Abstract
To identify common variants contributing to normal variation in two specific domains of cognitive functioning, we conducted a genome-wide association study (GWAS) of executive functioning and information processing speed in non-demented older adults from the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) consortium. Neuropsychological testing was available for 5429–32 070 subjects of European ancestry aged 45 years or older, free of dementia and clinical stroke at the time of cognitive testing from 20 cohorts in the discovery phase. We analyzed performance on the Trail Making Test parts A and B, the Letter Digit Substitution Test (LDST), the Digit Symbol Substitution Task (DSST), semantic and phonemic fluency tests, and the Stroop Color and Word Test. Replication was sought in 1311-21860 subjects from 20 independent cohorts. A significant association was observed in the discovery cohorts for the single-nucleotide polymorphism (SNP) rs17518584 (discovery P-value = 3.12 × 10−8) and in the joint discovery and replication meta-analysis (P-value = 3.28 × 10−9 after adjustment for age, gender and education) in an intron of the gene cell adhesion molecule 2 (CADM2) for performance on the LDST/DSST. Rs17518584 is located about 170 kb upstream of the transcription start site of the major transcript for the CADM2 gene, but is within an intron of a variant transcript that includes an alternative first exon. The variant is associated with expression of CADM2 in the cingulate cortex (P-value = 4 × 10−4). The protein encoded by CADM2 is involved in glutamate signaling (P-value = 7.22 × 10−15), gamma-aminobutyric acid (GABA) transport (P-value = 1.36 × 10−11) and neuron cell-cell adhesion (P-value = 1.48 × 10−13). Our findings suggest that genetic variation in the CADM2 gene is associated with individual differences in information processing speed.
INTRODUCTION
Cognitive function broadly refers to multiple dissociable, but inter-correlated cognitive domains, including memory, language, executive function, processing speed and visuospatial ability. Unimpaired cognitive abilities are an important determinant of quality of life. Impairment of cognitive abilities is seen with dementia, bipolar disorder, schizophrenia and attention deficit hyperactivity disorder.1–3 Among the cognitive domains, processing speed is considered a fundamental process, reflecting the speed at which cognitive operations are performed.4,5 Executive function reflects higher-order cognitive capabilities, presumably mediated by the frontal lobes, including response inhibition, attention, cognitive flexibility and planning.6
In addition to domain-specific variance, processing speed and executive function are also, in part, explained by an individual's general cognitive ability. The same holds true for the genetic variance of performance on individual cognitive tests.7 The estimated heritability of general cognitive ability from twin studies of intelligence ranges from approximately 50 to 80%, and appears to increase with age.8 The general intelligence construct g has a heritability of around 29%, for example, observed in a genome-wide association study (GWAS) based on the GCTA procedure.9,10 Heritability of performance on tests within individual cognitive domains has been estimated from 12 to 68%11–14 for processing speed and 16–63%13,15,16 for executive function.
Of note, there is also considerable covariation between cognitive domains.17,18 There is, for example, debate as to whether processing speed is merely one of the cognitive domains, or whether processing speed has a more unique role as a fundamental process underlying variation in more complex cognitive traits as well as in cognitive aging.19,20
Identifying variants that influence quantitative variation in processing speed and executive function may provide insight into the normal variation in these important cognitive functions, and may ultimately increase our understanding of diseases that disrupt these cognitive domains.
Although various genes have been identified as potential candidates affecting different dimensions of cognitive function, prior studies have yielded inconsistent results.21 Candidate gene meta-analyses have shown associations of the apolipoprotein E (APOE) gene22 and the DTNBP1 (dystrobrevin binding protein 1) gene23 to general cognitive ability, although these findings do not meet the current standard of genome-wide significance (P-value between 0.01 and 0.05 for APOE, P-value = 0.003 for DTNBP1). Linkage analyses of executive function tasks have identified regions on chromosomes 2q, 5q, 11q, 13q and 14q.24–26 To our knowledge, there are currently five published GWAS on processing speed and executive traits in adults.5,27–30 Processing speed was suggestively associated with several loci, of which the TRIB3 (tribbles homolog 3) gene was the strongest and biologically most interesting.5 For executive function, one study30 identified a genome-wide significant association (P-value = 4.32 × 10−8) of a single-nucleotide polymorphism (SNP) in the WDR72 gene (chromosome 15) for a cognitive test similar to the Stroop interference test. The WDR72 gene has also been associated with kidney function.31 Prior studies of executive function and processing speed have been limited by small sample sizes (700 and 4000 subjects), resulting in limited power, and by application of lenient significance thresholds. Replication of prior findings has been lacking both across and within cognitive domains.
In this study, we performed a large-scale meta-analysis to identify genetic variants associated with executive function and information processing speed combining GWAS from multiple cohorts of non-demented middle-aged and older adults from the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) consortium.32
MATERIALS AND METHODS
Study populations
The discovery phase included 20 cohorts contributing to one or more test (N per test = 5429–32 070) (Supplementary Table 1). The number of discovery cohorts and subjects varied based on the availability of each test. Each cohort had extensive phenotypic data on one or more traits, and genome-wide SNP data available. Details for each cohort are given in Supplementary Table 1. Subjects aged 45 and older who were free of stroke and dementia and of European ancestry were eligible for the study.
Twenty replication cohorts (total N = 1311–21 860) (Supplementary Table 1) were selected based upon comparability of the study populations to the discovery cohorts, and availability of genotype data, and these cohorts were invited to share data from a GWAS run according to the same protocols. Applying the same inclusion criteria for age (≥45 years) and the absence of stroke or dementia, we also included cohorts of African-American ancestry (N = 1004–3164 depending on the trait) in the replication phase, partly to evaluate whether our findings could be extrapolated to persons of other ethnicities. Data from the discovery and replication studies were further meta-analyzed. Additional details are provided in the Supplementary Material. Each participant provided informed consent and all studies were approved by their local Institutional Review Boards.
Executive function and processing speed tests
Each cohort included some tests of executive function and/or processing speed. Test batteries differed across cohorts. Tests of executive function included the Trail Making Test part B (Trail B), Stroop card 3 (color word card) and tests of phonemic (letters, see Supplementary Material) and semantic (animals) fluency. Tests of processing speed included Trail Making Test part A (Trail A), Stroop card 2 (color card), the Digit-Symbol Substitution Test (DSST), Letter-Digit Substitution Task (LDST), the Letter Digit Coding Test (LDCT) and the Symbol Digit Modalities Test (SDMT). All tests were administered in a standardized manner by an investigator unaware of any genetic information on the subjects. Details for the test administration and raw scores are given in Supplementary Table 1 and in the Supplementary Methods. Because the nature of the task and procedures for DSST, LDST, LDCT and SDMT were so similar, these measures were combined (that is, treated as the same test; referred to hereafter as DSST/LDST) in analysis. To assess the validity of combining these measures into a single meta-analysis, we conducted a substudy to determine the correlation between these measures in 102 volunteers not included in the meta-analysis cohorts. Details of the substudy are provided in the Supplementary Material, section 6 and Supplementary Table 5.
Genotyping and imputation
Genotyping was independently performed by each cohort using commercially available arrays ranging from the Illumina 300 and 610 k to the Affymetrix GeneChip SNP Array 6.0 (Supplementary Table 2). Each cohort applied standard quality control filtering before genetic imputations. These filters included a SNP call rate of at least 90%, sample call rate of at least 92%, minor allele frequency of at least 0.01, and Hardy–Weinberg deviation P-value of at most 10−3 (Supplementary Table 3). Genetic data imputations were performed in each cohort using HapMap II CEPH (Utah residents with ancestry from northern and western Europe) as the reference panel, with the exceptions of ARIC and GENOA African-American cohorts that imputed their genetic data using both HapMap II CEPH and YRI (Yoruba in Ibadan) populations as a reference.33 Details on genotyping and imputations are provided in Supplementary Tables 2–4.
