Summary
The bistably expressed K-state of Bacillus subtilis is characterized by two distinct features; transformability and arrested growth when K-state cells are exposed to fresh medium. The arrest is manifested by a failure to assemble replisomes and by decreased rates of cell growth and rRNA synthesis. These phenotypes are all partially explained by the presence of the AAA+ protein ComGA, which is also required for the binding of transforming DNA to the cell surface and for the assembly of the transformation pilus that mediates DNA transport. We have discovered that ComGA interacts with RelA and that the ComGA-dependent inhibition of rRNA synthesis is largely bypassed in strains that cannot synthesize the alarmone (p)ppGpp. We propose that the interaction of ComGA with RelA prevents the hydrolysis of (p)ppGpp in K-state cells, which are thus trapped in a non-growing state until ComGA is degraded. We show that some K-state cells exhibit tolerance to antibiotics, a form of type 1 persistence, and we propose that the bistable expression of both transformability and the growth arrest are bet-hedging adaptations that improve fitness in the face of varying environments, such as those presumably encountered by B. subtilis in the soil.
Keywords: competence, persistence, ppGpp, ComGA, RelA
Introduction
The state of competence for transformation in Bacillus subtilis is activated by the transcription factor ComK and exhibits two distinct features compared with most other characterized transformable bacteria; it is bistably expressed in a minority of the cells in a clonal population and the expressing cells are growth-arrested. Because ComK also activates the expression of several dozen genes not needed for transformation (Berka et al., 2002, Hamoen et al., 2002, Ogura et al., 2002), we refer to the ComK ON condition as the K-state (KS) (Berka et al., 2002). In the present study we ask two questions concerning the KS; what causes the growth arrest and how might the bistable co-expression of both transformability and the growth arrest confer fitness advantages?
KS regulation is complex. Briefly, positive autoregulation, noise and cooperativity in expression from the comK promoter (Maamar et al., 2007, Süel et al., 2007) collaborate with temporally regulated comK basal expression (Mirouze et al., 2012, Leisner et al., 2007) to cause the bifurcation into two subpopulations, consisting of KS and non-KS cells. In laboratory strains of B. subtilis, KS cells comprise about 15% of the total population.
It has been known for decades that competence for transformation is accompanied by arrested growth and cell division, as a result of which KS cells tolerate exposure to penicillin G (Nester & Stocker, 1963, Johnsen et al., 2009). Two ComK-induced proteins, ComGA and Maf (Butler et al., 1993), are known to be involved in the inhibition of cell division in KS cells (Briley et al., 2011b, Haijema et al., 2001). ComGA prevents FtsZ-ring (Z-ring) formation and probably DNA replication, while Maf redundantly blocks cell division at a later step (Briley et al., 2011b). ComGA is a membrane associated AAA+ (ATPases associated with diverse cellular activities) protein, which not only inhibits cell division but also plays a role in the binding of transforming DNA to the cell and is required for the construction of the competence-associated pseudopilus needed for DNA transport (Briley et al., 2011a). Maf is an inhibitor of cell division (Butler et al., 1993) and is a nucleotide pyrophosphatase, with orthologs present in bacteria, yeast and even humans (Tchigvintsev et al., 2013). The role, if any, of this pyrophosphatase activity in the ability of Maf to inhibit cell division is not clear.
Here we show that ComGA contributes to the inhibition of rRNA transcription and of cell growth in the KS. We present evidence that these activities of ComGA are due to its ability to bind RelA, probably inhibiting hydrolysis of the alarmone (p)ppGpp. The emergence of cells from the KS is accompanied by the degradation of ComGA. The non-growing status of KS cells when placed in fresh growth medium, their tolerance for antibiotics and the involvement of (p)ppGpp suggest that this state brings about a form of type 1 persistence, defined as antibiotic tolerance during the emergence from stationary phase (Balaban et al., 2004). We show that tolerance due to the KS extends to antibiotics other than penicillin G and makes a major contribution to the total antibiotic tolerance of cultures emerging from stationary phase. We argue that the bistable expression of transformability and the growth arrest are both adaptations to growth in an unpredictably fluctuating environment, such as that encountered in the wild by this soil-inhabiting bacterium.
Results
KS cells are delayed in the resumption of cell division
To more completely describe the previously reported (Briley et al., 2011b, Haijema et al., 2001) growth arrest phenotypes of KS cells, we performed time-lapse microscopy with cells incubated on agarose pads containing fresh competence growth medium. Fig. 1A shows image frames from an outgrowth experiment in which a KS cell was identified by the expression of a comG promoter fusion to the CFP coding sequence (PcomG-CFP). Although the KS cell was initially indistinguishable morphologically from the non-KS cells, it soon followed a distinct trajectory. The non-KS cells elongated and divided after a relatively short delay, while the KS cell shown in the figure did so after an extended lag, showing evidence of division only at 150 minutes. The kinked cell shape, which was evident in Fig. 1A after 45 minutes is typical of KS cells, perhaps indicative of a cell wall defect. Although the division times of both KS and non-KS cells in several experiments were broadly distributed, the non-KS cells typically divided after 45–60 minutes, while the majority of KS cells divided between 110–180 minutes after exposure to fresh growth medium.
Fig. 1.
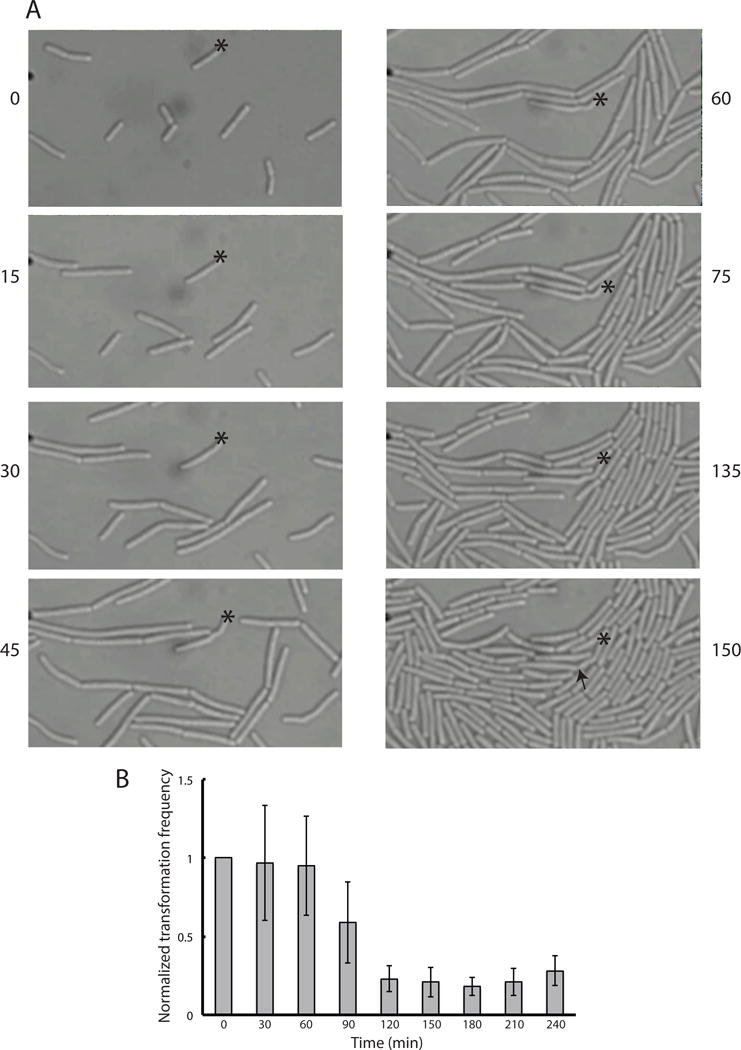
KS cells are delayed in growth. (A) Representative images from a time-lapse microscopy experiment during the reversal of the KS. An asterisk marks a single KS cell of strain BD5810, identified by use of a comG-cfp promoter fusion. The arrow indicates the first detectable indication of cell division. The elapsed time of each frame, in minutes, is given. (B) The transformation frequency to leucine prototrophy of strain BD2899 was measured during emergence from the KS. The values are normalized to the first time point. Error bars represent standard deviations.
If the non-KS cells were to divide before the KS cells, the transformation frequency would be expected to decrease and to reach a constant value when the KS cells begin to divide, presumably at the same rate as cells that had never been in the KS. To test this, cells at the time of maximal KS expression (T2) were incubated with transforming DNA for 30 minutes and then treated with DNase to destroy extracellular DNA. The cells were then diluted into fresh medium and at 30-minute intervals aliquots were plated for total viable count and for transformation to leucine prototrophy. The transformation frequency decreased after 60 minutes and reached a constant lower value after 120–150 minutes (Fig. 1B). These data confirm that the non-KS cells divide earlier, causing the transformation frequency to decline until the KS cells (and thus the transformants) begin to divide as well. The division time in this medium is about 25 minutes, from which we can infer that the transformed cells begin to divide 2–3 division times after the non-KS cells. The lag inferred from the time-lapse experiment ranged from 2–5 generations in agreement with the experiment shown in Fig. 1B. Note that the transformation frequency in Fig. 1B decreased 4–5-fold from 60 to 180 minutes, in reasonable agreement with this conclusion. Thus the data in Fig. 1 confirm the division delay of KS cells and show that during the delay, non-KS cells undergo two or three divisions.
KS cells grow slowly during outgrowth
During the time-lapse experiments we frequently observed that KS cells were not only delayed in division but also grew in length more slowly than non-KS cells. We measured the lengths of several KS (Fig. 2, blue lines) and non-KS (black lines) cells during outgrowth. KS cells were identified by their ComK-CFP fluorescence. To correct for cell division, the lengths of daughter cells were summed to yield the total length derived from each cell identified at the start of the experiment. Although the growth rates were quite heterogeneous, the KS cells typically grew more slowly than the non-KS cells. These data were modeled (Fig. S1) using a general linearized model (GLM), which has been used before to analyze differences in bacterial growth (Nelder & Wedderburn, 1972, Schaffner, 1998, Lindqvist, 2006). Based on this model we can say with at least 95% confidence that the predicted average change in cell length for the wild-type KS cells is significantly lower than for the non-KS cells.
Fig. 2.
