Abstract
Genome sequences encoding DUF1220 protein domains show a burst in copy number among anthropoid species and especially humans, where they have undergone the greatest human lineage-specific copy number expansion of any protein coding sequence in the genome. While DUF1220 copy number shows a dosage-related association with brain size in both normal populations and in 1q21.1-associated microcephaly and macrocephaly, a function for these domains has not yet been described. Here we provide multiple lines of evidence supporting the view that DUF1220 domains function as drivers of neural stem cell proliferation among anthropoid species including humans. First, we show that brain MRI data from 131 individuals across 7 anthropoid species shows a strong correlation between DUF1220 copy number and multiple brain size-related measures. Using in situ hybridization analyses of human fetal brain, we also show that DUF1220 domains are expressed in the ventricular zone and primarily during human cortical neurogenesis, and are therefore expressed at the right time and place to be affecting cortical brain development. Finally, we demonstrate that in vitro expression of DUF1220 sequences in neural stem cells strongly promotes proliferation. Taken together, these data provide the strongest evidence so far reported implicating DUF1220 dosage in anthropoid and human brain expansion through mechanisms involving increasing neural stem cell proliferation.
Background
Comparison of DUF1220 copy number among >40 mammalian species indicates that DUF1220 has undergone a major copy number expansion specifically among the anthropoid suborder of primates (monkeys, apes and humans). Among anthropoids, the most striking increase is found in humans, where DUF1220 shows the greatest evolutionary copy number expansion of any protein coding sequence in the human genome (>270 copies in human; 160 of which are unique to human; chimp=125, macaque=30 and mouse/rat=1)[1, 2, 3]. This burst in copy number shows a close correlation with both brain size and neuron number specifically among primates and thus fits well with the recent proposal that primate brain expansion has proceeded via a unique evolutionary path of neuronal scaling[4, 5].
The majority of DUF1220 copies map to 1q21 and DUF1220 copy number loss and gain have been implicated in 1q21.1-associated microcephaly and macrocephaly, respectively[6, 7]. DUF1220 copy number was also shown to be significantly associated with brain gray matter volume in a non-disease population[4] . Lastly, we have recently shown that increases in dosage of the CON1 DUF1220 clade exhibit a linear correlation with increased severity of each of the three primary symptoms associated with autism, a disease linked to brain overgrowth (manuscript submitted). Given these findings that link DUF1220 dosage to human brain expansion, we utilized three independent but complementary approaches in an effort to gain new insight into the function of DUF1220 domains during human brain development.
Materials and Methods
MRI Subjects
Magnetic resonance images (MRI) were obtained from a total of 193 adult primates including humans (Homo sapiens, n = 11, all adult males), chimpanzees (Pan troglodytes, n = 127, 53 males and 72 females, mean age = 27.52, s.d. = 10.35), gorillas (Gorilla gorilla, n =15, 11 males and 4 females, mean age = 32.88, s.d.=14.49), orangutans (Pongopygmaeus, n = 12, 7 males and 5 females, mean age = 22.69, s.d.=13.87)), gibbons (Hylobateslar, n = 3, 1 male and 2 females, mean age = 17.00, s.d. = 8.28 ), baboons (Papioanubis, n = 4, 2 males and 2 females, mean age = 7.23, s.d. = 2.11), and rhesus monkeys (Macacamulatta, n = 21, 16 males and 5 females, mean age = 9.09, .d.= 2.32). For the humans, chimpanzees, gibbons, and rhesus monkeys, all the MRI scans were obtained in vivo while a combination of in vivo (IV) and post-mortem (PM) scans made up the baboon (2 PM, 2 IV), orangutan (4 IV, 8 PM), and gorilla (2 IV, 13 PM) samples. All PM brains were collected following death by natural causes of the subjects. Thus, no subjects were sacrificed for the purposes of this study. For all species, the scanning procedures were performed under the guidelines of state and federal laws, the U.S. Department of Health and Human Services and institutional animal care and use committees.
