Abstract
Association studies have shown and continue to show a substantial amount of success in identifying links between multiple single nucleotide polymorphisms (SNPs) and phenotypes. These studies are also believed to provide insights toward identification of new drug targets and therapies. Albeit of all the success, challenges still remain for applying and prioritizing these associations based on available biological knowledge. Along with single variant association analysis, genetic interactions also play an important role in uncovering the etiology and progression of complex traits. For gene-gene interaction analysis, selection of the variants to test for associations still poses a challenge in identifying epistatic interactions among the large list of variants available in high-throughput, genome-wide datasets. Therefore in this study, we propose a pipeline to identify interactions among genetic variants that are associated with multiple phenotypes by prioritizing previously published results from main effect association analysis (genome-wide and phenome-wide association analysis) based on a-priori biological knowledge in AIDS Clinical Trials Group (ACTG) data. We approached the prioritization and filtration of variants by using the results of a previously published single variant PheWAS and then utilizing biological information from the Roadmap Epigenome project. We removed variants in low functional activity regions based on chromatin states annotation and then conducted an exhaustive pairwise interaction search using linear regression analysis. We performed this analysis in two independent pre-treatment clinical trial datasets from ACTG to allow for both discovery and replication. Using a regression framework, we observed 50,798 associations that replicate at p-value 0.01 for 26 phenotypes, among which 2,176 associations for 212 unique SNPs for fasting blood glucose phenotype reach Bonferroni significance and an additional 9,970 interactions for high-density lipoprotein (HDL) phenotype and fasting blood glucose (total of 12,146 associations) reach FDR significance. We conclude that this method of prioritizing variants to look for epistatic interactions can be used extensively for generating hypotheses for genome-wide and phenome-wide interaction analyses. This original Phenome-wide Interaction study (PheWIS) can be applied further to patients enrolled in randomized clinical trials to establish the relationship between patient’s response to a particular drug therapy and non-linear combination of variants that might be affecting the outcome.
Keywords: PheWAS, PheWIS, genetic interactions, Epistasis, ENCODE, Roadmap Epigenome, Pharmacogenomics, Clinical Trials, Annotations, prior biological knowledge
1. Introduction
Investigating the precise response of antiretroviral therapies given to patients is an important area of research. Previous studies have discovered interesting single gene effects as well as genetic interaction effects associated with response to anti-retroviral medications1, 2 in the AIDS Clinical Trials Group (ACTG) data (https://actgnetwork.org/). A recently published Phenome-wide association study (PheWAS)2 showed a number of variants associated with a list of 27 highly curated and transformed (for normal distribution) phenotypes collected in baseline model of AIDS clinical trials3, 4. Thus, this unique clinical trials dataset and the analyses performed earlier provide a backbone for performing epistatic interactions analyses among variants and genes that might be associated with multiple drug response phenotypes.
A wealth of data are being generated from speedy advancements in genotyping and sequencing technologies, thus providing opportunities to investigate not only single gene effects but also non-linear combined genetic effects of these variants. Genome wide association studies (GWAS) have been proven to detect many SNPs associated with multiple diseases or traits. These variants discovered by GWAS can only explain small proportion of genetic risk corresponding to the problem of “missing heritability”5. One conceivable explanation of missing heritability is the existence of genetic interactions or epistasis5 and the evidence for genetic interactions has been observed in both humans and model organisms6. Efficient identification of epistatic interactions is also an important biological problem because unlike GWA studies, gene-gene interaction studies are not yet fully equipped to produce reproducible results most importantly due to the combinations of pairwise models that are generated from each individual study. Additionally, testing for two or multi-way interactions still remains a challenge due to overhead of computing resources and also due to correction for false positives for each test performed. Thus, filtration of variants based on prior biological knowledge is used frequently in the search for epistasis7. Many studies have shown that filtration of variants based on strong and marginal main effects as determined by the data can be useful in detecting interactions8. Combining the main effect filtration method along with filtration based on prior-biological knowledge has also been proven to increase the power to detect epistatic interactions9–11.