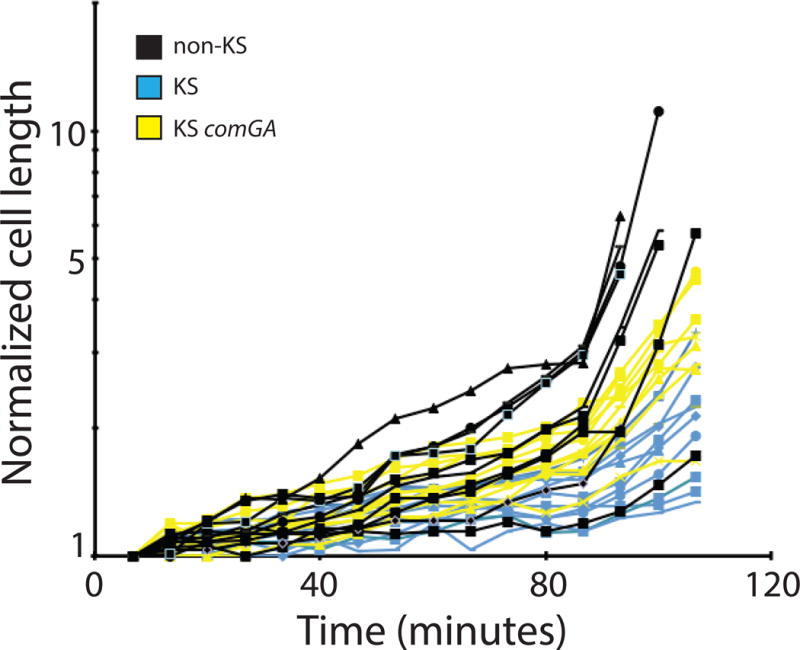
KS cells are delayed in cell elongation. ComGA contributes to this delay. KS cells were identified using YFP or CFP fusions to the promoter of comG. Cell lengths were measured at the indicated times. The blue lines represent data from KS comGA+ cells (BD5810) and the yellow lines from KS comGA (BD6757) cells. The black lines indicate the cell lengths of non-KS cells derived from both strains, which were mixed before placing them on the agarose pads to ensure that the cells were exposed to identical conditions. Because several non-KS cells divided during the course of the experiment, the lengths of daughter cells were added together. Each line represents one such lineage. All the measurements were normalized to the cell lengths at the start of the experiment and plotted logarithmically.
ComGA contributes to the growth defect of KS cells
We have shown previously that KS cells do not form Z-rings, while inactivation of comGA reverses this block. Although comGA loss-of-function mutants form Z-rings, they do not complete division because Maf, which is overexpressed in KS cells, inhibits cytokinesis at a stage after Z-ring formation (Briley et al., 2011b). As a consequence, comGA maf+ KS cells are somewhat filamented. The inactivation of comGA also contributes to filamentation by partially reversing the growth defect of KS cells (Fig. 2, yellow lines). Most of the comGA KS cells grew more rapidly than wild-type KS cells, although not as rapidly as non-KS cells. Furthermore the predicted mean growth rate for comGA KS cells also appears intermediate between those of KS and non-KS cells, though there is a modest overlap between the non-KS and comGA KS cell 95% confidence intervals (Fig. S1). Because the bypass due to inactivation of comGA is apparently not complete, another KS specific product probably contributes to the reduced rate of growth. It is possible that the inhibition of Z-ring formation by ComGA is related to its ability to reduce the rate of cell growth, so that cells are delayed in reaching a critical length for Z-ring formation.
DNA replication is inhibited in the KS, due in part to ComGA
We have reported previously that DAPI-stained KS cells retain the appearance of stationary phase cells for more than an hour before showing microscopic evidence of nucleoid segregation, while in non-KS cells the centrally located nuclear bodies split into daughter nucleoids earlier (Haijema et al., 2001). This was interpreted as most likely indicating a block in KS DNA replication. To confirm this, we monitored replisome assembly using DnaX-YFP, which revealed a dramatic difference in the replication status of KS and non-KS cells. The presence of a DnaX-YFP fluorescent dot indicates that replisome assembly has taken place and that replication may be underway (Lemon & Grossman, 2000). The non-KS cells imaged 90 minutes after dilution into fresh medium nearly all have at least one dot, while the KS cells possess few dots at this time (Fig. 3A(i)). This is shown quantitatively in Fig. 3B, where 77% of the KS cells lack even a single DnaX dot 90 minutes after resuspension in fresh growth medium. In contrast, this number is only 2% for the non-KS cells, in which 98% contain one or more dots at that time, indicating that replisomes have not only formed but that the replication forks have separated enough to be resolvable.
Fig. 3.
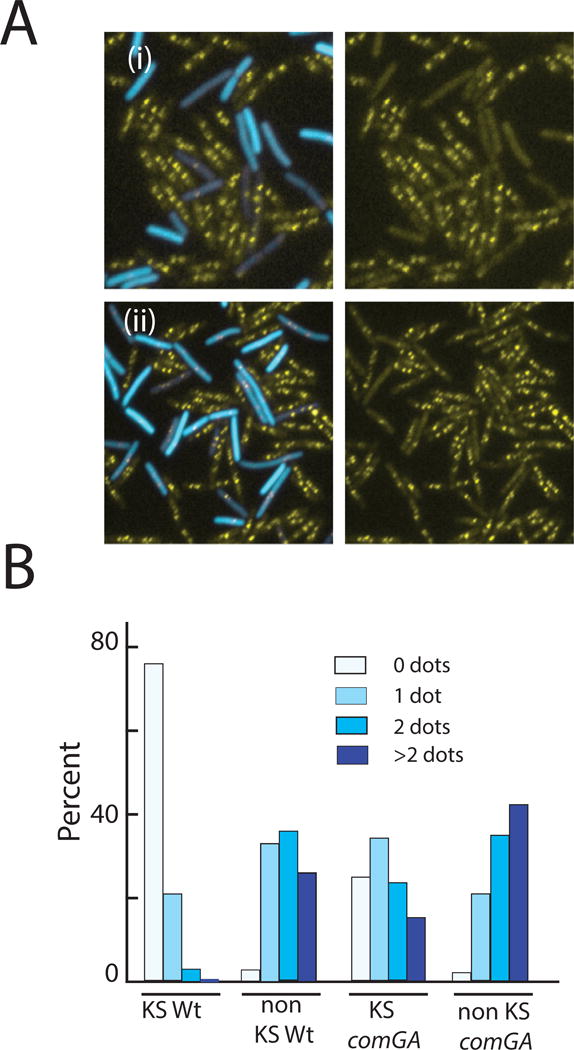
KS cells are arrested in replisome assembly. KS cells were identified using a PcomG-CFP promoter fusion using strains that also carried a DnaX-YFP fusion. Panel A(i) shows a representative image from a comGA+ strain (BD5820) and A(ii) from an isogenic comGA strain (BD6027). The histogram in Panel B summarizes dot-frequencies in KS and non-KS cells of the comGA+ (BD5820) and comGA (BD6027) strains, 90 minutes after the dilution of T2 cultures into fresh medium. A minimum of 200 cells was counted for each measurement.
A partial bypass of replisome assembly by comGA inactivation was observed (Fig. 3A(ii) and Fig. 3B). At 90 minutes after dilution, 75% of the comGA KS cells have at least one dot, compared to more than 99% of the non-KS cells (p<0.0001, V=0.3317), where V is Cramér’s V, a measure of the magnitude of the effect as described in Experimental procedures. The partial nature of the reversal is evident when the numbers of cells with 2 or more dots are compared (fewer than 40% for the comGA KS cells and 78% for the non-KS cells (p<0.0001, V=0.3766)). It appears that a KS-specific factor other than ComGA contributes to the inhibition of replisome assembly.
KS cells exhibit decreased staining with propidium iodide
We have previously noticed that KS cells exhibit weak propidium iodide (PI) staining compared to non-KS cells (Haijema et al., 2001). To further explore this, a culture at T2 was diluted into fresh medium and samples were taken at various times, mixed with PI and examined microscopically on polylysine-coated slides, which permeabilizes them for PI (Strahl & Hamoen, 2010). When cells were stained prior to dilution into fresh growth medium, no consistent difference between the staining of KS and non-KS cells could be discerned (Fig. S2). Only after exposure to fresh medium did a difference appear, as shown in Fig. 4A(i) for cells stained at 90 minutes after dilution. Indeed KS cells can be reliably identified by their weak PI staining. These differences were confirmed by flow cytometry (Fig. 4C). For this, samples were mixed with PI at the indicated times after dilution of a T2 culture into fresh medium and the cells were permeabilized by the addition of 0.1% Triton X-100. KS cells were identified from their PcomG-YFP fluorescence and the median PI intensities of KS and non-KS cells were measured. In agreement with the impression from microscopy (Fig. S2), the median intensities of KS and non-KS cells were similar at the time of dilution. With time, the intensities of the non-KS cells increased rapidly and that of the KS cells did so much more slowly. In this experiment a striking difference between KS and non-KS staining was evident even at 120 minutes.
Fig. 4.
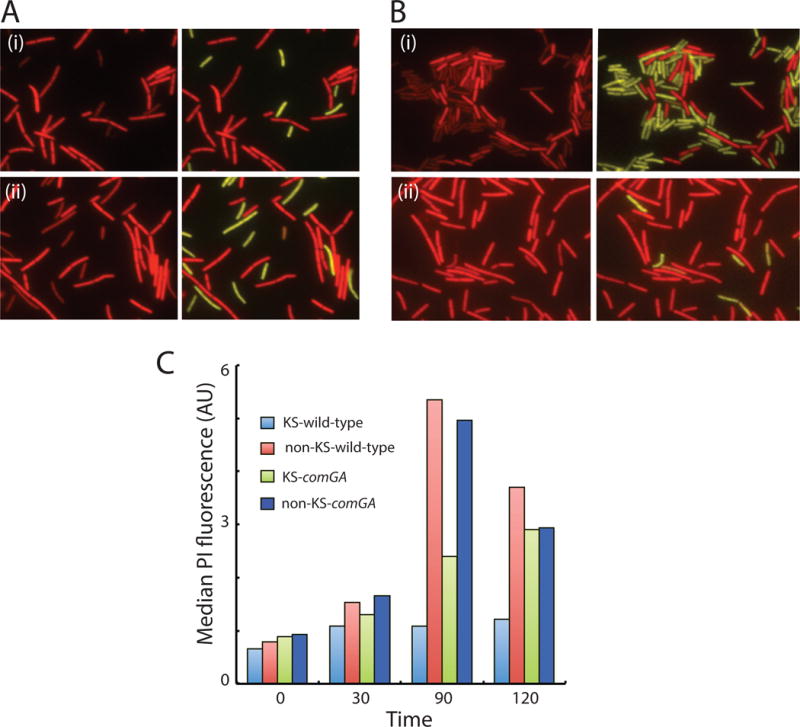
KS cells stain poorly with PI. Panels A(i) and A(ii) show PI staining of comGA+ and comGA strains (BD6011 and BD6757 respectively) 90 minutes after dilution into fresh medium. Panel B(i) shows a similar experiment using strain BD6854, which is comGA+ rok while panel B(ii) shows results using a comGA+ rok strain which also contains knockout mutations in relA yjbM and ywaC (BD7409). Panel C summarizes fluorescence intensity measurements for PI at the indicated times, measured by flow cytometry. The strains used for this experiment were the same as for panel A. Competent cells were identified by PcomGA-yfp expression for all experiments.
