Abstract
The biochemical mechanism by which mutations in nucleotide-binding oligomerization domain containing 2 (NOD2) cause Blau syndrome is unknown. Several studies have examined the effect of mutations associated with Blau syndrome in vitro, but none have looked at the implication of the mutations in vivo. To test the hypothesis that mutated NOD2 causes alterations in signaling pathways downstream of NOD2, we created a Nod2 knock-in mouse carrying the most common mutation seen in Blau syndrome, R314Q (corresponding to R334Q in humans). The endogenous regulatory elements of mouse Nod2 were unaltered. R314Q mice showed reduced cytokine production in response to i.p. and intravitreal muramyl dipeptide (MDP). Macrophages from R314Q mice showed reduced NF-κB and IL-6 responses, blunted phosphorylation of MAPKs, and deficient ubiquitination of receptor-interacting protein 2 in response to MDP. R314Q mice expressed a truncated 80 kDa form of NOD2 that was most likely generated by a posttranslational event since there was no evidence for a stop codon or alternative splicing event. Human macrophages from 2 patients with Blau syndrome also showed a reduction of both cytokine production and phosphorylation of p38 in response to MDP, indicating that both R314Q mice and cells from patients with Blau syndrome show reduced responses to MDP. These data indicate that the R314Q mutation when studied with the Nod2 endogenous regulatory elements left intact is associated with marked structural and biochemical changes that are significantly different from those observed from studies of the mutation using over-expression, transient transfection systems.
Introduction
The innate immune system consists of several families of pattern-recognition receptors (PRRs) capable of recognizing conserved constituents of microbial pathogens and triggering inflammatory responses. Nucleotide-binding oligomerization domain containing 2 (Nod2) is a nucleotide-binding and leucine-rich repeat-containing (NLR) family member that recognizes peptidoglycan fragments from bacterial cell walls containing muramyl dipeptide (MDP) (1, 2). Nod2 is composed of 3 domains: a C-terminal leucine-rich repeat (LRR) domain, which is essential for its MDP-sensing ability; a central nucleotide binding and oligomerization domain (NOD), which is important for ATP-dependent self-oligomerization; and two N-terminal caspase recruitment domains (CARD) that participate in protein-protein interactions and induction of subsequent intracellular signaling responses (3). Following recognition of MDP, Nod2 activates the transcription factors NF-kB and MAPKs via well characterized pathways leading to inflammatory responses and release of antimicrobial molecules [reviewed in (4)]. Nod2 plays a pivotal role in host defense in the recognition of bacterial pathogens and single-stranded RNA viruses, induction of autophagy and maintaining homeostasis with commensal bacteria [reviewed in (5)].
The importance of Nod2 in human health is further underscored by the fact that mutations in NOD2 are associated with the chronic inflammatory disorders Crohn’s disease and Blau syndrome (6–8). Given the prevalence of Crohn’s disease and the availability of clinical material to study, the role of Nod2 in this disorder has been extensively studied. NOD2 mutations linked to Crohn’s disease are clustered in the LRR region of the protein and several hypotheses regarding the mechanism of disease have been examined [reviewed in (9)]. A current paradigm proposes that loss of Nod2 function, either in controlling the gut microbiome or regulating TLR responses, is the underlying cause of Crohn’s disease. The inability of MDP to activate forms of Nod2 carrying Crohn’s disease-associated mutations has been observed both in vitro using cells transiently transfected with mutant forms of Nod2 and in macrophages prepared from patients with the disease (2, 10, 11).
In contrast, much less is understood about the mechanism by which mutations in NOD2 cause Blau syndrome, a rare autosomal dominant disorder characterized by granulomatous inflammatory arthritis, dermatitis and uveitis (12). Mutations associated with Blau syndrome are located in the NOD domain of NOD2 and at least 17 different mutations have been identified (12, 13). Transient transfection assays performed in vitro using plasmids with powerful promoters that overexpress NOD2 have found that mutations associated with Blau syndrome cause excessive NF-κB and MAPK activation compared to the wild-type form of NOD2, and this has led to the hypothesis that Blau syndrome is the result of a gain of function of Nod2 (14). However, this gain of function hypothesis has not been supported when clinical specimens from patients with Blau syndrome were analyzed (12, 15–17). PBMCs were not found to have spontaneous release of cytokines, nor were inflammatory responses observed when PBMCs were stimulated with MDP. Moreover, the degree of enhanced Nod2 function seen in vitro does not correlate with the severity of the disease in patients (16). In order to clarify the mechanism by which Blau-syndrome associated mutations alter the function of Nod2, we sought to develop a model not based on over-expression of the gene in vitro.
Here, we created a Nod2 knock-in (KI) mouse by homologous recombination. The mice will be referred to as R314Q mice since they carry one of the most common genetic variants (R314Q, the ortholog of R334Q in humans) associated with Blau syndrome. The regulatory elements for Nod2 expression were not altered by this approach. In this article we show that the R314Q mutation does not lead to a gain of function of NOD2. Rather, the mutation leads to a truncated form of NOD2 and altered cytokine and intracellular signaling responses to MDP both in vivo and in vitro. Macrophages from patients with Blau syndrome also showed reduced cytokine production and intracellular signaling in response to MDP.
Materials and Methods
Construction of the R314Q-Nod2 knock-in mice
A bacterial artificial chromosome (Children’s Hospital Oakland Research Institute, Oakland, CA) containing the murine Nod2 genomic locus was modified by recombineering to create the specific point mutation (G to A change) within codon 314 of Nod2 leading to an arginine (R) to glutamine (Q) amino acid change (codon number as per UniProtKB/Swiss-Prot: Q8K3Z0.1, the 1020 amino acid long isoform of murine NOD2). A second round of recombineering was performed to insert a neomycin resistance gene (Neo) flanked by FLIPPASE recognition target (FRT) sequences in a region of intron 3 showing low homology across several species. A BglII restriction site was placed 5′ to the first FRT site for use in screening. Subcloning from the mutated BAC clone was performed in pBluescript to create a construct with the point mutation, neomycin resistance and “homology arms” 4 kB on either side of the mutation (Fig. 1A, targeting vector). Using the University of Michigan Transgenic Animal Model Core Facility, the construct was electroporated into ES cells and cloned on soft agar containing the selection agent, G418. Successful homologous recombination creating an intermediate locus (5.1 kb) was distinguished from the wild type locus (3.8kb) by Southern blotting with BglII digestion and the indicated probe (Δ) (Fig. 1A). A clone containing the mutation was used for blastocyst injection and ES cell-mouse chimeras were produced. After additional breeding to ensure germline transmission, offspring were bred on an Flp recombinase strain to remove the Neo gene detected by the presence of a 3.1 kb BglII fragment (Fig. 1A, final locus), and then for 10 generations on C57BL/6j (Jackson Laboratory, Bar harbor, ME) to create R314Q heterozygous mice (+/m). Wild type (+/+) litter mates were used as controls for all studies. Brother-sister mating was used to create homozygous R314Q mice (m/m). The genotypes of the final lines were confirmed by Southern blot analysis (Fig. 1B) and sequence analysis of genomic DNA (Fig. 1C). Genotyping of +/+, +/m and m/m lines routinely performed by PCR screening of genomic DNA amplified by PCR (Taq DNA Polymerase with Thermopol Buffer, New England BioLabs) using primer: CATCAGAAATTGGCAGTGGA and primer: GCAGGAATATAGCCGGGAAC. The primers span intron 3, giving a wild type amplicon that results in a 221bp fragment and a mutant amplicon that results in a 254bp fragment due to the presence of the FRT sequence remaining after FLP-FRT recombination. Amplicons were analyzed by electrophoresis in a 3% agarose gel. Genotypes were also confirmed every third generation by Southern blot analysis of genomic DNA digested with BglII, electrophoresed in 1% agarose, transferred to a nylon membrane (Roche), crosslinked, then hybridized with a digoxigenin labeled probe and developed as per the protocol recommended by the manufacturer (Roche). The digoxigenin labeled probe corresponds to a region of exon 4 in Nod2 (Fig. 1A, top) and was generated by PCR (primer sequence available upon request). The R314Q locus yields a 3.1 kb restriction fragment while the wild type locus produces a 3.8 restriction fragment (Fig. 1B). NOD2 deficient mice (−/−) were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were maintained under specific pathogen-free (SPF) conditions at the Portland VA Medical Center (PVAMC). All animal studies were performed under protocols approved by the Institutional Animal Care and Use Committee of the PVAMC.