The Roadmap Epigenome has provided high-resolution genome wide interaction maps based on the chromatin accessibility, histone modifications, DNA methylation and mRNA expression across 127 epigenomes12, 13. These data can be used as a great resource of prior biological information for filtering variants based on the activity of the genomes as defined by chromatin states14. Annotations of variants associated with disease traits from the NHGRI GWAS Catalog15 have shown that 81% of variants associated with a disease can be annotated into one of the functional regulatory elements using ENCODE data where functional here refers to any biochemical activity as identified from at least one of the cell lines from ENCODE 12, 14. Roadmap epigenome data is collected from an even larger list of epigenomes and thus provide an extensive and more detailed map of regulatory activity of the genome.
In this study, we intended to use this extensive knowledge about regulatory elements as criteria to filter variants based on their functional activity before performing interaction testing rather than the more traditional approach of prioritizing variants based on their activity after conducting analysis. This will reduce the multiple testing burden and increase interpretability. In the remaining sections, we explain our proposed analytic pipeline for Phenome-wide interaction study (PheWIS), its application to the pre-treatment ACTG datasets, and a series of highly significant gene-gene interactions associated with baseline clinical variables. We show that combination of biological knowledge and main effect filtering provides a high-throughput, comprehensive pipeline to address the architecture of complex traits. This method can clearly be applied to patients from on-treatment imminent clinical trial data to generate hypothesis for epistatic gene-gene interactions that could influence drug response and treatment design.
2. Materials and Methods
2.1 Genotype and Phenotype data
ACTG data from treatment-naïve patients has been previously reported16–20. We used the same dataset as described in the pilot PheWAS conducted on ACTG data that consisted of 27 pre-treatment laboratory measurements (shown in supplementary table 1 at ritchielab.psu.edu/publications/supplementary-data/psb-2016/phewis) that have been normalized by appropriate transformations. From all 27 phenotypes, 26 were used as independent variables and one phenotype (CD4 T-cell counts) was used as a covariate due to its known confounding effect in HIV patients21. This dataset consisted of 2547 genotyped participants which were imputed in three phases based on a separate immunogenomics project22. Phase I and II were combined together (Discovery dataset), which consisted of 1366 samples and Phase III consisted of 1181 samples (Replication dataset) as described in detail in pilot PheWAS2. Supplementary Table 2 Lists the information on samples used in both the discovery and replication dataset along with the demographic information on these samples.
2.2 Annotation and Filtration of variants
The pilot PheWAS analysis reported 10,584 variants that replicated at p-value <0.01 with the same direction of effect across two datasets. We took all of these variants that passed the replication criteria in the pilot PheWAS and annotated them using Biofilter23. Biofilter is a unified framework that consists of data from multiple resources such as KEGG, GENCODE, RegulomeDB, etc. We added Roadmap Epigenome posterior probability data for 25 chromatin states averaged across all 127 epigenomes as a new source to Biofilter. We annotated variants with the help of Biofilter by specifying the Roadmap Epigenome as the single source to be used to annotate variants in order to remove any redundancy from similar sources such as RegulomeDB24 or HaploReg25 which also contain data from ENCODE project12.
We used the 25-state chromatin models data published on the Roadmap epigenome website (http://egg2.wustl.edu/roadmap/web_portal/imputed.html#chr_imp). The roadmap epigenome posterior probability raw data is a map of the human genome where the genome is divided into 200 base pair regions (chunks) and thus there are 15,478,375 total numbers of chunks of the genome (for human genome build 37) for which probabilities for each 25 states are provided. We combined posterior probabilities from 127 epigenomes (tissues/cell types) in Roadmap Epigenome data by doing an average across all values to calculate posterior probability of each state for each 200bp region. State with highest probability was then assigned to each region. Careful investigation of these data suggested that many consecutive chunks are annotated as the same chromatin states. Thus we dynamically combined chunks together to yield a larger contiguous region of the genome, thereby reducing the total number of chunks. In order to combine the consecutive chunks, we used a rule of 80% where the two chunks were combined and annotated as the same state if the probability of the same state in consecutive chunk is 80% or greater.