Image Collection and Procedure
This study was opportunistic in terms of availability of in vivo and post-mortem MRI scans. Thus, as noted above, the magnets and scanning protocols were not identical in all species. This presents some limitations for certain comparative analyses because variation in magnet strength and/or scanning protocol can influence the signal strength and sensitivity in contrast between grey matter, white matter and CSF. Moreover, shrinkage in tissue due to fixatives can result in some distortion in size of various structures. However, in all of the comparative analyses of CS surface area and depth, the individual values were adjusted for whole brain measures taken from the same scan. Thus, inherent differences in grey matter, white matter and CSF due to the scanning protocols or magnet strength were standardized within individuals when quantifying the surface area and depth of the CS.
For in vivo MRI scans in all species except humans, subjects were first immobilized by ketamine injection as appropriate for the species and subsequently anaesthetized with propofol (chimpanzees), or isofluorane (rhesus and bonnet macaques). The subjects remained anaesthetized for the duration of the scans as well as the time needed to transport them between their home cage and the imaging facility (total time ~ 2 h, MRI acquisition time ranging from 36–60 minutes). All scans were examined at the time of acquisition and any image with artifact was excluded in the subsequent image processing. After completing MRI procedures, the subjects were temporarily housed in a single cage for 6–12 h to allow the effects of the anesthesia to wear off, after which they were returned to their home cage. The archived MRI data were transferred to a PC running BrainVisa software for post-image processing. To provide an unbiased collection of human subjects, a heterogeneous sample set (n = 11) was randomly assembled from the BrainVisa database. All MR images were previously processed through the software before including them in our analyses. As such, the scans originated from many different scanners and protocols over the span of 20 years using an approximate gradient echo protocol (inversion time = 500 ms, pulse repetition = 10 ms, echo time = 2 ms, and a 256 x 256 matrix). For the post-mortem scanning, either 4.7 or 7T magnets were used and T2-weighted images were collected in the transverse plane using a gradient echo protocol (pulse repetition = 22.0 s, echo time = 78.0 ms, number of signals averaged = 8–12, and a 256 × 192 matrix reconstructed to 256 × 256).
Image Processing
The pipeline of process used to obtain the measures in this study were derived from a pipeline initially dedicated to the human brain and freely distributed as a BrainVISA toolbox (http://brainvisa.info). The human-dedicated pipeline has been used previously for at least 5000 different subjects. Some tuning of this pipeline was required to account for specificities of the nonhuman primate anatomy as well as the different protocols used to acquire the images in the in vivo and post-mortem brains. The pipeline processing steps proceeded in the following manner: First, correction of the spatial inhomogeneties of the signal, which prevent direct association between the signal intensity and the nature of the tissue, were performed. The estimation of the spatially smooth bias field used to restore the signal intensity was performed via minimization of the signal entropy. After correction, each tissue intensity distribution was stable across the brain. Second, automatic analysis of the signal histogram and mathematical morphology was then used to compute a binary mask of the brain. This approach is built on the fact that the brain is surrounded by dark areas corresponding to skull and cerebrospinal fluid (CSF). Therefore, once the range of intensities corresponding to brain tissue had been defined by histogram analysis, brain segmentation mainly amounts to splitting the connections with external structures like the optical nerves. For the nonhuman primate anatomy, some specific tuning had to be applied relative to the human-dedicated processing performed by BrainVISA. Specifically, for some primate species, the largest object in the image after splitting connections turned out to be the muscles, at least in the case of the in vivo scans. Hence, in order to reliably select the brain, we had to introduce an additional constraint relative to the localization of the brain in the middle of the head. Further, for the post-mortem MRI scans, we had to invert the intensities corresponding to grey and white matter in order for BrainVISA to run properly. Once the brain mask had been defined, the mask was split into three parts corresponding to hemispheres and cerebellum.