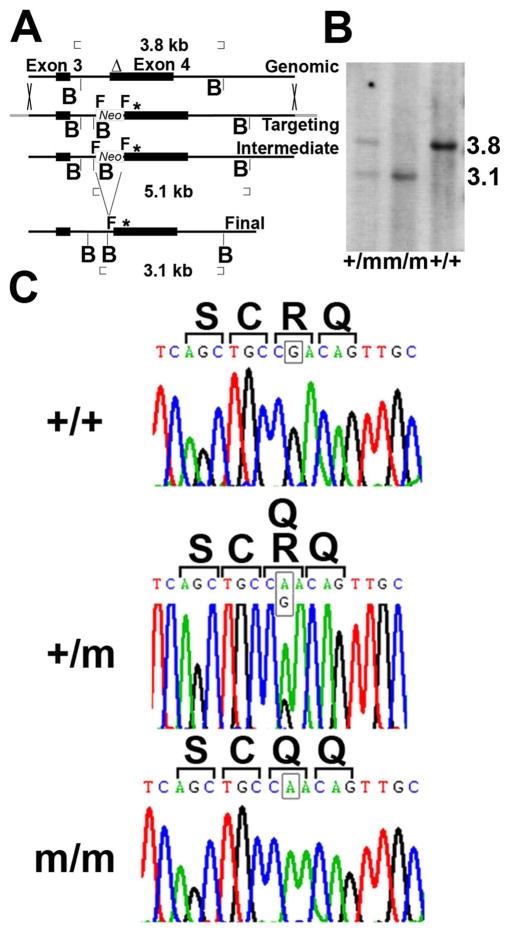
Figure 1.
Generation of R314Q Nod2 knock-in mice. (A) Schematic of genomic, targeting, intermediate and final loci described in materials and methods. Δ, probe for Southern blotting; B, BglII site; * point mutation; F, FRT; Neo, neomycin resistance gene. (B) Southern blot analysis of genomic DNA digested with BglII and hybridized with the probe (Δ) shown in A. +/+, wild type; m/m, homozygous mutant; +/m, heterozygous mutant. (C) Chromatograms from sequence analysis of exon 4 from genomic DNA from +/+, +/m and m/m mice confirming the G to A base change (see squares in each panel) leading to a codon change from R to Q.
Real time and reverse transcriptase (RT) PCR
RNA was isolated (RNeasy, Qiagen, Valencia, CA) from bone marrow derived macrophages (BMDM) and cDNA was synthesized (Superscript VILO, Invitrogen, Grand Island, NY). Real Time PCR for Nod2 and β-actin (Actb) mRNA levels were then performed with iQ Sybr-Green Supermix (BioRad, Hercules, CA) and analyzed according to comparative CT method using Actb as the endogenous control plus comparing the range of expression to untreated cells from wild type mice. RT-PCR to screen for splice variants was also performed by analyzing cDNA from +/+, +/m, and m/m macrophages and looking for changes in PCR size indicating a deletion/truncation (sequence of all primers are available upon request). Nod2 cDNA prepared from macrophages of m/m mice was also fully sequenced and compared to wild type Nod2 from C57BL/6 mice to assess for any changes other than the G to A change intentionally introduced.
Immunoblotting
Cell lysates were prepared in RIPA lysis buffer (50 mM Tris-HCl, pH =7.5, 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% Deoxycholate, 2 mM Na4P2O7, 5 mM NaF, 1 mM Na3VO4) plus proteinase inhibitor cocktails (Roche). SDS-PAGE and protein transfer to PVDF membrane was performed using the manufacturer’s recommended protocols (Invitrogen, Grand Island, NY). Immunoblotting was performed with antibodies to mouse IκBα, p-ERK, p-JNK, mouse p38, mouse p-p38 (Cell Signaling, Danvers, MA), mouse β-actin (Abcam, Cambridge, MA), ubiquitin, RIP2 (Santa Cruz Biotechnology, Santa Cruz CA), and mouse NOD2 (clone 26mNOD2 Ebioscience, San Diego, CA) in PBS plus 5% non-fat milk. HRP conjugated secondary antibodies (Santa Cruz Biotechnology) were detected by ECL (Pierce, Rockford, IL).
Macrophage generation and stimulation
Murine macrophages were generated from bone marrow cells with 30% L929 conditioned media as previously described (18). After 7 days, BMDM were treated overnight with either 100 μg/ml polyinosinic-polycytidylic acid (poly(I:C)) (InvivoGen, San Diego, CA), 10 μg/ml MDP (Bachem), or for 6 hours with 10 ng/ml IFN-γ. Human macrophages were generated from viably frozen PBMCs (obtained under a protocol approved by the Institutional Review Board of the DuPont Hospital for Children and viably frozen until use) from two patients with familial Blau syndrome (a mother and son carrying an R334W mutation). The mother (Blau-1), age 42, and son (Blau-2), age 15, had onset of disease at ages 2 and 1, respectively. Blau-1 had been treated with infliximab in the past and was on methotrexate and adalimumab to control eye disease and arthritis. Blau-2 had been on Anakinra in the past and was on daily Methylprednisolone, Azathioprine and Cyclosporine to control active eye disease. His arthritis was not active. Control macrophages were from viably frozen PBMCs from healthy individuals. PBMCs were incubated in RPMI 1640 containing 20% type AB Normal Human Serum (Atlanta Biologicals, Flowery Branch, GA) and 100 ng/ml GM-CSF (R&D Systems) for 5 days. After 5 days, nonadherent cells were discarded and the attached cells were treated with poly(I:C) (100 μg/ml), MDP (100 μg/ml) or LPS (100 ng/ml, Escherichia coli O55:B5 (Sigma-Aldrich),) for cytokine studies or MDP alone (100 μg/ml) for MAPK activation as described in Fig. 5.
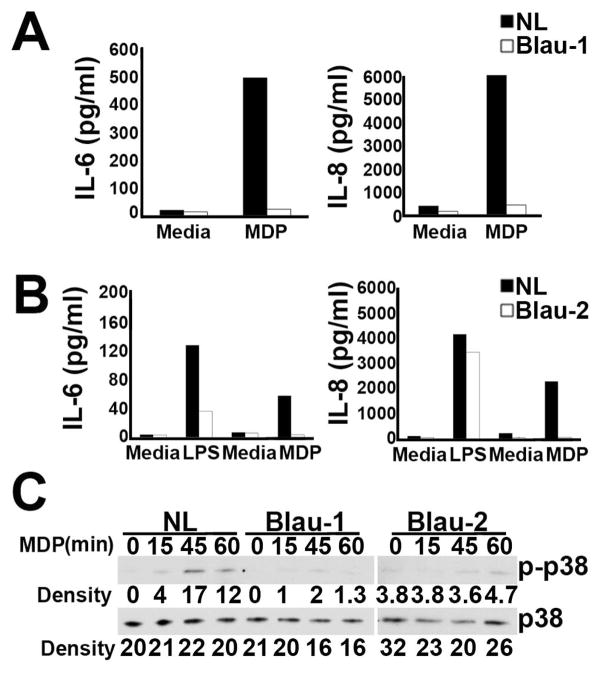
Figure 5.
Monocyte-derived macrophages (MDM) from patients with Blau syndrome have reduced responses to MDP. (A) IL-6 and IL-8 levels in supernatants of MDM from a healthy control (NL) and Blau patient 1 (Blau-1) pre-treated overnight in poly(I:C) followed by treatment for an additional 24 hours in media alone or media supplemented with MDP. (B) IL-6 and IL-8 levels in supernatants of MDM from a different healthy control (NL) and Blau patient 2 (Blau-2) pre-treated overnight in media alone (for subsequent study with LPS) or media supplemented with poly(I:C) followed by treatment for an additional 24 hours in media alone, or media supplemented with LPS or MDP. (C) MDM from a healthy control and Blau patients 1 and 2 were treated with MDP from 0–60 minutes (indicated at the top of each lane) and lysates were analyzed by immunoblot with antibodies for the phosphorylated and non-phosphorylated forms of p38. Blau-1 and Blau-2 were analyzed in different experiments. Densitometry readings of p-p38 and p38 bands are indicated below each lane.