It is known that PI stains nucleic acids. Ribosomal RNA (rRNA) makes the major contribution by mass to the total pool of nucleic acid and so the weak staining of KS cells could be explained by a deficit in rRNA content, perhaps due to a decreased rate of rRNA synthesis in KS cells compared to that in non-KS cells. However, PI also stains DNA and as noted above, KS cells are arrested in DNA replication. Because most of the rRNA genes are located near the replication origin it is possible that the earlier resumption of DNA replication in non-KS cells increases the copy number of rRNA genes and hence the rRNA content. To determine the role of DNA replication in the staining difference between KS and non-KS cells, a T2 culture of a strain expressing PcomG-yfp was diluted into fresh medium with and without 6-(p-hydroxyphenylazo)-uracil (HPura), an inhibitor of DNA replication (Brown, 1971). As described above (Figs. 4C and S2), the non-KS and KS cells (identified by YFP fluorescence) exhibited similar PI staining at T2 (not shown). After 90 minutes, the DAPI staining of both the KS and non-KS cells in the culture incubated with HPura was noticeably weaker than that of the uninhibited cells (Fig. S3, panels A and B) as expected. Despite the inhibition of DNA replication, the PI staining of the KS cells was still markedly weaker than that of the non-KS cells. Thus, the difference in PI staining cannot be due exclusively to differences in DNA content or to gene dosage and may be due instead to an inhibition of rRNA synthesis in KS cells.
Next, we investigated the role of ComGA in the KS-specific deficit in PI staining. In the comGA culture (Fig. S3C) there was still a difference in PI staining between KS and non-KS cells, although the difference appears to be smaller than in the wild-type culture (Fig. S3A). This result is consistent with the data presented in Fig. 4A(ii) and Fig. 4C, showing that the absence of ComGA partially reverses the staining difference between KS and non-KS cells. In addition, in the presence of HPura, there was little or no difference in PI staining between the two cell types (Fig. S3D). These data are consistent with a failure of the rRNA content of KS cells to increase from the level found in stationary phase due to the combined inhibitory effects of ComGA on rRNA synthesis and to the failure of KS cells to rapidly resume DNA replication. As shown above, the latter failure is due in part to ComGA. We conclude that KS cells are delayed in cell division, grow slowly, are delayed in the assembly of replisomes and probably have a lower content of rRNA than non-KS cells after a period of incubation in new growth medium. Furthermore it appears that ComGA contributes to these phenotypes. If KS cells are indeed rRNA deficient, the cell growth and cell division delays of KS cells could be caused by a low rate of protein synthesis.
ComGA inhibits rRNA synthesis
To test whether ComGA does inhibit rRNA operon transcription, we used a fusion of the gene encoding firefly luciferase to the P1 promoter of rrnB. The rRNA operons of B. subtilis are each driven by two promoters, P1 and P2, and in all of them P1 is susceptible to inhibition by the alarmone (p)ppGpp (Natori et al., 2009). Strains with a P1rrnB-luc fusion in wild-type, comGA and comK backgrounds were grown in a plate reader equipped for luminometry. Because only about 15% of the cells in a culture normally enter the KS, the three strains were made mutant for rok, which encodes a repressor of comK (Hoa et al., 2002). In this mutant about 80% of the cells in cultures of the two comK+ strains will express KS genes, and the plate reader experiment will approximately reflect rrnB transcription in the KS cells. Growth in competence medium was followed until the cultures reached T2, and then each culture was diluted 50-fold into pre-warmed competence medium and optical density and light output were measured at intervals (Fig. 5A). The rrnB transcription rates increased with markedly different kinetics in the three strains. In the comK strain (green), which reflects the behavior of non-KS cells, the rate rose rapidly. In the comK+ comGA+ strain (blue) the rate remained low for about two hours and thereafter rose more rapidly. This comparison indicates that KS cells are indeed delayed in the resumption of rrnB transcription. Interestingly, the rate in the comK+ comGA culture (red) was higher than in the wild-type culture, but still lower than in the comK strain (non-KS cells). Because 20% of the cells in the comK+ comGA+ and comK+ comGA cultures are not in the KS, the rates of rrnB expression in the KS subpopulations in these cultures must be somewhat lower than measured. Thus, the differences between the rates in these two cultures and that of the comK culture are even greater than depicted in Fig. 5. The order of growth resumption of the three cultures paralleled the transcription rates. Because this experiment was performed in a strain with a polar Tn917 insertion in comGA, we determined if any other comG gene product was needed for the delay in rrnB transcription. An isogenic strain with a polar insertion of Tn917 in the second gene of the comG operon was used. No relief of the KS inhibition of rrnB transcription was observed (Fig. S4) indicating that the only comG product needed for the inhibition of rrnB transcription was ComGA. rrnB is not the only rRNA operon subject to KS regulation. ComGA also inhibits the transcription of the rrnD operon during outgrowth from the KS and the transcriptional profiles of rrnD in wild-type, comK and comGA strains are quite similar to those of rrnB (Fig. S5).
Fig. 5.
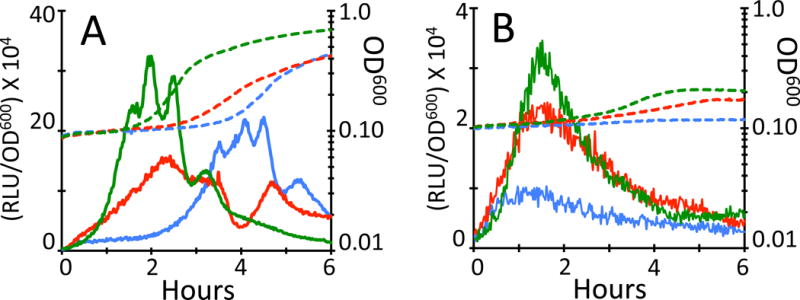
ComGA inhibits rrnB transcription during emergence from the KS. comGA+ (BD7069, blue), comGA (BD7106, red) and comK (BD7152, green) strains, all carrying luciferase fusions to the promoter of rrnB as well as a loss-of-function rok mutation, were grown in the presence of luciferin in a plate reader. Panel A shows results in the absence and B in the presence of HPura (100 μM). Growth (dashed lines) and light output (solid lines) were followed at 1.5-minute intervals. Light output values (RLU, Relative Luminescence Units) were corrected for OD600 and plotted on a logarithmic scale. Note that the ordinate scales in panels A and B differ by a factor of 10.
As described above for the PI staining phenotype of KS cells, a delayed increase in rrnB operon copy number may contribute to the decrease in the activity of the P1-rrnB promoter in KS cells. This is likely to be a less important effect than for total rRNA synthesis, because rrnB is not ori proximal. To test the magnitude of the copy number effect, we repeated the experiment shown in Fig. 5A in the presence of HPura (Fig. 5B). During the course of this experiment the wild-type strain exhibited almost no increase in growth, whereas the comK and comGA strains showed small increases (dashed lines), consistent with the reversal of cell growth inhibition when comGA was inactivated (Fig. 2). In the absence of DNA replication, the difference in transcription rate between the comK and comGA strains was quite small (Fig. 5B), and in two other replicate experiments was not detected at all (not shown). Thus, the partial restoration of rrnB transcription in the comGA strain in the absence of HPura (Fig. 5A) is most likely due to a lower gene copy number even when comGA is inactivated. This is consistent with the partial effect of comGA inactivation on replisome assembly (Fig. 3). The maximum transcription rates in the presence of HPura are ten-fold lower than without, suggesting that even in KS cells, increased rrnB copy number contributes substantially to the transcriptional activity after about an hour of outgrowth.
Considered together, these data suggest that the persistent weak PI staining of KS cells most likely reflects a low rate of rRNA synthesis, typical of bacterial cells in stationary phase (Piir et al., 2011, Aviv et al., 1996). Although initially the KS and non-KS cells have similar levels of PI staining (Figs. 4C and S2), the staining of non-KS cells increases rapidly as does the rate of rRNA synthesis in these cells. In contrast, the rate of rRNA synthesis in KS cells remains low for about 2.5–3 hours (Fig. 5A), and accordingly their staining by PI remains weak for a longer time (Figs. 4 and S3). It appears that in KS cells, the inhibition of DNA replication contributes to the lower rate of rRNA synthesis, by preventing an increase in the copy number of rRNA operons. Finally, ComGA contributes to the inhibition of total rRNA synthesis in two ways; by an effect on the rRNA operon promoters and by inhibiting DNA replication, thereby reducing rRNA operon gene dosage.
ComGA inhibits rRNA transcription in the absence of other ComK products
We next asked if the expression of comGA in the absence of other ComK-dependent proteins is sufficient to decrease the rate of rrnB transcription. For this experiment, comGA was expressed from the isopropyl ß-D-1 thiogalactopyranoside (IPTG) inducible Phyperspank (Phs) promoter in a strain carrying the P1rrnB-luc fusion (Fig. S6). The strains were grown in LB medium where KS genes are not expressed and then diluted into competence medium containing luciferin, with and without inducer (Fig. 6A). In both conditions, the apparent rate of rrnB transcription initially rose, as luciferin entered the cells. When comGA was induced, the rate of rrnB transcription rapidly decreased after a brief delay (blue solid line (Fig. 6A)). In the absence of inducer (red solid line) the rate climbed further and was sustained for a while (with oscillations), presumably to adjust the rRNA content to a level appropriate for the new growth rate, until the cells approached stationary phase, at which point the rate decreased. The growth of the induced culture was slightly retarded beginning at about 1.5 hours (blue dashed line), suggesting that it was limited by the rate of rRNA synthesis, which was clearly decreased in the presence of IPTG about an hour earlier. This demonstrates that ComGA can inhibit rRNA synthesis in the absence of other KS gene products. We have already shown that the elimination of ComGA by mutation only partially bypasses the KS phenotypes and so it is no surprise that the inhibition is not as severe as that seen in the context of the KS. Importantly, and as noted, these data strongly suggest that the delay in rRNA synthesis is a cause of the growth delay rather than the reverse, because the inhibition of rrnB transcription evident in Fig. 6A precedes the difference in growth rate between the induced and uninduced cultures by about one hour.
Fig. 6.