To get an estimate of the total number of regions for each chromatin state in a genome-wide study, we choose to look at approximately 5M variants from Illumina Omni5 platform as that is one of the largest genotyping chips. Table 2 provides an overall estimate of each chromatin state and the total number of regions combined dynamically for all variants genotyped on Illumina Omni5 chip (http://www.illumina.com/products/humanomni5-quad_beadchip_kit.html). We picked the Omni5 chip to show a large number of variants that can be covered with their respective chromatin states from the genotyping chips available. To get a better overview of variants on genotyping chips that are known to be associated with a disease using NHGRI GWAS catalog15, we also mapped these variants on Omni 5 chip to GWAS catalog (accessed May 2014) using Library of Knowledge Integration (LOKI) database in Biofilter and looked at how all variants in each state are associated with one or more disease from GWAS catalog. Table 2 also represents the number of times each chromatin state is represented in the NHGRI GWAS catalog as being associated with a disease.
Table 2.
Estimate of chromatin states from Illumina Omni5 genotyping chip and number of chromatin states in variants mapping to GWAS catalog that are associated with a disease. Here each 200 base pair region of the genome is combined together dynamically when the next region is represented as same state with at least 80% posterior probability.
| State | Description | #Occurrences in
Omni5 Chip |
#Occurrences in NHGRI GWAS Catalog |
|---|---|---|---|
| S1 | Active TSS | 6803 | 18 |
| S2 | Promoter Upstream TSS | 21901 | 51 |
| S3 | Promoter Downstream TSS 1 | 22854 | 65 |
| S4 | Promoter Downstream TSS 2 | 9007 | 24 |
| S5 | Transcribed 5' preferential | 90330 | 175 |
| S6 | Strong transcription | 42687 | 102 |
| S7 | Transcribed 3' preferential | 225664 | 449 |
| S8 | Weak transcription | 207773 | 404 |
| S9 | Transcribed Regulatory (Prom/Enh) | 15920 | 47 |
| S10 | Transcribed 5' preferential and Enh | 15170 | 40 |
| S11 | Transcribed 3' preferential and Enh | 9022 | 25 |
| S12 | Transcribed weak Enhancer | 19313 | 46 |
| S13 | Active Enhancer 1 | 7318 | 23 |
| S14 | Active Enhancer 2 | 6947 | 24 |
| S15 | Active Enhancer Flank | 10350 | 23 |
| S16 | Weak Enhancer 1 | 8878 | 25 |
| S17 | Weak Enhancer 2 | 18104 | 44 |
| S18 | Primary H3K27ac possible Enhancer | 895 | 5 |
| S19 | Primary DNAase | 19959 | 48 |
| S20 | ZNF genes and repeats | 9211 | 11 |
| S21 | Heterochromatin | 32644 | 40 |
| S22 | Poised Promoter | 3808 | 12 |
| S23 | Bivalent Promoter | 12285 | 35 |
| S24 | Repressed Polycomb | 69906 | 189 |
| S25 | Quiescent/Low | 3289868 | 5565 |
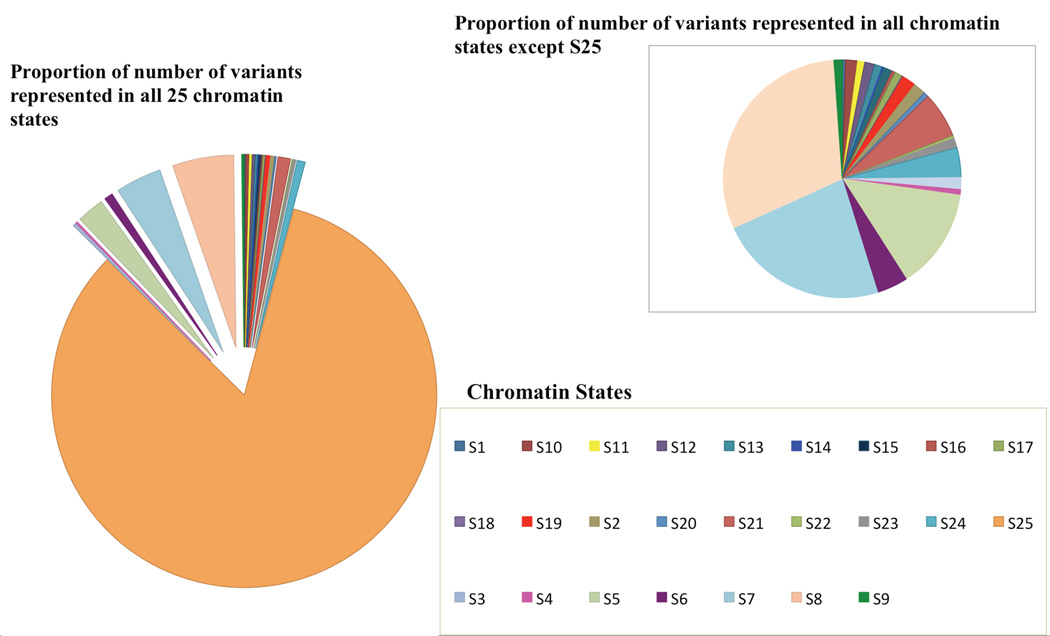
10,584 variants from the pilot PheWAS were annotated using the same approach described above. Figure 1 shows the proportion of variants in each of the 25 states. To filter these variants based on the activity of each region (corresponding to chromatin states), we removed any variants that fell in Chromatin State 25 (Quiescent/Low State) because as described in Roadmap epigenome, predominantly most of the inactive regions fall under quiescent state (approximately 40% of inactive region) and this state is represented on an average in 68% of the genome13. This annotation followed by filtration step resulted in 1773 variants that were further considered for association testing.