Genome-wide association analysis
Each cohort performed a linear regression model of test scores against the dosage of coded alleles for each SNP testing an additive effect of the genetic variants. Skewness and kurtosis were evaluated before the analyses. A detailed description of the screening for latent population substructure in each cohort is given in part 5 of the Supplementary Material and Supplementary Table 3. Where appropriate, principal components or family-specific methods such as mixed model score were applied to correct for familial relationships (Supplementary Material, part 5).
We used two models of association analysis; with and without education level as a covariate in addition to age, gender and other study specific confounders, for example, study site, familial relations or population substructure. This is because there is a dynamic, two-way relationship between cognitive function and level of education. The ability to score high on cognitive tests is influenced by background familiarity with, for example, the numbers and alphabet (Trails tests) or a large vocabulary (fluency tests), which are typically acquired during one's formal education. Estimates are that, even in the Netherlands (10%) and the USA (22%), functional illiteracy is common among adults aged 16–65.34,35 Conversely, there is a genetic correlation between education and cognitive ability.36
Meta-analysis
The meta-analysis was performed in METAL.37 For all tests except the Stroop test and LDST/DSST, meta-analysis was performed using the inverse variance method. For the Stroop test and LDST/DSST, a sample size weighted meta-analysis was performed because of the differences in the test methodology and measurement units that impeded the pooling of the beta coefficients. The z-statistic was weighted by the effective sample size (sample size × (observed dosage variance/expected dosage variance)) for each SNP. Genomic control was applied within each cohort before meta-analysis. The meta-analyses were restricted to autosomal SNPs common to all studies for each neurocognitive test.
Additional analyses
Expression quantitative trait locus analyses
All variants with a discovery P-value < 5 × 10−6 were analyzed further to test whether they were associated with RNA expression. For this, we used the Genotype Tissue expression portal (GTex, Broad Institute, Boston, MA, USA; http://www.gtexportal.org/home/) to assess the variants for their influence on expression of their closest genes in brain tissue.38 We also performed expression quantitative trait locus (eQTL) experiments in 138 human hippocampal cell lines obtained in vivo from patients undergoing surgery for treatment-resistant epilepsy. Details on the methods of this functional follow-up study are given in the Supplementary Material. Cognitive tests are never completely limited to a single cognitive domain. Even in executive function and processing speed tests, working memory (and other capabilities) may have a role. For example, one needs to remember the assignment and which words were already used in fluency tasks, and those who are better able to passively learn the key in the LDST or DSST should likewise produce a higher (better) score. Thus, some hippocampal involvement might be expected along with the predominant frontal processes of executive function and processing speed. Hence we used the hippocampal cell lines to identify associations between whole-genome SNP (Illumina Human 660 W array) and RNA expression data (Illumina HumanHT-12v3). The Bonferroni corrected level of eQTL significance for a given SNP can be assumed to be 0.05/n of genes for a given SNP and a given test, leading to a P-value of 0.05/20 000 = 2.5 × 10−6 for the hypothesis-free, genome-wide search for eQTL associations.
Gene network and functional prediction analysis
A gene network analysis was performed using the Gene Network database (http://www.genenetwork.nl/genenetwork, by Groningen University, The Netherlands) (Fehr-mann et al., manuscript in preparation), which incorporates gene expression data from 77 840 human, mouse and rat Affymetrix microarrays from the Gene Expression Omnibus.39,40 Using principal components analysis on the probeset correlation matrices, so-called transcriptional components were identified that describe major biological pathways. We combined these data into a multi-species gene network with 19 997 unique human genes. Predictions of gene function were made using biological databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG, www.genome.jp/kegg),41 Gene Ontology (GO, www.geneontology.org) Database42 and Reactome pathway database (www.reactome.org).43 Additionally, we consulted the GTex website for tissue expression data on the genes of interest. More detailed information on the gene network approach is given in the Supplementary Material.
RESULTS
Phenotypes
Analyses in an independent group of 102 volunteers showed correlations between LDST, DSST and SDMT above 0.8 (P-value < 0.01). After partialling out age, correlations remained above 0.7 (P-value < 0.01). (Supplementary Table 6 and Supplementary Figure 1). We therefore deemed it appropriate to combine these three tests into one sample size-weighted meta-analysis.
GWAS and eQTL analysis
Baseline characteristics of the study populations and mean test results for each trait are provided in Supplementary Table 1. Supplementary Figure 2 shows the quantile-quantile plots for the discovery-phase GWAS meta-analysis of each trait. No inflation due to hidden substructure or cryptic relatedness was observed for any cohort.
In the meta-analysis for processing speed (LDST/DSST tests), only one genome-wide significant association was observed at an intronic variant (rs17518584, P-value = 3.12 × 10−8 in the model adjusted for age, sex and education and P-value = 6.25 × 10−7 in the model adjusted for age and sex) in the gene encoding cell adhesion molecule 2 (CADM2) on chromosome 3 (Supplementary Figure 3 and Table 1). This gene is also known as SYNCAM2 (synaptic cell adhesion molecule 2). This variant explains 0.05% (age, sex and education-adjusted model) of the variance in scores on the LDST/DSST in the Rotterdam Study, one of the largest population-based cohorts.
Table 1.