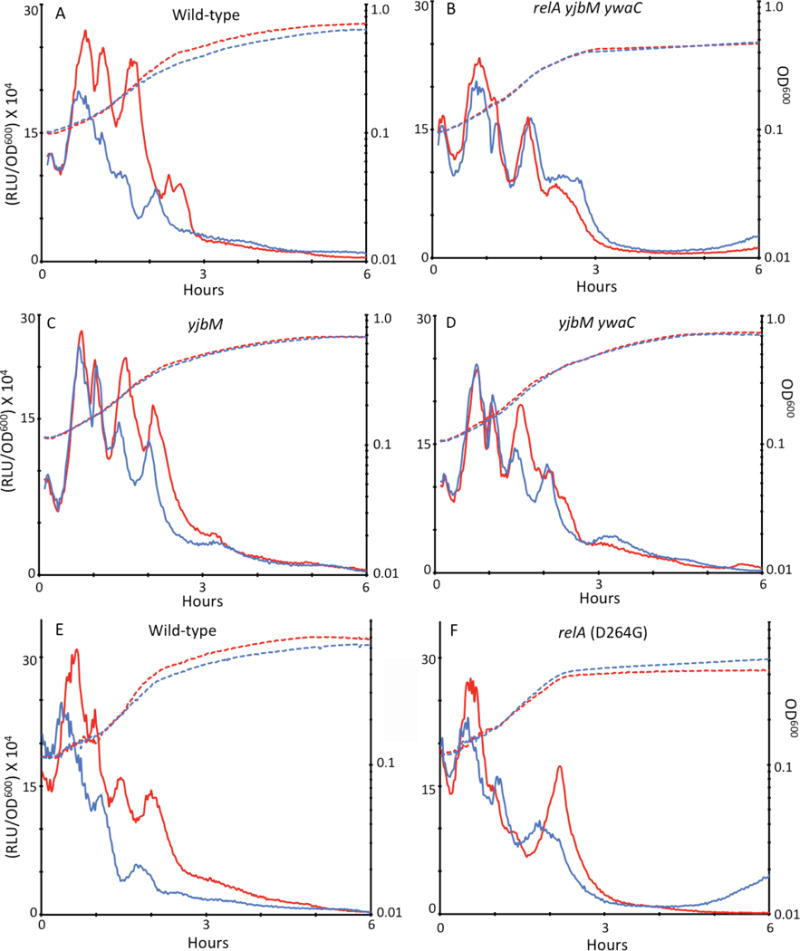
ComGA slows rrnB transcription during growth independently of other KS proteins and by a (p)ppGpp-dependent mechanism. In all four panels the dashed lines represent growth (OD600) plotted on a logarithmic scale and the solid lines show light output from the rrnB-luc construct, corrected for OD600. Blue and red lines show results obtained in the presence and absence of IPTG, respectively. The strains used were as follows: (A) Phs-comGA (BD6851), (B) Phs-comGA relA ywaC yjbM (BD6876), (C) Phs-comGA yjbM (BD6870), (D) Phs-comGA ywaC yjbM (BD6871), (E) Phs-comGA (BD6851) and (F) Phs-comGA relA D264G (BD7573). The data in panels A–D were obtained in one experiment and those in panels E and F in another.
These data reinforce the following working model. As cells approach the stationary phase their ribosome content decreases. Indeed in Escherichia coli, about half of the ribosomes are degraded (Piir et al., 2011) and the rate of rRNA transcription decreases in both E. coli and B. subtilis as growth slows (Aviv et al., 1996, Mirouze et al., 2011). This happens in all the cells and accordingly the PI staining of KS and non-KS cells are similar at T2. When diluted into fresh growth medium, rRNA synthesis rapidly resumes in non-KS cells but is retarded in KS cells, due to the effect of ComGA on rRNA operon transcription and of the combined action of ComGA and an unknown factor on rRNA operon copy number.
ComGA interacts with RelA
To better understand the role of ComGA, we characterized its in vivo protein interactions in the KS using immunoaffinity purification and mass spectrometry. A functional, C-terminally MYC-tagged ComGA (Hahn et al., 2005) was placed at the native locus, as described in Experimental procedures. Protein complexes containing ComGA-MYC were isolated from rok cells grown to T2. Since ComGA is a peripheral membrane protein (Chung et al., 1998), we performed replicate experiments using different lysis conditions, one optimized for enrichment of cytoplasmic proteins (Buffer A), the other to improve the isolation of membrane associated proteins (Buffer B). Immunopurified proteins were identified by MS/MS using a nano-LC-ESI coupled directly to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) as described. In general, more total proteins were co-isolated when using Buffer A than with Buffer B (Fig. 7A). Importantly, several competence proteins known to interact with ComGA were recovered as anticipated (Hahn et al., 2005, Kramer et al., 2007), confirming that the isolation conditions preserved protein-protein interactions that had been independently established (Table S2). An interesting cytoplasmic protein that was newly identified as prominently interacting with ComGA-MYC was the (p)ppGpp synthetase RelA (Fig. 7B). Although RelA was more efficiently isolated with Buffer A, consistent with its cytosolic location, the ComGA association with RelA was retained using Buffer B, despite its higher salt and detergent concentrations, suggesting that the interaction is strong. This association was of obvious interest because of the well-known ability of (p)ppGpp to inhibit rRNA synthesis and DNA replication (Magnusson et al., 2005, Potrykus & Cashel, 2008, Dalebroux & Swanson, 2012, Wang et al., 2007).
Fig. 7.
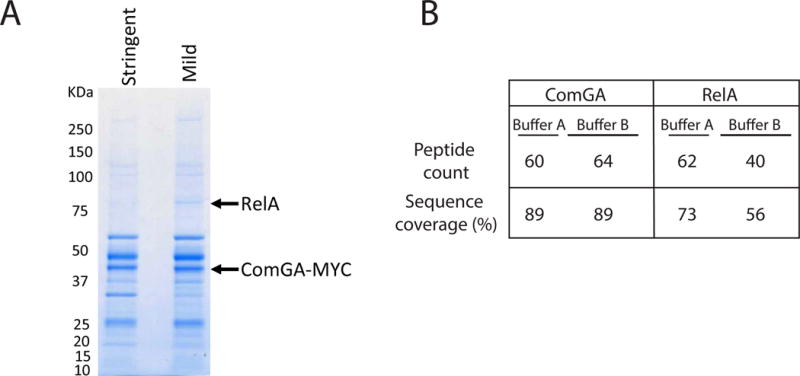
ComGA is associated with RelA in vivo. Cells from a culture expressing a functional ComGA-MYC fusion, were broken and immunoprecipitated using anti-MYC antiserum. Two buffer conditions were used as explained in the text. Interacting proteins were visualized by SDS-PAGE (A) and excised bands were further examined by mass spectrometry. In panel A the positions of RelA and ComGA are indicated. Selected mass spectrometry results are summarized in panel B.
Although these data strongly suggest that ComGA is in a complex with RelA they do not demonstrate direct binding. To address this, we expressed GST-ComGA and RelA separately and together in E. coli, using the pQLink system (Scheich et al., 2007). ComGA was tagged at its N-terminus with GST, followed by a PreScission protease cleavage site. While the single expressing strains were readily obtained, it was not possible to obtain a strain expressing both GST-ComGA and the B. subtilis RelA, even in the absence of inducer (IPTG), unless an E. coli lacIQ host was used. Even in a lacIQ background, colonies grown on LB agar containing IPTG were tiny, while those of strains expressing GST-ComGA or RelA individually were of normal size. This synthetic phenotype strongly suggested an interaction between RelA and ComGA, perhaps resulting in an expansion of the (p)ppGpp pool. E. coli cells expressing either GST-ComGA, RelA or both were lysed, and the resulting extracts were clarified by centrifugation. Although most of each induced protein was insoluble, enough was present in the soluble fraction to permit further analysis. The supernatants from all three cultures were passed over a glutathione column. When the B. subtilis RelA protein alone was expressed, it appeared in the flow-through (F) and wash (W) fractions and no further RelA protein was released following on-column treatment with PreScission protease (C), showing that RelA did not interact non-specifically with the glutathione resin (Fig. 8A). When GST-ComGA alone was expressed, ComGA was eluted only after cleavage by PreScission protease, as expected (Fig. 8B). When the extract from cells expressing both RelA and GST-ComGA was loaded on the column, proteins of the correct sizes for both RelA (~80 kDa) and cleaved ComGA (~40 kDa) were released by protease treatment, showing that RelA was retained on the column only due to its association with GST-ComGA.
Fig. 8.
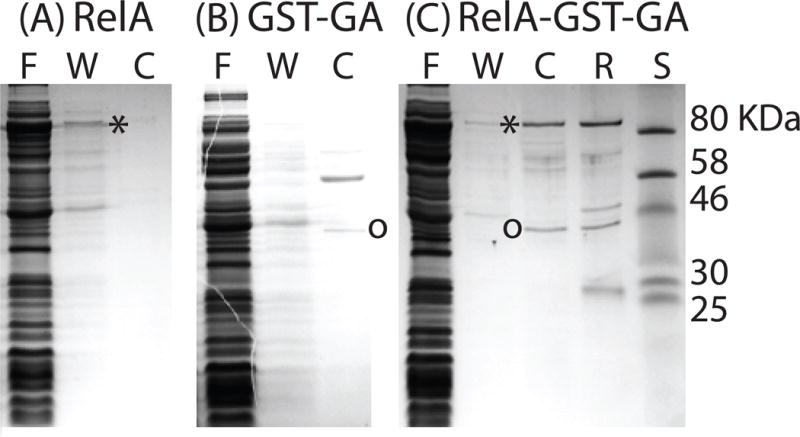
ComGA binds to RelA from B. subtilis (RelABsu). Three E. coli strains, expressing RelABsu (A), GST-ComGA (B) and both proteins (C) were grown, lysed and the extracts were passed over a glutathione column. Proteins were eluted following on-column cleavage with PreScission protease and the various fractions were analyzed by SDS-PAGE. “F”, “W” and “C” refer to flow through, wash and cleaved fractions respectively. “R” refers to the material remaining on the resin and the lane labeled “S” contains molecular weight standards. Asterisks and circles indicate the locations of RelABsu and of ComGA respectively. The identity of RelABsu was confirmed by mass spectrometry.
To verify the identities of the two eluted proteins, and to exclude the possibility that the 80 kDa protein in Fig. 8C was the RelA protein from E. coli, the bands were excised from the gel and subjected to tryptic digestion. The resulting peptides were analyzed by mass spectrometer, and spectra were searched against the B. subtilis and E. coli databases for protein identification. The predominant protein present in the 80 kDa band was B. subtilis RelA, with 53 unique peptides identified, and 54% sequence coverage. The main protein in the 40 kDa band was ComGA, with 87 unique peptides and 87% sequence coverage. Although traces of E. coli protein contaminants were present, no other B. subtilis proteins were identified, as expected. These experiments independently confirmed the association of ComGA with the B. subtilis RelA protein that had been shown by co-immunoprecipitation and demonstrated that the association is due to direct binding.
The ability of ComGA to inhibit rRNA synthesis is bypassed in a (p)ppGpp null mutant
Since ComGA associates with RelA we next considered whether the ComGA-associated phenotypes may be due to an increase in the intracellular pool of (p)ppGpp, as suggested by the synthetic slow growth phenotype of E. coli co-expressing ComGA and the B. subtilis RelA. The B. subtilis RelA protein possesses (p)ppGpp hydrolase activity as well as weak synthetase activity, like its close ortholog from Streptococcus equisimilis (Mechold et al., 1996). ComGA binding to RelA may inhibit its hydrolase activity or stimulate the synthesis of (p)ppGpp. In either case, if the relevant ComGA phenotypes were due to its ability to increase the (p)ppGpp pool, these phenotypes might be bypassed in a strain that cannot synthesize this molecule. To construct a (p)ppGpp° strain (null for (p)ppGpp synthesis), it is necessary to inactivate not only RelA, but also two other synthetases, encoded by ywaC and yjbM (Nanamiya et al., 2008, Srivatsan et al., 2008). It is worth noting that the phenotype of a relA single knockout was not investigated because such a strain is slow growing, rapidly accumulates suppressors that inactivate ywaC and yjbM and because the frequency of KS cells is markedly reduced (not shown). The reason for this is unknown and decreased competence expression in a relA strain has been reported previously (Inaoka & Ochi, 2002).