Figure 1.
Distribution of all 25 chromatin states in 10,584 SNPs from the pilot PheWAS study (on left) and the proportions of variants used in PheWIS (on right)
2.3 Statistical Analysis
To test for pairwise interactions among 1773 annotated variants in both discovery and replication datasets, all variants were encoded as additive where risk incurred by heterozygous alternate allele is half the risk incurred by homozygous alternate alleles. We ran linear regression where a reduced model consisted of main effects of all variants adjusted by covariates and a full model consisted of main effects and an interaction term for each pairwise SNP-SNP model adjusted by covariates. A likelihood ratio test was conducted to obtain the significance of the interaction effect above and beyond the main effect of each variant. Below is the mathematical description for the reduced and full model:
| (1) |
| (2) |
| (3) |
We used PLATO (http://ritchielab.psu.edu/software/plato-download) to conduct PheWIS in both discovery and replication datasets where all 26 phenotypes were calculated simultaneously for each pairwise interaction model. We adjusted the analysis by age, gender, CD4 T-cell count (square root) and first 5 principal components (to account for genetic ancestry). We also calculated Bonferroni and FDR based corrected p-values26, 27 for each model tested. Here the models are adjusted for all pairwise combination of variants and all phenotypes (40,842,828 tests). We ran the regression analyses separately for discovery and replication datasets and then looked for each pairwise combination of SNPs associated with the same phenotype to determine if results were replicating across the two independent datasets.
3. Results
Annotation of all 10,584 variants from the pilot PheWAS analysis showed that the majority of variants represent state 25 (S25; Quiescent/Low) as shown in Figure 1.Variants detected from GWAS are highly enhanced in regulatory regions as illustrated in Table 2 where a large number of variants are represented in all 25 states but the majority of variants associated with a disease represent the most inactive state “S25”. Since a large proportion of variants known to be associated from GWA studies only represent small proportion of genetic risk28 and one of the biggest challenges is in understanding the role of the majority of these variants29. Therefore, prioritizing variants based on the affect that they can impose on gene regulation is a crucial step in understanding the associations between variants and phenotypes. We aimed this study to focus on only variants that are represented in more active states (with state 1 being the most active and state 25 being the least active) with the potential for a larger proportion of variance to be explained by these variants. A total of 50,798 SNP-SNP pair and phenotype results replicate at p-value<0.01. In order to adjust for multiple testing burden and to reduce false positives, we required replication between the two datasets based on Bonferroni adjusted p-value and False Discovery Rate (FDR) adjusted p-value26, 27, 30. A total of 2,176 results replicate for just one phenotype (fasting glucose) based on Bonferroni based correction and 12,146 results replicated for two phenotypes: fasting glucose and high density lipoprotein (HDL), for FDR based correction of p-values. We used Biofilter to again annotate the position of these variants with chromatin states and then further annotate each SNP from SNP-SNP pairs with genes. SNPs are annotated as genes where the position of a SNP falls within gene boundaries. Therefore, more than one SNP can be annotated to same genes. Table 3 presented the distribution of variants from Bonferroni and FDR based results for each of the 24-chromatin states. We also looked at the expression of top genes in various tissues using GTEx portal31. For HDL results, we looked for expression in adipose and liver tissue and for fasting glucose; we looked for expression in the pancreas.
