Abstract
Translation of the 5.7 kb luteovirus genome is controlled by the 3’ untranslated region (UTR). Base pairing between regions of the 3’ UTR and sequences kilobases upstream is required for cap-independent translation and ribosomal frameshifting needed to synthesize the viral replicase. Luteoviruses produce subgenomic RNAs, which can serve as mRNA, but one sgRNA also regulates translation initiation in trans. As on all viruses, the 3’ and 5’ ends contain structures that are presumed to facilitate RNA synthesis. This review describes the structures and interactions of Barley yellow dwarf virus RNA that facilitate the complex interplay between the above events and result in a successful virus infection. We also present surprising results on the apparent lack of need for some subgenomic RNAs for the virus to infect cells or whole plants. In summary, the UTRs of luteoviruses are highly complex entities that control and fine-tune many key events of the virus replication cycle.
1. Introduction
The genomic RNA of a positive strand RNA virus must serve both as genome, i.e. the template for replication and production of progeny genomes, and as the messenger RNA for production of viral proteins. These events are conflicting and thus switching from one process to the other must be highly regulated. Because translation and replication both begin at ends of the viral RNAs, the 5’ and 3’ ends play crucial roles in regulating both processes. They do so by interacting with different host and viral proteins, macromolecular complexes and organelles at different stages of the replication cycle. To achieve this, most RNA structures at the ends are probably dynamic, changing in conformation as their roles change. Viruses in the Luteovirus genus and related viruses in the Tombusviridae family (tombusvirids) have become well-characterized examples of such control by RNA structural rearrangements (Nicholson and White, 2014).
2. Genus Luteovirus and the Luteoviridae
The Luteoviridae family includes the Luteovirus, Polerovirus and Enamovirus genera. (The term luteovirus refers only to members of genus Luteovirus, and not other members of the Luteoviridae family.) Members of the Luteoviridae are defined by their properties of persistent, circulative transmission by aphids, and confinement to the phloem in the plant (except for the enamovirus). These properties are conferred by the structural and movement proteins of the viruses, which share sequence similarity throughout the Luteoviridae family. However, based on sequences of the replicase genes, transcriptional and translational control signals, and RNA termini, luteoviruses are more closely related to tombusvirids (particularly the Alphanecrovirus, Betanecrovirus and Dianthovirus genera), than to poleroviruses and enamoviruses, which in turn are more similar to genus Sobemovirus (no family assigned) (Miller et al., 2002). Of particular relevance to this review, the 5’ ends of the RNAs of luteoviruses and tombusvirids are unmodified, while the 5’ ends of the genomes of poleroviruses, enamoviruses and sobemoviruses are covalently attached to a small viral protein (VPg). The 3’ ends of luteovirus and tombusvirid genomes terminate in CCC, while the polerovirus genome ends in GU. Luteoviruses contain GU[G/A]AAG at or near the 5’ terminus, whereas the 5’ end of the polerovirus (but not enamovirus) genome begins with ACAAAA (with occasional substitution of a non-terminal A with G). Thus, both positive and negative strand synthesis of polerovirus RNA initiates with AC. Poleroviruses and enamoviruses are clearly distinct from luteoviruses in their polymerase genes and in the RNA sequences recognized by the polymerase. Thus many aspects of this review apply more to tombusvirids and the closely related umbraviruses than to poleroviruses or enamoviruses. (See also the article in this issue by Anne Simon on the ends of RNAs of viruses in the Carmovirus genus of the Tombusviridae).
This review will focus mainly on the barley yellow dwarf viruses (BYDVs). According to the ICTV, there are five BYDVs in genus Luteovirus, BYDV-PAV, BYDV-PAS, BYDV-MAV, BYDV-kerII, BYDV-kerIII (Domier, 2012). Two others, BYDV-SGV and BYDV-GPV remain unassigned to a genus. Some viruses formerly known as BYDV, have been reclassified as poleroviruses. These include Cereal yellow dwarf virus-RPV (CYDV-RPV), CYDV-RPS, Maize yellow dwarf virus and Wheat yellow dwarf virus. These will not be discussed in this review because they have completely different terminal sequences, and end modification (VPg), and lack known translation enhancer elements.
BYDVs and the above yellow dwarf poleroviruses comprise the most economically important and widespread viruses of small grain cereals (Lister and Ranieri, 1995). BYDV-PAV is the most common of these viruses and will be the focus of this review. BYDV-PAV is by far the best-characterized luteovirus, and has become a paradigm of translational control in particular, owing to the plethora of exquisite translational control mechanisms it (and presumably other luteoviruses) employs (Fig. 1). Much of this translation is controlled by sequences in the untranslated regions (UTRs) the viral RNA. These regions also control genome replication and transcription of subgenomic RNAs (Fig. 1). Here, we will describe how ends of genomic and subgenomic RNAs control these events in cis and in trans.
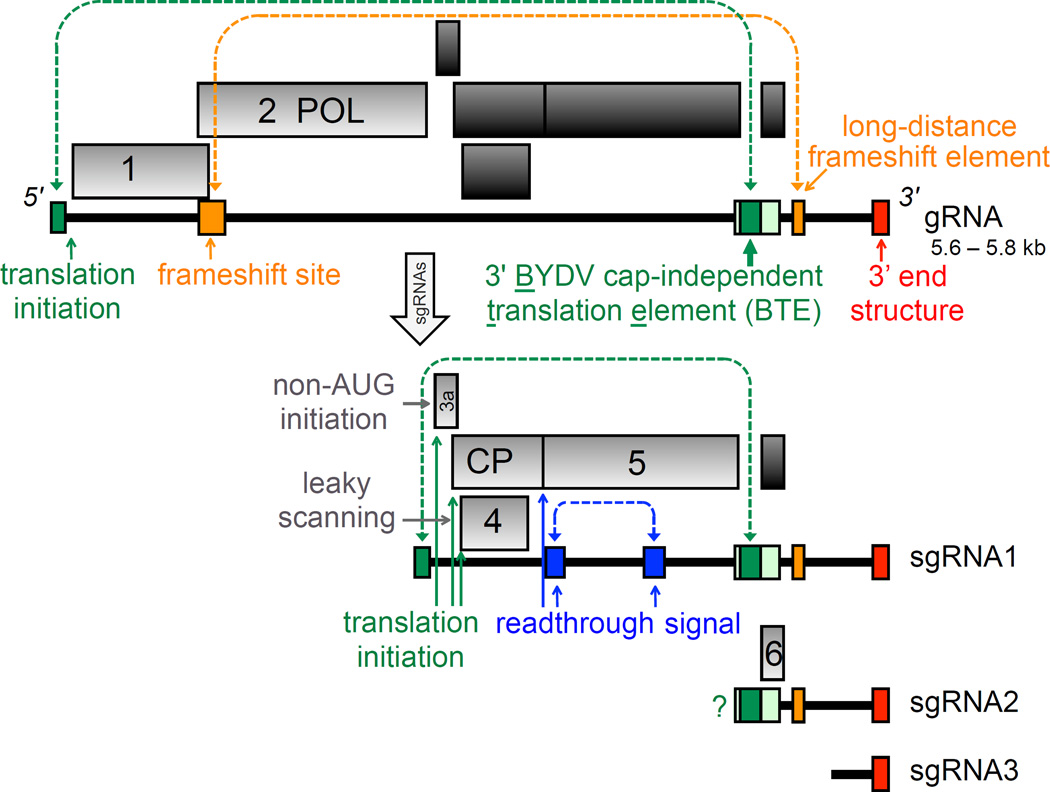
Fig. 1.
Genome organization of BYDVs. Boxes in light gray are translated from the RNA (bold line) underneath. Dark gray boxes indicate ORFs not translated from that RNA. Genomic (virion) RNA (gRNA) is at the top, and the three subgenomic RNAs (sgRNA) are generated in infected cells, with sgRNA1 serving as mRNA for four proteins. sgRNA2 is a trans-regulator of translation and possible mRNA for ORF 6. Question mark indicates that translation of ORF 6 is predicted, but its product, P6 has not been detected in infected cells. Colored boxes indicate locations of cis-acting translational control elements. Dark green indicates minimal BTE, while light green includes additional base pairing needed for function in vivo. Dashed lines indicate known (blue, orange) or predicted (blue) long-distance base pairing that controls the indicated translation event. POL: RNA-dependent RNA polymerase, CP: coat protein.