Indeed, ComGA requires the presence of (p)ppGpp for its inhibitory effect on P1rrnB transcription (Fig. 6B). (p)ppGpp is also needed for the usual KS inhibition of PI staining (Fig. 4B(ii)), where it was restored to an intermediate level in the (p)ppGpp° strain, compared to that of cells that are capable of synthesizing this molecule (Fig. 4B(i)). Although a quantitative comparison of the staining restoration accomplished by elimination of (p)ppGpp synthesis with that achieved in a comGA knockout was not attempted, the levels appear to be approximately similar.
To investigate the contributions of the three (p)ppGpp synthetases, we also tested the effect of ComGA overproduction in the absence of yjbM (Fig. 6C) or of both yjbM and ywaC (Fig. 6D). In both of these backgrounds, the effects of comGA induction early in growth were eliminated (compare with Fig. 6A), although effects on rrnB transcription were still evident after about 1.5 hours during exponential growth and as the cultures approached stationary phase. ComGA cannot inhibit transcription early in growth when YjbM is absent, so this synthetase must be most active at that time, consistent with the results of transcriptional analysis of yjbM expression (Nanamiya et al., 2008). The elimination of ywaC has little effect, consistent with the reported dependence of this gene on SigW for its expression and its induction in response to cell wall stress (Cao et al., 2002). The small effect that its absence does impose is evident only during the stationary phase (compare Fig. 6C and D). These data suggest that YjbM is active as a synthetase primarily during early growth and that YwaC is active in stationary phase. The inhibition by ComGA when RelA is the only source of (p)ppGpp takes place during exponential growth (compare Fig. 6B with 6D), suggesting that the synthetase activity of this protein is active at this time, perhaps as the cells sense the depletion of nutrients. These results also hint that ComGA decreases the hydrolase activity of RelA instead of augmenting its ability to produce (p)ppGpp. If ComGA increased the production of (p)ppGpp by RelA, we might expect ComGA production to show an effect early in growth in the yjbM or yjbM ywaC mutants.
The ability of ComGA to inhibit rrnB transcription does not require the (p)ppGpp synthetase activity of RelA
If the effects of ComGA on rrnB transcription were due to its ability to augment the synthetase activity of RelA, the D264G relA mutation should eliminate the ComGA imposed inhibition of rrnB transcription. This mutation, which inactivates the synthetase activity without affecting the hydrolase activity (Nanamiya et al., 2008, Natori et al., 2009), was introduced at the relA locus, so that the mutant protein was expressed under native control. In the mutant strain, a clear difference in rrnB transcription can be seen in the presence and absence of inducer (Fig. 6F). This difference strongly suggests that the effect of ComGA does not depend only on the synthetase activity of RelA and that the hydrolysis of (p)ppGpp by RelA is therefore inhibited by ComGA. The magnitude of this difference is less than that observed with the wild-type strain (compare Fig. 6, panels E and F), as expected because the total level of (p)ppGpp is likely to be decreased in the mutant strain. Interestingly, there is no difference between the induced and uninduced cultures during exponential growth, suggesting that RelA is the major source of (p)ppGpp at this time, as inferred above, based on the comparison between Fig. 6B and 6D. Although this experiment supports an effect of ComGA on the hydrolysis of (p)ppGpp by RelA, it does not exclude the possibility that the ability of RelA to synthesize (p)ppGpp is also affected.
ComGA is degraded during the reversal of the KS
Whatever its mode of action, ComGA must be inactivated for cells to fully emerge from the KS. To determine if this inactivation is accomplished by degradation, we monitored ComGA levels in cultures of KS cells after dilution into fresh medium by Western blotting. By 90 minutes after dilution into fresh medium, ComGA is undergoing degradation and its concentration in the KS cells is decreasing rapidly just when the cells resume growth (Fig. 9). It is important to note that in this culture, the non-KS cells are dividing, which means that non-KS proteins would likely increase in amount as cell growth occurs, and thus would not serve as good comparisons for the behavior of ComGA. For the same reason, loading an equal mass of total protein on each lane would cause a misleading apparent decrease in ComGA as the KS cells are diluted by growth of the non-KS cells. For this reason, lysates derived from a constant volume of culture were loaded on each lane, so that the Western blot signals reflect the total amount of each protein per unit volume. We also analyzed the stability of the competence protein Maf, as it too is specifically expressed in KS cells. Remarkably, Maf is not degraded and in fact is seen to increase in amount with time. Although Maf is a potent inhibitor of cell division (Briley et al., 2011b, Butler et al., 1993), it is clearly present during the time that KS cells divide. How the effects of Maf are circumvented remains the subject of future work. In contrast to this, ComGA is degraded by an unknown mechanism as the KS cells initiate growth and division.
Fig. 9.

ComGA is removed by degradation. Equal volume aliquots were taken from a KS culture at the indicated times after resuspension in fresh competence medium of a culture grown to T2. The samples were analyzed by Western blotting on separate gels with anti-ComGA or anti-Maf antisera as described in Experimental procedures.
The KS imposes a form of type I persistence
It has been shown that transformants are tolerant of penicillin G in the KS, as would be expected from their slowed growth (Johnsen et al., 2009, Nester & Stocker, 1963). It was postulated that the decreased growth rate of KS cells confers an advantage during rare episodes of exposure to antibiotics or to other conditions that injure growing cells (Johnsen et al., 2009). We have extended this finding to kanamycin (Kan), an aminoglycoside antibiotic and to oxolinic acid, a quinolone, demonstrating that the antibiotic tolerance of KS cells is not restricted to penicillin G. The earlier experiments compared the penicillin G sensitivities of transformants, as a surrogate for competent cells, with that of total colony forming units. Because KS cells may be heterogeneous with respect to transformability, we instead compared the killing kinetics of total colony forming units of wild-type and comK strains, until plateaus were reached, in order to measure the ComK-dependent and independent tolerant populations. The tolerant (persistent) population for both antibiotics was approximately 10-fold higher for the wild-type cultures than for the comK mutant (Fig. 10). This indicates that about 90% of the total persistence in the wild-type cultures was dependent on the KS. However, because approximately 15% of the cells in this culture were in the KS, while the persistent population comprised a very small fraction (~ 10−4) of the total colony forming units, the vast majority of KS cells were killed by Kan and by oxolinic acid. This indicates that the KS is heterogeneous for persistence, consistent with the elongation rate heterogeneity shown in Fig. 2, suggesting that cells with slower elongation rates may be more antibiotic tolerant. A distinction has been made between type II persistence, which is manifested within populations of growing cells, and type I persistence, which is exhibited by cells that experience a prolonged lag when emerging from stationary phase (Balaban et al., 2004, Fridman et al., 2014). Our data show that some KS cells exhibit type I persistence.
Fig. 10.
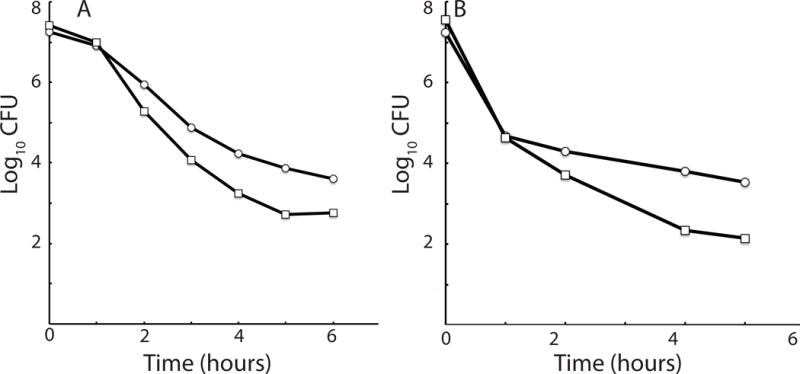
KS cells exhibit type 1 persistence. Cultures of the wild-type strain (BD630, ○) and its isogenic comK derivative (BD5004, □) were grown to T2 and then diluted into competence medium containing 10 μg/ml of oxolinic acid (A) or into LB medium containing kanamycin (10 μg/ml). (B). Samples were taken for determination of surviving colony-forming units (CFU) at hourly intervals.
Discussion
The role of ComGA in growth arrest
The B. subtilis KS is characterized by a decreased rate of rRNA synthesis (Figs. 4 and 5), slowed cell elongation (Fig. 2), delayed DNA replication (Fig. 3) and the absence of Z-rings (Haijema et al., 2001). Fig. 11 summarizes these effects and our current understanding of the roles of ComGA and Maf (Briley et al., 2011b) in establishing these disparate growth defects. ComGA interacts with RelA, thereby increasing the level of (p)ppGpp, thereby inhibiting rRNA synthesis. ComGA also inhibits the assembly of replisomes by an unknown mechanism. The elevated level of (p)ppGpp induced by ComGA binding to RelA may also inhibit DNA replication after replisome assembly, by interfering with primase activity (Wang et al., 2007). The inhibition of replication further decreases rRNA synthesis in KS cells by preventing an increase in rRNA operon copy number, particularly because most rRNA operons are ori proximal. Thus, ComGA contributes to the inhibition of rRNA synthesis both by its interaction with RelA and by down-regulating the assembly of replisomes. The resulting deficit in rRNA then plausibly restrains the rate of cell growth and hence the rate of Z-ring formation. Maf, another ComK-dependent protein, inhibits cell division at a step subsequent to the formation of Z-rings (Briley et al., 2011b, Butler et al., 1993).
Fig. 11.
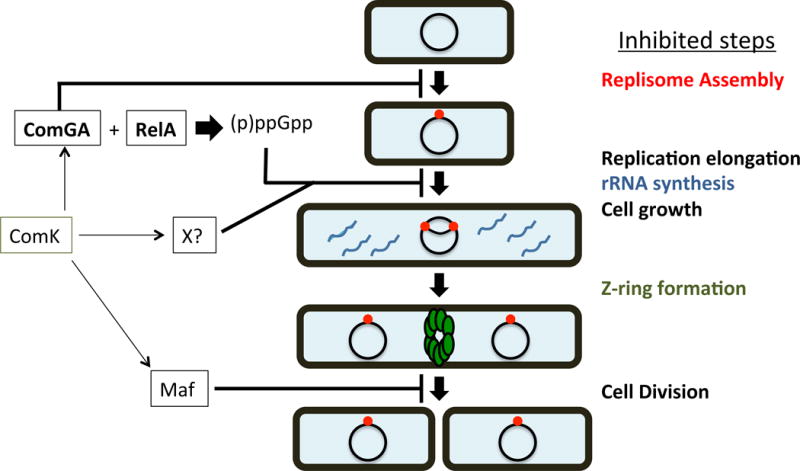
The growth arrest during the KS. Inhibited steps are listed to the right. ComK activates the expression of at least 3 factors required for the growth arrest of KS cells, as follows. (i) ComGA inhibits replisome assembly in a (p)ppGpp-independent fashion, and also directly interacts with RelA to increase the pool of (p)ppGpp in the cell. The increased (p)ppGpp inhibits both replication elongation and rRNA synthesis. (ii) An unidentified factor, dependent on ComK for its synthesis (X?), also inhibits rRNA synthesis. (iii) Maf redundantly inhibits cell division at a step following Z-ring formation (Butler et al., 1993, Briley et al., 2011b). Arrows depict positive effects and lines ending in perpendiculars show negative effects, summarizing data from this work and from previous studies (Wang et al., 2007).