SiRNA
Approximately 4 × 106 BMDM from +/+, +/m, m/m, and −/− mice were resuspended in 100 μl sterile OptiMem (Invitrogen) plus 200 picomoles of ON-TARGETplus Mouse NOD2 SiRNA (Thermo Scientific, #257632) or SiGenome Non-Targeting SiRNA Pool #1 (Thermo Scientific). After gently mixing, the cells were pulsed in a Gene Pulser II electroporation apparatus (BioRad) (400 volts and 150 μF resistance) and cultured for 2 days. BMDM were then treated with poly(I:C) for 24 hours, lysed and then analyzed by Western blot for levels of NOD2 and β-actin protein.
Mass Spectrometry
HEK293 monolayers were transfected with pcDNA3 plasmid (Invitrogen) containing mouse Nod2 cDNA, cultured for 24 hours, lysed and electrophoresed in a 4–12% gradient polyacrylamide gel. A slice of the gel corresponding to the 116 kDa region was excised and analyzed further by the Proteomics Shared Resource at Oregon Health & Science University. Gel slices were reduced with dithiothreitol, alkylated with iodoacetamide and digested overnight with trypsin (19). Samples were then dried, dissolved in 5% Formic Acid, and analyzed by LC-MS/MS (MS2) using an Orbitrap Fusion mass spectrometer (Thermo Scientific) to collect MS2 spectra in data-dependent mode, 75 μm × 50 cm Easy Spray C18 column, 2–30% acetonitrile gradient over 100 min in a mobile phase containing 0.1% formic acid, and 0.3 μl/min flow rate. 22 unique peptides belonging to NOD2 were identified by the program Sequest using a database of mouse protein sequences downloaded from the Swiss-Prot database, and a list of 70 corresponding mass to charge (m/z) values for these peptides was created. This served as the reference library for further analysis of NOD2 expressed in BMDM from +/+ and m/m mice. BMDM from mice of each genotype were lysed, electrophoresed, and gel slices from the 116 kDa (for +/+) and 80 kDa (for m/m) were excised. Tryptic digests from these slices were then screened for the presence of the 22 observed NOD2 peptides using the Orbitrap Fusion under identical chromatography conditions, but in targeted MS2 mode to increase sensitivity for NOD2 peptide detection. Data analysis was performed using the reference library containing the 70 MS2 data assigned to a digest of NOD2 standard and Skyline software (http://www.ncbi.nlm.nih.gov/pubmed/20147306). The fragment ions and retention times for each peptide in the reference library were then compared to the MS2 spectra generated by the targeted MS2 analysis of digest from BMDM from +/+ and m/m mice.
TUBE assay
Evaluation of ubiquitinated proteins was performed using tandem ubiquitin binding entities (TUBE) containing GST tags as per the manufacturer’s instructions (Life Sensor, Malvern, PA). Protein lysate from MDP treated (0–120 minutes) BMDM isolated from +/+, +/m, and m/m mice were used as starting material and were further precipitated with glutathione sepharose (Sigma, St. Louis, MO) to isolate polyubiquitinated proteins, electrophoresed for analysis by immunoblotting with anti-ubiquitin and anti-RIP2 antibodies.
Transient transfection of HEK293T cells
HEK293T cells (CRL-11268, American Type Culture Collection, Manassas, VA) were transiently transfected with empty pcDNA3.1(+) or pcDNA3.1(+) containing the mouse Nod2 cDNA using the TransIT-293 transfection reagent according to manufacturer’s instructions (Mirus Bio, Madison, WI). After transfection, HEK cells were incubated at 37°C for 24 hours, lysed and analyzed by immunoblotting.
Histological evaluation and fundus exams
Eye and joint tissues were harvested at the indicated times and prepared for sectioning and staining with H&E as previously reported (20, 21). Ocular fundus exams were performed under inhalation of isoflurane anesthesia using topical endoscopic fundus imaging (TEFI) as previously described (22–24).
Near-infrared fluorescence imaging
The cathepsin activity within joints was monitored by near-infrared (NIR) fluorescence imaging as previously described (25). Mice were injected with 2 nmole/150 μl volume of ProSense (ViseEN Medical, Woburn, MA). Imaging was performed with the Li-Cor-Biosciences Pearl Imager imaging system with a mousePod attachment. Images were analyzed with LiCor software.
MDP-injection in vivo
Adult mice (8–12 wk old) were administered an i.p. injection of 300 μg/mouse of MDP (Bachem) and serum IL-6, MIP2α, and KC levels were measured 3 hours after injection by ELISA (R&D Systems, Minneapolis, MN). For uveitis experiments, each eye was intravitreally administered either MDP 100 μg/eye (2 μl) or Saline (2 μl) according to previously established protocols (26). KC cytokine levels in the ocular fluid were measured 5 hours after MDP administration.
Statistical analysis
Data are represented as mean +/− standard deviation (SD). Statistically significant differences were determined by a two-tailed Student t test and were considered significant when p values were <0.05.
Results
R314Q-KI mice are refractive to MDP challenge in vivo
In order to better understand the mechanism by which mutations of NOD2 cause alterations in NOD2-signaling pathways, we constructed a KI mouse wherein the most frequent mutation observed in Blau syndrome (R334Q) was inserted into the Nod2 locus of C57BL/6J mice at the orthologous location (R314Q) in exon 4. An important aspect of the homologous recombination approach taken here was that this allowed the endogenous regulatory elements of the mouse Nod2 gene to remain intact and unaltered (Fig. 1A). Southern blot analysis of genomic DNA confirmed the successful recombination of the WT allele (“+”) and mutated (“m”) allele as detected by the 3.8 and 3.1 kb fragments, respectively (Fig. 1B). As shown in Fig. 1C, genetic sequencing further identified the introduction of the point mutation (G to A) within codon 314 of the Nod2 gene, which lead to an arginine (R) to glutamine (Q) amino acid substitution. The KI mice were backcrossed 10 generations onto C57BL/6J background and survived normally and showed the expected Mendelian inheritance pattern for the mutation. We did not observe any gross abnormalities of the skin, inflammatory changes in the eyes, or cathepsin activation indicative of arthritis in mice that were housed in SPF conditions and examined up to 36 weeks of age (supplemental Fig. 1). The cellularity of peripheral blood (white blood cells, lymphocytes, mononuclear cells, granulocytes) as well as albumin, alkaline phosphatase, alanine aminotransferase, amylase, blood urea nitrogen, calcium, creatinine, globulin, glucose, potassium, sodium, phosphorus, total bilirubin, total protein were comparable in +/+, +/m and m/m mice (data not shown).
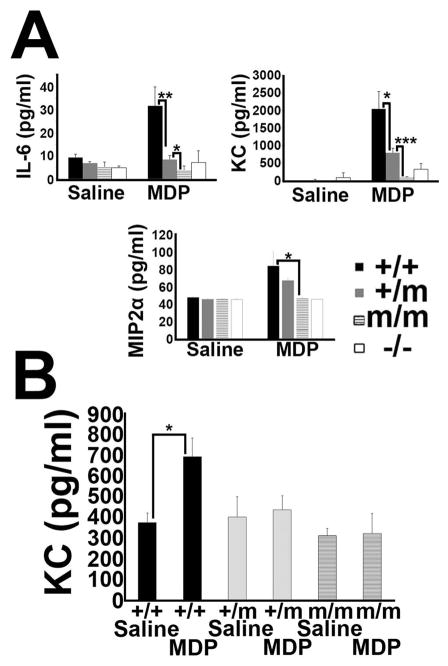
In order to study the effect of the R314Q mutation at the systems level, we tested the in vivo inflammatory response of KI mice to an MDP challenge. Compared to WT mice which responded to MDP with a 3 fold increase in IL-6, the addition of MDP to KI mice did not alter basal IL-6 levels (Fig. 2A). NOD2 knock-out (KO) mice also did not respond to MDP. A similar pattern was observed for KC (murine IL-8 homologue) and MIP2α, other commonly measured indicators of MDP-induced NOD2 activation (27, 28). We did not observe any basal changes in the levels of IL-6, MIP2α, and KC amongst the 3 genotypes, which argues against a gain of function induced by R314Q as measured by a systemic response in vivo. These results indicate that the R314Q-NOD2 mutation results in a loss of function of NOD2 to respond to MDP in vivo.