3. Translational control
3.1 Control of translation by the 3’ end in cis
The first step upon entry of the virion into the cell is disassembly and release of the RNA from the virion. The mechanism for this is virtually unknown for plant icosahedral viruses, although it has been proposed to be cotranslational for turnip crinkle virus (TCV) (species Turnip crinkle virus, genus Carmovirus, family Tombusviridae (Bakker et al., 2012). As with all positive sense RNA viruses, translation of the genomic RNA must occur prior to RNA replication because translation is necessary to produce the viral RNA-dependent RNA polymerase (RdRp) required for genome replication. Initiation of luteovirus translation requires non-canonical signals in the RNA because the RNA of luteoviruses (and all twelve genera of the Tombusviridae) lack both the m7G(5’)ppp(5’)N cap structure at the 5’ end and the poly(A) tail at the 3’ end. These terminal modifications are required for significant translation initiation on host mRNAs because they interact with translation initation factors that recruit the ribosome to the mRNA (reviewed by (Jackson et al., 2010)). A key rate-limiting factor is eIF4F, which is a heterodimer consisting of the cap-binding protein eIF4E, and the scaffolding and scanning protein, eIF4G (Patrick and Browning, 2012). On host mRNAs, eIF4E binds the 5’ cap, bringing eIF4G to the 5’ end of the mRNA. Poly(A) binding protein (PABP) simultaneously binds the poly(A) tail and eIF4G. Then eIF3, which is bound to the 43S ribosome pre-initiation complex, brings this complex to the 5’ end of the mRNA via by simultaneously binding eIF4G. Owing to their lack of a 5’ cap and a poly(A) tail, luteoviruses and tombusvirids harbor a cap-independent translation element (CITE) located near the 5’ end of the 3’ UTR. These RNA structures, usually around 100 nt long, replace the need for a 5’ cap, and like the cap, they bind eIF4F. Various CITEs bind either eIF4E or eIF4G, but bind most tightly to the heterodimer, eIF4F (Gazo et al., 2004; Nicholson et al., 2010; Treder et al., 2008; Wang et al., 2009).
There are about eight known classes of RNA structures that serve as 3’ CITEs (Miras et al., 2014; Simon and Miller, 2013). These include the BYDV-like CITE (BTE) in the luteoviruses, dianthoviruses, and alpha- and betanecroviruses (Shen and Miller, 2004a; Wang et al., 2010), a Y-shaped structure in tombusviruses (Fabian and White, 2006), the panicum mosaic virus-like CITE (PTE) in the panicoviruses and some carmo- and umbraviruses (Batten et al., 2006; Wang et al., 2009), the T-shaped element in TCV and related viruses (Simon, this issue and (Gao et al., 2012), an I-shaped element in various Tombusviridae including Melon necrotic spot virus (MNSV) (Nicholson et al., 2010; Nicholson et al., 2013), an ill-defined bulged stem-loop in Satellite tobacco necrosis virus (STNV) (Satellite tobacco necrosis virus, genus Alpha/beta necrovirus, family Tombusviridae) (van Lipzig et al., 2002) and a few other tombusvirids, and a 50 nt sequence known only in the Xiang strain of cucurbit aphid-borne yellows polerovirus (CABYV, Cucurbit aphid-borne yellows virus, genus Dianthovirus, family Tombusviridae) and one resistance-breaking strain of MNSV (Miras et al., 2014). The distribution of these elements does not always correlate with phylogenetic relationships of the viruses, suggesting recent and frequent recombination among the 3’ UTRs of tombusvirids (Simon and Miller, 2013). Where studied, it appears that all of them bind a surface of eIF4F with affinity that appears to be higher than binding of capped mRNA to eIF4F (Banerjee and Goss, 2014; Gazo et al., 2004; Nicholson et al., 2010; Treder et al., 2008; Wang et al., 2011). Upon binding of eIF4F, the 40S ribosomal subunit is recruited, either to the 3’ CITE directly (Yuan et al., 2012) or possibly to the 5’ UTR after the factor-bound CITE has base-paired with the 5’ UTR and placed eIF4F in the proximity of the 5’ end of the RNA (Rakotondrafara et al., 2006).
In most cases, the 3’ CITE base pairs to the 5’ UTR of the genomic RNA or subgenomic mRNA1 (sgRNA1) in order to deliver the factors and possibly the 40S ribosomal subunit which then scans to the 5’-terminal AUG (usually) where protein synthesis ensues (Nicholson et al., 2010). This base pairing occurs as a kissing stem-loop interaction between five bases in the terminal loop of stem-loop III (SL-III) in the BTE and five bases in a stem-loop in the 5’ UTR (Guo et al., 2001). In BYDV genomic RNA this is the fourth stem-loop from the 5’ end, shortly upstream of the start codon (Fig. 2A). The BTE also mediates cap-independent translation of sgRNA1 by base pairing to the stem-loop at the extreme 5’ end of sgRNA1. In all other luteoviral genomic RNA and subgenomic RNA1s, the BTE also base pairs to the 5’-proximal stem-loop.
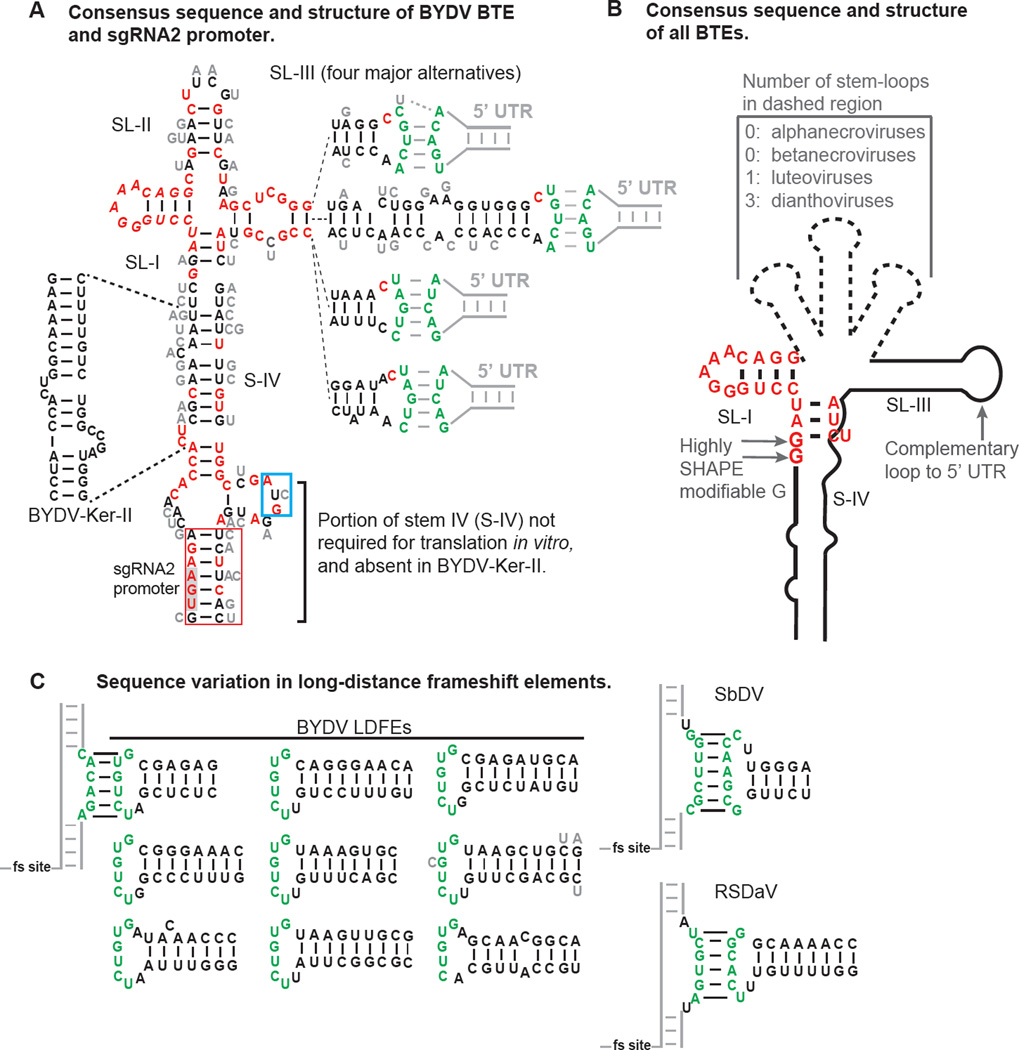
Fig. 2.
