Abstract
Defining protein-protein contacts is a challenging problem and cross-linking is a promising solution. Here, we present a case of mitochondrial single strand binding protein Rim1 and helicase Pif1, an interaction first observed in immuno-affinity pull-down from yeast cells using Pif1 bait. We found that only the short succinimidyl-diazirine cross-linker or formaldehyde captured the interaction between recombinant Rim1 and Pif1. In addition, Pif1 needed to be stripped of its N-terminal and C-terminal domains, and Rim1's C-terminus needed to be modified for the cross-linked product to become visible. Our report is an example of a non-trivial analysis, where a previously identified stable interaction escapes initial capture with cross-linking agents and requires substantial modification to recombinant proteins and fine-tuning of the mass spectrometry-based methods for the cross-links to become detectable.
We used high resolution mass spectrometry to detect the cross-linked peptides. A 1:1 mixture of 15N and 14N-labeled Rim1 was used to validate the cross-links by their mass shift in the LC-MS profiles. Two sites on Rim1 were confirmed: 1) the N-terminus, and 2) the K29 residue. Performing cross-linking with a K29A variant visibly reduced the cross-linked product. Further, K29A-Rim1 showed a five-fold lower affinity to single stranded DNA compared to wild-type Rim1. Both the K29A variant and wild type Rim1 showed similar degrees of stimulation of Pif1 helicase activity. We propose structural models of the Pif1-Rim1 interaction and discuss its functional significance. Our work represents a non-trivial protein-protein interface analysis and demonstrates utility of short and non-specific cross-linkers.
Keywords: Protein cross-linking, Protein-protein interaction, Protein-DNA interaction, Mass spectrometry (MS), DNA-helicase
Introduction
Pif1 is a highly conserved eukaryotic helicase [1], which has nuclear and mitochondrial forms [2], and is involved in maintenance of telomeric, ribosomal, and mitochondrial DNA. Its function is important for suppressing genomic instability associated with non-canonical DNA structures and stalled replication forks [3,4]. Also, it is essential for normal mitochondrial function in the presence of DNA damaging agents [5]. Despite its importance, Pif1's mechanism of action, its targets and protein interaction partners in mitochondria are poorly defined. Recently, we discovered that Pif1 interacts strongly with the mitochondrial single-stranded DNA binding protein (SSB), Rim1, and provided a detailed functional characterization of this interaction [6]. Rim1 is essential for respiratory growth [7]. Using unwinding assays we have demonstrated that Rim1 stimulates the Pif1 helicase activity in vitro. Apart from our report [6], the Pif1-Rim1 interaction is not mentioned in any other literature and is absent from major protein-protein interaction (PPI) databases. In fact, when we queried PPI databases (PINA database [8], iRefIndex database [9], and the STRING database [10]), Rim1 was found only amongst the second degree neighbors of Pif1 in yeast. In humans, the closest structural homolog of Rim1 is mtSSB, which in found only within the third degree neighbors. Table 1 compares the network properties of Pif1 and Rim1 (mtSSB in human) in yeast and human PPI networks. It follows that Pif1 is more functionally important in the yeast PPI network compared to human PPI network based on the scoring of protein networks. Pif1 scores higher by several fold in degree and betweenness (betweenness measures the extent to which a vortex lies on paths between other vertices) centralities, and also the higher number of second degree neighbors. Based on centrality measures, Rim1/mtSSB is more functionally important than Pif1 in both organisms. In the human PPI network, mtSSB is more central than Rim1 in the yeast PPI (as judged by higher betweenness). Because of the clear importance of this interaction for mtDNA maintenance, we decided to capture the interaction between recombinant proteins and to determine amino acid residues at the interaction interface.
Table 1.
