oinuma K, Takaya N, Kondo A. Simultaneous saccharification and fermentation of kraft pulp by recombinant Escherichia coli for phenyllactic acid production. Biochem Eng J. 2014;88:188–194. doi: 10.1016/j.bej.2014.04.014. [DOI] [Google Scholar]
- 95.Kawaguchi H, Teramura H, Uematsu K, Hara KY, Hasunuma T, Hirano K, Sazuka T, Kitano H, Tsuge Y, Kahar P, Niimi-Nakamura S, Oinuma KI, Takaya N, Kasuga S, Ogino C, Kondo A. Phenyllactic acid production by simultaneous saccharification and fermentation of pretreated sorghum bagasse. Bioresour Technol. 2015;182:169–178. doi: 10.1016/j.biortech.2015.01.097. [DOI] [PubMed] [Google Scholar]
- 96.Nijkamp K, van Luijk N, de Bont JAM, Wery J. The solvent-tolerant Pseudomonas putida S12 as host for the production of cinnamic acid from glucose. Appl Microbiol Biotechnol. 2005;69:170–177. doi: 10.1007/s00253-005-1973-7. [DOI] [PubMed] [Google Scholar]
- 97.Noda S, Miyazaki T, Miyoshi T, Miyake M, Okai N, Tanaka T, Ogino C, Kondo A. Cinnamic acid production using Streptomyces lividans expressing phenylalanine ammonia lyase. J Ind Microbiol Biotechnol. 2011;38:643–648. doi: 10.1007/s10295-011-0955-2. [DOI] [PubMed] [Google Scholar]
- 98.Kaneko T, Matsusaki M, Hang TT, Akashi M. Thermotropic liquid-crystalline polymer derived from natural cinnamoyl biomonomers. Macromol Rapid Comm. 2004;25:673–677. doi: 10.1002/marc.200300143. [DOI] [Google Scholar]
- 99.Kaneko D, Kinugawa S, Matsumoto K, Kaneko T. Terminally-catecholized hyper-branched polymers with high performance adhesive characteristics. Plant Biotechnol. 2010;27:293–296. doi: 10.5511/plantbiotechnology.27.293. [DOI] [Google Scholar]
- 100.Huang Q, Lin YH, Yan YJ. Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol Bioeng. 2013;110:3188–3196. doi: 10.1002/bit.24988. [DOI] [PubMed] [Google Scholar]
- 101.Eo SM, Oh SJ, Tan LS, Baek JB. Poly(2, 5-benzoxazole)/carbon nanotube composites via in situ polymerization of 3-amino-4-hydroxybenzoic acid hydrochloride in a mild poly (phosphoric acid) Eur Polym J. 2008;44:1603–1612. doi: 10.1016/j.eurpolymj.2008.03.024. [DOI] [Google Scholar]
- 102.Liu SP, Xiao MR, Zhang L, Xu J, Ding ZY, Gu ZH, Shi GY. Production of l-phenylalanine from glucose by metabolic engineering of wild type Escherichia coli W3110. Process Biochem. 2013;48:413–419. doi: 10.1016/j.procbio.2013.02.016. [DOI] [Google Scholar]
- 103.Suzuki H, Ohnishi Y, Furusho Y, Sakuda S, Horinouchi S. Novel benzene ring biosynthesis from C3 and C4 primary metabolites by two enzymes. J Biol Chem. 2006;281:36944–36951. doi: 10.1074/jbc.M608103200. [DOI] [PubMed] [Google Scholar]
- 104.Kawaguchi H, Sasaki K, Uematsu K, Tsuge Y, Teramura H, Okai N, Nakamura-Tsuruta S, Katsuyama Y, Sugai Y, Ohnishi Y, Hirano K, Sazuka T, Ogino C, Kondo A. 3-Amino-4-hydroxybenzoic acid production from sweet sorghum juice by recombinant Corynebacterium glutamicum. Bioresour Technol. 2015;198:410–417. doi: 10.1016/j.biortech.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 105.Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. Generation of a bacterium with a 21 amino acid genetic code. J Am Chem Soc. 2003;125:935–939. doi: 10.1021/ja0284153. [DOI] [PubMed] [Google Scholar]