Secondary structures that regulate luteovirus translation. A. Secondary structure and base variations of BYDV BTEs. The 5’ end of the sequence shown is the 5’ end of sgRNA2. Sequence variation in fifty-one isolates of BYDV species is indicated, with invariant bases in red. Alternative bases found at each position are shown in gray beside each base. There are four major forms of the distal end of stem-loop III (SL-III), with each indicated separately, connected by dashed lines to the proximal end of SL-III. Green bases form kissing stem-loop base pairing with the stem-loop in the 5’ UTR indicated by gray lines. The sequence in the red box is sufficient for production of sgRNA2 by BYDV-PAV6. UGA (gray shading) is stop codon of ORF 5. AUG in blue box is the ORF 6 start codon. B. Consensus structure of known BTEs, which includes all viruses in the Luteovirus, Dianthovirus and Necrovirus genera. Note that the RCNMV BTE has no known loop complementary to the 5’ UTR. The first or second guanosine residue (but not both) in the 17 nt conserved sequence is highly modified by SHAPE probing in BTEs of all genera, indicating that it is not base paired and may protrude from the structure in an unusual way (Kraft et al., 2013). (Two of the fifty-one isolates contain an A at position 2 of the 17 nt conserved sequence. Their translation efficiency is unknown.) C. Base pairing of the long-distance frameshift elements (LDFE) in the 3’ UTR of luteoviruses with a bulge adjacent to frameshift site. Sequences found in 51 isolates of five BYDV species contain one of the nine structures shown. Note the lack of sequence conservation in the stem, but conservation of the loop that base pairs to the bulged helix, CACAGA, which is invariant in all 51 isolates adjacent to the frameshift site (fs site). One isolate (gray C, Genbank accession no. EU332331) has a mismatch, and another (BYDV-Ker-II, accession no. NC_021481, bottom right) has only five bases of complementarity. Different sequences participate in long-distance base pairing in SbDV and RSDaV. No obvious LDFE was identified in the 3’ UTR of BLRV.
There does not seem to be a requirement for a specific sequence in the kissing stem-loops, as they can be altered and still facilitate translation as long as complementarity is retained. For infectious clone BYDV-PAV6, the kissing stem-loop base pairing sequence UGUCA:UGACA between the BTE and 5’ UTR[WAM1] has been determined experimentally. Analysis of genome sequences of fifty-one BYDV isolates representing several species (BYDV-GAV, BYDV-PAV, BYDV-PAS and BYDV-KerII), suggests that a subset, GACA:UGUC, may be sufficient, and that UAGUC:GACUA also may facilitate translation (Fig. 2A). This is not surprising, as other sequences are predicted to base pair in kissing stem-loops of Soybean dwarf virus (Soybean dwarf virus (SbDV) (Soybean dwarf virus, genus Luteovirus, family Luteoviridae), and Bean leafroll luteovirus (BLRV) (genus Luteovirus, family Luteoviridae) and the alpha- and betanecroviruses (Meulewaeter et al., 2004; Shen and Miller, 2004a). In fact, the complementary bases need not be located within the BTE or in kissing stem-loops. The BYDV 3’BTE is capable of facilitating cap-independent translation when added to the 3’ UTR of a reporter construct containing 5’ and 3’ UTRs of dengue virus flanking a luciferase ORF (Rakotondrafara et al., 2006). A ten base cyclization sequence in the 5’ end of dengue (and other flavivirus) RNA base pairs to a complementary sequence in the 3’ UTR (Khromykh et al., 2001). This long-distance base pairing is sufficient to replace the need for the kissing stem-loop base pairing present in BYDV RNA, and allows the BTE in the dengue virus reporter 3’ UTR to facilitate cap-independent translation (Rakotondrafara et al., 2006).
An apparent exception to the rule for long-distance base pairing to the 5’ UTR appears to occur in translation mediated by the BTE of Red clover necrotic mosaic virus (Red clover necrotic mosaic virus, genus Dianthovirus, family Tombusviridae, RCNMV). RNA1 of RCNMV and other dianthoviruses has a remarkable BTE (called 3’TE-DR1) consisting of six helices (five stem-loops and one stem connecting to the rest of the genome) radiating from the central hub (Kraft et al., 2013; Mizumoto et al., 2003). Stem-loop I (SL-I) includes most of the conserved 17 nt sequence present in all BTEs, so any of the other stem-loops are candidates for functional homologs to SL-III in the BYDV BTE which base pairs to the 5’ UTR. However, mutations in the 5’ UTR predicted to disrupt base-pairing to the 3’TE-DR1 did not affect translation in cowpea protoplasts. In contrast, mutation or deletion of any of the 5’ UTR loops including predicted 3’TE-DR1-complementary sequences did prevent translation in tobacco BY2 protoplasts, but this translation could not be restored by compensating mutations in the 3’ BTE (3’TE-DR1) (Sarawaneeyaruk et al., 2009). Thus there are clear host-specific differences in translational control of RCNMV RNA. Moreover, unlike with BYDV, the authors identified a role for the 5’-proximal stem-loop of the 5’ UTR of RCNMV RNA1 in translation enhancement (Sarawaneeyaruk et al., 2009). Disruption by mutagenesis of a predicted kissing loop interaction between the STNV translation enhancer domain (TED) and the 5’ UTR did not significantly affect its ability to stimulate cap-independent translation (Meulewaeter et al., 1998). Thus, some 3’ CITEs may find ways to interact with the 5’ UTR without dependence on Watson-Crick base pairing.
The 3’ CITE of luteoviruses, the BTE, binds eIF4G (Kd for the BYDV BTE = 177 nM) on which it depends to facilitate translation (Banerjee and Goss, 2014; Treder et al., 2008). eIF4E is not necessary for BTE-mediated translation, although it enhances the activity of eIF4G by about 25% in a wheat germ translation extract. Congruent with this observation, the C-terminal half of eIF4G is sufficient to facilitate BTE-mediated translation (Kraft et al., 2013). This truncation lacks the eIF4E binding domain, as well as the PABP binding domain, neither of which are necessary. This result indicates that the region(s) of eIF4G that provide high affinity binding to the BTE are also located in the C-terminal half of the protein. An RNA-binding site located between the eIF4E binding site and the MIF4G region of eIF4G which is required to facilitate ribosome scanning, is suspected to play a role, as deletion of this region prevents eIF4G from restoring cap-independent translation in an eIF4F-depleted wheat germ extract (Kraft et al., 2013).
How the structure of the 3’ BTE attracts high affinity binding of eIF4G, thus stimulating translation is unclear. Phylogenetic comparisons of BTE structures reveal three to six helices protruding from a central hub (Wang et al., 2010) (Fig. 2B). There are usually several unconserved, non-paired bases at this junction region. Mutation of most of these bases to each of the three other possible bases greatly reduced activity of the BYDV BTE (Guo et al., 2000; Kraft et al., 2013). Phylogenetic comparison of BTEs from 51 BYDV isolates (representing five BYDV species) confirms that Watson-Crick paired bases, and non-paired [WAM3]bases tolerate little variation at the helical junction (Fig. 2A). Moreover, deletion of SL-II to make the BYDV BTE resemble that of the alpha/betanecroviruses which lacks a structural homolog to SL-II, also destroyed BTE activity (Guo et al., 2000). Thus there are key interactions around the core hub region with no obvious sequence or structure that are necessary for BTE function. Fortunately, we now have a better understanding of the interaction with eIF4G. RNA footprinting experiments using SHAPE probing to simultaneously determine effect of eIF4G binding on BTE structure and to identify those bases protected from modification by eIF4G binding revealed that eIF4G binds SL-I and certain other bases around the hub, including the conserved bases at the proximal end of SL-III (Fig. 2A). eIF4G binding did not affect overall BTE structure (Kraft et al., 2013). Thus, an induced fit binding mechanism is unlikely. SL-I resembles the boxB RNA of bacteriophage lambda, which is bound by the N protein to terminate transcription in an interaction well understood at atomic resolution (Legault et al., 1998). Both the BTE and boxB contain a 5 nt tetraloop fitting the motif GNRNA, so eIF4G may bind the BTE via similar interactions as N on boxB.