Network properties of Rim1 and Pif1 in yeast and human PPI network.
| Property | Yeast | Human |
|---|---|---|
| Rim1 (mtSBB) degree neighbor relative to Pif1 | 2 | 3 |
| Number of shortest paths between Pif1 and Rim1 | 9 | 27 |
| Pif1, degree centrality | 89 | 9 |
| Pif1, betweenness | 2006 | 9 |
| Pif1, second degree neighbors | 4574 | 896 |
| Rim1 (mtSSB), degree centrality | 160 | 42 |
| Rim1 (mtSSB), betweenness | 11295 | 24102 |
Chemical cross-linking mass-spectrometry (CXMS) is used for establishing identities of interacting proteins, mapping of protein-protein interaction (PPI) interfaces, and as a structure analysis tool [11,12]. In practice, however, the CXMS analysis is difficult, especially on the large scale: 1) not all interactions are captured due to the possibility that the modifiable amino acid residues (e.g. lysines) may not be in close proximity; 2) cross-linked peptides might have poor ionization properties because positively charged lysine side chains have been modified and, as a consequence, have lower intensity in the presence of linear peptides; 3) overall abundance of cross-linked peptides is low compared to non-cross-linked ones; 4) abundance of peptides representing inter-protein cross-links is still lower compared to intra-protein cross-links. Thus far these challenges have been addressed by 1) testing a panel of cross-linkers of various lengths and chemo-selectivities (reviewed in [12]); 2) using heavy and light version of cross-linker to distinguish cross-linked peptides from unmodified peptides using the isotopic mass shift (for an example, see reference [13]); 3) using 18O incorporation to label peptide C-termini [14,15] (cross-linked peptides will have at least two C-termini corresponding to the 8 Da mass difference between 16O and 18O forms); 4) using affinity handles, click chemistry conjugation, and MS-labile groups to enrich and detect the cross-linked species [16-18]; 5) using chromatography and acquisition conditions, which exploit the higher average length and charge of the cross-linked species [19,20].
In our recent review article, we argued that short, broadly specific cross-linkers, could yield more coverage of an interactome compared to long, chemo-selective cross-linkers, and could allow mapping of individual protein-protein interaction interfaces [12]. We also noted major analytical challenges that need to be solved for such a strategy to be successful. One such challenge is the increase in computational expense if one allows for non-specific cross-linking (i.e. insertion of a cross-link at any amino acid residue). Another challenge is related to the inaccessibility of affinity handles and/or MS-cleavable groups due to the short length of the cross-link. For the analysis of the interaction between a pair of proteins, it is relatively easy to design a cross-link search algorithm, which uses a pair-wise combination of peptides to constrain the masses of possible cross-linked precursors (reviewed in [12]) . In the current report we use StavroX [21] as part of our strategy to identify cross-linked peptides and to define the position of the cross-linking sites between Pif1 and Rim1. We use a short cross-linker based on NHS-diazirine chemistry (succinimidyl 4,4’-azipentanoate, SDA) to capture the interaction. SDA is a 3.9 Å-short cross-linker, which cross-links free amino-group groups on one protein (via succinimidyl reaction) to any amino-acid on the other protein (via UV-driven decomposition of the diazirine to a reactive carbene). Recently, Gomez et al. used the 13.5 Å-long, cleavable NHS-diazirine cross-linker, SDAD to study cross-linking of model peptides and equine myoglobin [22]. As expected, the NHS-diazirine captured more interactions compared to the lysine-to-lysine cross-linker of the similar length. Similarly, diazirine-labeled amino acid analogues [23] in combination with high-resolution mass spectrometry have been successfully used to map protein-protein interactions at zero-length [24]. Aside from these papers the descriptions of NHS-diazirine cross-linking chemistry and practical applications of SDA-derived cross-links has been scarce. In addition to the detailed characterization of the Rim1-Pif1 interaction, our current report provides a methodology, applicable to difficult-to-detect cross-linked protein pairs.
Methods
Materials
The following materials were purchased from ThermoFisher Scientific or its subsidiaries: HPLC-grade acetonitrile, formic acid, HEPES, Tris, NaCl, EDTA, BSA, MgCl2, SDS, KOH, β-mercaptoethanol, acrylamide, bisacrylamide, formamide, xylene cyanol, bromphenol blue, urea, glycerol, SDA cross-linker, formaldehyde, DSS, Gel-Code blue stain, and Zeba-Spin Desalting columns for buffer exchange. ATP, poly(dT), Sephadex G-25, zymolyase T20, and protease inhibitor cocktail for use with fungal and yeast extracts were obtained from Sigma. 15N-ammonium chloride was obtained from Chembridge Isotopes. [γ-32P]ATP was obtained from Perkin-Elmer Life Sciences. All the DNA oligonucleotides were obtained from Integrated DNA Technologies (IDT), purified using denaturing polyacrylamide gel electrophoresis, and quantified by UV absorbance at 260 nm. Epoxy (M270) Dynabeads and pre-cast 5-15% gradient gels were purchased from Life Technologies.