SNPs with P-value < 10−6 in original meta-analysis
| Trait | SNP | Chr | Position | Gene | Feature | A1 | A2 | Average EAF |
Discovery |
Replication |
Combined |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of subjects |
Age, sex adjusted | Age, sex, education adjusted |
No. of subjects |
Age, sex adjusted | Age, sex, education adjusted |
Age, sex adjusted | Age, sex, education adjusted |
|||||||||||||||
| Tests with summary values in beta (SE) | Beta (SE) | P-value | Beta (SE) | P-value | Beta (SE) | P-value | Beta (SE) | P-value | Beta (SE) | P-value | Beta (SE) | P-value | ||||||||||
| Trail A | rs9514964 | 13 | 109679886 | MY016 | ntron | A | G | 0.17 | 5429 | 0.048 (0.009) | 3.01E-07 | 0.044 (0.009) | 2.35E-06 | 9553 | −0.006 (0.007) | 0.37 | 0.007 (0.007) | 0.32 | 0.029 (0.007) | 1.55E-04 | 0.020 (0.006) | 3.03E-04 |
| rs9559465 | 13 | 109683883 | MY016 | ntron | A | G | 0.84 | −0.048 (0.010) | 3.30E-07 | −0.043 (0.009) | 2.59E-06 | 0.006 (0.008) | 0.43 | −0.007 (0.008) | 0.36 | −0.035 (0.008) | 1.58E-04 | −0.021 (0.006) | 2.35E-04 | |||
| rs1230154 | 4 | 99988659 | METAP1, ADH5 | ntergenic | T | C | 0.71 | −0.038 (0.007) | 7.69E-07 | −0.036 (0.008) | 1.64E-06 | 0.003 (0.006) | 0.63 | 0.001 (0.006) | 0.93 | −0.030 (0.007) | 9.34E-04 | −0.013 (0.005) | 4.83E-03 | |||
| Trail B | rs11082233 | 18 | 37265119 | PIK3C3, KC6 | ntergenic | T | C | 0.11 | 6210 | 0.073 (0.013) | 6.95E-08 | 0.061 (0.013) | 2.28E-06 | 10817 | −0.012 (0.010) | 0.25 | −0.017 (0.010) | 0.09 | 0.020 (0.008) | 1.73E-02 | 0.0123 (0.0079) | 0.12 |
| Phonemic fluency | rs6583634 | 8 | 143546590 | BAI1 | ntron | T | C | 0.15 | 13 454 | 1.046 (0.209) | 5.75E-07 | 0.914 (0.194) | 2.47E-06 | 12734 | −0.162 (0.230) | 0.48 | −0.009 (0.164) | 0.96 | 0.498 (0.155) | 1.00E-03 | 0.376 (0.125) | 0.0027 |
| rs10481393 | 8 | 143556168 | BAI1 | ntron | T | C | 0.15 | 1.050 (0.210) | 5.96E-07 | 0.914 (0.195) | 2.78E-06 | 0.068 (0.479) | 0.88 | −0.015 (0.183) | 0.937 | 0.891 (0.193) | 3.69E-06 | 0.420 (0.134) | 0.0016 | |||
| Semantic fluency | rs6587905 | 1 | 61093597 | C1orf87, NFIA | ntergenic | T | C | 0.16 | 6383 | 0.664 (0.129) | 2.80E-07 | 0.656 (0.126) | 2.09E-07 | 21860 | 0.091 (0.099) | 0.36 | 0.138 (0.077) | 0.071 | 0.303 (0.079) | 1.16E-04 | 0.280 (0.065) | 1.90E-05 |
| rs10115337 | 9 | 88711574 | LOC392358, GAS1 | ntergenic | T | C | 0.18 | −0.494 (0.100) | 8.02E-07 | −0.478 (0.098) | 9.88E-07 | −0.058 (0.079) | 0.46 | −0.083 (0.059) | 0.159 | −0.224 (0.062) | 2.88E-04 | −0.189 (0.050) | 1.65E-04 | |||
| rs10780801 | 9 | 88710941 | LOC392358, GAS1 | ntergenic | T | C | 0.82 | 0.493 (0.100) | 8.18E-07 | 0.477 (0.976) | 1.01E-06 | 0.055 (0.079) | 0.49 | 0.081 (0.059) | 0.166 | 0.223 (0.062) | 3.19E-04 | 0.189 (0.050) | 1.79E-04 | |||
| Tests with summary values in Z-scores | Z-score | P-value | Z-score | P-value | Z-score | P-value | Z-score | P-value | Z-score | P-value | Z-score | P-value | ||||||||||
| Stroop | 12 866 | 6381 | ||||||||||||||||||||
| LDST/DSST | rs664154 | 6 | 19099538 | RP1-239K6.1 | ntergenic | T | C | 0.24 | 32 070 | 5.22 | 1.84 E-07 | 4.79 | 1.67E-06 | 1311 | −0.96 | 0.34 | −1.06 | 0.29 | 5.03 | 5.01E-07 | 4.57 | 4.90E-06 |
| rs17518584 | 3 | 85687613 | CADM2 | ntron | T | C | 0.64 | 4.98 | 6.25 E-07 | 5.54 | 3.12E-08 | 2.23 | 0.03 | 1.95 | 0.05 | 5.43 | 5.56E-08 | 5.92 | 3.28E-09 | |||
| rs2734839 | 11 | 112791700 | DRD2 | ntron | T | C | 0.61 | 4.99 | 5.92 E-07 | 4.86 | 1.18E-06 | 0.55 | 0.58 | 0.20 | 0.84 | 5.13 | 2.95E-07 | 4.91 | 9.18E-07 | |||
| rs2118666 | 2 | 222808767 | PAX3 | ntron | T | G | 0.46 | −4.95 | 7.33E-07 | −4.25 | 2.17E-05 | 0.10 | 0.92 | −0.07 | 0.94 | −4.95 | 7.45E-07 | −4.26 | 2.08E-05 | |||
Abbreviations: A1, effect allele; A2, reference allele; Beta, beta coefficient of effect allele; Chr, Chromosome; EAF, effect allele frequency; SE, standard error of beta coefficient; LDST/DSST, Letter-Digit or Digit-Symbol Substitution Task; SNP, single-nucleotide polymorphism.
Additional information was sought for all SNPs with a discovery P-value of below 5 × 10−6 and a minor allele frequency of >0.05. At this phase, 7 SNPs were analyzed for LDST/DSST, 15 SNPs for the Stroop test, 12 SNPs for Trail A, 17 SNPs for Trail B, 9 SNPs for letter fluency and 34 SNPs for semantic fluency. Of all the loci tested in the independent cohorts, including both subjects of European and African-American ancestries, a nominally significant association in the same direction as in the discovery analyses was observed only for the CADM2 variant rs17518584 with LDST/DSST (nominal P-value = 0.03 for meta-analyses of results in the additional cohorts, adjusting for age and sex, and nominal P-value = 0.05 for meta-analysis of results after adjusting for age, sex and education). Meta-analysis of discovery and replication cohorts yielded genome-wide significant evidence for association of rs17518584 in CADM2 with scores for the LDST/DSST processing speed tests, in the fully-adjusted model that included age, sex and education (P-value = 3.28 × 10−9) (Table 1). The findings of the individual studies (both discovery and replication) are shown in Table 2.
Table 2.
Effect size and direction of effects across cohorts for rs17518584
| Study | Test | Time (seconds) | Coded allele | Non-coded allele | Beta Coeff./SE (age, sex adjusted) | P-value | Beta Coeff./SE (age, sex, education adjusted) | P-value |
|---|---|---|---|---|---|---|---|---|
| Discovery | ||||||||
| ARIC | DSST53 | 90 | T | C | 0.235 (0.157) | 1.34 × 10−1 | 0.275 (0.146) | 5.97 × 10−2 |
| CHS | 90 | T | C | 0.317 (0.387) | 4.12 × 10−1 | 0.469 (0.358) | 1.90 × 10−1 | |
| GENOA | 90 | T | C | 0.503 (0.550) | 3.61 × 10−1 | 0.553 (0.534) | 3.02 × 10−1 | |
| ORCADES | 90 | T | C | 1.516 (1.101) | 1.68 × 10−1 | 1.541 (1.054) | 1.44 × 10−1 | |
| AGES | DSST54 | 90 | T | C | 0.682 (0.282) | 1.56 × 10−2 | 0.808 (0.259) | 1.84 × 10−3 |
| Korcula | 120 | T | C | -0.350 (0.967) | 7.18 × 10−1 | −0.161 (0.838) | 8.48 × 10−1 | |
| Split | 60 | T | C | 0.495 (0.664) | 4.56 × 10−1 | 0.549 (0.588) | 3.51 × 10−1 | |
| Vis | 120 | T | C | 0.725 (1.155) | 5.31 × 10−1 | 0.880 (1.089) | 4.19 × 10−1 | |
| Health ABC | DSST55 | 90 | T | C | 0.448 (0.452) | 3.22 × 10−1 | 0.458 (0.432) | 2.89 × 10−1 |
| LBC 1921 | DSST56 | 120 | T | C | 0.755 (1.178) | 5.22 × 10−1 | 0.804 (1.140) | 4.80 × 10−1 |
| LBC 1936 | 120 | T | C | 1.074 (0.630) | 8.84 × 10−2 | 0.776 (0.611) | 2.04 × 10−1 | |
| ASPS | LDST57 | 60 | T | C | 0.850 (0.600) | 1.55 × 10−1 | 0.545 (0.580) | 3.44 × 10−1 |
| RS | 60 | T | C | 0.176 (0.174) | 3.12 × 10−1 | 0.224 (0.165) | 1.76 × 10−1 | |
| RS2 | 60 | T | C | 0.370 (0.212) | 8.08 × 10−2 | 0.329 (0.205) | 1.09 × 10−1 | |
| RS3 | 60 | T | C | 0.648 (0.216) | 2.72 × 10−3 | 0.599 (0.208) | 3.91 × 10−3 | |
| MAP | SDMT58 | 90 | T | C | −1.214 (0.530) | 2.15 × 10−2 | −1.193 (0.520) | 2.14 × 10−2 |
| ROS | 90 | T | C | 1.154 (0.462) | 1.22 × 10−2 | 1.218 (0.450) | 6.58 × 10−3 | |
| PROSPER | LDCT59 | 60 | T | C | 0.336 (0.155) | 3.04 × 10−2 | 0.303 (0.148) | 4.07 × 10−2 |
| Replication | ||||||||
| BLSA | DSST53 | 90 | T | C | −0.119 (0.663) | 8.58 × 10−1 | −0.155 (0.678) | 8.20 × 10−1 |
| MAS | DSST55 | 90 | T | C | 1.651 (0.586) | 4.81 × 10−3 | 1.425 (0.576) | 1.33 × 10−2 |
Abbreviations: Coeff, coefficient; DSST, Digit Symbol Substitution Test; LDCT, Letter Digit Coding Test; LDST, Letter-Digit Substitution Task; SDMT, Symbol Digit Modalities Test; SE, standard error of beta coefficient; Model 1: adjusted for age and gender; Model 2: adjusted for age, gender and education.