After a mask has been defined for each hemisphere, a negative mold of the white matter was computed. The outside boundary of this mold results from a 5-mm “closing” of the masked hemisphere. Here, “closing” is an operation of Mathematical Morphology used to analyze shapes: the mask of the hemisphere is first “dilated” then “eroded” which results in deleting the folds that are less than 5mm wide. The inside boundary is the grey/white interface computed with topology preserving deformations assuring the spherical topology of the mold. Imposing the actual topology of the cortex to the mold prevents the detection of spurious folds resulting from noisy data. The mold is finally skeletonized in order to detect the cortical fold as crest surfaces of the 3D MR image located inside the mold. Skeletonization is another standard technique in Mathematical Morphology. An object is eroded until losing its thickness: for example, a door would become a flat 2D surface or a ball with a cavity would become a sphere. The crest surfaces stem from a morphological watershed process iteratively eroding the 3D mold from the lightest intensities to the darkest intensities. Topological constraints guarantee that the resulting surfaces have no holes. The end result is a set of topologically elementary surfaces located along the darkest part of the fold corresponding to CSF. These elementary surfaces are split further when a deformation of the deepest part of the fold indicates the presence of a buried gyrus. The clues allowing the detection of buried gyri are embedded in the curvature of the grey/white interface. Indeed, a buried gyrus leads to a horse saddle shape in the depth of the grey/white interface, which results in a negative Gaussian curvature providing these clues. Finally, the cortical folds comprising the sulci are presented in the 3D visualization graph. .
Global Cortical Measures
Using BV, we calculated hemispheric grey and white matter volumes (mm3) and overall gyrification index (GI) for each subject. To calculate the grey (GM) and white matter (WM) volumes, the program used the grey/white interface generated in the sulci extraction pipeline process to generate a cerebral spinal fluid (CSF) interface, providing the total volumes in each hemisphere. The cerebellum and related brain stem structures were excluded from calculations in total GM and WM volume. As has been done in many previous studies, the overall gyrification index (GI) was determined by measuring the surface area sulcal internal contours of the cortex and dividing it by the total surface area for each hemisphere, which BV calculates (see Bogart et al., 2013[8]).
Human Fetal Brain Tissue Samples
Fetal tissue samples were collected from spontaneous abortions from 2009 to 2013, formalin fixed and paraffin embedded for archival at the University of Colorado Hospital. Samples with no abnormal brain related diagnosis were screened for intact cortical/neural tube tissue. Samples were collected under COMIRB protocol 09-0455.
LNA probes for in situ hybridization
Two non-overlapping oligo sequences (A and B) for human NBPF were identified 5’-ATCCAGCAGCTCCCTGCTGA-3’ and 5’-TCTTGCAAGACTTCAGGCCCTT-3’. Locked nucleic acids (LNA) were designed into the sequence resulting in RNA Tm’s of 84°C for both probes. We also included a scramble LNA probe and a miR-126 LNA probe (See Jørgensen methods). The LNA oligos were 6-carboxyfluorescein (FAM)-labeled at the 5’- and 3’-ends (double-FAM labeled probes) and obtained from Exiqon, Vedbæk, Denmark.
In situ hybridization
In situ hybridization was performed using a Tecan in situ hybridization instrument (Tecan, Männedorf, Switzerland) essentially as described elsewhere [10] . In brief, 6-μm thick tissue sections were pre-digested with proteinase-K (25μg/ml for 8 minutes at 37°C). In situ hybridization was performed by incubating the two double-FAM labeled NBPF LNA probes mixed or separately at 40 or 60 nM diluted in Exiqon hybridization buffer at 57°C for 1 hour. Double-FAM labeled scramble (60 nM) and miR-126 (at 60 nM) probes were used as negative and positive controls, respectively. After stringent washes in SSC buffers, the sections were incubated with alkaline phosphatase – conjugated anti-FAM (1:800, Roche, Mannheim, Germany). Slides were developed in 4-nitro-blue tetrazolium (NBT) and 5-brom-4-chloro-3’-Indolyl-phosphate (BCIP) substrate (Roche) for 60 minutes resulting in a dark-blue precipitate. Slides were counter stained with nuclear fast red (Vector Laboratories, Burlingname, CA) if not otherwise stated.