Yeast strains and growth conditions
S. cerevisiae BY4741 parent strain and PIF1::TAP-HIS3 BY4741 strain (C-terminal TAP-tag) were grown in YPG medium (1% yeast extract, 2% bacto-peptone, 3% glycerol) until the mid-log phase. The cells were harvested by centrifugation and frozen as pellets using liquid nitrogen.
Yeast mitochondria isolation
Yeast mitochondria was isolated from the BY4741 strain grown on YPG medium until the mid-log phase using the spheroplast/zymolyase method according to [25]. The yeast cells were harvested by centrifugation for 5 min at 3000 × g, and the cell pellets were resuspended in DTT buffer, 2 mg per g (wet weight) cells (100 mM Tris/H2SO4 (pH 9.4), 10 mM dithiothreitol) followed by incubation in a shaker at 70 rpm for 20 min, 30°C. Next, the cells were collected by centrifugation for 5 min at 3000×g, and resuspended in Zymolyase buffer, 7 ml per g (wet weight) cells (20 mM potassium phosphate (pH 7.4), 1.2 M sorbitol) and pelleted again. The cell pellets were resuspended in the Zymolyase buffer for the second time, and Zymolyase-20T) 5 mg of Zymolyase-20T per g (wet weight) cells) was added to the cell suspension. The cells with Zymolyase were incubated at 70 rpm in a shaker for 30 min at 30°C. Next, the spheroplasts were pelleted by centrifugation for 8 min at 2200 × g at 4°C and resuspended in the ice-cold homogenization buffer (10 mM Tris/HCl (pH 7.4), 0.6 M sorbitol, 1 mM EDTA, 0.2% (w/v) BSA), 6.5 ml per g (wet weight) cells. The sheroplasts were homogenized in a glass homogenize with 15 strokes followed by removal of the unbroken cells, nuclei, and large debris by centrifuging for 5 min at 1500 × g at 4°C. Next, the resulting supernatant was centrifuged for 5 min at 3000 × g at 4°C, and then for 15 min at 12,000 × g at 4°C. The resulting pellet – crude mitochondrial fraction - was resuspended in 3 ml of ice-cold SEM buffer (10 mM MOPS/KOH (pH 7.2), 250 mM sucrose, 1 mM EDTA). The crude mitochondrial fraction was purified further using the sucrose step gradient (60%, 32%, 23%, 15%) by Centrifugation in a Beckman SW41 Ti swinging-bucket rotor for 1 h at 134,000 × g (33000 rpm) at 4°C. The band at the 60-32 interface was collected and the pure mitochondria were pelleted at 10,000 × g for 30 min at 4°C.
Affinity purification of Pif1-interacting proteins from yeast
From the frozen yeast pellet, the Pif1-interacting proteins were precipitated using a protein A antibody conjugated to epoxy M270 Dynabeads as described in [6,26]. Briefly, yeast cells were harvested by centrifugation and resuspended in IP buffer (20 mM HEPES, pH 7.4, 2 mM MgCl2, 300 mM NaCl, 0.1 % tween-20, with protease inhibitor cocktail added), 5 ml per g (wet weight) cells. The suspension sonicated for 10 min using bench-top Polytron homogenizer at maximum speed and rotated at 4°C for 1 hr. Cell debri were removed by centrifugation at 3000 × g, and the supernatant mixed with the protein A antibody-conjugated M270 Dynabeads followed by 4-hr incubation at 4°C, with gentle rotation. The beads were collected by magnet and washed 5 times with the IP buffer. The beads were boiled with the SDS gel loading buffer for 5 min, and the proteins were resolved by SDS-PAGE.
Affinity purification of ssDNA-interacting proteins
Biotinylated oligonucleotides (T15-BioTEG, 200 pmol) were attached to M280 Streptavidin Dynabeads (1 mg) in 5 mM Tris, 0.5 EDTA, and 150 mM KCl, pH 7.5 (buffer R) and washed three times with buffer R to remove the unbound species. Next, the beads were resuspended in the 20 mM Tris, pH 7.5, 300 mM KCl, 0.1% Tween with protease inhibitor cocktail added (buffer P). Cellular lysate was added and the mixture was rotated at 4°C for 4 hrs. Next, the beads were collected and washed 5 times with the buffer P.