Table 3.
Occurrences of Bonferroni and FDR corrected results in all 24 chromatin states
| State | Description | #Occurrences in Bonferoni corrected results |
#Occurrences in FDR corrected results |
|---|---|---|---|
| S1 | Active TSS | 1 | 4 |
| S2 | Promoter Upstream TSS | 6 | 15 |
| S3 | Promoter Downstream TSS 1 | 3 | 14 |
| S4 | Promoter Downstream TSS 2 | 3 | 7 |
| S5 | Transcribed 5' preferential | 33 | 118 |
| S6 | Strong transcription | 5 | 24 |
| S7 | Transcribed 3' preferential | 38 | 183 |
| S8 | Weak transcription | 80 | 292 |
| S9 | Transcribed Regulatory (Prom/Enh) | 2 | 4 |
| S10 | Transcribed 5' preferential and Enh | 2 | 10 |
| S11 | Transcribed 3' preferential and Enh | 6 | 7 |
| S12 | Transcribed weak Enhancer | 1 | 11 |
| S13 | Active Enhancer 1 | 6 | 12 |
| S14 | Active Enhancer 2 | 0 | 2 |
| S15 | Active Enhancer Flank | 2 | 9 |
| S16 | Weak Enhancer 1 | 1 | 1 |
| S17 | Weak Enhancer 2 | 0 | 6 |
| S18 | Primary H3K27ac possible Enhancer | 1 | 2 |
| S19 | Primary DNAase | 2 | 8 |
| S20 | ZNF genes and repeats | 2 | 5 |
| S21 | Heterochromatin | 7 | 34 |
| S22 | Poised Promoter | 0 | 1 |
| S23 | Bivalent Promoter | 4 | 20 |
| S24 | Repressed Polycomb | 10 | 27 |
We also mapped all SNP-SNP pairs to genes using Biofilter. Bonferroni significant results consisted of 212 unique genes that were mapped to 66 genes and FDR-based significant results consisted of 690 unique SNPs that represent 245 unique genes. Details of all replicated results can be found in supplementary material online (supplementary table 3 and 4 at ritchielab.psu.edu/publications/supplementary-data/psb-2016/phewis).
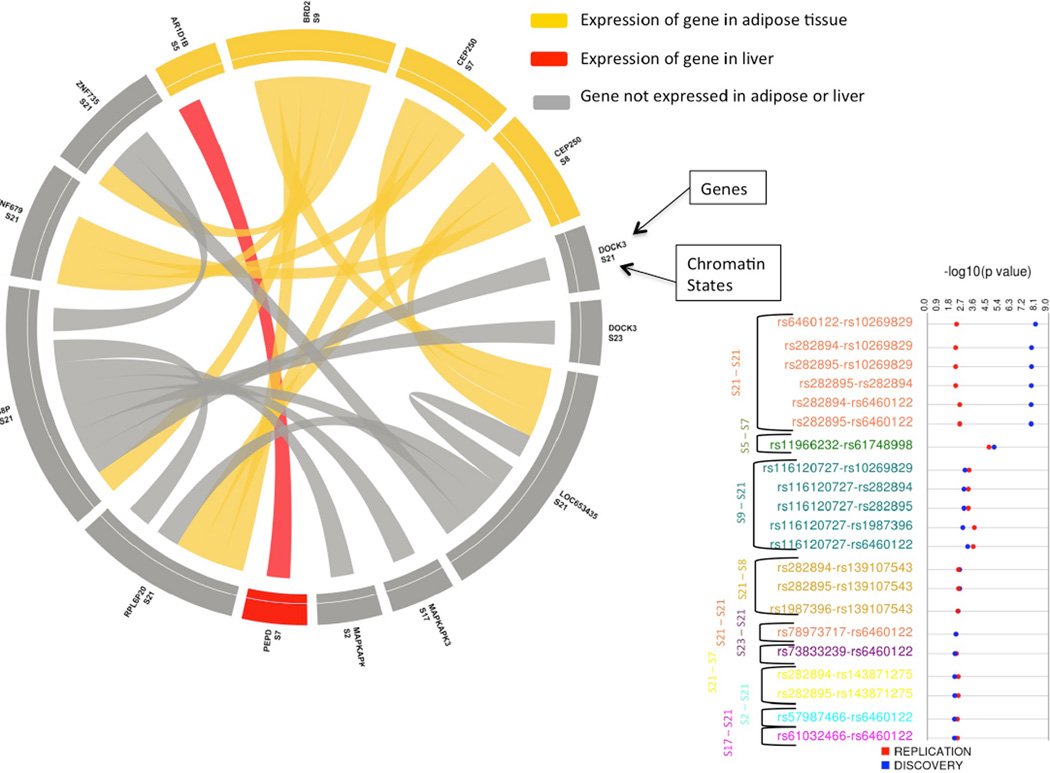
Figure 2 represents the top 30 results for fasting glucose that are less than Bonferroni corrected p-value 0.01. Each SNP-SNP pair and their corresponding genes are shown along with −log10 (p-value) track for both Discovery and Replication datasets. Interactions among the specific chromatin states that the SNP falls under are shown on the right side. Six unique gene-gene pairs are also expressed in the pancreas. Figure 3 shows a circular plot for HDL providing the interaction between the SNPs in the genes and the states that the SNPs represent. The genes are colored based on the tissue that they are expressed in. Figure 3 also represents the FDR corrected p-values for each SNP-SNP interaction pairs. For details on all results that were replicated, please refer to supplementary material online (supplementary table 3 and 4 at ritchielab.psu.edu/publications/supplementary-data/psb-2016/phewis)
Figure 2.
Synthesis-view plot (http://visualization.ritchielab.psu.edu/synthesis_views/plot) illustrating interactions among top 30 SNP-SNP pair for fasting glucose phenotype. Different color for text corresponds to the combination of chromatin states that SNP-SNP pairs are mapped to as represented on the right axis.
Figure 3.