Several lines of evidence lead to our conclusion that ComGA and RelA interact. First, the co-immunoprecipitation experiment in B. subtilis strongly suggests that these proteins associate, either directly or indirectly. Second, the co-purification of RelA with ComGA after expression in E. coli confirms the association and shows that it requires no additional B. subtilis protein and is thus a direct interaction. Third, the synthetically deleterious phenotype exhibited by the co-expressing E. coli strain supports the existence of an in vivo interaction. Finally, the bypass of the inhibition of rRNA synthesis by ComGA in the (p)ppGpp° strains provides indirect support for the involvement of RelA in the inhibitory action of ComGA.
The discovery that the inhibition of rRNA synthesis by ComGA is bypassed in (p)ppGpp° strains and that ComGA binds to RelA, suggest a simple scenario to explain the role of ComGA in the KS. As growth slows and cells enter the stationary phase, the (p)ppGpp pool expands in response to nutritional limitation, contributing to a slowdown in growth and in rrn transcription (Paul et al., 2004, Gralla, 2005, Potrykus & Cashel, 2008, Srivatsan & Wang, 2008). This is expected to happen in both KS and non-KS cells and indeed a sharp downturn in rrnB transcription occurs in stationary phase B. subtilis, which is much less apparent in a (p)ppGpp° background (Mirouze et al., 2011). When stationary phase cells encounter conditions conducive to the resumption of growth, particularly an ample supply of nutrients, the (p)ppGpp pool must decrease before growth can resume. The B. subtilis RelA protein is likely to be a (p)ppGpp hydrolase, like its close ortholog in Streptococcus equisimilis (Mechold et al., 1996). We propose that the binding of ComGA to RelA inhibits its (p)ppGpp hydrolase activity, so that stationary phase KS cells are trapped in a slow-growth state until ComGA is degraded. This is consistent with the marked growth defect of a relA strain, in which (p)ppGpp continues to be synthesized by YjbM and YwaC (Nanamiya et al., 2008, Srivatsan et al., 2008) but presumably cannot be degraded. We propose that the sustained presence of (p)ppGpp in KS cells contributes to a decreased rate of rRNA synthesis and a decreased rate of cell growth. As noted above, this in turn may delay the formation of Z-rings. We have attempted the direct measurement of (p)ppGpp in cells emerging from stationary phase but the amount in KS cultures was below the level that could be reliably quantified (unpublished results). We prefer the hypothesis that the effect of ComGA on RelA is to inhibit its hydrolase activity rather than to increase the rate of (p)ppGpp synthesis. The failure of the relAD164G mutation to prevent the inhibitory effect of ComGA supports this idea (Fig. 6). As noted above, further support derives from the inability of ComGA to inhibit rrnB transcription early in growth, when YjbM is absent but RelA is present (compare Fig. 6A and 6C).
Orthologs of ComGA are encoded in the genomes of nearly all known transformable bacteria, and ComGA-like proteins are nearly universally required for transformation. Particularly in B. subtilis, ComGA is a remarkable protein with many roles in the KS. Its conserved role seems to be in the assembly of the competence pilus and it has been shown to be required for the binding of transforming DNA to the cell surface and for the internalization of this DNA (Briley et al., 2011a). Remarkably, only B. subtilis has been shown to undergo a prolonged growth arrest when competent. It appears that ComGA has been recruited in B. subtilis for a unique function in addition to its roles in transformation (Briley et al., 2011a). Similarly maf, encoding a highly conserved protein present in both prokaryotes and eukaryotes, has been recruited to the ComK regulon, where its only known role is to inhibit cell division. The considerable redundancy in growth-inhibiting pathways (Fig. 11) suggests that the semi-dormancy manifested by the K-state enhances fitness. We next address the biological rational for the growth arrest and for its bistable expression.
Why are both the growth arrest and transformability expressed bimodally?
Only B. subtilis has been reported to exhibit the bimodal expression of transformability and as just noted, only B. subtilis has coupled transformation to a growth arrest. For example, Neisseria gonorrhoeae is constitutively competent during growth (Biswas et al., 1977). Haemophilus influenzae becomes transformable when deprived of critical nutrients (MacFadyen et al., 2001) but no evidence has been reported that competent cells are growth-inhibited, nor has its competence been shown to express in a subpopulation. All the cells in a Vibrio cholerae culture express competence when grown under homogeneous conditions (Lo Scrudato & Blokesch, 2012) and no growth arrest behavior has been reported. In S. pneumoniae, although all the cells in a culture appear to become competent (Martin et al., 2010), evidence has been reported for a transient growth arrest, and its competence has been likened to the SOS state of other bacteria (Dagkessamanskaia et al., 2004). What are the advantages and disadvantages of the bimodal expression of both the growth arrest and of transformability and why might these features associate in B. subtilis during the KS? As noted above, at least one other unknown KS-specific gene product contributes to the growth inhibition and Maf aids in the cell division delay of KS cells, underscoring the important contribution the arrest must make to the fitness of B. subtilis. We propose that both of these two unique aspects of the KS have evolved as an adaptation for survival in a variable environment, like that in the soil. Setting aside the specific nature of the fitness advantages conferred by the bimodal expression of transformability and of the growth arrest, several theoretical studies have shown that stochastic switching to a new gene expression state can be advantageous in the face of environments that change infrequently with relatively lengthy intervening periods of stasis (Kuwahara & Soyer, 2012, Kussell & Leibler, 2005, Thattai & van Oudenaarden, 2004). Bimodal gene expression of the KS, like the sessile-motile switch of B. subtilis (Kearns & Losick, 2005), may be viewed as particular examples of such bet-hedging.
Although these theoretical studies are quite relevant, they are not informative about the specific nature of the fitness advantages conferred by the growth arrest and by the bimodal expression of DNA uptake. The growth arrest by itself clearly imposes a fitness burden on KS cells. In fact, it was shown that in direct competition experiments, comK+ cells were at a growth disadvantage compared to comK cells in a medium in which the KS is expressed (Johnsen et al., 2009). This fitness cost must therefore be balanced in nature against an advantage conferred by the KS decrease in growth rate. We have previously suggested that the growth delay may function to permit genome repair after the massive recombination that can occur after transformation (Briley et al., 2011b), and this may indeed contribute to fitness. On the other hand, if this were true we might expect a similar delay in resuming growth in all transformable bacteria. It has been shown that KS cells are antibiotic tolerant (Johnsen et al., 2009, Nester & Stocker, 1963) and we have extended these observations in this study (Fig. 10). It is interesting that the stress-responsive regulatory pathway that regulates competence in S. mutans, also leads to the formation of antibiotic-tolerant cells (Leung & Levesque, 2012). The KS may therefore represent a form of persistence that confers tolerance to antibiotics and other insults that preferentially affect growing cells. For example, in heterogeneous populations, slow growing yeast cells are relatively resistant to heat killing (Levy et al., 2012). The KS cells that are the subjects of the present study resemble type 1 persisters because they are formed in stationary phase and display a prolonged lag before resuming growth. In this model, the frequency of KS cells must have been set by selection in accordance with the probability that rare antibiotic-like stress conditions occur. The concept that the stochastic expression of persistence offers an advantage to bacteria in the face of fluctuating environments is well established (Balaban et al., 2004, Kussell et al., 2005, Acar et al., 2008).
Why might the fitness advantage conferred by the ability to take up new genes also be restricted to a subpopulation? Transformation, a widespread bacterial phenotype (Lorenz & Wackernagel, 1994) is commonly proposed to confer a fitness advantage due to the rare acquisition of beneficial alleles, although other hypotheses have been advanced (Redfield, 1988, Redfield, 1993, Redfield et al., 1997). A recent theoretical study has considered the advantages of sex, or at least of recombination (Nowak et al., 2014). It was suggested that recombination provides a continual survival advantage to cells adapting in the face of competition in rapidly changing environments, a situation in accord with the Red Queen hypothesis (Van Valen, 1973). Recombination can bring advantageous mutations together and would have a robust effect in the case of fitness enhancing interactions between genes (positive epistasis). However, in a constant environment, recombination may eventually pose a fitness cost by disrupting advantageous combinations of alleles. Based on these considerations, it is possible that the bistable expression of transformability may offer a bet-hedging solution just as the bimodal expression of persistence has been postulated to do, with the frequency of KS cells once more set by the rate of environmental change. By bistably expressing the ability to recombine, a population may have the short term benefit derived in the face of environmental change and simultaneously be buffered against the disruptive effect of recombination. By coupling both the growth arrest and transformability to the bistable expression of comK, evolution may have arrived at an economical way to regulate these phenotypes. Other well-studied transformable bacteria may have evolved different strategies to confront environmental vagaries.
It is notable that persistence in other bacteria has also been linked to the accumulation of (p)ppGpp (Korch et al., 2003, Dahl et al., 2003, Sureka et al., 2008, Nguyen et al., 2011, Maisonneuve et al., 2013, Amato et al., 2013, Maisonneuve & Gerdes, 2014, Mwangi et al., 2013, Kim et al., 2013). Although (p)ppGpp has been classically studied in the context of drastic amino acid deprivation, it is increasingly apparent that it plays a variety of often subtle regulatory role (Potrykus & Cashel, 2008, Liu et al., 2015, Gaca et al., 2015). Its involvement in situations as diverse as the B. subtilis KS and persistence in E. coli, Mycobacterium tuberculosis and Pseudomonas probably represent cases of convergent evolution, in which the growth rate regulating alarmone has been recruited to perform analogous tasks by different mechanisms.
Experimental procedures
Strains construction and growth of bacteria
B. subtilis strains listed in Table S1 are derived from the 168 derivative BD630. All strain constructions were carried out by transformation (Albano et al 1987) or transduction with bacteriophage PBS1 with selection for the appropriate antibiotics at the following concentrations: Kan (5 μg/ml), chloramphenicol (5 μg/ml), erythromycin (5 μg/ml), tetracycline (20 μg/ml), phleomycin (1 μg/ml), spectinomycin (100 μg/ml) and ampicillin (Amp) (100 μg/ml). Molecular constructs were created using standard molecular biological methods. The particulars are described in the Supporting Information. Liquid cultures of B. subtilis were grown with shaking at 37 °C in LB or competence medium (Albano et al., 1987) and growth was monitored by either measurement of turbidity in a Klett colorimeter or by OD600 in an Envision 2104 plate reader (Perkin-Elmer).