Image Processing
Pairs of images (DUF1220 probe and Scramble probe) were obtained at identical exposures on a Nikon i80, and adjusted for levels by the same values between pairs in ImageJ. Each image was split into component color channels, red, green and blue, and the green channel was discarded. A representative measurement window large enough to span several cells, but not including damaged tissue or the ventricular surface (because of edge artifacts), was defined and applied to all four images in the same manner via the ROI Manager. In the event that the measurement window had to be moved in order to avoid the ventricular surface or damaged tissue in one image, it was done so identically for both channels. Mean gray values were then measured and recorded, and the gray value images were then rotated and scaled as necessary for presentation in figure 2. Blue channel values are primarily Nuclear Fast Red, and were thus used to control for non-specific staining differences, while the red channel primarily measures in situ probe (BCIP) signal. In situ values were therefore defined as red channel difference minus blue channel difference for each pair of images (Supplemental Fig. 2). A negative value thus indicates a stronger probe signal in the Scramble sample than in the test sample.
Figure 2.
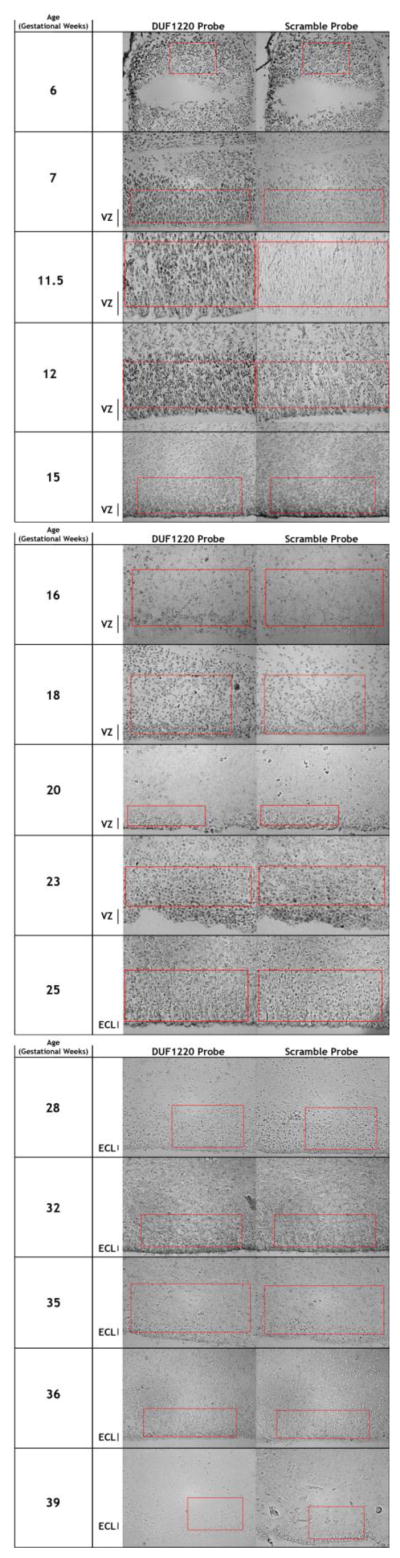
Time course of human fetal brain tissue samples showing differences in DUF1220 (left column) and Scramble (right column) probe signals. Telencephalic samples are presented at 40X, and regions used for probe quantification are indicated by red boxes. Estimates for the ventricular and subventricular zones by cell density are indicated to the left. Developmental time is in gestational weeks (G.W.). VZ: ventricular zone. ECL: ependymal cell layer.
Live Cell Imaging
H9 derived human embryonic stem cells (Gibco #N7800-100) were plated at approximately 60–70% coverage in a six well plate (Falcon #353046) pre coated with CellSTART CTS (Gibco #10142-01) and cultured at 37 degrees with 5% CO2 in complete media (KnockOut DMEM/F-12 (Gibco #12660-012) supplemented with 1% GlutaMAX -I (Gibco #35050-061), 2μg per 100 mL of Recombinant FGF-Basic (Gibco #PHG0024), 2μg per 100 mL of Recombinant EGF (Gibco #PHG0314) and 0.2% serum (Gibco #A10508-01)). After 24 hours, cells were switched to minimal media (KnockOut DMEM/F-12 supplemented with 1% GlutaMAX -I and 0.2% serum) and transfected with a synthetic NBPF15 construct (OriGene #RC212377 ) or a mock construct (OriGene #PS100001), using TurboFectin 8.0 (OriGene #TF81001) according to manufacturer’s specifications. Briefly, 3μL of TurboFectin 8.0 per well was added to 100 μL of Opti-MEM I Reduced Serum Medium (Gibco #31985-070) per well, gently mixed by pipetting, and incubated at room temperature for 5 minutes. Plasmid DNA was then added (1μg per well), gently mixed by pipetting, and incubated at room temperature for 30 minutes. Transfection mixture was then added to each well drop-wise. Images were collected in a 4 x 4 array in an Incucyte ZOOM (Essen Bioscience) once every 4 hours. Incucyte software was trained to identify cell coverage on an image of the cells. Raw numbers from this processing definition were plotted and a first degree polynomial curve was fitted to them with R in order to define the growth period in the NBPF15-transfected cells. Original images used to plot cell coverage were also manually inspected for morphological differences that might confound the coverage metric, such as actively apoptotic cells or floating cells no longer adhered (Supplemental Fig. 5), and no such differences were detected.