DNA binding
Polarization of fluorescein labeled ssDNA was measured to determine the binding affinity of wild type and mutant Rim1 at 25°C in 25 mM HEPES pH 7.5, 50 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 2 mM β-ME, and 0.1 mg/mL BSA. A solution containing 1 nM of 3’-fluorescein T70 (3’F-T70) was incubated with increasing concentrations of Rim1. Fluorescence polarization values for the experiment were collected using a PerkinElmer Life Sciences Victor3V 1420 with excitation and emission wavelengths set to 485 nm and 535 nm, respectively. Fluorescence polarization was converted to anisotropy and plotted versus concentration of SSB using KaleidaGraph. The data was fit to the Hill equation to obtain a Hill Coefficient and K0.5 value.
Multiple turnover DNA unwinding
Partial duplex substrate 70T30bp was prepared and radiolabeled on the displaced strand as described (27). All concentrations listed are after mixing. The unwinding experiments were performed at 25°C in a buffer containing 25 mM HEPES pH 7.5, 50 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 2 mM β-ME, and 0.1 mg/mL BSA. Reactions contained 2 nM DNA substrate and 5 mM ATP and were initiated upon the addition of 100 nM Pif1 and 60 nM unlabeled displaced strand to trap the unwound loading strand. Unwinding experiments in the presence of SSB proteins were performed by preincubating 100 nM Rim1 protein (tetramer) with a mixture containing 2 nM substrate and 5 mM ATP for 5 minutes prior to addition of 100 nM Pif1 and 60 nM unlabeled displaced strand. At the desired times, aliquots of the reaction mixture were transferred to the quench solution (200 mM EDTA, 0.6% SDS, 0.1% bromphenol blue, 0.1% xylene cyanol, 6% glycerol, and 112 μM T70). The role of the T70 was to sequester proteins after the reaction. The substrate and ssDNA product were resolved on a 20% native polyacrylamide gel. Radiolabeled substrate and product were detected using a Typhoon Trio PhosphorImager (GE Healthcare) and quantified using ImageQuant software. The amount of product formed over time was plotted using KaleidaGraph and fit to a single exponential.
Recombinant proteins
Recombinant helicase domain of Pif1, full-length Pif1, and Rim1-C-terminal 6×His proteins used in this work were purified as described in [6]. K29A Rim1-C-terminal 6×His mutant was purified from E. coli, using the same isolation protocols as for wild-type Rim1, established in [6]. Throughout the article, we call Rim1-C-terminal 6×His as wild-type Rim1, and K29A Rim1-C-terminal −6×His as K29A Rim1. The proteins without 6×His tag as labeled as “no-tag”, when appropriate.
15N labeled Rim1
To metabolically label Rim1, E. coli expressing Rim1 was grown on media prepared with 15N-ammonium chloride as the sole source of nitrogen [28]. The 15N-labeled Rim1 was purified according to the previously established protocol [6]. Briefly, M9 media for E.Coli growth was prepared with 15N-ammonium chloride. E. coli strain BL21 was transformed with the the Rim1 - 6×His construct. The cells were grown to OD600 0.8 in the 15N media and the production of the Rim1 was cold-induced with IPTG at 20°C. The cells were harvested after 12 hr of incubation at 20°C, lysed in a microfluidizer, and the Rim1 protein was isolated using Talon Metal Affinity Column.
Cross-linking reactions
Prior to the cross-linking experiments, recombinant proteins purified from E. coli were thawed on ice, and using the Zeba Spin Desalting columns, the storage buffer was exchanged to the cross-linking reaction buffer (25 mM HEPES, 100 mM NaCl, 10% glycerol, pH 7.5). In all of the cross-linking reactions, the final concentrations of Pif1 (monomer) and Rim1 (tetramer) were 2.5 μM and 3 μM. The following panel of cross-linkers was used in this work: disuccinimidyl suberate (DSS), 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide (EDC), formaldehyde, and succinimidyl 4,4’-azipentanoate (SDA). The cross-linking was performed as follows:
Formaldehyde
Recombinant Rim1 and Pif1 were mixed in 28 μL of the cross-linking buffer and incubated for 15 minutes at 30°C. Next, 2 μL of appropriately diluted formaldehyde stock was added to result in 0.1 to 1% final formaldehyde concentrations. The reactions were carried out for an additional hour at 30°C and quenched by addition of 10 μL of 4× SDS loading buffer.