When examining association signals for loci previously identi-fied in genetic analyses of cognitive function, rs7412 and rs429358 used to genotype the APOE ε4 allele associated with general cognition22 were not present in the analyzed SNP sets. However, a single SNP proxy for APOE ε4 (rs4420638)44 was associated with the LDST/DSST (P-value = 2.11 × 10−4) in the fully adjusted model. We found no effect of additional adjustments for rs4420638 on the association of rs17518584 with the LDST/DSST in the Rotterdam study cohorts. There was no evidence for association of SNPs in any of the other tested candidate genes that have previously been associated with executive and processing speed functions. (DTNBP1: lowest P-value = 0.019 for rs2619522 for the Stroop test, TRIB3: lowest P-value = 0.155 for phonemic fluency, WDR72 lowest P-value = 0.157 for LDST/DSST in our GWAS meta-analysis for the fully adjusted model). Also, regions for which linkage was reported (2q, 5q, 11q, 13q and 14q) were not genome-wide significant. The strongest association was seen in the 11q.25 region in which we found a SNP rs2734839 to be suggestively associated with performance on the LDST/DSST (P-value = 4.39 × 10−7 for the age- and sex-adjusted model and 3.08 × 10−6 for the model adjusted for age, sex and education). Rs2734839 is an intronic SNP in the DRD2 gene encoding the D2 subtype of the dopamine receptor.
Bioinformatics analysis
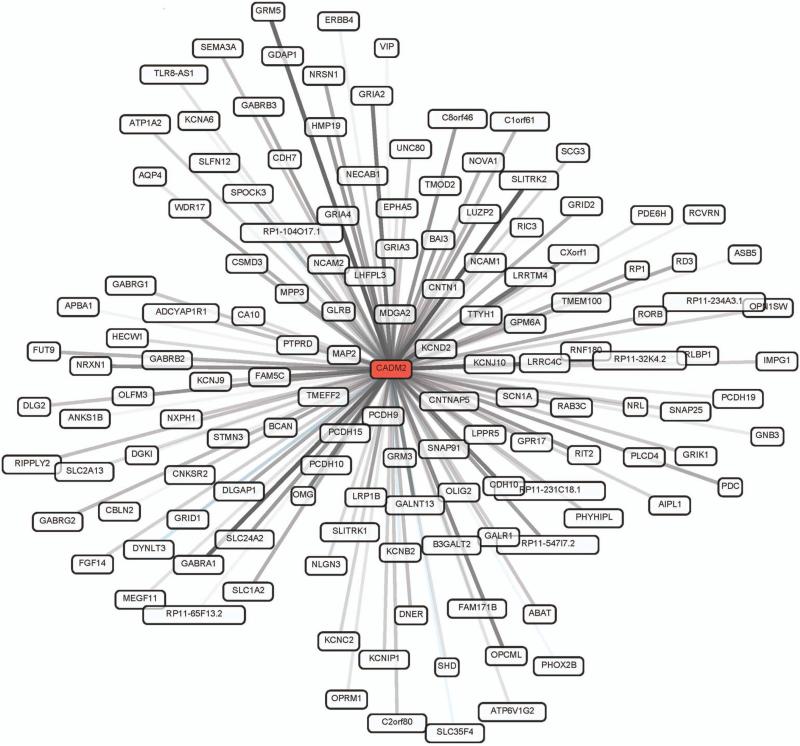
The GTex tissue expression data (www.broadinstitute.org/gtex) show that CADM2 is expressed more abundantly in different areas of the brain than in any other tissue, and most specifically in the frontal and anterior cingulate cortex (Supplementary Figure 4). In the GTex eQTL analysis, the top hit from the LDST/DSST GWAS, rs17518584, showed association with RNA expression levels of CADM2 in the cingulate cortex (P-value = 4 × 10−4), general hemispheral cortex (P-value = 0.004), substantia nigra (P-value = 0.007), frontal cortex (P-value = 0.02) and cerebellum (P-value = 0.03). No significant cis or trans associations were identified in the ex-vivo hippocampal data. Gene network analysis shows that CADM2 is significantly expressed in human brain (P-value for expression in cerebral cortex = 2.47 × 10−171, area under the curve (AUC) = 0.99; P-value for expression in the prefrontal cortex specifically = 1.29 × 10−31, AUC = 1.0; P-value for expression in the hippocampus = 8.2 × 10−36, AUC = 0.99). The gene network analyses suggest that CADM2 is likely involved in biological processes that include the glutamate signaling pathway (P-value = 7.22 × 10−15), gamma-aminobutyric acid (GABA) transport (P-value = 1.36 × 10−11) and neuron cell-cell adhesion (P-value = 1.48 × 10−13). CADM2 shows strong positive co-expression with several genes involved in the GABA and other glutamate neurosignaling pathways, such as GABA A receptor alpha 1 and beta 2 (GABRA1, GABRB2) and glutamate receptor metabotropic 5 (GRM5). Further, there is strong positive co-expression with many members of the voltage-gated potassium channel group including KCNJ9, KCNJ10, KCNB2 and KCNC, and with the OPCML (opioid binding protein/cell adhesion molecule-like) gene and EPHA5 (EPH receptor A5) (Figure 1). Supplementary Table 7 and Supplementary Figure 4 provide the expression data in neuronal tissues. As expected, Gene Network and GTex show that DRD2 is predominantly expressed in the pituitary (GTex reads per kilobase per million (RPKM) = 50), putamen (gene network AUC = 1, P-value = 5 × 10−12, GTex RPKM = 40), substantia nigra (gene network AUC = 0.98, P-value = 4 × 10−15, GTex RPKM = 4) and nucleus accumbens (GTex RPKM = 28). However, the intronic variant rs2734839 is not associated with expression at nominal significance in any region reported (cingulate, frontal or general cortex, amygdala, caudate nucleus, nucleus accumbens, putamen, substantia nigra, cerebellum, hippocampus, hypothalamus, pituitary or spinal cord).
Figure 1.
Gene network plot for CADM2, based on biological processes. Gray lines indicate positive co-expression and blue lines indicate negative co-expression, with the density of the line reflecting the strength of the co-expression.