Statistical Methods
Associations of DUF1220 copy number with mean MRI measured volumetric brain regions across primate lineages were obtained from t-tests from four linear regression analyses. Cell proliferation rates were analyzed with a polynomial regression. A first degree polynomial curve was fit to cell proliferation at hourly time points in order to define the growth period in the NBPF15-transfected cells. An interaction term formally testing differing slopes between mock and NBPF was included and was highly significant (p<0.0001). R version 2.15.1 (http://cran.r-project.org/) was used for statistical analyses.
Results
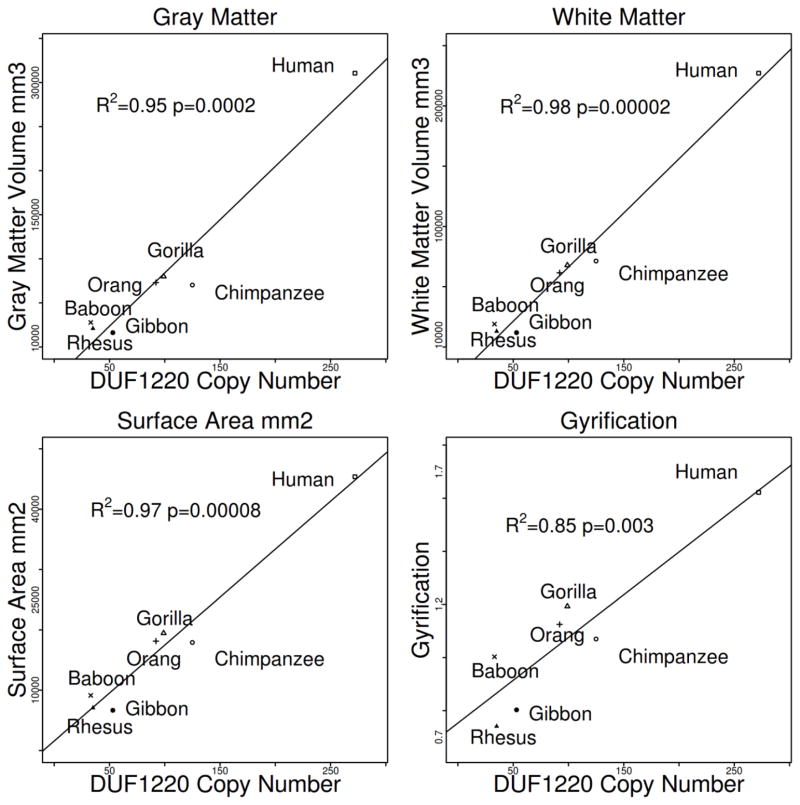
Our first approach utilized mean volumetric brain MRI measurements of 131 primates representing 7 species including humans. Significant correlations were identified between DUF1220 copy number and increases in brain gray matter volume (R2 = 0.95, F = 91.98 1, 5 df, p = 1.3 x 10−4), surface area (R2 = 0.97, F = 138.1 1, 5 df, p = 7.8 x 10−5), gyrification (R2 = 0.85, F = 27.22 1, 5 df, p = 2.0 x 10−3) and white matter volume (R2 = 0.98, F = 239.2 1, 5 df, p = 2.1 x 10−5) (Fig.1). In contrast, no significant association was found between DUF1220 copy number and body size (p > 0.1) (Supplemental Fig. 1). These findings expand upon an earlier comparison which showed a strong correlation between DUF1220 copy number and both overall brain size and estimated cortical neuron number across primate lineages[4] .