Figure 2.
In vivo responses to MDP. (A) Serum IL-6, MIP2α, and KC levels 3 hours after i.p. administration of 300 μg of MDP in +/+, +/m, m/m and −/− mice (3 mice each genotype, repeated three times with comparable result). Results are given as means +/− SD. *:p<0.05; **:p<0.01; ***p<0.001 (B) KC cytokine levels in the eye of +/+, +/m, and m/m mice 5 hours after intravitreal injection of MDP or saline (5 mice each genotype and treatment). Results are given as means +/− SD. *:p<0.05
To further examine the consequences of the R314Q mutation on inflammatory responses in vivo, mice were administered an intra-ocular injection of MDP (Fig. 2B), which we had previously shown elicits an acute ocular inflammation and measurable cytokine response (26). In WT (+/+) mice, MDP injection results in a significant increase in KC production compared to control, saline-injected eyes (Fig. 2B). In contrast to the WT-mice, +/m and m/m mice failed to respond to MDP through production of KC (or IL-6, data not shown here), thereby indicating an inability to respond to MDP.
The R314Q-NOD2 mutation results in loss of macrophage responsiveness to MDP challenge
We further sought to investigate whether the loss of MDP responsiveness at the systemic level coincides with functional changes at the cellular level. Poly(I:C) primed BMDM were stimulated with MDP and the production of IL-6 was measured amongst the 3 genotypes and KO mice (Fig. 3A). Because the level of NOD2 expression is relatively low in BMDM, poly(I:C) treatment was used in these and subsequent studies to enhance NOD2 and RIP2 expression and augment NOD2 signaling (29). In response to MDP, a significant increase in IL-6 production was observed in WT cells, and this was absent in negative control NOD2 KO cells, thereby indicating the NOD2-specificity for the MDP-induced IL-6 response. Interestingly, MDP-triggered IL-6 production was significantly reduced in a gene-dosage dependent effect. Indeed, the MDP-induced IL-6 production in m/m cells was completely abrogated and was indistinguishable from the cellular response of NOD2 KO cells.
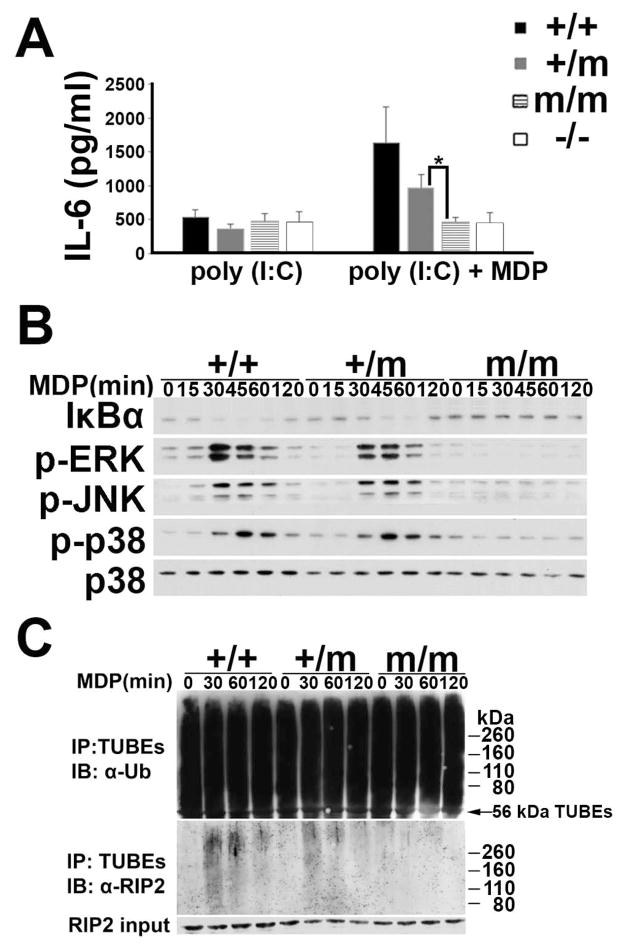
Figure 3.
BMDM from R314Q-Nod2 mice have reduced responses to MDP. (A) BMDM from +/+, +/m, m/m and −/− mice were treated with poly(I:C) alone or poly(I:C) and MDP for 24 hours and IL-6 measured in supernatants (3 mice each genotype). Error bars represent SD. Results are representative of 3 separate experiments. *:p<0.05 (B) BMDM from +/+, +/m and m/m were treated with MDP from 0–120 minutes (indicated at the top of each lane) and lysates were analyzed by immunoblot with antibodies to monitor the amount of IκBα, and the degree of phosphorylation (p) of ERK, JNK and p38. Antibodies to p38 were used as a control for the amount of protein loaded. (C) BMDM from +/+, +/m, and m/m were treated with MDP from 0–120 minutes (indicated at the top of each lane). TUBE-GST protein complexes were used to precipitate all polyubiquitinated proteins from lysates of BMDMs and subsequent immunoblotting was performed using anti-ubiquitin (top) or anti-RIPs (middle). The bottom panel is an immunoblot with anti-RIP2 antibodies on lysates prior to treatment with TUBEs. IP: immunoprecipitation, IB: Immunoblot
To assess whether the decrease in cytokine production in KI-cells was due to impaired intracellular signaling responses, we examined MDP-induction of the NF-κB and MAPK pathways in BMDMs (Fig. 3B). Western blot analysis detected diminished expression of IκBα in the WT cells within 30 minutes, which ultimately recovered 120 minutes later. Consistent with the heterogenous expression of Nod2, a disappearance of IkBa was observed in +/− cells; albeit it was slightly delayed (i.e. it took 45 minutes). BMDMs of m/m mice did not respond to MDP as indicated by the unaltered expression of IκBα. The lack of responsiveness to MDP in m/m cells was further observed for MDP-induction of the MAPK pathway. MDP-stimulation resulted in increased expression level of the active, phosphorylated forms of the signaling mediators ERK, JNK and p38, which occurred within 30 minutes in WT cells. This response was impaired and absent in m/+ and m/m cells, respectively.
One of the most immediate and measurable intracellular responses of MDP-induced NOD2 activity is the ubiquitination of the kinase RIP2, which is necessary for downstream signal transduction events in the NF-kB and MAPK pathways (30). Using tandem ubiquitin binding entities (TUBE), which are protein complexes that bind cellular polyubiquitinated proteins and protect them from de-ubiquitination or proteasomal degradation (31), we evaluated whether the R314Q mutation altered MDP-induced RIP2 ubiquitination. As shown in Fig. 3C, anti-ubiquitin immunoblotting showed the abundance of ubiquitinated proteins in all 3 genotypes, indicating that the TUBE protein complexes were successful in isolating polyubiquitinated proteins from the BMDM (Fig 3C, top panel). However, immunodetection specifically for the ubiquitinated form of RIP2 showed different amounts of polyubiquitination amongst the 3 different genotypes (Fig 3C, middle panel). RIP2 is detected over a broad molecular weight range because the diverse array of ubiquitin linkages that can occur creates a modified population of RIP2 protein of varying sizes (32). The amount of RIP2 in the lysates of all genotypes prior to pull down with TUBEs was comparable (Fig. 3C, bottom panel). The level of polyubiquitinated RIP2 was the highest in BMDM from +/+ mice, reduced lysates prepared from +/m mice, and not observed in lysates from m/m, even at the later time points. These results show that NOD2 in BMDM prepared from +/m and m/m mice have an impaired ability to activate RIP2 in response to MDP, which is as a function of copy number of the mutation. Collectively, these data indicate that introduction of Blau-point mutation results in an altered form of NOD2 that exhibits impaired cellular responses to MDP involving RIP2 initiated signal transduction of NF-kB and MAPK pathways and diminished IL-6 production.