Statistical analysis
All statistical analyses were performed using Stata/SE 13.1. Differences between the proportions of DnaX dots were determined using 2×2 contingency tables and all p-values were Bonferroni corrected. Cramér’s V (V) is a measure of effect size, which is calculated from the chi-square. Values run from 0 to 1. V values of 0.1, 0.3 and 0.5 are taken to indicate small, medium and large effects respectively (Abbott & McKinney, 2013).
The large heteroscedasticity in the cell length data prevented the use of a simple linear regression analysis of cell length versus time. A previously utilized method to deal with growth curves that exhibit such characteristics is to employ a GLM (Nelder & Wedderburn, 1972, Schaffner, 1998, Lindqvist, 2006). The logarithm of the normalized cell length was plotted versus time in minutes. A GLM assuming a gamma distribution of the lengths, utilizing an identity link function, and setting the y-intercept to 0 was found to appropriately model the errors for all functions (p < 0.001). The GLM for the data displayed in Fig. 2 is shown in Fig. S1.
Antibiotic tolerance
B. subtilis was grown to T2 in competence medium and then diluted 20 fold into pre-warmed competence medium containing either 10 μg/ml of oxolinic acid or into LB containing 10 μg/ml of Kan. At hourly intervals 10 ml was removed from the growing culture, centrifuged and resuspended in 10 ml of medium without antibiotic, and viable counts were determined.
Microscopy
Cells were imaged on an upright Nikon Eclipse 90i microscope outfitted with an Orca ER Digital Camera (Hamamatsu) with a Nikon Tirf 1.45 NA Plan Neoflur 100 oil immersion objective. Semrock Optical filter sets were used for fluorescence detection. The Volocity software package 6.3 was used for image acquisition. Live cells were imaged on 1% agarose pads made up in competence medium. Time-lapse studies were performed on agarose pads in a Bioptechs chamber (Micro-Environmental Systems) held at 37 °C. Strains BD5810 and BD6757 were grown individually to competence and then diluted 20-fold into a single tube containing fresh competence medium. 1 μl of this mixed culture was placed on an agarose pad made up in competence medium in the chamber and mounted on the microscope. Images were collected every 5 min for the first hour and every 15 min for the next hour. When required, samples were stained for 5 min with 2 μg/ml PI + 0.1% Triton X100 on poly L-lysine-coated slides, washed 1X with PBS (81 mM Na2HPO4 + 24.6 mM NaH2PO4 + 100 mM NaCl) and then mounted in Slowfade (Molecular Probes).
Flow cytometry
Cells were grown to T2, diluted 50-fold into fresh pre-warmed competence medium and incubated further at 37 °C. Samples of 1 ml were taken immediately and at 30, 60, 90 and 120 minutes after dilution. The cells were fixed in 3.2% paraformaldehyde and stored at 4 °C until use. Prior to cytometric analysis the cells were concentrated 2-fold in PBS and stained with a final concentration of 2 μg/ml PI in 0.1% Triton X100. Fluorescence was measured on a BD LSRII flow cytometer (BD Bioscience) operating a solid-state laser at 488 nm. Data were collected using a 530/30 filter for YFP and a 575/26 filter for PI. Photomultiplier voltage was set at 599 for YFP and 477 for PI. For each sample at least 10,000 events were counted and the data were collected and analyzed using BD FACSDiva 6 software. The analysis was carried out by the Rutgers New Jersey Medical School Flow Cytometry and Immunology Core Laboratory.
Western blot analysis
Western blotting was carried out essentially as described (Hahn et al., 2005). Cells were grown to T2 and diluted 50-fold into fresh competence medium. Equal volume aliquots (25 ml) were taken at the indicated times. Cells were recovered by centrifugation and resuspended in 200 μl of 50 mM NaCl, 25% sucrose (w/v), 5 mM MgCl2, 50 mM Tris-HCl, pH 8.0 containing 300 μg/ml of lysozyme. After incubation at 37 °C for 5 minutes, 50 μl of 5X cracking buffer (225 mM Tris-HCl, pH 8.0, 5% SDS, 50% glycerol, 1% ß-mercaptoethanol and 0.05% bromphenol blue) was added and 10 μl was loaded on each lane of a 10 % Tricine SDS-polyacrylamide gel (Schagger & von Jagow, 1987). Anti-Maf and anti-ComGA antisera, raised in rabbits, were used at a dilution of 1:1,000 and 1:5,000 respectively, and the secondary antibodies (horseradish peroxidase conjugated goat anti-rabbit, Kirkegaard & Perry Laboratories, Inc.) were used at a dilution of 1:10,000.
Protein affinity purification
Single transformants of ED1751, ED1752 or ED1758 were inoculated into 25 ml of LB broth containing 100 μg/ml ampicillin and grown overnight at 37 °C with vigorous shaking. A liter of LB containing 100 μg/ml Amp was inoculated with 20 ml of each overnight culture and shaken vigorously at 37 °C. At an OD600 of 0.5–0.7, IPTG was added to a final concentration of 500 μM and the cultures were incubated at 25° C for an additional 3 hours. The cultures were then centrifuged for 15 min at 5000 × g and the pellets were frozen at −80 °C until use.
Protein purification of GST tagged proteins was carried out essentially as described previously (Carabetta et al., 2013). Briefly, pellets were thawed on ice and resuspended in 20 ml lysis buffer (50 mM sodium phosphate, pH 7.5, 300 mM NaCl, 10 mM tris(2-carboxyethyl)phosphine (TCEP) + 1 tablet EDTA-free protease inhibitor capsule (Thermo-Fisher Scientific). Lysis was accomplished with 3 passages through an Avestin EmulsiFlex-C5 cell disrupter at an operating pressure of 12,000 psi (Neiditch & Hughson, 2007). The lysate was centrifuged for 60 minutes at 14,000 × g at 4°C. The supernatant was mixed with 2 ml pre equilibrated glutathione-Super flow resin (Clontech) and gently rotated for 1 h at 4°C. It was then loaded on a column and the flow through was collected. The column was washed with 20 volumes of lysis buffer minus the protease inhibitor. To elute the protein, 2 ml (1 column volume) of lysis buffer was added to the resin and 800 μl of a 1:10 dilution of PreScission Protease (GE Healthcare) in lysis buffer was added to the slurry and incubated overnight at 4°C. The flow through was collected and the column was washed with 1 column volume of buffer, collected in the same tube. For analysis, the samples were electrophoresed on 10% Tricine SDS polyacrylamide gels.
Isolation of ComGA and analysis of its interactions by mass spectrometry
Cryogenic cell lysis, conjugation of magnetic beads, immunopurification of ComGA-containing complexes, and mass spectrometric analyses were carried out as described previously, with minor modifications (Cristea et al., 2005, Carabetta et al., 2010, Greco et al., 2012, Carabetta et al., 2013, Mann et al., 2013, Joshi et al., 2013). ComGA-MYC (BD3798) was expressed in a rok background so that ~80% of the cells would be in the KS. Cells were grown to the KS in liquid competence medium, pelleted and rapidly frozen in liquid nitrogen as described (Cristea et al., 2005, Carabetta et al., 2013). Since ComGA is a peripheral membrane protein, two buffers were selected for analysis. The first (Buffer A) was optimized to efficiently isolate cytoplasmic proteins: 20 mM HEPES pH 7.4, 100 mM CH3CO2K, 2 mM MgCl2, 0.1% tween-20 (v/v), 1 mM ZnCl2, 1 mM CaCl2, 0.2% Triton X-100, 150 mM NaCl, 10 μg/μl DNaseI, 1:100 protease inhibitor cocktail (Sigma), and 2 mg/ml phenylmethylsulphonyl fluoride (PMSF). For the isolation of the membrane-bound fraction of interacting partners, a similar buffer (Buffer B) was used with the following changes: the NaCl and Triton X-100 concentrations were increased to 250 mM and 1%, respectively, DNaseI was omitted, and 0.5% sodium deoxycholate was added. Anti-MYC antibodies (AbCam) were conjugated to magnetic M270 Epoxy Dynabeads (Life Technologies) at a concentration of 10 μg of antibody per mg of beads. ComGA-containing complexes were eluted directly into LDS-PAGE sample loading buffer (Life Technologies), resolved on a 4–12% NuPAGE bis-tris precast gradient gel (Life Technologies), prepared for in-gel digestion with trypsin, and peptides were recovered as described previously (Carabetta et al., 2013). Interacting proteins were identified by nLC-MS/MS on a Dionex Ultimate 3000 RSLC coupled to an LTQ-Orbitrap Velos ETD mass spectrometer (Thermo Fisher Scientific, San Jose, CA), and data was analyzed as described (Carabetta et al., 2013, Joshi et al., 2013).
Luminometry
Luciferase assays were preformed as previously described (Mirouze et al., 2011) Briefly, strains were grown in LB for approximately 2 hrs., harvested and resuspended in competence medium to an OD600 of 2. This suspension was then further diluted 20-fold in competence medium and 200 μl was added to each well of a 96 well clear bottomed, black plate (Corning) containing luciferin (Caliper Life Sciences) to yield a final concentration of 4.7 nM. The plates were incubated at 37 °C with agitation in a Perkin Elmer Envision 2104 Multilabel Reader equipped with an enhanced sensitivity photomultiplier for luminometry. The plate lids were heated to 38°C to prevent condensation. Relative Luminescence Units (RLU) and OD600 were measured at 1.5 min intervals. After 5 hours of growth, at which point T2 had been reached, the plate reader was stopped and the cells were diluted 50-fold into an empty well containing fresh competence medium and luciferin. The plate reader was switched on and the RLUs and OD600 measurements were continued for another 9 hours.
Acknowledgments
We thank the members of our respective laboratories for advice and discussion. We thank Alan Grossman, Katherine Lemon and David Rudner for providing strains and Ken Briley for making the Phyperspank-comGA construct. We also thank Tilia Tanner and Craig Beisel for advice with the statistical analyses. Finally, we thank George Wright of GLSynthesis Inc. for his kind gift of HPura. The work was supported by NIH RO1GM057720 in the DD lab and by NIH DP1DA026192 and R21HD073044-01A1 in the IMC lab. AT was supported in part by the New Jersey Health Foundation.