Figure 1.
Correlation between DUF1220 genomic copy number and MRI-determined gray matter volume, white matter volume, surface area, and gyrification among indicated primate species.
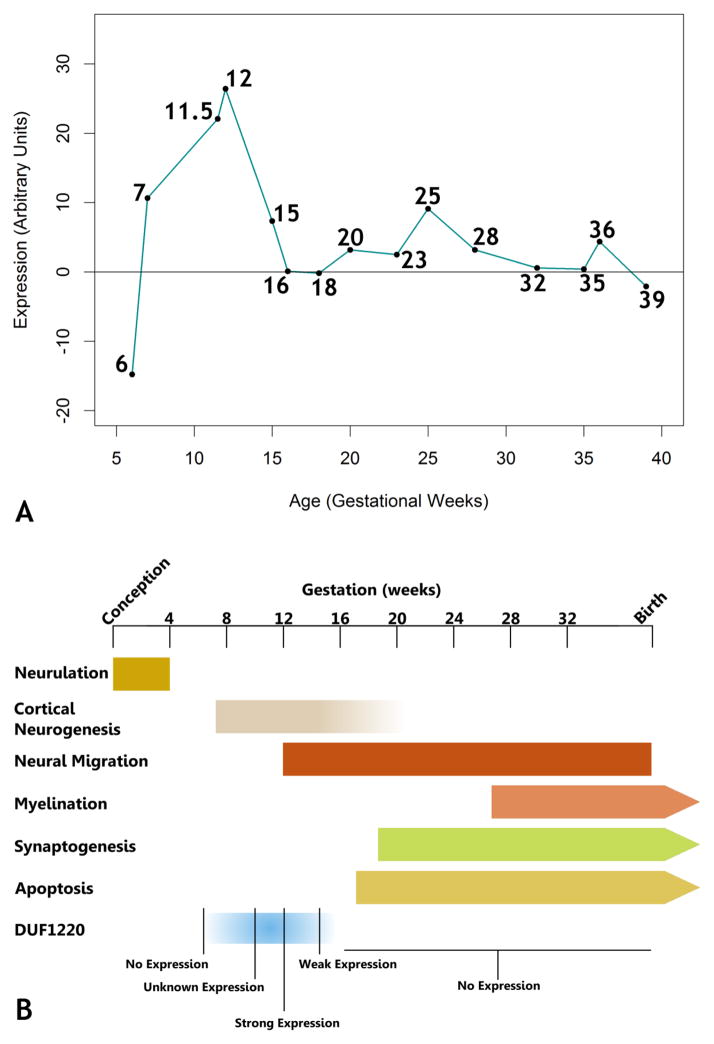
Secondly, we sought to determine whether expression of DUF1220 was localized to brain regions undergoing neural stem cell expansion. To address this we utilized in situ hybridization to investigate DUF1220 expression in human fetal brain tissue samples collected from spontaneous and induced abortions spanning virtually the entirety of human fetal brain development (gestational weeks (G.W.) 6 – 39). We show that in human brain HLS-type DUF1220 domains are highly expressed at the RNA level predominantly in the ventricular and subventricular zones, the primary sites for production of new excitatory neurons (Fig. 2). In addition, we show that the expression overlaps with the cortical neurogenic window but not earlier or later in development. While no signal was detected at week 6 or 7 (Fig. 3), expression was detected in the G.W. 11.5 and 12 samples, during which the ventricular zone is largest (Fig. 2), where the strongest signal is present predominantly in the dorso-lateral ventricular zone. A much fainter signal is detected at week 15, and no real signal is detected after this point.
Figure 3.
A: Timecourse of DUF1220 RNA expression. In situ hybridization values were plotted by gestational age, showing peak expression at gestational weeks 11 and 12. B: A timeline of human fetal brain development indicating general developmental events, including DUF1220 expression. Strong DUF1220 expression is seen by 11.5 gestational weeks, tapers off to lower expression by week 15, and is gone by week 18, indicating strong expression by at least mid-neurogenesis. In addition to this timing, strong DUF1220 probe signal in the ventricular zone also suggests a possible effect on neurogenesis. After references 15, 16 and 17.