The R314Q mutation results in the conversion of NOD2 from full length protein to a smaller form
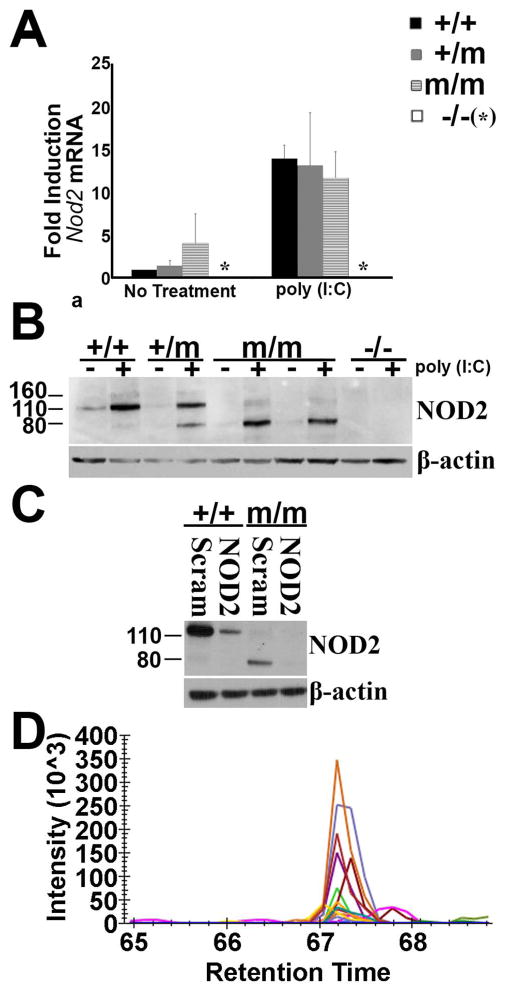
To assess whether the abnormal response to MDP observed both in vivo and in vitro for R314Q mice was the result of alterations in transcriptional and/or translational control of Nod2 at the cellular level, studies were performed on BMDM prepared from WT and KI mice. To determine whether the point mutation alters Nod2 transcription, the levels of mRNA expression were analyzed by quantitative real-time PCR. As shown in Fig. 4A, the expression levels of Nod2 in the baseline state were comparable amongst all 3 genotypes. Stimulation with poly(I:C) elicited a robust induction of Nod2 mRNA expression, and this gene induction was not significantly altered by the point mutation. As expected, the mRNA expression of Nod2 was non-detectable in BMDMs prepared from NOD2 KO mice. This data indicates that the introduction of the R314Q mutation does not impair the transcriptional control of Nod2.
Figure 4.
The R314Q mutation results in the conversion of NOD2 from full length protein to a smaller form. (A) Nod2 mRNA levels present in BMDM from +/+, +/m, m/m, and −/− mice cultured in media alone (no treatment) or media supplemented with poly(I:C) for 24 hours. Transcript levels were determined in cDNA by quantitative-RT PCR and expressed as fold induction compared to β-actin for each condition. Error bars represent SD. Results are representative of 3 separate experiments. (B) Western blot analysis with anti-NOD2 antibody on lysates from BMDM of +/+, +/m, m/m, and −/− mice cultured in media alone (−) or media supplemented (+) with poly(I:C) for 24 hours. Two examples (different BMDM preparations) are shown for m/m mice. Results are representative of 3 separate experiments. (C) si-RNA knock down of NOD2. Immunoblot of lysates from poly(I:C)-primed BMDM of +/+ or m/m mice which were electroporated with either nonspecific scrambled control siRNA (Scram) or NOD2 specific siRNA. Blots ere developed with anti-NOD2 antisera or anti-β-actin as a loading control. (D) Mass spectrometry analysis of proteins eluted from gel slices from the 80 kDa region following electrophoresis of lysates from BMDM of m/m mice. Tryptic digests were analyzed by targeted LC-MS2 for the presence of NOD2 peptides. A peptide corresponding to residues 724–739 of mouse NOD2 in the 80 kDa band is shown. Each line on the chromatogram represents the intensity of one of the fragment ions also observed when a digest of NOD2 standard was analyzed.
To investigate whether a change in protein expression occurs as a consequence of the R314Q mutation, NOD2 protein levels were measured in BMDM in the resting state or upon induction with poly(I:C). Immunoblotting revealed the expected full length 116 kDa form of NOD2 in cells from WT cells (+/+) (Fig. 4B), which was further increased upon stimulation with poly(I:C). NOD2 expression was appropriately absent in cells prepared from control NOD2 KO mice (−/−), indicating the antibody specificity of this assay. In contrast, BMDMs from KI mice (+/m or m/m) did not show constitutive NOD2 expression in the naïve state. Indeed the m/m cells lacked the full length NOD2 and instead showed the appearance of a smaller, 80 kDa protein that reacted with the anti-NOD2 antibody. The smaller 80 kDa band was further increased by stimulation with poly(I:C). Consistent with their heterozygous genotype, the BMDMs from +/m mice showed expression of both the 116 and 80 kDa forms of putative NOD2 upon induction with poly(I:C). In line with prior reports that NOD2 is upregulated by IFN-γ (33), we found increased expression of 116 kDa form of NOD2 in +/+ BMDMs when stimulated with IFN-γ (Supplemental Fig. 2A). Again, the full-length form of NOD2 was absent from both m/m cells and NOD2 KO cells and instead generation of the 80 kDa band in m/m or m/+ cells was confirmed. Of note, a longer exposure of the blot revealed a faint 80 kDa form of NOD2 in WT-cells. This raises the possibility that the 80 kDa form may be generated normally when NOD2 is upregulated and that the R314Q mutation promotes the conversion to the 80 kDa form of NOD2. We have further confirmed the specificity of the anti-NOD2 antibody used in these studies using HEK293 cells transiently transfected with a plasmid carrying mouse Nod2 cDNA (Supplemental Fig. 2B). These data indicate that despite normal transcription, the point mutation results in loss of full-length NOD2 protein.
To further investigate generation of the putative 80 kDa form of NOD2 that occurs in KI mice, knock down experiments were performed in BMDMs prepared from WT (+/+) versus mutant mice (m/m) using short interfering RNA (siRNA). Compared to the scrambled, control siRNA, the specific siRNA treatment resulted in significant reduction in full-length WT-NOD2 (Fig. 4C, left) and also the 80 kDa form in the mutant BMDCs (Fig 4C, right). These data demonstrate that post-transcriptional gene silencing by siRNA effectively knocked down both forms of NOD2, thereby indicating that introduction of R314Q mutation results in a unique and smaller form of the NOD2 protein.
The identity of the mutant 80 kDa form of NOD2 was further evaluated by mass spectrometry (Fig. 4D). Analysis of tryptic digests from gel slices cut from the 116 kDa region of lysates from BMDM of +/+ mice identified two peptides - GFSEEGIQLYLRK (amino acids 426–438) and SLYEMQEEQLAQEAVR (amino acids 724–739) of NOD2 defined by m/z ratios and fragment ions created from the reference library of overexpressed NOD2. The peptide SLYEMQEEQLAQEAVR was also identified in gel slices from the 80 kDa region of lysates of BMDM isolated from m/m mice, thereby identifying NOD2 in the 80 kDa region. Other spectra matched too few ions to be able to identify their source. While the mass spectrometry results indicate that the 80 kDa region contained a modified form of the full length 116 kDa NOD2 protein, the yield of tryptic fragments was insufficient to generate spectra throughout the entire length of the protein to provide information on the exact residues that were missing from the 80 kDa form.
Since Nod2 splice variants have been described that lead to truncated forms of NOD2 protein (34), mRNA from BMDM of +/+, +/m and m/m mice were analyzed for evidence of splice variants by PCR. DNA fragments of the predicted size were obtained that were comparable in all genotypes and smaller sized fragments, that would have been indicative of altered exon splicing, were not observed (Supplemental Fig. 2C). To rule out the possibility of introduction of a premature stop codon, Nod2 mRNA from BMDM of m/m mice was reverse transcribed and sequenced. The resulting sequence was exactly as predicted (data not shown), with the only difference from wild type being the G to A change that we intentionally introduced to create the R314Q mutation. Collectively, these data indicate that the R314Q mutation results in a post-translational modification of NOD2, wherein the full-length protein is converted to a smaller form of NOD2.