References
- Abbott M, McKinney J. Understanding and applying research design. John Wiley & Sons, Inc; Hoboken, N.J: 2013. p. xix.p. 425. [Google Scholar]
- Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40:471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- Amato SM, Orman MA, Brynildsen MP. Metabolic control of persister formation in Escherichia coli. Mol Cell. 2013;50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Giladi H, Oppenheim AB, Glaser G. Analysis of the shut-off of ribosomal RNA promoters in Escherichia coli upon entering the stationary phase of growth. FEMS Microbiol Lett. 1996;140:71–76. doi: 10.1111/j.1574-6968.1996.tb08317.x. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, Cui X, Sloma A, Widner W, Dubnau D. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol Microbiol. 2002;43:1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x. [DOI] [PubMed] [Google Scholar]
- Biswas GD, Sox T, Sparling PF. Factors affecting genetic transformation of Neisseria gonorrhoeae. J Bacteriol. 1977;129:983–992. doi: 10.1128/jb.129.2.983-992.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briley K, Jr, Dorsey-Oresto A, Prepiak P, Dias MJ, Mann JM, Dubnau D. The secretion ATPase ComGA is required for the binding and transport of transforming DNA. Mol Microbiol. 2011a;81:818–830. doi: 10.1111/j.1365-2958.2011.07730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briley K, Jr, Prepiak P, Dias MJ, Hahn J, Dubnau D. Maf acts downstream of ComGA to arrest cell division in competent cells of B. subtilis. Mol Microbiol. 2011b;81:23–39. doi: 10.1111/j.1365-2958.2011.07695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NC. Inhibition of bacterial DNA replication by 6-(p-hydroxyphenylazo)-uracil: differential effect on repair and semi-conservative synthesis in Bacillus subtilis. J Mol Biol. 1971;59:1–16. doi: 10.1016/0022-2836(71)90409-8. [DOI] [PubMed] [Google Scholar]
- Butler YX, Abhayawardhane Y, Stewart GC. Amplification of the Bacillus subtilis maf gene results in arrested septum formation. J Bacteriol. 1993;175:3139–3145. doi: 10.1128/jb.175.10.3139-3145.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Kobel PA, Morshedi MM, Wu MF, Paddon C, Helmann JD. Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J Mol Biol. 2002;316:443–457. doi: 10.1006/jmbi.2001.5372. [DOI] [PubMed] [Google Scholar]
- Carabetta VJ, Silhavy TJ, Cristea IM. The response regulator SprE (RssB) is required for maintaining poly(A) polymerase I-degradosome association during stationary phase. J Bacteriol. 2010;192:3713–3721. doi: 10.1128/JB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta VJ, Tanner AW, Greco TM, Defrancesco M, Cristea IM, Dubnau D. A complex of YlbF, YmcA and YaaT regulates sporulation, competence and biofilm formation by accelerating the phosphorylation of Spo0A. Mol Microbiol. 2013;88:283–300. doi: 10.1111/mmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YS, Breidt F, Dubnau D. Cell surface localization and processing of the ComG proteins, required for DNA binding during transformation of Bacillus subtilis. Mol Microbiol. 1998;29:905–913. doi: 10.1046/j.1365-2958.1998.00989.x. [DOI] [PubMed] [Google Scholar]
- Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- Dagkessamanskaia A, Moscoso M, Henard V, Guiral S, Overweg K, Reuter M, Martin B, Wells J, Claverys JP. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol. 2004;51:1071–1086. doi: 10.1111/j.1365-2958.2003.03892.x. [DOI] [PubMed] [Google Scholar]
- Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE., 3rd The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Swanson MS. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: the intricacies of (p)ppGpp metabolism and Its control of bacterial homeostasis. J Bacteriol. 2015;197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla JD. Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol Microbiol. 2005;55:973–977. doi: 10.1111/j.1365-2958.2004.04455.x. [DOI] [PubMed] [Google Scholar]
- Greco TM, Miteva Y, Conlon FL, Cristea IM. Complementary proteomic analysis of protein complexes. Methods Mol Biol. 2012;917:391–407. doi: 10.1007/978-1-61779-992-1_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell. 2005;122:59–71. doi: 10.1016/j.cell.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema BJ, Hahn J, Haynes J, Dubnau D. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol Microbiol. 2001;40:52–64. doi: 10.1046/j.1365-2958.2001.02363.x. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Smits WK, de Jong A, Holsappel S, Kuipers OP. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 2002;30:5517–5528. doi: 10.1093/nar/gkf698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa TT, Tortosa P, Albano M, Dubnau D. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol Microbiol. 2002;43:15–26. doi: 10.1046/j.1365-2958.2002.02727.x. [DOI] [PubMed] [Google Scholar]
- Inaoka T, Ochi K. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J Bacteriol. 2002;184:3923–3930. doi: 10.1128/JB.184.14.3923-3930.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen PJ, Dubnau D, Levin BR. Episodic selection and the maintenance of competence and natural transformation in Bacillus subtilis. Genetics. 2009;181:1521–1533. doi: 10.1534/genetics.108.099523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Mwangi M, Chung M, Milheirico C, de Lencastre H, Tomasz A. The mechanism of heterogeneous beta-lactam resistance in MRSA: key role of the stringent stress response. PLoS One. 2013;8:e82814. doi: 10.1371/journal.pone.0082814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- Kramer N, Hahn J, Dubnau D. Multiple interactions among the competence proteins of Bacillus subtilis. Mol Microbiol. 2007;65:454–464. doi: 10.1111/j.1365-2958.2007.05799.x. [DOI] [PubMed] [Google Scholar]
- Kussell E, Kishony R, Balaban NQ, Leibler S. Bacterial persistence: a model of survival in changing environments. Genetics. 2005;169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- Kuwahara H, Soyer OS. Bistability in feedback circuits as a byproduct of evolution of evolvability. Mol Syst Biol. 2012;8:564. doi: 10.1038/msb.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner M, Stingl K, Radler JO, Maier B. Basal expression rate of comK sets a ‘switching-window’ into the K-state of Bacillus subtilis. Mol Microbiol. 2007;63:1806–1816. doi: 10.1111/j.1365-2958.2007.05628.x. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. Movement of replicating DNA through a stationary replisome. Mol Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- Leung V, Levesque CM. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J Bacteriol. 2012;194:2265–2274. doi: 10.1128/JB.06707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012;10:e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist R. Estimation of Staphylococcus aureus growth parameters from turbidity data: characterization of strain variation and comparison of methods. Appl Environ Microbiol. 2006;72:4862–4870. doi: 10.1128/AEM.00251-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Bittner AN, Wang JD. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol. 2015;24C:72–79. doi: 10.1016/j.mib.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Scrudato M, Blokesch M. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet. 2012;8:e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFadyen LP, Chen D, Vo HC, Liao D, Sinotte R, Redfield RJ. Competence development by Haemophilus influenzae is regulated by the availability of nucleic acid precursors. Mol Microbiol. 2001;40:700–707. doi: 10.1046/j.1365-2958.2001.02419.x. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, Castro-Camargo M, Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- Mann JM, Carabetta VJ, Cristea IM, Dubnau D. Complex formation and processing of the minor transformation pilins of Bacillus subtilis. Mol Microbiol. 2013;90:1201–1215. doi: 10.1111/mmi.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B, Granadel C, Campo N, Henard V, Prudhomme M, Claverys JP. Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol Microbiol. 2010;75:1513–1528. doi: 10.1111/j.1365-2958.2010.07071.x. [DOI] [PubMed] [Google Scholar]
- Mechold U, Cashel M, Steiner K, Gentry D, Malke H. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J Bacteriol. 1996;178:1401–1411. doi: 10.1128/jb.178.5.1401-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze N, Desai Y, Raj A, Dubnau D. Spo0A~P imposes a temporal gate for the bimodal expression of competence in Bacillus subtilis. PLoS Genet. 2012;8:e1002586. doi: 10.1371/journal.pgen.1002586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze N, Prepiak P, Dubnau D. Fluctuations in spo0A transcription control rare developmental transitions in Bacillus subtilis. PLoS Genet. 2011;7:e1002048. doi: 10.1371/journal.pgen.1002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi MM, Kim C, Chung M, Tsai J, Vijayadamodar G, Benitez M, Jarvie TP, Du L, Tomasz A. Whole-genome sequencing reveals a link between beta-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb Drug Resist. 2013;19:153–159. doi: 10.1089/mdr.2013.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- Natori Y, Tagami K, Murakami K, Yoshida S, Tanigawa O, Moh Y, Masuda K, Wada T, Suzuki S, Nanamiya H, Tozawa Y, Kawamura F. Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient in (p)ppGpp synthetase genes, relA, yjbM, and ywaC. J Bacteriol. 2009;191:4555–4561. doi: 10.1128/JB.00263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch MB, Hughson FM. The regulation of histidine sensor kinase complexes by quorum sensing signal molecules. Methods Enzymol. 2007;423:250–263. doi: 10.1016/S0076-6879(07)23011-3. [DOI] [PubMed] [Google Scholar]
- Nelder JA, Wedderburn RWM. Generalized linear models. Journal of the Royal Statistical Society Series A (General) 1972;135:370–384. [Google Scholar]
- Nester EW, Stocker BAD. Biosynthetic latency in early stages of deoxyribonucleic acid transformation in Bacillus subtilis. J Bacteriol. 1963;86:785–796. doi: 10.1128/jb.86.4.785-796.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak S, Neidhart J, Szendro IG, Krug J. Multidimensional epistasis and the transitory advantage of sex. PLoS Comput Biol. 2014;10:e1003836. doi: 10.1371/journal.pcbi.1003836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Yamaguchi H, Kobayashi K, Ogasawara N, Fujita Y, Tanaka T. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J Bacteriol. 2002;184:2344–2351. doi: 10.1128/JB.184.9.2344-2351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Piir K, Paier A, Liiv A, Tenson T, Maivali U. Ribosome degradation in growing bacteria. EMBO Rep. 2011;12:458–462. doi: 10.1038/embor.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Redfield RJ. Evolution of Bacterial Transformation: Is sex with dead cells ever better than no sex at all? Genetics. 1988;119:213–221. doi: 10.1093/genetics/119.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RJ. Genes for breakfast: the have-your-cake-and-eat-it-too of bacterial transformation. J Hered. 1993;84:400–404. doi: 10.1093/oxfordjournals.jhered.a111361. [DOI] [PubMed] [Google Scholar]
- Redfield RJ, Schrag MR, Dean AM. The evolution of bacterial transformation: sex with poor relations. Genetics. 1997;146:27–38. doi: 10.1093/genetics/146.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner DW. Predictive food microbiology Gedanken experiment: why do microbial growth data require a transformation? Food Microbiology. 1998;15:185–189. [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Scheich C, Kummel D, Soumailakakis D, Heinemann U, Bussow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- Sureka K, Ghosh B, Dasgupta A, Basu J, Kundu M, Bose I. Positive feedback and noise activate the stringent response regulator Rel in Mycobacteria. PLoS One. 2008;3:e1771. doi: 10.1371/journal.pone.0001771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchigvintsev A, Tchigvintsev D, Flick R, Popovic A, Dong A, Xu X, Brown G, Lu W, Wu H, Cui H, Dombrowski L, Joo JC, Beloglazova N, Min J, Savchenko A, Caudy AA, Rabinowitz JD, Murzin AG, Yakunin AF. Biochemical and structural studies of conserved Maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides. Chem Biol. 2013;20:1386–1398. doi: 10.1016/j.chembiol.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A. Stochastic gene expression in fluctuating environments. Genetics. 2004;167:523–530. doi: 10.1534/genetics.167.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. A new evolutionary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]