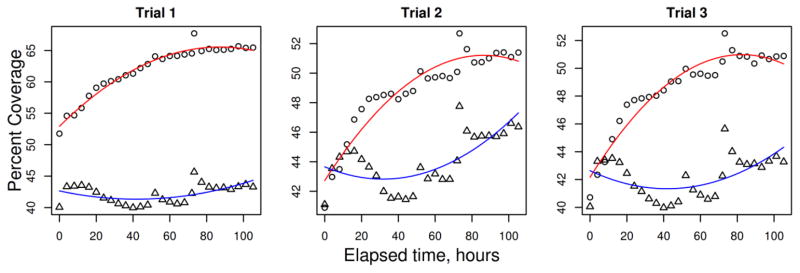
Finally we used transient transfection to test the ability of DUF1220 domains to influence proliferation in human neural stem cells in vitro. Compared to a mock transfection (empty vector), transfection of H9-derived neural stem cells with a DUF1220-encoding gene (NBPF15 cDNA) showed a greater rate of increase in cell coverage that lasts approximately 90 hours (Fig. 4). More specifically, NPBF15-transfected cells show an immediate increase in coverage within the first day, and continue to grow approximately 10%, while mock transfected cells begin to decrease within the first day, and increase only 3–4% over the duration of the proliferative burst.
Figure 4.
Total cell coverage plotted over time. Cultures of H9-derived human embryonic stem cells were transfected with NBPF15 (6 DUF1220 domains; circles) or an empty mock construct (triangles) at time = 0 and imaged every three hours. Cellular coverage was measured for each time point and plotted over time, and a curve was fitted to each experiment (NBPF15 in red and mock in blue).
Discussion
We have previously demonstrated a strong correlation between DUF1220 copy number and both brain size and estimated cortical neuron number across anthropoid primate species[4], but sought to identify more specific features of the brain that correlate with DUF1220 copy number in an effort to identify a developmental function. The four features we identified, gray matter volume, surface area, gyrification and white matter volume, are most easily explained by a model of brain expansion in which DUF1220 domains promote an increase in neuron number. Very little data describing the spatio-temporal expression pattern of DUF1220-containing genes existed, however, so we undertook a series of in situ hybridizations in a developmental time course to address this. We find that expression appears to spike preferentially in the ventricular zone at approximately G.W. 11.5/12, corresponding to roughly 70 days post conception. This may suggest an expression that corresponds to the onset of superficial layer neurogenesis (e.g. layers II/III). Although a slight bump in expression is detected later in G.W. 25, the higher values appear to be due to non-specific “streaking” (most likely an edge artifact, Fig. 2), as opposed to the punctuate pattern indicative of a true RNA signal. This streaking also appears in the blue channel (data not shown), and is therefore not likely to be real signal.
These data suggest that DUF1220 domains are expressed at the right time and place to influence neural stem cell proliferation in humans, and are associated with exaggerated neural stem cell proliferation in anthropoid primates. In order to test the hypothesis that DUF1220 domains can promote neural stem cell proliferation, we exogenously expressed a construct bearing 6 DUF1220 domains in human neural stem cells, which showed an immediate burst of proliferation, compared to little or no change in a mock transfection. Our primate brain MRI results and ventricular zone expression data suggest a possible DUF1220-driven brain expansion via the horizontal growth of the cortex, though the mechanism by which this occurs is not known. Although expression of DUF1220 at the ventricular zone is consistent with expansion of neural stem cells, and therefore consistent with hypotheses such as the radial unit hypothesis[11], expression in the sub-ventricular zone is also consistent with a mechanism of intermediate progenitor expansion[12]. Though more work will be required to investigate possible mechanisms of cellular expansion in order to determine the relevance of DUF1220 to either or both of these possibilities, the work presented here, when taken together, provide the strongest evidence to date that DUF1220 domains have influenced anthropoid brain expansion via increased neural stem cell proliferation.