Macrophages from patients with Blau syndrome demonstrate loss of responsiveness to MDP
To compare the observations obtained in mutant KI mice with clinical disease, we sought to examine the cellular response of patients with Blau syndrome. PBMCs obtained from 2 different patients with Blau syndrome (with mutation at orthologous location R334Q) were used to generate peripheral blood derived macrophages (PBDM) Comparable numbers of PBDM were generated from both patients as well as healthy controls. Experiments were carried out as above, wherein macrophages primed with poly(I:C) were challenged with MDP, and IL-6 and IL-8 production were measured (Fig. 5A, B). In contrast to PBDM from healthy controls, which produced both IL-6 and IL-8 in response to MDP stimulation, PBDM from both of the patients with Blau syndrome demonstrated a complete loss of IL-6 and IL-8 production (Blau-1, Fig. 5A; Blau-2, Fig. 5B). Differences in spontaneous release of IL-6 or IL-8 (i.e. media control) were not observed, which is contrary to the gain of function model theorized for Blau syndrome. Consistent with prior reports using PBMCs of Blau patients (15, 17), we confirmed that the PBDMs used here were capable of responding to LPS as measured by production of IL-6 (Fig. 5B), indicating that the NOD2 mutation does not impair cellular capacity to respond to other PRR ligands and rules out indirect effects of immunosuppressive therapy. Investigation of the MDP-triggered MAPK pathway activation in PBDM of Blau patients, showed that the phosphorylation of p38, quantitated by densitometry, was absent or severely blunted in both patients (Fig. 5C). Studies of NF-κB activation, measured by disappearance of IκBα, using PBDM from Blau-1 showed no evidence of NF-κB activation (data not shown), further indicating impaired intracellular signaling.
Discussion
We created the R314Q KI mouse with the goal of studying the function of a variant of Nod2 carrying the most common Blau-syndrome associated mutation. The data here demonstrate that R314Q KI mice, wherein Nod2 was endogenously expressed under the control of its normal regulatory elements, exhibited impaired NOD2 activation to MDP. The macrophage response to MDP was impaired in that RIP2 activation was reduced, which resulted in mitigated responses within the MAPK and NF-κB pathways and diminished cytokine production. Such abated cellular inflammatory responses were paralleled in vivo in that KI mice were refractory to MDP challenge. MDP-induced cytokine production systemically as well as locally within the eye was absent. In contrast to previous studies based on transient transfection assays, we did not find any evidence for the spontaneous activation of NF-κB or enhanced responses to MDP. PBDMs from 2 different patients with Blau syndrome exhibited the same phenotype in that MDP-induced cytokine and intracellular signaling responses involving MAPK and NF-κB were impaired. These data are all consistent with the possibility that Blau syndrome, like Crohn’s disease, is also a disease mediated by loss of function of NOD2. While Blau syndrome is not associated with gut inflammation, arthritis and uveitis can occur in Crohn’s disease and both diseases are associated with granulomatous inflammation.
Even though macrophages from R314Q mouse showed a comparable biochemical response to challenge with MDP as did macrophages from patients, the mice did not develop a spontaneous clinical phenotype. This is comparable to other genetic manipulations of Nod2, including Nod2 KO mice (35) and mice engineered to contain a mutation found in Crohn’s disease (28). It is possible that the transcriptome response in humans that causes Blau syndrome does not trigger the same orthologs in B6 mice and the mice can’t respond to the NOD2 mutation in the same way as a human carrying the mutation (36). Human diseases caused by loss of function mutations in NLR family members may be more difficult to model in mice than gain of function mutations. Our R314Q KI model showing a loss of function of NOD2 and no spontaneous disease is in contrast to well understood gain of function genetic disorder involving the NLR family member, NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3). Mutations in NLRP3 cause cryopyrin-associated periodic syndrome (CAPS), a gain of function autosomal dominant disorder associated with spontaneous activation of the NLRP3 inflammasome, excessive IL-1β release and a uniform response in patients to IL-1β inhibitors (37). This phenotype has readily been reproduced in KI mouse models where autoactivation of the inflammasome was demonstrated (38, 39). Since mutations in Blau syndrome and CAPS are found at analogous positions in their respective NLR family member genes, it has been inferred that they would have a comparable mechanism of action (40, 41). However, in contrast to patients with CAPS, patients with Blau syndrome do not spontaneously release cytokines from PBMCs. Our R314Q model, in contrast to NLRP3 KI models, also does not show spontaneous activation of cytokines triggered by the NOD2 pathway.
We have expressed wild type and murine R314Q mutated NOD2 in HEK cells using expression vectors and in murine J774 macrophages using retroviral vectors to produce stable transformants (data not shown). Western blot analysis of lysates from these cells do not show the 80 kDa truncated form of NOD2. It is possible in artificial over-expression systems that the cellular processes responsible for the 80 kDa form of NOD2 are not functional or are only expressed in cell types different from these cell lines used because of their ease of in vitro culture and manipulation. The lack of NOD2 processing when mutant NOD2 is over-expressed in HEK cells may also account for the discrepancy between the gain-of-function attributed to mutant NOD2 in HEK cells and the lack of response to MDP observed in patients with Blau syndrome. We have initiated studies of PBDM to investigate whether the conversion process of full length Nod2 to the 80 kDa form is observed in patients with Blau syndrome. However, as previously described by others (43), interpreting these studies is difficult because of the quality and specificity of antisera or antibodies currently available. Using a reagent from ProSci that had been used by others to detect human Nod2 (42), multiple irresolvable bands were identified in the 80–120 kDa section of western blots from PBDM of healthy controls as well as patients Blau-1 and -2 (data not shown) such that a definitive conclusion about the disappearance of full length Nod2 or appearance of an 80 kDa truncated form could not be made and will require further study. The heterozygous nature of Blau syndrome creates a further challenge for experiments using western blot analysis. Since patients express a normal 116 kDa form of Nod2, detecting the disappearance of full length Nod2 on a western blot would require interpreting relative intensities of bands compared to healthy individuals, unlike the homozygous R314Q mutant mice created by breeding where the disappearance of full length NOD2 is readily appreciated.
One mechanism by which we believe that mutant NOD2 results in loss of responsiveness to MDP is through the post-translational modification and conversion of the full-length NOD2 to a smaller form of NOD2 that lacks the ability to sense MDP. The structure of the 80 kDa form of NOD2 is under further study. The commercial anti-mouse NOD2 monoclonal antibody used here was created by immunizing rats with two separate large peptide fragments (NOD2 amino acids 150–450 and 481–750) but the exact epitope was not elucidated. Furthermore, this antibody did not work in our hands for immunoprecipitation studies requiring us to analyze whole cell lysates to study the structure. While our work with mass spectrometry did identify one peptide that allowed us to conclude that the 80 kDa region of electrophoresis gels of lysates from BMDM from m/m did contain NOD2, the approach did not yield enough material to be quantitative and provide information of which residues or domain of the NOD2 protein was altered. The structure of the 80kDa mutant form of NOD2 is currently under further study. Nonetheless, it is interesting to consider whether the 80kDa form has a yet-to-be determined function. For example, if the 80 kDa form showed auto-inhibitory capability for NOD2 oligomerization, like NOD2-S (34), or antagonism for MDP-induced activation of NOD2, like NOD2-C2 (43), this could explain the loss of response to systemic administration of MDP seen in R314Q +/m mice and the absent response to MDP seen in macrophages from Blau patients who still carry a normal copy of NOD2. It has been shown that the truncated form of NOD2 caused by the 3020insC mutation found in Crohn’s disease does result in a new function that may be pathogenic – the inhibition of basal levels of IL-10 release by interfering with phosphorylation of hnRNP-A1 (44). Interestingly, this new function was only observed in humans, not mice, indicating an important difference between mouse and human orthologues of NOD2.