DISCUSSION
The discovery phase of our study yielded genome-wide significance for an association of processing speed (LDST/DSST) with rs17518584. In silico replication in additional independent cohorts yielded nominally significant support for association. In the combined discovery-replication analysis, the model adjusted for age, sex and education resulted in genome-wide significant association between rs17518584 and LDST/DSST performance. The variant is associated with CADM2 expression, at least at nominal significance.
Rs17518584 is located about 170 kb upstream of the transcription start site of the major transcript for the CADM2 gene, but is within an intron of a variant transcript that includes an alternative first exon. Variants in the CADM2 gene have been previously associated with body mass index,37 but not with cognitive function. The gene has been studied as a candidate for autism spectrum disorders,45 and showed a suggestive association with scores on the persistence items on a personality scale.46 The protein is likely involved in long-term signal depression and potentiation and neuroactive ligand-receptor interaction (http://www.genome.jp/kegg/), and is a member of the immunoglobulin superfamily. The gene encodes a neuronal adhesion molecule that has been shown to be widely expressed in the developing and adult brain in laboratory mice,47 as well as in human and rat brain tissue, with highest expression AUCs for the prefrontal cortex. In the GTex analyses, rs17518584 significantly influenced CADM2 expression levels in the brain, most specifically in the cingulate cortex. This finding is of interest in the background of diffusion tensor imaging experiments that have shown an association between fractional anisotropy in the cingulum and performance on executive and processing speed tasks.48,49 The co-expression data from gene network analysis also revealed some interesting links to Alzheimer's disease (Figure 1). OPCML has been associated with a variety of cognitive domains in a follow-up of linkage regions for Alzheimer's disease.50 EPHA5 is a member of the same family of ephrin receptors as the EPHA1 gene, which is associated with Alzheimer's disease.51,52
No significant cis or trans associations with gene expression were identified in the experiments using ex-vivo hippocampal tissue from epilepsy patients. However, both GTex and Gene Network bioinformatics sources show that CADM2 is expressed in normal hippocampal tissue (obtained postmortem).
We identified only one genome-wide significant variant explaining a small part of the variance in performance on the LDST/DSST (R2 = 0.005). These findings should be interpreted in light of the sample size studied. The number of persons analyzed varied considerably between outcomes, with 5555–32 900 subjects per individual test. This variability is a consequence of the nature of the CHARGE consortium, which uses the data previously collected in the individual cohorts, long before the GWAS era. As a consequence, there were differences in the test batteries administered in each cohort, and some tests were available in a larger number of cohorts than others. Limitations in sample size may have prevented us from finding variants (false negatives) associated with some outcomes. To minimize the detection of false positives, we sought replication in independent studies.
This may also explain, in part, why rs17518584 was identified only in LDST/DSST, which had the largest discovery sample. The strongest association for this SNP in other traits was in letter fluency, with a P-value of 0.076 in the discovery meta-analysis, with all studies showing a positive effect of the T allele (as in LDST/DSST). This may reflect the influence of processing speed on fluency performance previously described in the literature.18 Our analyses lacked power to address genes with small effects affecting tests for which we had only a limited sample size. The lack of test-specific power should also be taken into account when interpreting the lack of association with candidate genes beyond APOE (DTNBP1, TRIB3 and WDR72). There was only one other intronic variant (rs2734839) in the DRD2 gene, encoding the D2 subtype of the dopamine receptor, that approached genome-wide significance. The bioinformatic analyses did not support a functional effect of the expression of the gene in the substantia nigra. This variant therefore remains to be replicated. A further limitation is that the cognitive phenotypes were based on single assessments and thus we were unable to determine genetic effects on age-related cognitive decline. Despite some limitations, this is the first study to discover and replicate a genetic variant involved in processing speed.
The present report provides the most comprehensive meta-analysis of processing speed and executive function GWAS to date. We found a genome-wide significant association between widely-used tests of processing speed (LDST/DSST) and a SNP in the CADM2 gene, which is involved in glutamatergic and GABA-ergic transmission, in middle-aged and older non-demented adults. This gene is a candidate for autism and personality, but based on the pathway and expression analyses it may also be relevant to a broad range of neuropsychiatric diseases including dementias.
Supplementary Material
ACKNOWLEDGMENTS
3CS: The work was made possible by the generous participation of the control subjects and their families. This work was supported by the National Foundation for Alzheimer's disease and related disorders, the Institut Pasteur de Lille, the Centre National de Génotypage, Inserm, FRC (fondation pour la recherche sur le cerveau) and Rotary. This work has been developed and supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant (Development of Innovative Strategies for a Transdisciplinary approach to ALZheimer's disease). JCL was funded by the MEDIALZ Project (Grant 11001003) financed by ERDF (European Regional Development Fund) and Conseil Régional Nord Pas de Calais. The Three-City Study was performed as part of a collaboration between the Institut National de la Santé et de la Recherche Médicale (Inserm), the Victor Segalen Bordeaux II University and Sanofi-Synthélabo. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study was also funded by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, MGEN, Institut de la Longévité, Agence Française de Sécurité Sanitaire des Produits de Santé, the Aquitaine and Bourgogne Regional Councils, Fondation de France and the joint French Ministry of Research/INSERM ‘Cohortes et collections de données biologiques’ program. Lille Génopôle received an unconditional grant from Eisai.
AAA: We thank the cohort participants and team members who contributed to this study. Phenotype collection and DNA extraction were supported by the Wellcome Trust, the British Heart Foundation and the Chief Scientist Office of the Scottish Executive. The AAA Trial was performed and the database is maintained by members of the University of Edinburgh Molecular Epidemiology Research Group in the Centre for Population Health Sciences. We also thank staff at the Wellcome Trust Clinical Research Facility in Edinburgh where some of the research clinics and genotyping were undertaken.
AGES: Aging Gene-Environment Susceptibility-Reykjavik Study: The research has been funded by NIA Contract N01-AG-12100 with contributions from NEI, NIDCD and NHLBI, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association) and the Althingi (the Icelandic Parliament).
ARIC: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung and Blood Institute Contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C), R01HL70825, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute Contract U01HG004402; and National Institutes of Health Contract HHSN268200625226C. We thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
ASPS: We thank Ing. Johann Semmler and Irmgard Pölzl for creating the DNA bank and for supervising the quality managementof the biobanking and DNA analyses.The ASPS is funded by the Austrian Science Fond (FWF) Grant Number P20545-P05 and P13180.
BLSA: Baltimore Longitudinal Study of Aging (BLSA): The Baltimore Longitudinal Study of Aging is supported by the Intramural Research Program of the NIH, National Institute on Aging.
Cardiovascular Health Study: This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, and R01HL120393 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629, R01AG15928, R01AG20098, and R01AG027058 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHSNHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CROATIA-Korcula: The CROATIA-Korcula study was funded by grants from the Medical Research Council (UK), European Commission Framework 6 project EUROSPAN (Contract No. LSHG-CT-2006-018947) and Republic of Croatia Ministry of Science, Education and Sports research grants to IR (108-1080315-0302). We would like to acknowledge the invaluable contributions of the recruitment team in Korcula, the administrative teams in Croatia and Edinburgh and the people of Korcula. The SNP genotyping for the CROATIA-Korcula cohort was performed in Helmholtz Zentrum München, Neuherberg, Germany.
CROATIA-Split: The CROATIA-Split study is funded by grants from the Medical Research Council (UK), European Commission Framework 6 project EUROSPAN (Contract No. LSHG-CT-2006-018947) and Republic of Croatia Ministry of Science, Education and Sports research grants to IR (108-1080315-0302). We would like to acknowledge the invaluable contributions of the recruitment team in Split, the administrative teams in Croatia and Edinburgh and the people of Split. The SNP genotyping for the CROATIA-Split cohort was performed by AROS Applied Biotechnology, Aarhus, Denmark.