Recent evidence has suggested that primate brain expansion is primarily due to unique scaling of neuronal constituents[5]. Specifically, gains in brain mass in the primate species are associated with far greater increases in neuron number than are found in other lineages. Consistent with this, the evolutionary expansion of DUF1220 copy number, and the NBPF gene family in which they primarily exist13, is confined to the anthropoid suborder of primates and closely parallels increases in neuron number among these species[2, 4]. Indeed, the work presented here provides additional support to the view that human evolutionary brain expansion is primarily a neuron number-driven phenomenon, and points to increases in DUF1220 domain dosage as a possible key driver behind this process.
Supplementary Material
References
- 1.Popesco M, Maclaren E, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff G, Sikela J. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313(5791):1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 2.O’Bleness M, Dickens C, Dumas L, Kehrer-Sawatzki H, Wyckoff G, Sikela J. Evolutionary history and genome organization of DUF1220 protein domains. G3. 2012;2(9):977–986. doi: 10.1534/g3.112.003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumas L, Sikela J. DUF1220 domains, cognitive disease, and human brain evolution. Cold Spring Harbor Symposia on Quantitative Biology. 2009;74:375–382. doi: 10.1101/sqb.2009.74.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumas L, O’Bleness M, Davis J, Dickens C, Anderson N, Keeney J, Jackson J, Sikela M, Raznahan A, Giedd J, Rapoport J, Nagamani S, Erez A, Brunetti-Pierri N, Sugalski R, Lupski J, Fingerlin T, Cheung S, Sikela J. DUF1220-domain copy number implicated in human brain-size pathology and evolution. American Journal of Human Genetics. 2012;91(3):444–454. doi: 10.1016/j.ajhg.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herculano-Houzel S. The human brain in numbers: a linearly scaled-up primate brain. Frontiers in Human Neuroscience. 2009 doi: 10.3389/neuro.09.031.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunetti-Pierri N, Berg J, Scaglia F, Belmont J, Bacino C, Sahoo T, Lalani S, Graham B, Lee B, Shinawi M, Shen J, Kang S, Pursley A, Lotze T, Kennedy G, Lansky-Shafer S, Weaver C, Patel A. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nature Genetics. 2008;40(12):1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mefford H, Sharp A, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney V, Crolla J, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers E, de Leeuw N, Eichler E. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. New England Journal of Medicine. 2008;359(16):1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogart S, Bennett A, Shapiro S, Reamer L, Hopkins W. Different early rearing experiences have long-term effects on cortical organization in captive chimpanzees (Pan troglodytes) Developmental Science. 2013 doi: 10.1111/desc.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jørgensen S, Baker A, Møller S, Nielsen B. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods. 2010;52(4):375–381. doi: 10.1016/j.ymeth.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen B, Jørgensen S, Fog J, Søkilde R, Christensen I, Hansen U, Brünner N, Baker A, Møller S, Nielsen H. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clinical and Experimental Metastasis. 2011;28(1):27–38. doi: 10.1007/s10585-010-9355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends in Neurosciences. 1995;18(9):383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- 12.Noctor S, Martínez-Cerdeño V, Kriegstein A. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. The Journal of Comparative Neurology. 2008;508(1):28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandepoele K, Van Roy N, Staes K, Speleman F, van Roy F. A novel gene family NBPF: Intricate structure generated by gene duplications during primate evolution. MolBiolEvol. 2005;22:2265–2274. doi: 10.1093/molbev/msi222. [DOI] [PubMed] [Google Scholar]
- 14.Rilling J, Insel T. The primate neocortex in comparative perspective using magnetic resonance imaging. Journal of Human Eution. 1999;37(2):191–223. doi: 10.1006/jhev.1999.0313. [DOI] [PubMed] [Google Scholar]
- 15.Bayer S, Altman J, Russo R, Zhang X. Timetables of neurogenesis in the human brain based on experimentally determined patterns in the rat. Neurotoxicology. 1993;14:83–144. [PubMed] [Google Scholar]
- 16.Howdeshell K. A model of the development of the brain as a construct of the thyroid system. Environ Health Perspect. 2002;110:337–348. doi: 10.1289/ehp.02110s3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tau G, Peterson B. Normal development of brain circuits. Neuropsychopharmacology. 2010;35:147–168. doi: 10.1038/npp.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.