Circular plot representing interactions of SNP-SNP pair combined based on the genes and the chromatin states represented for HDL phenotype. Yellow color corresponds to the expression of gene in adipose tissue, red color corresponds to expression of gene in liver tissue and grey color corresponds to expression on gene in neither adipose nor liver tissues. Lines show the interactions between the variants in the genes and corresponding states. On right, showing a synthesis view plot where FDR p-values of both discovery and replication dataset for each pair SNP-SNP interactions representing unique gene and chromatin state is represented. Color for SNP-SNP pair corresponds to different combinations of interactions among chromatin states
4. Discussion
This study presents a pilot Phenome-wide Interaction study (PheWIS), which is the first of its kind, in the AIDS Clinical Trials Group data. With the help of statistical methods to detect genetic interactions associated with one or multiple phenotypes, we showed significant interactions for SNPs mapped to different chromatin states. The purpose of this study is aimed at mimicking the regulatory genetic networks by showing how interactions between two different chromatin states impacted by genetic variants are associated with a trait. In this paper, we used a-priori biological information from Roadmap Epigenome data to test for variants that represent active chromatin states. Among the top associations with Bonferroni p-value<0.01 are the interactions between SEH1L gene and RCL1 gene to be associated with fasting glucose. Interactions between these two genes are represented by two top-most SNP-SNP interaction pair as shown in Figure 2. In these interactions, the three-chromatin states represented are S3 (Promoter Downstream TSS 1), S5 (Transcribed 5’ preferential) and S8 (Weak Transcription), which suggests interactions among transcribed regions that could be of potential interest. SEH1L gene participates in the regulation of glucose transport process (GO:0010827) and functional studies in yeast have shown that growth of yeast on glucose media requires function RCL132. PheWIS aims at identifying interactions among variants above and beyond the main effects of individual variant. Thus, with this approach we are able to identify several known and novel interactions that could not be identified with PheWAS alone.
The majority of interactions in the FDR corrected results for HDL show interactions among chromatin state 21 (S21; Heterochromatin) and other states. In Roadmap epigenome data, heterochromatin state is mostly represented by constitutive heterochromatin and heterochromatin state is highly tissue specific13. Since in this analysis, we combined data from all cell lines to represent all 25 chromatin states, nothing can be said about the heterochromatin in adipose or liver cell lines. Thus, suggesting that in the future, more work would be required to look at these polymorphic regions based on the tissue that phenotype is affecting or the tissue using which the study samples are collected. For the HDL PheWIS results, one potential interesting interaction is between ARID1B and PEPD genes. Peptidase D (PEPD) and ARID1B genes have been known to be associated with HDL33–35. Both of these genes are highly expressed in adipose tissue with PEPD being also highly expressed in liver.