CROATIA-Vis: The CROATIA-Vis study was funded by grants from the Medical Research Council (UK) and Republic of Croatia Ministry of Science, Education and Sports research grants to IR (108-1080315-0302). We would like to acknowledge the staff of several institutions in Croatia that supported the field work, including but not limited to The University of Split and Zagreb Medical Schools, the Institute for Anthropological Research in Zagreb and Croatian Institute for Public Health. The SNP genotyping for the CROATIA-Vis cohort was performed in the core genotyping laboratory of the Wellcome Trust Clinical Research Facility at the Western General Hospital, Edinburgh, Scotland.
ERF: This study was financially supported by the Netherlands Organization for Scientific Research (NWO), the Internationale Stichting Alzheimer Onderzoek (ISAO), the Hersenstichting Nederland (HSN) and the Centre for Medical Systems Biology (CMSB) in the framework of the Netherlands Genomics Initiative (NGI) and by the Russian Foundation for Basic Research (RFBR). We thank the participants from the Genetic Research in Isolated Populations, Erasmus Rucphen Family, who made this work possible. Also, we thank Petra Veraart for collecting all genealogical data.
FHS: This work was supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278) and Grants (U01 HL096917 and R01 HL093029). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This study was also supported by grants from the National Institute of Neurological Disorders and Stroke (NS17950) and the National Institute of Aging (U01 AG049505, AG033193, AG008122, AG16495). The content is solely the responsibility of the authors and does not necessarily represent the official views of NINDS, NHLBI, NIA, NIH or AHA. Dr. Debette is a recipient of a Chaire d'Excellence grant from the Agence National de la Recherche and a grant from the Leducq Foundation.
GENOA: Support for the Genetic Epidemiology Network of Arteriopathy (GENOA) was provided by the National Heart, Lung and Blood Institute (HL054464, HL054457, HL054481, HL071917 and HL87660) and the National Institute of Neurological Disorders and Stroke (NS041558) of the National Institutes of Health. Genotyping was performed at the Mayo Clinic (S.T.T., Mariza de Andrade, Julie Cunningham) and was made possible by the University of Texas Health Sciences Center (Eric Boerwinkle, Megan L Grove-Gaona). We would also like to thank the families that participated in the GENOA study.
GS: We are grateful to the GS Executive Committee Professors Blair H. Smith, David J. Porteous, Sandosh Padmanabhan and Dr. Lynne J. Hocking, and all the families who took part, the general practitioners and the Scottish School of Primary Care for their help in recruiting them, and the whole Generation Scotland team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, healthcare assistants and nurses. Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates [CZD/16/6] and the Scottish Funding Council [HR03006].
HBCS: We thank all study participants as well as everybody involved in the Helsinki Birth Cohort Study. Helsinki Birth Cohort Study has been supported by grants from the Academy of Finland, the Finnish Diabetes Research Society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Signe and Ane Gyllenberg Foundation, University of Helsinki, Ministry of Education, Ahokas Foundation, Emil Aaltonen Foundation, Juho Vainio Foundation and Wellcome Trust (Grant Number WT089062).
Health ABC: This research was supported by NIA Contracts N01AG62101, N01AG62103 and N01AG62106. The genome-wide association study was funded by NIA Grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract Number HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
Hunter: The authors would like to thank the men and women participating in the HCS as well as all the staff, investigators and collaborators who have supported or been involved in the project to date. The cohort was made possible with support from the University of Newcastle's Strategic Initiative Fund, the Vincent Fairfax Family Foundation and the Hunter Medical Research Institute.
InCHIANTI: The Invechhiare in Chianti (InCHIANTI) Study was supported as a targeted project (ICS 110.1RS97.71) by the Italian Ministry of Health, by the US National Institute on Aging (Contracts N01[AG]916413, N01[AG] 821336, 263 MD 9164 13, and 263 MD 821336), and, in part, by the Intramural Research Program, National Institute on Aging, National Institutes of Health.
LBC1921/LBC1936: We thank the cohort participants and team members who contributed to these studies. Phenotype collection in the Lothian Birth Cohort 1921 was supported by the BBSRC, The Royal Society and The Chief Scientist Office of the Scottish Government. Phenotype collection in the Lothian Birth Cohort 1936 was supported by Research Into Ageing (continues as part of Age UK The Disconnected Mind project). Genotyping of the cohorts was funded by the UK Biotechnology and Biological Sciences Research Council (BBSRC). The work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (G0700704/84698). Funding from the BBSRC, Engineering and Physical Sciences Research Council (EPSRC), Economic and Social Research Council (ESRC) and MRC is gratefully acknowledged.
MAS: We would like to acknowledge and thank the Sydney MAS participants and the Research Team for their contributions and assistance. We would like to specifically acknowledge the support and contributions of Professor Henry Brodaty (Chief Investigator), Dr Simone Reppermund (Study Co-ordinator), Professor Peter Schofield, Dr Arezoo Assareh and Dr John Kwok to this work. DNA was extracted by Genetic Repositories Australia, an Enabling Facility supported by NHMRC Grant 401184. DNA sample preparation was undertaken in the laboratory of Professor Peter Schofield and Dr John Kwok, Neuroscience Research Australia, with the assistance of Dr Arezoo Assareh. Genotyping was performed by the Ramaciotti Centre, University of New South Wales. Sydney MAS is supported by the Australian National Health & Medical Research Council Program Grants 350833 and 568969. Karen Mather is supported by the Capacity Building Grant 568940. Nicola Armstrong is supported by the NHMRC Project Grant 525453.
Hippocampal eQTL: The hippocampal gene expression study was supported by the German Federal Ministry of Education and Research (BMBF) through the Integrated Genome Research Network (IG) MooDS (Systematic Investigation of the Molecular Causes of Major Mood Disorders and Schizophrenia, under the auspices of the National Genome Research Network plus (NGFNplus)).
NHS: This study was supported by research Grants CA87969, CA49449, HL34594, U01HG004399, DK058845, CA65725, CA67262, CA50385, 5UO1CA098233, EY09611, EY015473, HG004728, HL35464, CA55075, CA134958 and DK070756 from the National Institutes of Health. The genotyping was partly supported by an unrestricted grant from Merck Research Laboratories. Dr Sun is supported by career development award K99HL098459 from the National Heart, Lung and Blood Institute. Supported in part by NIH.
ORCADES: ORCADES was supported by the Chief Scientist Office of the Scottish Government, the Royal Society, the MRC Human Genetics Unit, Arthritis Research UK and the European Union framework program 6 EUROSPAN project (Contract No. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. We would like to acknowledge the invaluable contributions of Lorraine Anderson and the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney.
PROSPER: The PROSPER study was supported by an investigator initiated grant obtained from Bristol-Myers Squibb. Prof. Dr JW Jukema is an Established Clinical Investigator of the Netherlands Heart Foundation (Grant 2001 D 032). Support for genotyping was provided by the seventh framework program of the European commission (Grant 223004) and by the Netherlands Genomics Initiative (Netherlands Consortium for Healthy Aging Grant 050-060-810).
RS: The generation and management of GWAS genotype data for the Rotterdam Study is supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating the GWAS database, and Karol Estrada and Maksim V. Struchalin for their support in creation and analysis of imputed data. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission 49 (DG XII), and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. Dr Ikram was supported by a ZonMW Veni Grant: 916.13.054.