Upon binding the BTE, eIF4G may then recruit additional factors including eIF3 which brings in the 40S ribosomal subunit. Whether ribosome recruitment occurs directly to the BTE or to the 5’ UTR after the BTE base pairs to the 5’ UTR allowing it to “deliver” eIF4G to the 5’ end is unknown. A six base conserved sequence that overlaps with SL-I, GAUCC, is complementary to ribosomal 18S RNA at the position homologous to the site of the anti-Shine-Dalgarno sequence in Escherichia coli. Thus, it was proposed that the BTE may base pair to the 18S rRNA to recruit the ribosome, in addition to eIF4G-mediated recruitment (Wang et al., 1997). In this case, the 40S ribosomal subunit would bind the BTE directly and be delivered to the 5’ end directly. Owing to the known requirement for scanning from the 5’ end (Guo et al., 2001; Rakotondrafara et al., 2006) (Sarawaneeyaruk et al., 2009), we conclude that the ribosome is not delivered directly to the start codon.
A stem-loop structure in the 3’ UTR about 200 nt downstream of the BTE is required for ribosomal frameshifting, which occurs almost 4000 bases upstream to facilitate translation of the RdRp (Fig. 1). This stem-loop, called the long-distance frameshift element (LDFE), contains a loop (UCUGUG) that base-pairs to a bulge (CACAGA) in a large bulged stem-loop immediately downstream of the frameshift site, to form a complex pseudoknot that is required for the low level (around 1–2% frameshifting) needed for expression of the RdRp and thus replication (Barry and Miller, 2002). Sequence comparisons of the 51 BYDV isolates reveal that, except for one Chinese BYDV-PAV isolate and BYDV-KerII (in which only five bases are complementary), the long distance base pairing sequence does not vary (Fig. 2C). In contrast, there seem to be no constraints on the sequences of the stem of the LDFE, and it varies from 6 bp in the isolate in which it was discovered, to 9 base pairs (Fig. 2C). This differs from the BTE which shows more conservation in base paired regions around the helical junction region, but tolerated differences in the long-distance base pairing sequence. However, the long-distance base pairing sequences of the LDFEs of other luteoviruses, such as SbDV and rose spring dwarf-associated virus (RSDaV, Rose spring dwarf-associated virus, genus Luteovirus, family Luteoviridae) differ from that of BYDV (Fig. 2C) (Barry and Miller, 2002; Salem et al., 2008). Similar long-distance base pairing, spanning thousands of bases, between an LDFE in the 3’ UTR and a bulged stemloop adjacent to the frameshift site was shown to be required for frameshifting by the dianthovirus RCNMV (Tajima et al., 2011).
Programmed readthrough of the CP ORF stop codon to generate the CP-RTD protein is also predicted to be facilitated by long-distance base pairing with a downstream sequence (Brown et al., 1996). This mechanism also requires a (CCXXXX)8 repeat sequence in the RNA beginning about 8 nt downstream of the stop codon. Andy White’s lab has shown readthrough controlled by such long-distance base pairing in various tombusvirids (Cimino et al., 2011; Newburn et al., 2014). In BYDV sgRNA1, the long-distance base pairing is predicted to span “only” 700 nucleotides (Fig. 1). Because these control elements are not near the ends of a viral RNA, we will not discuss them further.
The signals for cap-independent translation and ribosomal frameshifting may be located thousands of bases downstream of the site of action in order to facilitate a switch from translation of the viral genome to replication of the viral genome. A viral RNA can’t be replicated and translated at the same time (Barton et al., 1999; Gamarnik and Andino, 1998). In the presence of translating ribosomes, it is highly unlikely that the ~100 kDa RdRp could displace the 3.2 million kDa 80S ribosome as it moves along the RNA synthesizing protiens. Thus translation must be stopped and ribosomes removed from the RNA before it can be replicated. As proposed previously (Barry and Miller, 2002; Miller and White, 2006), this may be achieved as follows. After the long-distance base pairing allows cap-independent translation and ribosomal frameshifting to produce the viral RdRp, the viral RdRp then proceeds to initiate negative strand synthesis at the 3’ end. As the RdRp moves in the 5’ direction on the template, the RdRp would encounter the LDFE and BTE, and disrupt their long-distance interactions with the frameshift site, and 5’ UTR, respectively. These disruptions would shut off translation far upstream and free the viral RNA of ribosomes. This, in turn, would permit complete copying of the viral RNA and RNA replication would proceed unfettered by the translational machinery.
3.2 Control of translation by the 3’ end in trans
BYDV generates three subgenomic RNAs in infected cells (Kelly et al., 1994; Koev et al., 1999). The largest, sgRNA1, is the mRNA from which the P3a, CP, P4 and CP-RTD proteins are translated (Fig. 1). The 5’ end of sgRNA2 maps to a region near the ORF 5 (CP-RTD) stop codon. It contains the BTE as its 5’ UTR and codes for the enigmatic ORF6. In ORF6, the first 15 codons or so show variation in the wobble position that supports selection for a functional protein, but the 3’ end of ORF 6 is the most variable region in the entire genome, often including a stop codon, so the encoded protein (P6) ranges from 4.3 to 7.2 kDa. ORF6 is not conserved in other luteoviruses (Domier et al., 2002; Yamagishi et al., 2003). Mutation of the start codon of ORF6, or fusion of ORF6 to a reporter gene prevented replication of BYDV-PAV RNA in oat protoplasts, however it isn’t clear whether these effects are due to a requirement for a functional P6 protein or due to disruption of key cis-acting signals in the RNA (Shen et al., 2006). Despite efforts, P6 protein was not detected by immunoblot in BYDV-infected protoplasts (Shen et al., 2006). However, Liu et al. (Liu et al., 2012) provided evidence that P6 of BYDV-GAV is a viral suppressor of RNA-mediated silencing. They showed that agrobacterium vector engineered to express P6, suppressed silencing of GFP in transgenic 16c Nicotiana benthamiana plants. However, the authors did not demonstrate expression of P6 protein (Liu et al., 2012).