There are few limitations in this study. Although after correcting for multiple testing based on Bonferroni and FDR methods, we identified many statistical interactions associated with two phenotypes; future research is required to understand these novel interaction associations. Next, all these results are based on treatment naïve patients enrolled in clinical trials, similar analysis in post-treatment quantitative phenotypes can help explore more associations that are linked to the side-effects presented by drugs as well as the benefits of the drug given to patients. Our approach is based on averaging across 127 epigenomes from Roadmap data to annotate regions of the genome. With this approach, we might have missed useful information on chromatin states that are specific to just one tissue type. Future studies can be focused on tissue specific annotation approach or a more comprehensive approach where annotations for an active region can be from any one tissue as well rather than average across all tissues. Lastly, we only excluded the variants that were mapped to state 25 from Roadmap epigenome data whereas future studies could also focus on excluding variants that are under represented in more than one states and only including the variants that map to states which are over-represented in our data.
5. Conclusions
We present the first phenome-wide SNP-SNP interaction study in a pharmacogenomics dataset. Though this study is on treatment naïve patients, it presents a great framework to look for statistical epistasis in a large number of phenotypes, which are collected post treatment. Most of the interactions associated with traits in this study are novel and would require more extensive future work to understand if any of these associations explain biological processes that are also linked to one or more phenotypes. Methods such as the one proposed for PheWIS will enable researchers to investigate more territory in the etiology of complex traits.
Supplementary Material
Acknowledgements
This project was supported by Award Number U01AI068636 from the National Institute of Allergy and Infectious Diseases and supported by National Institute of Mental Health (NIMH), National Institute of Dental and Craniofacial Research (NIDCR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. Grant support included AI077505, TR000445, AI054999 (DWH), HG006385, HL065962 (MDR).Study drugs were provided by Bristol-Myers Squibb Company (Princeton, NJ), Gilead Sciences (Gilead Sciences, Inc., Foster City, CA), GlaxoSmithKline, Inc (Research Triangle Park, NC), and Boehringer Ingelheim (Ridgefield, CT).Clinical Research Sites that participated in ACTG protocols ACTG 384, A5095, A5142, or A5202, and collected DNA under protocol A5128, were supported by the following grants from NIH National Institute of Allergy and Infectious Diseases (NIAID): AI069532, AI069484, AI069432, AI069450, AI069495, AI069434, AI069424, AI069439, AI069467, AI069423, AI069513, AI069477, AI069465, AI069419, AI069502, AI069474, AI069472, AI069501, AI069418, AI069494, AI069471, AI069511, AI069452, AI069428, AI069556, AI069415, AI032782, AI046376, AI046370, AI038858, AI034853, AI027661, AI025859, AI069470, AI027675, AI073961, AI050410, AI045008, AI050409, AI072626, AI069447, AI027658, AI027666, AI058740, and AI025868, and by the following grants from NIH National Center for Research Resources (NCRR): RR000046, RR000425, RR025747, RR025777, RR025780, RR024996, RR024160, RR023561, RR024156, RR024160, and RR024160.