Another topic of interest uncovered by our studies is the investigation of the mechanism by which the full-length form of NOD2 is converted to the mutant 80 kDa form. We have ruled out a premature stop codon in the mRNA and a splice variant. Several proteins have been shown to bind to Nod2 and negatively regulate downstream signaling events, including Erbin, CENTB1, AAMP, CAD, and JNKBP1 (45–50). These negative regulators are not thought to act by cleavage of Nod2. More recently, Nod2-binding partners were identified that regulated Nod2 pathways by controlling proteasome-mediated turnover of Nod2, including Hsp90, SOCS-3 and TRIM27 (33, 51). Nod2 degradation did not occur by these pathways in the presence of proteasomal inhibitors. We have conducted studies to investigate if full length NOD2 could be recovered in BMDM from +/m and m/m mice by a variety of inhibitors, including the calpain inhibitors ALLN and calpastatin, the proteasome inhibitors bortezomib and MG132, the pan caspase inhibitor z-vad.fmk, the cathepsin inhibitors L-006235, chymostatin and pepstatin A, and NH4Cl as an inhibitor of phagosome-lysosomal fusion (data not shown). All of these treatments, including some in combination, failed to restore the 116 kDa form of NOD2. If a specific enzyme is responsible for generating the 80 kDa form in the setting of the mutation, it remains to be identified. A recent study using a genome-wide siRNA screen for proteins regulating Nod2 and NF-kB signaling in HEK-293 cells found that while there were hundreds serving as general regulators of the NF-kB pathway, there were few that were negative regulators specific to Nod2 (52). If the faint 80 kDa band found in macrophages from wild type mice represents a normally occurring cleavage of NOD2 that is enhanced by the R314Q mutation, future studies to elucidate the mechanism would be an important advance in identifying additional negative regulators specific to NOD2.
Through the R314Q mouse we have identified a structural change in NOD2 when the ortholog of human NOD2 has been modified to carry one point mutation found in Blau syndrome. R314Q mice showed reduced cytokine production in vivo in response to MDP. Macrophages from R314Q mice and from patients from Blau syndrome showed reduced cytokine production and intracellular signaling of pathways normally activated by MDP. In contrast to analyses of Blau mutations done in transient transfection assays, spontaneous activation of the NF-κB pathway was not observed. These data provide new insights into how mutations associated with Blau syndrome could alter the function of NOD2.
Supplementary Material
Acknowledgments
This research was supported by the Department of Veterans Affairs Biomedical Laboratory Research and Development Service as well as NIH Training Grant #5-T32-AI07472.
We thank Thom Saunders of the University of Michigan Transgenic Animal Model Core Facility, Larry David of the Proteomics Shared Resource at Oregon Health & Science University for performing mass spectrometry and helpful discussions and Neal Copeland of the National Cancer Institute, Frederick, MD, for the gift of plasmids used in recombineering. We thank Emily E. Vance for her technical contributions.
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Girardin SE, I, Boneca G, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 2.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 3.Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 4.Jun JC, Cominelli F, Abbott DW. RIP2 activity in inflammatory disease and implications for novel therapeutics. J Leukoc Biol. 2013;94:927–932. doi: 10.1189/jlb.0213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw MH, Kamada N, Warner N, Kim YG, Nunez G. The ever-expanding function of NOD2: autophagy, viral recognition, and T cell activation. Trends Immunol. 2011;32:73–79. doi: 10.1016/j.it.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 7.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 8.Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 9.Strober W, Asano N, Fuss I, Kitani A, Watanabe T. Cellular and molecular mechanisms underlying NOD2 risk-associated polymorphisms in Crohn’s disease. Immunol Rev. 2014;260:249–260. doi: 10.1111/imr.12193. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Moran T, Swanson E, Julian C, Harris J, Bonen DK, Hedl M, Nicolae DL, Abraham C, Cho JH. Regulation of IL-8 and IL-1beta expression in Crohn’s disease associated NOD2/CARD15 mutations. Hum Mol Genet. 2004;13:1715–1725. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 11.Netea MG, Ferwerda G, de Jong DJ, Jansen T, Jacobs L, Kramer M, Naber TH, Drenth JP, Girardin SE, Kullberg BJ, Adema GJ, Van der Meer JW. Nucleotide-binding oligomerization domain-2 modulates specific TLR pathways for the induction of cytokine release. J Immunol. 2005;174:6518–6523. doi: 10.4049/jimmunol.174.10.6518. [DOI] [PubMed] [Google Scholar]
- 12.Rose CD, Martin TM, Wouters CH. Blau syndrome revisited. Curr Opin Rheumatol. 2011;23:411–418. doi: 10.1097/BOR.0b013e328349c430. [DOI] [PubMed] [Google Scholar]
- 13.Punzi L, Gava A, Galozzi P, Sfriso P. Miscellaneous non-inflammatory musculoskeletal conditions. Blau syndrome. Best Pract Res Clin Rheumatol. 2011;25:703–714. doi: 10.1016/j.berh.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Kanazawa N, Okafuji I, Kambe N, Nishikomori R, Nakata-Hizume M, Nagai S, Fuji A, Yuasa T, Manki A, Sakurai Y, Nakajima M, Kobayashi H, Fujiwara I, Tsutsumi H, Utani A, Nishigori C, Heike T, Nakahata T, Miyachi Y. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: common genetic etiology with Blau syndrome. Blood. 2005;105:1195–1197. doi: 10.1182/blood-2004-07-2972. [DOI] [PubMed] [Google Scholar]
- 15.Martin TM, Zhang Z, Kurz P, Rose CD, Chen H, Lu H, Planck SR, Davey MP, Rosenbaum JT. The NOD2 defect in Blau syndrome does not result in excess interleukin-1 activity. Arthritis Rheum. 2009;60:611–618. doi: 10.1002/art.24222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okafuji I, Nishikomori R, Kanazawa N, Kambe N, Fujisawa A, Yamazaki S, Saito M, Yoshioka T, Kawai T, Sakai H, Tanizaki H, Heike T, Miyachi Y, Nakahata T. Role of the NOD2 genotype in the clinical phenotype of Blau syndrome and early-onset sarcoidosis. Arthritis Rheum. 2009;60:242–250. doi: 10.1002/art.24134. [DOI] [PubMed] [Google Scholar]
- 17.Son S, Lee J, Woo CW, Kim I, Kye Y, Lee K, Lee J. Altered cytokine profiles of mononuclear cells after stimulation in a patient with Blau syndrome. Rheumatol Int. 2010;30:1121–1124. doi: 10.1007/s00296-009-1342-4. [DOI] [PubMed] [Google Scholar]
- 18.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, Schuler G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 19.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 20.Rosenzweig HL, Clowers JS, Nunez G, Rosenbaum JT, Davey MP. Dectin-1 and NOD2 mediate cathepsin activation in zymosan-induced arthritis in mice. Inflamm Res. 2011;60:705–714. doi: 10.1007/s00011-011-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kezic JM, Davey MP, Glant TT, Rosenbaum JT, Rosenzweig HL. Interferon-gamma regulates discordant mechanisms of uveitis versus joint and axial disease in a murine model resembling spondylarthritis. Arthritis Rheum. 2012;64:762–771. doi: 10.1002/art.33404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paques M, Guyomard JL, Simonutti M, Roux MJ, Picaud S, Legargasson JF, Sahel JA. Panretinal, high-resolution color photography of the mouse fundus. Invest Ophthalmol Vis Sci. 2007;48:2769–2774. doi: 10.1167/iovs.06-1099. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, Koch P, Chen M, Lau A, Reid DM, Forrester JV. A clinical grading system for retinal inflammation in the chronic model of experimental autoimmune uveoretinitis using digital fundus images. Exp Eye Res. 2008;87:319–326. doi: 10.1016/j.exer.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Furtado JMDM, Choi D, Lauer AK, Appukuttan B, Bailey ST, Rahman HT, Payne JF, Stempel JS, Mohs K, Powers MR, Yeh S, Smith JR. Imaging Retinal Vascular Changes in the Mouse Model of Oxygen-Induced Retinopathy. In. Trans Vis Sci Tech. 2012 doi: 10.1167/tvst.1.2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenzweig HL, Jann MJ, Vance EE, Planck SR, Rosenbaum JT, Davey MP. Nucleotide-binding oligomerization domain 2 and Toll-like receptor 2 function independently in a murine model of arthritis triggered by intraarticular peptidoglycan. Arthritis Rheum. 2010;62:1051–1059. doi: 10.1002/art.27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenzweig HL, Martin TM, Jann MM, Planck SR, Davey MP, Kobayashi K, Flavell RA, Rosenbaum JT. NOD2, the gene responsible for familial granulomatous uveitis, in a mouse model of uveitis. Invest Ophthalmol Vis Sci. 2008;49:1518–1524. doi: 10.1167/iovs.07-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carneiro LA, Magalhaes JG, Tattoli I, Philpott DJ, Travassos LH. Nod-like proteins in inflammation and disease. J Pathol. 2008;214:136–148. doi: 10.1002/path.2271. [DOI] [PubMed] [Google Scholar]