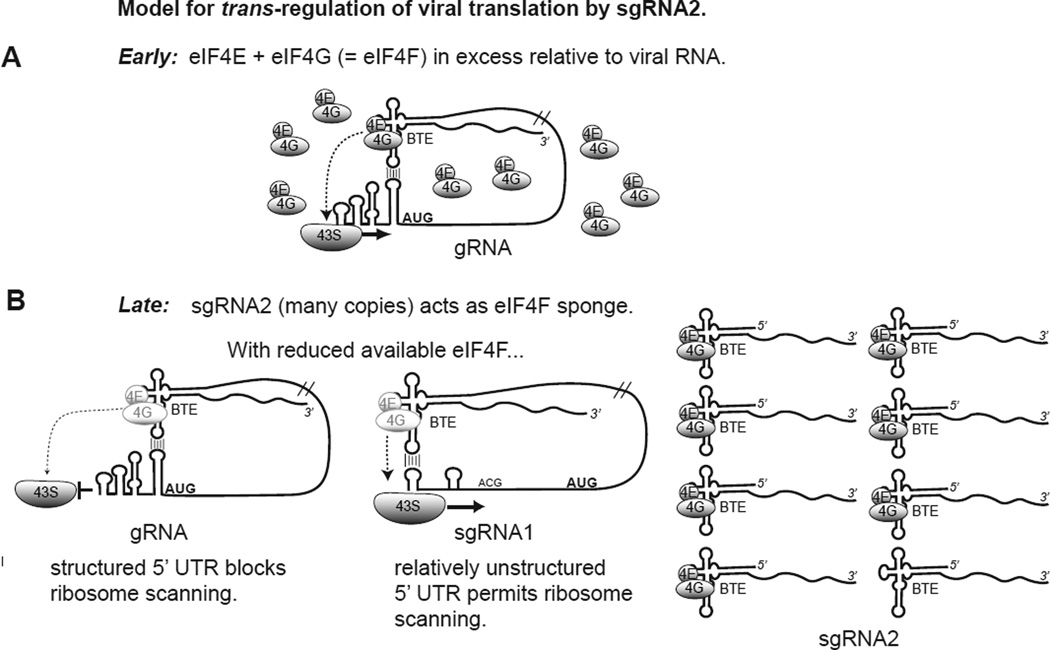
Independent of the role of P6, sgRNA2 RNA itself can act as a trans-regulator of viral translation (Fig. 3). sgRNA2 accumulates to a vast molar excess relative to the larger sgRNA1 and even the genome (Fig. 4A) (Shen and Miller, 2004b). By virtue of the BTE at its 5’ end, sgRNA2 effectively inhibits translation of capped mRNAs and other BTE-containing RNAs (Shen and Miller, 2004b) by binding eIF4G (Treder et al., 2008). In the presence of the viral genomic RNA and sgRNA1, sgRNA2 selectively inhibits translation of genomic RNA. This seems to be an elegant mechanism by which the virus switches from translating replicase from genomic RNA early in infection when no sgRNAs are present, to translation of the coat and movement protein genes later in infection, when sgRNA1 and sgRNA2 accumulate (Fig. 3). This differential translation of sgRNA1 in favor of genomic RNA is conferred by the 5’ end sequence of sgRNA1, as reporter mRNAs that differed only by containing either the genomic RNA or sgRNA1 5’ UTR showed the same differential translation in the presence of sgRNA2 (Shen et al., 2006). This differential translation appears to be due to the sequestration of eIF4G by sgRNA2, because, under normal translation conditions, it is not seen in the absence of sgRNA2 (i.e. genomic and sgRNA1 both translate equally). Under highly competitive conditions, such as high mRNA concentrations or in extracts with reduced levels of eIF4F, then the advantage for sgRNA1 is apparent (K. Treder, personal communication). This reduced dependence on eIF4G can be explained by the relative lack of secondary structure in the 5’ end of sgRNA1 compared to the highly structured 5’ UTR of genomic RNA (Pestova and Kolupaeva, 2002; Shen et al., 2006).
Fig. 3.
Trans-regulation model of BYDV gene expression. A. Early in infection, when few viral genomes are present, the BTE binds the eIF4G subunit of the key translation initiation factor, eIF4F and delivers it to 5’ end via long-distance base pairing to recruit (or deliver) the 43S ribosomal pre-initiation complex to the 5’ end. The relative excess of eIF4F in the cell facilitates ribosome scanning through the highly structured 5’ UTR. B. Late in infection, the vast excess of sgRNA2 binds much of the eIF4F pool, leaving fewer eIF4F complexes available for translation of gRNA and sgRNA1 (light shaded eIF4E/eIF4G). This allows structure in gRNA 5’ UTR to inhibit ribosome scanning, while 5’ UTR of sgRNA1 has less structure, making it less eIF4F-dependent (Pestova and Kolupaeva, 2002), thus permitting ribosome scanning and translation. Thus sgRNA2 accumulation serves as a switch to inhibit translation of the polymerase from gRNA, and favor translation of structural (CP, CP-RTD) and movement proteins (ORF3a, ORF4) from sgRNA1 (Fig. 1). Details and supporting data are in Shen et al. (Shen et al., 2006).
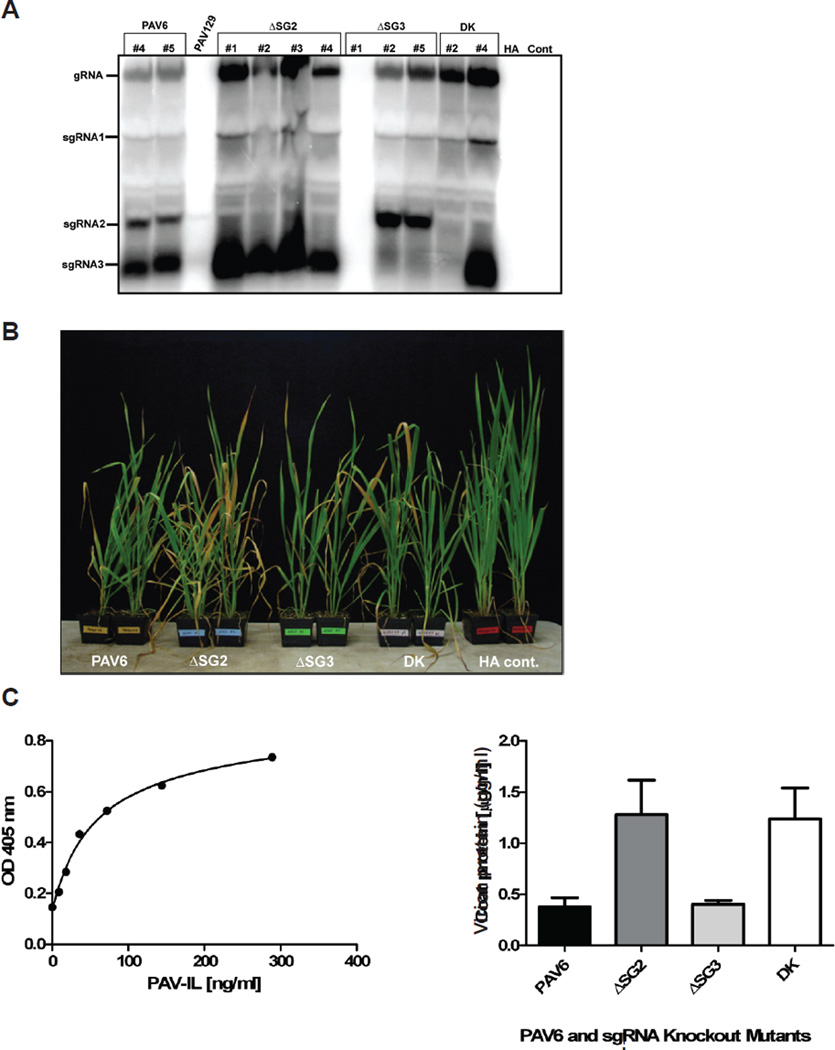
Fig. 4.
Infection of oat plants with BYDV-PAV6 mutants unable to synthesize sgRNA2 and/or sgRNA3. A. Northern blot hybridization of RNA from infected plants. Total RNA was isolated from plants infested with viruliferous aphids (Rhopalosiphum padi) containing the indicated mutant virus that showed symptoms approximately 7 weeks post-infestation (wpi). Aphids acquired virus purified from oat protoplasts 48 hr after transfection with infectious transcript containing the indicated mutation in PAV6. Mobilities of genomic and sgRNAs are identified on left side of blot. Specific plants that were tested in the study are identified by the numbers at the top of each lane. B. Comparison of virus symptoms induced by wild type BYDV-PAV6 and BYDV-PAV6 sgRNA knockout mutants. Plants were inoculated with viruliferous aphids containing the indicated mutant virus. Two plants taken from each group of infected plants were compared to plants infested with virus-free aphids (healthy aphid controls, HA cont.). BYDV-PAV6 and sgRNA knockout infected plants show similar symptoms while controls show no signs of infection by BYDV at ~7 wpi. C. ELISA detection of BYDV virions in infected oat (Clintland 64) plants. Total protein was extracted from infected plant tissue at 7 weeks post-infestation. Several leaves were pooled from each plant and four replicates were tested for each pooled leaf sample. Left: calibration curve of BYDV-PAV virion standard. Right: Total virion amounts in plants inoculated with the indicated mutant BYDV-PAV6 constructs.
The other luteoviruses SbDV and BLRV also generate a small sgRNA that corresponds in position to sgRNA3 of BYDV, but apparently not an sgRNA2 homologue (Domier et al., 2002; Yamagishi et al., 2003). However, viruses in the closely related genus, Dianthovirus do express an sgRNA2-like RNA with a similar trans-regulatory function. RCNMV generates a small 3’ co-terminal sgRNA (SR1f) containing the BTE near its 5’ end (Iwakawa et al., 2008). Like sgRNA2, this SR1f RNA inhibits cap-independent (viral) and cap-dependent (host) translation in trans. Whether it selectively controls translation of the other viral RNAs, as demonstrated for BYDV, has not been reported. SR1f also inhibits negative strand RNA[WAM6] synthesis, which has not been tested for BYDV.