References
- 1.Motsinger AA, et al. Multilocus genetic interactions and response to efavirenz-containing regimens: an adult AIDS clinical trials group study. Pharmacogenet. Genomics. 2006;16:837–845. doi: 10.1097/01.fpc.0000230413.97596.fa. [DOI] [PubMed] [Google Scholar]
- 2.Moore CB, et al. Phenome-wide Association Study Relating Pretreatment Laboratory Parameters With Human Genetic Variants in AIDS Clinical Trials Group Protocols. Open Forum Infect. Dis. 2014;2 doi: 10.1093/ofid/ofu113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holzinger ER, et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet. Genomics. 2012;22:858–867. doi: 10.1097/FPC.0b013e32835a450b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rotger M, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin. Pharmacol. Ther. 2007;81:557–566. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 5.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 6.Evans DM, Marchini J, Morris AP, Cardon LR. Two-stage two-locus models in genome-wide association. PLoS Genet. 2006;2:e157. doi: 10.1371/journal.pgen.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritchie MD. Using biological knowledge to uncover the mystery in the search for epistasis in genome-wide association studies. Ann. Hum. Genet. 2011;75:172–182. doi: 10.1111/j.1469-1809.2010.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, et al. Analysis pipeline for the epistasis search - statistical versus biological filtering. Front. Genet. 2014;5:106. doi: 10.3389/fgene.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L, et al. Knowledge-driven analysis identifies a gene-gene interaction affecting high-density lipoprotein cholesterol levels in multi-ethnic populations. PLoS Genet. 2012;8:e1002714. doi: 10.1371/journal.pgen.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grady BJ, et al. Use of biological knowledge to inform the analysis of gene-gene interactions involved in modulating virologic failure with efavirenz-containing treatment regimens in ART-naïve ACTG clinical trials participants. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 2011:253–264. doi: 10.1142/9789814335058_0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner SD, et al. Knowledge-Driven Multi-Locus Analysis Reveals Gene-Gene Interactions Influencing HDL Cholesterol Level in Two Independent EMR-Linked Biobanks. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0019586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbins GK, et al. Comparison of sequential three-drug regimens as initial therapy for HIV-1 infection. N. Engl. J. Med. 2003;349:2293–2303. doi: 10.1056/NEJMoa030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulick RM, et al. Triple-nucleoside regimens versus efavirenz-containing regimens for the initial treatment of HIV-1 infection. N. Engl. J. Med. 2004;350:1850–1861. doi: 10.1056/NEJMoa031772. [DOI] [PubMed] [Google Scholar]
- 18.Gulick RM, et al. Three- vs four-drug antiretroviral regimens for the initial treatment of HIV-1 infection: a randomized controlled trial. JAMA. 2006;296:769–781. doi: 10.1001/jama.296.7.769. [DOI] [PubMed] [Google Scholar]
- 19.Riddler SA, et al. Class-sparing regimens for initial treatment of HIV-1 infection. N. Engl. J. Med. 2008;358:2095–2106. doi: 10.1056/NEJMoa074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daar ES, et al. Atazanavir plus ritonavir or efavirenz as part of a 3-drug regimen for initial treatment of HIV-1. Ann. Intern. Med. 2011;154:445–456. doi: 10.1059/0003-4819-154-7-201104050-00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crampin A, Mwaungulu F, Ambrose L, Longwe H, French N. Normal Range of CD4 Cell Counts and Temporal Changes in Two HIVNegative Malawian Populations. Open AIDS J. 2011;5:74–79. doi: 10.2174/1874613601105010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International HIV Controllers Study et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pendergrass SA, et al. Genomic analyses with biofilter 2.0: knowledge driven filtering, annotation, and model development. BioData Min. 2013;6:25. doi: 10.1186/1756-0381-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gronostajski RM, Guaneri J, Lee DH, Gallo SM. The NFI-Regulome Database: A tool for annotation and analysis of control regions of genes regulated by Nuclear Factor I transcription factors. J. Clin. Bioinforma. 2011;1:4. doi: 10.1186/2043-9113-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y. Discovering the false discovery rate. J. R. Stat. Soc. Ser. B Stat. Methodol. 2010;72:405–416. [Google Scholar]
- 28.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visscher PM, Brown MA, McCarthy MI, Yang J. Five Years of GWAS Discovery. Am. J. Hum. Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhandari M, et al. The risk of false-positive results in orthopaedic surgical trials. Clin. Orthop. 2003:63–69. doi: 10.1097/01.blo.0000079320.41006.c9. [DOI] [PubMed] [Google Scholar]
- 31.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horn DM, Mason SL, Karbstein K. Rcl1 Protein, a Novel Nuclease for 18 S Ribosomal RNA Production. J. Biol. Chem. 2011;286:34082–34087. doi: 10.1074/jbc.M111.268649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Global Lipids Genetics Consortium et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Q-Z, et al. Sex-specific association of the peptidase D gene rs731839 polymorphism and serum lipid levels in the Mulao and Han populations. Int. J. Clin. Exp. Pathol. 2014;7:4156–4172. [PMC free article] [PubMed] [Google Scholar]
- 35.Govindaraju DR, et al. Genetics of the Framingham Heart Study Population. Adv. Genet. 2008;62:33–65. doi: 10.1016/S0065-2660(08)00602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.