- 28.Kim YG, Shaw MH, Warner N, Park JH, Chen F, Ogura Y, Nunez G. Cutting edge: Crohn’s disease-associated Nod2 mutation limits production of proinflammatory cytokines to protect the host from Enterococcus faecalis-induced lethality. J Immunol. 2011;187:2849–2852. doi: 10.4049/jimmunol.1001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YG, Park JH, Reimer T, Baker DP, Kawai T, Kumar H, Akira S, Wobus C, Nunez G. Viral infection augments Nod1/2 signaling to potentiate lethality associated with secondary bacterial infections. Cell Host Microbe. 2011;9:496–507. doi: 10.1016/j.chom.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao M, Scacheri PC, Marinis JM, Harhaj EW, Matesic LE, Abbott DW. ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr Biol. 2009;19:1255–1263. doi: 10.1016/j.cub.2009.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marblestone JG, Larocque JP, Mattern MR, Leach CA. Analysis of ubiquitin E3 ligase activity using selective polyubiquitin binding proteins. Biochim Biophys Acta. 2012;1823:2094–2097. doi: 10.1016/j.bbamcr.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malynn BA, Ma A. Ubiquitin makes its mark on immune regulation. Immunity. 2010;33:843–852. doi: 10.1016/j.immuni.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee KH, Biswas A, Liu YJ, Kobayashi KS. Proteasomal degradation of Nod2 protein mediates tolerance to bacterial cell wall components. J Biol Chem. 2012;287:39800–39811. doi: 10.1074/jbc.M112.410027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenstiel P, Huse K, Till A, Hampe J, Hellmig S, Sina C, Billmann S, von Kampen O, Waetzig GH, Platzer M, Seegert D, Schreiber S. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc Natl Acad Sci USA. 2006;103:3280–3285. doi: 10.1073/pnas.0505423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pauleau AL, Murray PJ. Role of nod2 in the response of macrophages to toll-like receptor agonists. Mol Cell Biol. 2003;23:7531–7539. doi: 10.1128/MCB.23.21.7531-7539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG Inflammation, and L. S. C. R. P. Host Response to Injury. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman HM, Throne ML, Amar NJ, Sebai M, Kivitz AJ, Kavanaugh A, Weinstein SP, Belomestnov P, Yancopoulos GD, Stahl N, Mellis SJ. Efficacy and safety of rilonacept (interleukin-1 Trap) in patients with cryopyrin-associated periodic syndromes: results from two sequential placebo-controlled studies. Arthritis Rheum. 2008;58:2443–2452. doi: 10.1002/art.23687. [DOI] [PubMed] [Google Scholar]
- 38.Brydges SD, Mueller JL, McGeough MD, Pena CA, Misaghi A, Gandhi C, Putnam CD, Boyle DL, Firestein GS, Horner AA, Soroosh P, Watford WT, O’Shea JJ, Kastner DL, Hoffman HM. Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity. 2009;30:875–887. doi: 10.1016/j.immuni.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng G, Zhang F, Fuss I, Kitani A, Strober W. A mutation in the Nlrp3 gene causing inflammasome hyperactivation potentiates Th17 cell-dominant immune responses. Immunity. 2009;30:860–874. doi: 10.1016/j.immuni.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albrecht M, Lengauer T, Schreiber S. Disease-associated variants in PYPAF1 and NOD2 result in similar alterations of conserved sequence. Bioinformatics. 2003;19:2171–2175. doi: 10.1093/bioinformatics/btg370. [DOI] [PubMed] [Google Scholar]
- 41.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 42.Brooks MN, Rajaram MV, Azad AK, Amer AO, Valdivia-Arenas MA, Park JH, Nunez G, Schlesinger LS. NOD2 controls the nature of the inflammatory response and subsequent fate of Mycobacterium tuberculosis and M. bovis BCG in human macrophages. Cell Microbiol. 2011;13:402–418. doi: 10.1111/j.1462-5822.2010.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramer M, Boeck J, Reichenbach D, Kaether C, Schreiber S, Platzer M, Rosenstiel P, Huse K. NOD2-C2 - a novel NOD2 isoform activating NF-kappaB in a muramyl dipeptide-independent manner. BMC Res Notes. 2010;3:224. doi: 10.1186/1756-0500-3-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noguchi E, Homma Y, Kang X, Netea MG, Ma X. A Crohn’s disease-associated NOD2 mutation suppresses transcription of human IL10 by inhibiting activity of the nuclear ribonucleoprotein hnRNP-A1. Nat Immunol. 2009;10:471–479. doi: 10.1038/ni.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald C, Chen FF, Ollendorff V, Ogura Y, Marchetto S, Lecine P, Borg JP, Nunez G. A role for Erbin in the regulation of Nod2-dependent NF-kappaB signaling. J Biol Chem. 2005;280:40301–40309. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- 46.Kufer TA, Kremmer E, Banks DJ, Philpott DJ. Role for erbin in bacterial activation of Nod2. Infect Immun. 2006;74:3115–3124. doi: 10.1128/IAI.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto-Furusho JK, Barnich N, Xavier R, Hisamatsu T, Podolsky DK. Centaurin beta1 down-regulates nucleotide-binding oligomerization domains 1- and 2-dependent NF-kappaB activation. J Biol Chem. 2006;281:36060–36070. doi: 10.1074/jbc.M602383200. [DOI] [PubMed] [Google Scholar]
- 48.Bielig H, Zurek B, Kutsch A, Menning M, Philpott DJ, Sansonetti PJ, Kufer TA. A function for AAMP in Nod2-mediated NF-kappaB activation. Mol Immunol. 2009;46:2647–2654. doi: 10.1016/j.molimm.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 49.Richmond AL, Kabi A, Homer CR, Marina-Garcia N, Nickerson KP, Nesvizhskii AI, Sreekumar A, Chinnaiyan AM, Nunez G, McDonald C. The nucleotide synthesis enzyme CAD inhibits NOD2 antibacterial function in human intestinal epithelial cells. Gastroenterology. 2012;142:1483–1492. e1486. doi: 10.1053/j.gastro.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lecat A, Di Valentin E, Somja J, Jourdan S, Fillet M, Kufer TA, Habraken Y, Sadzot C, Louis E, Delvenne P, Piette J, Legrand-Poels S. The c-Jun N-terminal kinase (JNK)-binding protein (JNKBP1) acts as a negative regulator of NOD2 protein signaling by inhibiting its oligomerization process. J Biol Chem. 2012;287:29213–29226. doi: 10.1074/jbc.M112.355545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zurek B, Schoultz I, Neerincx A, Napolitano LM, Birkner K, Bennek E, Sellge G, Lerm M, Meroni G, Soderholm JD, Kufer TA. TRIM27 negatively regulates NOD2 by ubiquitination and proteasomal degradation. PLoS One. 2012;7:e41255. doi: 10.1371/journal.pone.0041255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warner N, Burberry A, Franchi L, Kim YG, McDonald C, Sartor MA, Nunez G. A genome-wide siRNA screen reveals positive and negative regulators of the NOD2 and NF-kappaB signaling pathways. Sci Signal. 2013;6:rs3. doi: 10.1126/scisignal.2003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.