Recently much of the sequence that was thought to be the 172 nt 5’ UTR on BYDV-PAV sgRNA1 was discovered to encode a small ORF (Fig. 1). All luteoviruses and poleroviruses contain a 45 – 50 codon ORF (ORF3a) beginning with a non-AUG codon (AUU, AUA, ACG, or CUG depending on the virus) and terminating between the start codons of ORFs 3 (CP ORF) and ORF 4 (Smirnova et al., 2015).[WAM7] Owing to the lack of an AUG codon, ORF 3a is translated at low levels, so most ribosomes (40S subunits) do not initiate at the ORF3a start codon and continue to scan until they initiate translation at the ORF 3 (CP) or ORF4 start codon. Thus, for these ribosomes, the first 172 nt on sgRNA1 is effectively untranslated sequence. Whether the actual 64 nt 5’ UTR ending at the start codon of ORF3a is sufficient to confer the translational advantage to sgRNA1 is unknown, but this does not affect the interpretation of the observations that led to the trans-regulation model before ORF3a was discovered.
4. Subgenomic RNAs 2 and 3 are dispensible for infection
BYDV-PAV that lacked the ability to produce sgRNA2, via a point mutation in the sgRNA2 promoter, differentially affected the ability of reporter RNAs to translate in infected cells (Shen et al., 2006). However, this mutation did not appear to affect viral RNA accumulation in infected protoplasts. We took advantage of this phenomenon to demonstrate the role of sgRNA2 accumulation in reducing translation of other RNAs in trans (Shen et al., 2006). The lack of need for sgRNA2 for viral replication in individual cells led us to speculate that perhaps sgRNA2, as well as sgRNA3, are required only for infection of whole plants. We tested the effects of mutations that knock out synthesis of sgRNA2 and/or sgRNA3 on viral accumulation in whole plants.
Infectious clones of BYDV (PAV6) containing point mutations that knock out synthesis of sgRNA2 (ΔsgRNA2) or sgRNA3 (ΔsgRNA3) or both (DK) were transfected into protoplasts as described in Shen et al. (Shen et al., 2006). After 48 hr, virions were partially purified from the protoplasts, then fed to aphids (Rhopalosiphum padi) in a 20% sucrose solution through Parafilm membranes. After 24 hr acquisition time, aphids were placed on 7 day-old oat (Clintland 64) plants for 3 days at which point the aphids were removed with insecticide. At various times post-infestation, plants were examined for symptoms and tested for virus by northern blot hybridization and ELISA.
Surprisingly, inability to synthesize sgRNA2 or sgRNA3 or both RNAs had no negative effect on virus accumulation in plants. Northern blot hybridization showed high levels of accumulation of viral RNAs, except the deleted sgRNA (Fig. 4A). The retention of the point mutations were confirmed by sequencing. Plants inoculated with the sgRNA2/sgRNA3 knockouts also showed similar yellowing symptoms as when infected with wild type virus (Fig. 4B). ELISA assays revealed accumulation of more virus coat protein, suggesting presence of more virus particles, when sgRNA2 was absent due to the point mutations, but not when sgRNA3 alone was absent (Fig. 4C). Strikingly, occasionally a new sgRNA would appear, of a different size than either sgRNA2 or sgRNA3. For example, plant #4 inoculated with PAV6 mutant containing the double knockout (DK) of sgRNA2 and sgRNA3 gave rise to an abundant sgRNA slightly smaller than sgRNA3 (Fig. 4A).
We conclude that, despite its demonstrated ability to selectively inhibit translation of gRNA relative to sgRNA1, and also to inhibit translation of capped, nonviral mRNAs (Shen and Miller, 2004b), sgRNA2 is not required for the virus to replicate and spread within a plant in a reasonably efficient way, at least within the highly susceptible Clintland 64 oats used here. The mutant viruses lacking sgRNA2 and/or sgRNA3 may still be less fit than the wild type virus in a way that would be detectable only by direct competition with wild type virus. This would require co-infecting plants with wild type and and mutant viruses. It is also possible that sgRNAs 2 and 3 are needed only for infection of certain hosts. P6, or possibly sgRNA2 itself, has been reported to serve as a VSR but only in N. benthamiana which is not a natural host (Liu et al., 2012). Importantly, the sgRNA2 homologue of RCNMV, SR1f, which, like sgRNA2, inhibits translation in trans, also is not necessary for virus replication in cells or in plants (Iwakawa et al., 2008). However, SR1f is noncoding and the virus containing the point mutation that prevented SR1f synthesis accumulated to somewhat lower levels than wild type virus (Iwakawa et al., 2008).
5. Terminal structures required for RNA replication
5.1 3’-terminal structure of BYDV
For want of a cell-free replication system for luteoviruses, little is known about the structures required directly for RNA synthesis. Nor is the replicase binding site on luteovirus RNA known. It may be at the 3’ end where (−) strand synthesis begins or it could be far upstream, as in the case of tombusviruses (Pogany et al., 2005). However, the secondary structural requirements of the 3’-terminal 100 nt for accumulation of BYDV RNA have been determined. The sequence folds into three to four stem-loops, depending on the BYDV virus (Fig. 5). A striking feature is that the 3’-terminal bases are embedded in a helix that coaxially stacks with an internal helix: stem-loop 3 (Fig. 5) (Koev et al., 2002). We propose that this hides the 3’ end from exonucleases, as well as the viral replicase, because it may participate in a conformational switch to control RNA synthesis. In this embedded form, negative strand synthesis would be inhibited due to the inability of the replicase to access the 3’ end, and positive strand synthesis from the 3’ end of the negative strand would be favored. In a possible alternative conformation, the four 3’-terminal bases are exposed and single stranded, and this would allow recognition by the replicase for negative strand synthesis (Koev et al., 2002). Because this is a positive strand RNA virus, we speculate that the embedded conformation would be favored most of the time, favoring positive strand RNA accumulation. This conformation would also be favored during translation. We found that this terminal sequence also enhanced translation in vivo. Upon its removal, translation in vivo was reduced, and addition of a poly(A) tail restored efficient translation in vivo, consistent with a role for the 3’ end structure in protection from exonucleases (Shen and Miller, 2007).
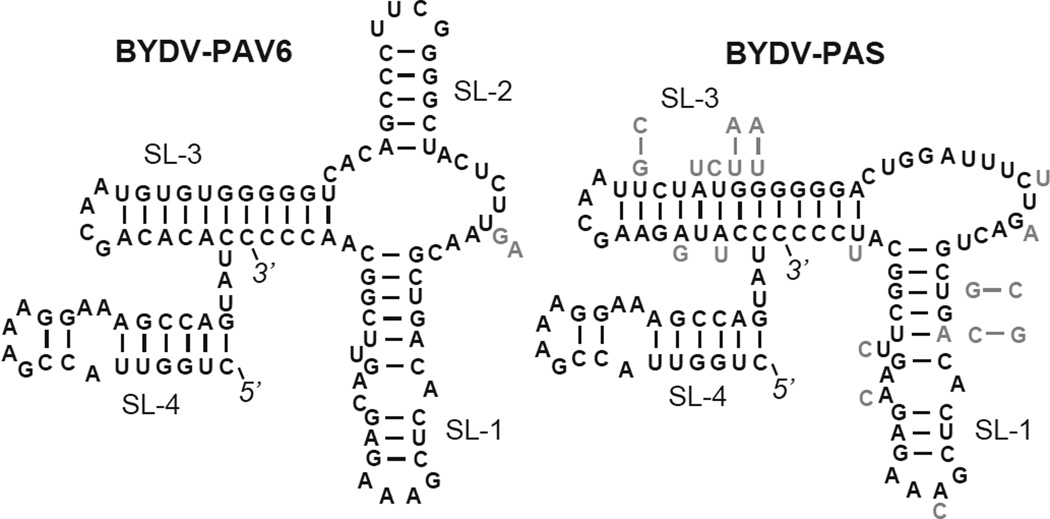
Fig. 5.