RUSH: Supported in part by NIA Grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, K08AG34290 and K25AG41906.
SHIP: SHIP is part of the Community Medicine Research net of the University Medicine of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania. The SHIP authors are grateful to Holger Prokisch and Thomas Meitinger (Helmholtz Zentrum München) for the genotyping of the SHIP-TREND cohort.
WGHS: The WGHS is supported by HL043851 and HL080467 from the National Heart, Lung and Blood Institute and CA047988 from the National Cancer Institute, the Donald W Reynolds Foundation and the Fondation Leducq, with collaborative scientific support and funding for genotyping provided by Amgen.
The study sponsors had no role in the study design, the collection, analysis, and interpretation of data; writing the report, or the decision to submit the report for publication.
Footnotes
AUTHOR CONTRIBUTIONS
Study concept and design were performed by CAIV, JB, SD, MS, AVS, JCB, GD, ST, CW, BHS, VG, JWJ, TBH, OLL, IF, JTB, GH, STT, KY, JvS, LHC, SLRK, DSK, PWS, DJS, JMS, LJH, GE, RS, RJS, AP, RFG, BAO, JIR, MMN, AH, RGJW, PAW, AGU, BMP, HJG, SB, DIC, LF, JRA, IR, CH, AFW, JFW, MF, DAB, IJD, MAI, LJL, ALF, SS, CMvD and THM. Acquisition of data was carried out by CAIV, MS, ST, KP, OP, LZ, PH, JK, JL, SMR, FG, MS, BHS, VG, JWJ, PLDeJ, TBH, OLL, JTB, MKJ, RA, RSNF, SH, STT, JvS, LHC, SLRK, DSK, WMM, GH, PWS, DCL, PR, AJG, AP, LJH, NJA, SMcL, DJP, JMS, GE, RS, RJS, NAK, AP, JGE, RFG, BAO, AB, JIR, MMN, AH, PES, PAW, AGU, BMP, HJG, SB, DIC, FG, KR, JFP, PSS, LF, JRA, CH, SC, LF, JFW, MF, DAB, MAI, SS, CMvD and THM. Statistical analysis and interpretation of the data were performed by CAIV, JB, SD, MS, AVS, JCB, GD, ST, JS, CW, LBC, YL, VV, MK, LZ, JK, JL, DL, COS, KM, VC, QS, LMR, CO, QY, SSM, NA, OS,AT, NK, RSNF, WZ, KY, KL, SLRK, EGH, TT, JW, NJA, LCP, RJS, NK, AP, YCH, LY, ALdeS, AB, MG, MMN, JCL, PSS, JRA, CH, JD, AJMDeC, IJD, MAI and THM. The manuscript was drafted by CAIV, JB, SD, MS, JCB, NA, HS, SS, CMvD and THM. Critical revision of the manuscript was performed by CAIV, JB, SD, MS, AVS, JCB, GD, ST, JS, CV, LBC, YL, VV, MK, KP, OP, LZ, PH, JK, JL, DL, COS, KAM, VC, QS, SMR, LMR, CO, MS, BHS, VG, QY, SSM, JWJ, PLDeJ, TBH, DCL, NA, LHC, OS, OLL, RS, AT, IF, NK, JTB, MJK, RA, RSNF, SH, MN, WZ, STT, KY, KL, JCvS, SLRK, DSK, WMM, GH, EGH, PWS, TT, DJS, JW, PR, AJG, AP, JMS, LJH, NJA, SMcL, JMS, LCP, GE, RJS, NAK, AP, YCH, JGE, AP, RFG, BAO, LY, ALDeS, AB, MG, JIR, MN, AH, PES, RGJW, BMB, PAW, AGU, BMP, HJG, SB, DIC, FG, KR, JCL, DJP, JFP, PSS, LF, JRA, IR, CH, AFW, JFW, SC, LF, HS, JD, AJMDeC, MF, DAB, ID, MAI, LJL, AF, SS, CMvD and THM. Funding was obtained by ST, SMR, BHS, VG, JWJ, PLDeJ, TBH, OLL, IF, JTB, MN, STT, JCvS, SLRK, GH, PWS, JMS, AP, JGE, BAO, JIR, MMN, AH, PES, RGJW, BMB, PAW, AGU, BMP, HJG, SB, DIC, FG, KR, PSS, JRA, IR, AFW, JFW, AJMDeC, DAB, IJD, MAI, LJL, ALF, SS, CMvD and THM.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Green MF. Cognitive impairment and functional outcome in schizophrenia and bipolar disorder. J Clin Psychiatry. 2006;67:3–8. discussion 36-42. [PubMed] [Google Scholar]
- 2.Seidman LJ, Biederman J, Weber W, Hatch M, Faraone SV. Neuropsychological function in adults with attention-deficit hyperactivity disorder. Biol Psychiatry. 1998;44:260–268. doi: 10.1016/s0006-3223(97)00392-2. [DOI] [PubMed] [Google Scholar]
- 3.Huntley JD, Howard RJ. Working memory in early Alzheimer's disease: a neuropsychological review. Int J Geriatr Psychiatry. 2009;25:121–132. doi: 10.1002/gps.2314. [DOI] [PubMed] [Google Scholar]
- 4.Salthouse TA. The processing-speed theory of adult age differences in cognition. Psychol Rev. 1996;103:403–428. doi: 10.1037/0033-295x.103.3.403. [DOI] [PubMed] [Google Scholar]
- 5.Luciano M, Hansell NK, Lahti J, Davies G, Medland SE, Raikkonen K, et al. Whole genome association scan for genetic polymorphisms influencing information processing speed. Biol Psychol. 2011;86:193–202. doi: 10.1016/j.biopsycho.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith EE, Jonides J. Storage and executive processes in the frontal lobes. Science (New York, NY) 1999;283:1657–1661. doi: 10.1126/science.283.5408.1657. [DOI] [PubMed] [Google Scholar]
- 7.Deary IJ, Penke L, Johnson W. The neuroscience of human intelligence differences. Nat Rev Neurosci. 2010;11:201–211. doi: 10.1038/nrn2793. [DOI] [PubMed] [Google Scholar]
- 8.Lee T, Henry JD, Trollor JN, Sachdev PS. Genetic influences on cognitive functions in the elderly: a selective review of twin studies. Brain Res Rev. 2010;64:1–13. doi: 10.1016/j.brainresrev.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Marioni RE, Davies G, Hayward C, Liewald D, Kerr SM, Campbell A, et al. Molecular genetic contributions to socioeconomic status and intelligence. Intelligence. 2014;44:26–32. doi: 10.1016/j.intell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright M, De Geus E, Ando J, Luciano M, Posthuma D, Ono Y, et al. Genetics of cognition: outline of a collaborative twin study. Twin Res. 2001;4:48–56. doi: 10.1375/1369052012146. [DOI] [PubMed] [Google Scholar]
- 12.Lee T, Mosing MA, Henry JD, Trollor JN, Lammel A, Ames D, et al. Genetic influences on five measures of processing speed and their covariation with general cognitive ability in the elderly: the older Australian twins study. Behav Genet. 2012;42:96–106. doi: 10.1007/s10519-011-9474-1. [DOI] [PubMed] [Google Scholar]
- 13.Cirulli ET, Kasperaviciute D, Attix DK, Need AC, Ge D, Gibson G, et al. Common genetic variation and performance on standardized cognitive tests. Eur J Hum Genet. 2010;18:815–820. doi: 10.1038/ejhg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker-Drob EM, Reynolds CA, Finkel D, Pedersen NL. Shared and unique genetic and environmental influences on aging-related changes in multiple cognitive abilities. Dev Psychol. 2014;50:152–166. doi: 10.1037/a0032468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee T, Mosing MA, Henry JD, Trollor JN, Ames D, Martin NG, et al. Genetic influences on four measures of executive functions and their covariation with general cognitive ability: the Older Australian Twins Study. Behav Genet. 2012;42:528–538. doi: 10.1007/s10519-012-9526-1. [DOI] [PubMed] [Google Scholar]