Secondary structures of the 3’ end of BYDV genomes, determined for BYDV-PAV6 (Genbank no. NC_004750) (Koev et al., 2002) and predicted here for 50 other luteoviruses. Twenty-two of the 50 BYDVs formed a structure identical in sequence to that of BYDV-PAV NC_004750 on the left, and 28 of the viruses/isolates formed the structure on the right with one less stem-loop. Sequence of BYDV-PAS (Genbank no. AF218798) is shown. Variant bases are shown in gray beside the particular base in the structure. When bases co-vary to conserve base pairing, the entire variant base pair is shown in gray beside or above the helix. Note that in all cases, the 3’ terminal nucleotides are base-paired in a helix that co-axially stacks with stem-loop 3 (SL-3).
In support of the structures that were based on four BYDV sequences and previous mutagenesis experiments (Koev et al., 2002), here we compare the 3’ ends of 51 isolates of five BYDV species. All of the sequences folded into one or the other of the two previously determined structures. Twenty-eight of the structures lack SL-2, while the remainder have the exact same sequence as the structure containing SL-2 (Fig. 5).
5.2. Stem-loop at the 5’ terminus
Compared to the 3’ UTR, the 5’ UTR appears to be less complex. As mentioned above, the 5’ end contains a stem-loop that kisses with the BTE to facilitate translation. But 5’ UTR structures required for replication of luteovirus RNA have not been determined. One common denominator is a modest, conserved stem-loop[WAM10] that begins just one nucleotide from the 5’ end of luteovirus genomic RNA and sgRNA1. The complementary strand would be predicted to form this stem-loop as well. This complementary stem-loop may serve as a replication origin for plus strand RNA synthesis. A similar stem-loop is found in many tombusvirids, and is required for RNA replication (Panavas et al., 2002). In addition, the plus strand version of the stem-loop may serve to protect against exonucleolytic degradation. An unanswered question for luteoviruses and the tombusvirids, is how their RNAs, lacking a 5’ cap or any other modification, evade Xrn4, the exonuclease that degrades uncapped mRNAs in plants. Perhaps this stem-loop has sufficient structure to block Xrn4. However, RCNMV takes advantage of cellular exonulease activity to generate the noncoding SR1f subgenomic RNA (Iwakawa et al., 2008). The exonuclease degrades RCNMV RNA1 from the 5’ end until it is blocked by a structure 400 nt from the 3’ end. The remaining 3’ fragment is the SR1f RNA (Iwakawa et al., 2008). The same mechanism generates a 3’ noncoding sgRNA of flaviviruses (Pijlman et al., 2008).
6. Conclusion
To summarize, viruses are complex systems with intricate regulatory processes controlled by the structures of the viral RNA and, in turn, its interaction with host and viral proteins. These structures and thus interactions must change with time over the course of the replication process as proteins and RNAs accumulate. Luteoviruses in particular display many elegant RNA-mediated regulatory processes. One such manifestation is the long-distance interactions required for translation that allow translation initiation and frameshifting to be sensitive to structural changes thousands of bases downstream. Another is the “sponging” of translation factor eIF4F by sgRNA2 as it accumulates to massive levels, which favors translation of late proteins from sgRNA1. Finally, local structural alternatives at the extreme 3’ terminus may control switching from negative to positive strand RNA synthesis and translation, although the data to support this hypothesis remain to be gleaned.
The complexity of these regulatory mechanisms, even in such a small genetic entity, makes them difficult to identify and predict. Hence, we are left with the puzzle of sgRNAs 2 and 3 which appear to be dispensible for infection of oat plants, giving symptoms and virus yield that differ little from wild type. However, many thought-to-be-indispensible genes from higher organisms have been deleted with little effect on apparent fitness. Most likely, sgRNAs 2 and 3 provide a slight competitive advantage that would be detected only by direct comparison in co-infections with wild type virus. Or perhaps the environmental conditions, or host genotype or species for which the sgRNAs 2 and 3 provide an advantage are different than in our system which employed highly susceptible oats (cv. Clintland 64) in growth chambers. The selective forces in the field would be indeed quite different. Many other questions remain to be answered, such as (i) how the ends of these unmodified RNAs deter exonucleolytic degradation, (ii) the replicase binding site(s) and requirements for plus and minus strand RNA synthesis, (iii) the mechanisms by which the subgenomic RNAs arise, and (iv) the sequences that interact with the coat protein for virion assembly.
Highlights.
Luteoviruses employ a plethora of noncanonical translation mechanisms.
Base pairing between the 3’ UTR and regions kilobases upstream controls translation.
A subgenomic RNA can regulate viral translation in trans.
Small subgenomic RNAs may be optional for virus replication.
Acknowledgments
This work was funded by NIH grant 5 R01067104-11 to WAM, and NIH grant F31 AI 56673 to JJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bakker SE, Ford RJ, Barker AM, Robottom J, Saunders K, Pearson AR, Ranson NA, Stockley PG. Isolation of an asymmetric RNA uncoating intermediate for a single-stranded RNA plant virus. J Mol Biol. 2012;417(1–2):65–78. doi: 10.1016/j.jmb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee B, Goss DJ. Eukaryotic initiation factor (eIF) 4F binding to barley yellow dwarf virus (BYDV) 3'-untranslated region correlates with translation efficiency. J Biol Chem. 2014;289(7):4286–4294. doi: 10.1074/jbc.M113.530329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JK, Miller WA. A-1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc Natl Acad Sci U S A. 2002;99(17):11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DJ, Morasco BJ, Flanegan JB. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J Virol. 1999;73(12):10104–10112. doi: 10.1128/jvi.73.12.10104-10112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batten JS, Desvoyes B, Yamamura Y, Scholthof KB. A translational enhancer element on the 3'-proximal end of the Panicum mosaic virus genome. FEBS Lett. 2006;580(11):2591–2597. doi: 10.1016/j.febslet.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Brown CM, Dinesh-Kumar SP, Miller WA. Local and distant sequences are required for efficient read-through of the barley yellow dwarf virus-PAV coat protein gene stop codon. J. Virol. 1996;70:5884–5892. doi: 10.1128/jvi.70.9.5884-5892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino PA, Nicholson BL, Wu B, Xu W, White KA. Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS pathogens. 2011;7(12):e1002423. doi: 10.1371/journal.ppat.1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier LL. Family Luteoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on the Taxonomy of Viruses. Amsterdam: Elsevier Academic Press; 2012. pp. 1045–1053. [Google Scholar]
- Domier LL, McCoppin NK, Larsen RC, D'Arcy CJ. Nucleotide sequence shows that Bean leafroll virus has a Luteovirus-like genome organization. J Gen Virol. 2002;83(Pt 7):1791–1798. doi: 10.1099/0022-1317-83-7-1791. [DOI] [PubMed] [Google Scholar]
- Fabian MR, White KA. Analysis of a 3'-translation enhancer in a tombusvirus: a dynamic model for RNA-RNA interactions of mRNA termini. RNA. 2006;12(7):1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12(15):2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Kasprzak W, Stupina VA, Shapiro BA, Simon AE. A ribosome-binding, 3' translational enhancer has a T-shaped structure and engages in a long-distance RNA-RNA interaction. Journal of virology. 2012;86(18):9828–9842. doi: 10.1128/JVI.00677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazo BM, Murphy P, Gatchel JR, Browning KS. A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3'-untranslated region of satellite tobacco necrosis virus. J Biol Chem. 2004;279(14):13584–13592. doi: 10.1074/jbc.M311361200. [DOI] [PubMed] [Google Scholar]
- Guo L, Allen E, Miller WA. Structure and function of a cap-independent translation element that functions in either the 3' or the 5' untranslated region. RNA. 2000;6:1808–1820. doi: 10.1017/s1355838200001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Allen E, Miller WA. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Mizumoto H, Nagano H, Imoto Y, Takigawa K, Sarawaneeyaruk S, Kaido M, Mise K, Okuno T. A viral noncoding RNA generated by cis-element-mediated protection against 5'->3' RNA decay represses both cap-independent and cap-dependent translation. J Virol. 2008;82(20):10162–10174. doi: 10.1128/JVI.01027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly L, Gerlach WL, Waterhouse PM. Characterisation of the subgenomic RNAs of an Australian isolate of barley yellow dwarf luteovirus. Virology. 1994;202(565):565–573. doi: 10.1006/viro.1994.1378. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J Virol. 2001;75(14):6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev G, Liu S, Beckett R, Miller WA. The 3'-terminal structure required for replication of barley yellow dwarf virus RNA contains an embedded 3' end. Virology. 2002;292:114–126. doi: 10.1006/viro.2001.1268. [DOI] [PubMed] [Google Scholar]
- Koev G, Mohan BR, Miller WA. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J. Virol. 1999;73:2876–2885. doi: 10.1128/jvi.73.4.2876-2885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft JJ, Treder K, Peterson MS, Miller WA. Cation-dependent folding of 3' cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with eIF4G. Nucleic Acids Res. 2013;41(5):3398–3413. doi: 10.1093/nar/gkt026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93(2):289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- Lister RM, Ranieri R. Distribution and economic importance of barley yellow dwarf. In: D'Arcy CJ, Burnett P, editors. Barley yellow dwarf: 40 years of progress. St. Paul: APS Press; 1995. pp. 29–53. [Google Scholar]
- Liu Y, Zhai H, Zhao K, Wu B, Wang X. Two suppressors of RNA silencing encoded by cereal-infecting members of the family Luteoviridae. J Gen Virol. 2012;93(Pt 8):1825–1830. doi: 10.1099/vir.0.042135-0. [DOI] [PubMed] [Google Scholar]
- Meulewaeter F, Danthinne X, Van Montagu M, Cornelissen M. 5'- and 3'-sequences of satellite tobacco necrosis virus RNA promoting translation in tobacco. Plant J. 1998;14(2):169–176. doi: 10.1046/j.1365-313x.1998.00104.x. [published erratum appears in Plant J 1998 Jul;15(1):153–4] [DOI] [PubMed] [Google Scholar]
- Meulewaeter F, van Lipzig R, Gultyaev AP, Pleij CW, Van Damme D, Cornelissen M, van Eldik G. Conservation of RNA structures enables TNV and BYDV 5' and 3' elements to cooperate synergistically in cap-independent translation. Nucleic Acids Res. 2004;32(5):1721–1730. doi: 10.1093/nar/gkh338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Liu S, Beckett R. Barley yellow dwarf virus: Luteoviridae or Tombusviridae? Mol. Plant Pathol. 2002;3:177–183. doi: 10.1046/j.1364-3703.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- Miller WA, White KA. Long distance RNA-RNA interactions in plant virus gene expression and replication. Ann. Rev. Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras M, Sempere RN, Kraft JJ, Miller WA, Aranda MA, Truniger V. Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 2014;202(1):233–246. doi: 10.1111/nph.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto H, Tatsuta M, Kaido M, Mise K, Okuno T. Cap-independent translational enhancement by the 3' untranslated region of red clover necrotic mosaic virus RNA1. J Virol. 2003;77(22):12113–12121. doi: 10.1128/JVI.77.22.12113-12121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburn LR, Nicholson BL, Yosefi M, Cimino PA, White KA. Translational readthrough in Tobacco necrosis virus-D. Virology. 2014;450–451:258–265. doi: 10.1016/j.virol.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Nicholson BL, White KA. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol. 2014;12(7):493–504. doi: 10.1038/nrmicro3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BL, Wu B, Chevtchenko I, White KA. Tombusvirus recruitment of host translational machinery via the 3' UTR. RNA. 2010;16(7):1402–1419. doi: 10.1261/rna.2135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BL, Zaslaver O, Mayberry LK, Browning KS, White KA. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J Virol. 2013;87(3):1872–1883. doi: 10.1128/JVI.02711-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T, Pogany J, Nagy PD. Analysis of minimal promoter sequences for plus-strand synthesis by the Cucumber necrosis virus RNA-dependent RNA polymerase. Virology. 2002;296(2):263–274. doi: 10.1006/viro.2002.1423. [DOI] [PubMed] [Google Scholar]
- Patrick RM, Browning KS. The eIF4F and eIFiso4F Complexes of Plants: An Evolutionary Perspective. Comp Funct Genomics. 2012;2012:287814. doi: 10.1155/2012/287814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16(22):2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell host & microbe. 2008;4(6):579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Pogany J, White KA, Nagy PD. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J Virol. 2005;79(8):4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotondrafara AM, Polacek C, Harris E, Miller WA. Oscillating kissing stem-loop interactions mediate 5' scanning-dependent translation by a viral 3'-cap-independent translation element. RNA. 2006;12(10):1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem NM, Miller WA, Rowhani AK, Golino DA, Moyne A-L, Falk BW. Rose spring dwarf-associated virus has RNA structural and gene-expression features like those of Barley yellow dwarf virus. Virology. 2008;375:354–360. doi: 10.1016/j.virol.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarawaneeyaruk S, Iwakawa HO, Mizumoto H, Murakami H, Kaido M, Mise K, Okuno T. Host-dependent roles of the viral 5' untranslated region (UTR) in RNA stabilization and cap-independent translational enhancement mediated by the 3' UTR of Red clover necrotic mosaic virus RNA1. Virology. 2009;391(1):107–118. doi: 10.1016/j.virol.2009.05.037. [DOI] [PubMed] [Google Scholar]
- Shen R, Miller WA. The 3' untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J Virol. 2004a;78(9):4655–4664. doi: 10.1128/JVI.78.9.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Miller WA. Subgenomic RNA as a riboregulator: negative regulation of RNA replication by Barley yellow dwarf virus subgenomic RNA 2. Virology. 2004b;327(2):196–205. doi: 10.1016/j.virol.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Shen R, Miller WA. Structures required for poly(A) tail-independent translation overlap with, but are distinct from, cap-independent translation and RNA replication signals at the 3' end of Tobacco necrosis virus RNA. Virology. 2007;358(2):448–458. doi: 10.1016/j.virol.2006.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, Rakotondrafara AM, Miller WA. Trans regulation of cap-independent translation by a viral subgenomic RNA. J Virol. 2006;80(20):10045–10054. doi: 10.1128/JVI.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AE, Miller WA. 3' cap-independent translation enhancers of plant viruses. Annu Rev Microbiol. 2013;67:21–42. doi: 10.1146/annurev-micro-092412-155609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Firth AE, Scheidecker D, Brault V, Reinbold C, Rakotondrafara AM, Chung BY, Miller WA, Ziegler-Graff V. A small non-AUG-initiated ORF in poleroviruses and luteoviruses is required for long-distance movement. PLoS Pathog. 2015 doi: 10.1371/journal.ppat.1004868. Under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima Y, Iwakawa HO, Kaido M, Mise K, Okuno T. A long-distance RNA-RNA interaction plays an important role in programmed-1 ribosomal frameshifting in the translation of p88 replicase protein of Red clover necrotic mosaic virus. Virology. 2011;417(1):169–178. doi: 10.1016/j.virol.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treder K, Pettit Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA. The 3' cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14(1):134–147. doi: 10.1261/rna.777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lipzig R, Gultyaev AP, Pleij CW, van Montagu M, Cornelissen M, Meulewaeter F. The 5' and 3' extremities of the satellite tobacco necrosis virus translational enhancer domain contribute differentially to stimulation of translation. RNA. 2002;8(2):229–236. doi: 10.1017/s1355838202018071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Browning KS, Miller WA. A viral sequence in the 3'-untranslated region mimics a 5' cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16(13):4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kraft JJ, Hui AY, Miller WA. Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology. 2010;402(1):177–186. doi: 10.1016/j.virol.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Parisien M, Scheets K, Miller WA. The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure. 2011;19(6):868–880. doi: 10.1016/j.str.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Treder K, Miller WA. Structure of a viral cap-independent translation element that functions via high affinity binding to the eIF4E subunit of eIF4F. J Biol Chem. 2009;284(21):14189–14202. doi: 10.1074/jbc.M808841200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi N, Terauchi H, Kanematsu S, Hidaka S. Characterization of the small subgenomic RNA of Soybean dwarf virus. Arch Virol. 2003;148(9):1827–1834. doi: 10.1007/s00705-003-0137-2. [DOI] [PubMed] [Google Scholar]
- Yuan X, Shi K, Simon AE. A local, interactive network of 3' RNA elements supports translation and replication of Turnip crinkle virus. Journal of virology. 2012;86(8):4065–4081. doi: 10.1128/JVI.07019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]