- 16.Sleegers K, de Koning I, Aulchenko YS, van Rijn MJ, Houben MP, Croes EA, et al. Cerebrovascular risk factors do not contribute to genetic variance of cognitive function: the ERF study. Neurobiol Aging. 2007;28:735–741. doi: 10.1016/j.neurobiolaging.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Finkel D, Reynolds CA, McArdle JJ, Hamagami F, Pedersen NL. Genetic variance in processing speed drives variation in aging of spatial and memory abilities. Dev Psychol. 2009;45:820–834. doi: 10.1037/a0015332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elgamal SA, Roy EA, Sharratt MT. Age and verbal fluency: the mediating effect of speed of processing. Can Geriatr J. 2011;14:66–72. doi: 10.5770/cgj.v14i3.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deary IJ, Spinath FM, Bates TC. Genetics of intelligence. Eur J Hum Genet. 2006;14:690–700. doi: 10.1038/sj.ejhg.5201588. [DOI] [PubMed] [Google Scholar]
- 20.Deary IJ, Ritchie SJ. 10 quick questions about processing speed. Br Acad Rev. 2014;24:4. [Google Scholar]
- 21.Payton A. The impact of genetic research on our understanding of normal cognitive ageing: 1995–2009. Neuropsychol Rev. 2009;19:451–477. doi: 10.1007/s11065-009-9116-z. [DOI] [PubMed] [Google Scholar]
- 22.Wisdom NM, Callahan JL, Hawkins KA. The effects of apolipoprotein E on non-impaired cognitive functioning: A meta-analysis. Neurobiol Aging. 2009;32:63–74. doi: 10.1016/j.neurobiolaging.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JP, Burdick KE, Lencz T, Malhotra AK. Meta-analysis of genetic variation in DTNBP1 and general cognitive ability. Biol Psychiatry. 2010;68:1126–1133. doi: 10.1016/j.biopsych.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buyske S, Bates ME, Gharani N, Matise TC, Tischfield JA, Manowitz P. Cognitive traits link to human chromosomal regions. Behav Genet. 2006;36:65–76. doi: 10.1007/s10519-005-9008-9. [DOI] [PubMed] [Google Scholar]
- 25.Rommelse NN, Arias-Vasquez A, Altink ME, Buschgens CJ, Fliers E, Asherson P, et al. Neuropsychological endophenotype approach to genome-wide linkage analysis identifies susceptibility loci for ADHD on 2q21.1 and 13q12.11. Am J Hum Genet. 2008;83:99–105. doi: 10.1016/j.ajhg.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almasy L, Gur RC, Haack K, Cole SA, Calkins ME, Peralta JM, et al. A genome screen for quantitative trait loci influencing schizophrenia and neurocognitive pheno-types. Am J Psychiatry. 2008;165:1185–1192. doi: 10.1176/appi.ajp.2008.07121869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seshadri S, DeStefano AL, Au R, Massaro JM, Beiser AS, Kelly-Hayes M, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med Genet. 2007;8:S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cirulli ET, Kasperaviciute D, Attix DK, Need AC, Ge D, Gibson G, et al. Common genetic variation and performance on standardized cognitive tests. Eur J Hum Genet. 2010;18:815–820. doi: 10.1038/ejhg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, et al. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18:4650–4661. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeBlanc M, Kulle B, Sundet K, Agartz I, Melle I, Djurovic S, et al. Genome-wide study identifies PTPRO and WDR72 and FOXQ1-SUMO1P1 interaction associated with neurocognitive function. J Psychiatr Res. 2012;46:271–278. doi: 10.1016/j.jpsychires.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International HapMap Consortium Frazer KA, Ballinger DG Cox DR, Hinds DA Stuve LL, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirsch IS, Jungeblut A, Jenkins L, Kolstad A. Adult literacy in America. National Center for Education Statistics. U.S. Department of Education Office of Educational Research and Improvement; Washington, DC, USA: 2002. [Google Scholar]
- 35.Fourage D, Houtkoop W, Van der Velden R. Laaggeletterdheid in Nederland Utrecht/'s-Hertogenbosch. Expertisecentrum Beroepsonderwijs; The Netherlands: Sep, 2011. Report No.: Contract No.: ISBN/EAN 978-94-6052-039-6. [Google Scholar]
- 36.Calvin CM, Deary IJ, Webbink D, Smith P, Fernandes C, Lee SH, et al. Multivariate genetic analyses of cognition and academic achievement from two population samples of 174,000 and 166,000 school children. Behav Genet. 2012;42:699–710. doi: 10.1007/s10519-012-9549-7. [DOI] [PubMed] [Google Scholar]
- 37.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 45.Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R, et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet. 2012;131:565–579. doi: 10.1007/s00439-011-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Service SK, Verweij KJ, Lahti J, Congdon E, Ekelund J, Hintsanen M, et al. A genome-wide meta-analysis of association studies of Cloninger's Temperament Scales. Transl Psychiatry. 2012;2:e116. doi: 10.1038/tp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas LA, Akins MR, Biederer T. Expression and adhesion profiles of SynCAM molecules indicate distinct neuronal functions. J Comp Neurol. 2008;510:47–67. doi: 10.1002/cne.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peters BD, Ikuta T, Derosse P, John M, Burdick KE, Gruner P, et al. Age-related differences in white matter tract microstructure are associated with cognitive performance from childhood to adulthood. Biol Psychiatry. 2013;75:248–256. doi: 10.1016/j.biopsych.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasson E, Doniger GM, Pasternak O, Tarrasch R, Assaf Y. White matter correlates of cognitive domains in normal aging with diffusion tensor imaging. Front Neurosci. 2013;7:32. doi: 10.3389/fnins.2013.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P, et al. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet. 2007;81:17–31. doi: 10.1086/518720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wechsler D. WAIS-R Manual. Psychological Corporation; Cleveland, OH, USA: 1981. [Google Scholar]
- 54.Wechsler D. The Measurement and Appraisal of Adult Intelligence Manual for the Wechsler Adult Intelligence Scale. Psychological Corporation; New York, NY, USA: 1955. [Google Scholar]
- 55.Wechsler D. Wechsler Adult Intelligence Scale – Third Edition (WAIS III) Psychological Corporation; San Antonio, TX, USA: 1997. [Google Scholar]
- 56.Wechsler D. WAIS-IIIUK Administration and Scoring Manual London. Psychological Corporation; UK: 1998. [Google Scholar]
- 57.Jolles J, Houx PJ, Van Boxtel MPJ, Ponds RWHM. The Maastricht Aging Study: Determinants of Cognitive Aging. Neuropsych Publishers; Maastricht, Netherlands: 1995. [Google Scholar]
- 58.Smith A. Symbol Digit Modalities Test Manual - Revised. Western Psychological Services; Los Angeles, CA, USA: 1992. [Google Scholar]
- 59.Houx PJ, Shepherd J, Blauw GJ, Murphy MB, Ford I, Bollen EL, et al. Testing cognitive function in elderly populations: the PROSPER study. PROspective Study of Pravastatin in the Elderly at Risk. J Neurol Neurosurg Psychiatry. 2002;73:385–389. doi: 10.1136/jnnp.73.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.