SDA
To a 2× stock of the protein to be reacted at the first step in the cross-linking buffer (5 μM in the case of Pif1, 6 μM in the case of Rim1) SDA stock solution in DMSO was added (50 μM SDA and 0.5% DMSO final concentrations). The NHS reaction was carried out at 25°C for 30 min followed by addition of 1M Tris, pH 8.0 (50 mM final Tris) followed by an additional incubation at 25°C for 5 min. Next, to remove the unreacted cross-linker, the buffer was exchanged with the fresh cross-linking buffer using Zeba Spin Desalting columns. 90-95% percent protein recovery was verified by Bradford assay. For the second step of the SDA-cross-linking reaction, an equal volume of the 2× protein stock to be reacted was added to the NHS-reacted mix. UV-cross-linking was performed in Stratalinker 1800 using coated bulbs (365 nm emission maximum) at a 2 cm distance from the bulbs, in black, flat-bottomed 96-well plates. 4× SDS buffer was added to the reactions and reaction products were resolved by 5-15% gradient SDS-PAGE.
DSS
Recombinant Pif1 and Rim1 were mixed in the cross-linking buffer at a 1:1.2 ratio. A 100 mM stock of DSS was prepared in DMSO and added to the reaction mixture, to a final concentration of 2.5 mM. The reaction was incubated at room temperature for 40 minutes, with rotation. Next, the reaction was quenched addition of 1M Tris, pH 8.0 (50 mM final Tris). Next, to remove the unreacted cross-linker, the buffer was exchanged with fresh cross-linking buffer using Zeba Spin Desalting columns. 4 × SDS loading buffer was added to the sample, and the reaction products were resolved by 5-15% gradient SDS-PAGE.
EDC
The two-step protocol, suggested by the manufacturer (Thermo) was used for the EDC cross-linking reaction. Rim1 was prepared in the activation buffer (100 mM MES, 100 mM NaCl, pH 6), and EDC was added to a final concentration of 2 mM. Next, N-hydroxysuccinimide was added to a final concentration of 5 mM. The reaction mixture was incubated for 15 min at room temperature and quenched by the addition of 20 mM beta-mercaptoethanol (final concentration). The buffer was exchanged to cross-linking buffer using Zeba Spin Desalting columns and Pif1 was added to the mixture. The reaction was incubated for 1 hour at 30°C. Next, 10 mM of hydroxylamine was added to 10 mM (final concentration) followed by 5 min incubation at 30°C. The excess reagents were removed by the second exchange into cross-linking buffer. 4 × SDS loading buffer was added to the sample, and the reaction products were resolved by 5-15% gradient SDS-PAGE.
RPLC-LTQ-Orbitrap mass spectrometry. Large-scale protein identification
The conditions and acquisition settings listed in this section were used for the analysis of TAP-tagged Pif1 purification, ssDNA-interacting proteins, and mitochondrial proteome analyses. Protein gel bands were excised and subjected to in-gel trypsin digestion by the following procedure. Gel slices were destained in 50% methanol, 100 mM ammonium bicarbonate for 1 hr, followed by reduction in 10 mM Tris[2-carboxyethyl]phosphine and alkylation in 50 mM iodoacetamide. Gel slices were then dehydrated in acetonitrile, followed by addition of 100 ng porcine trypsin in 100 mM ammonium bicarbonate and incubation at 37°C for 12-16 hours. Peptide products were then acidified in 0.1% formic acid. Tryptic peptides were separated on a 100 × 0.075 mm reverse phase column using a nanoAcquity UPLC system (Waters). Peptides were eluted using a 30 min gradient from 98:2 to 40:60 buffer A:B ratio. [Buffer A = 0.1% formic acid, 0.05% acetonitrile; buffer B = 0.1% formic acid, 75% acetonitrile.] Eluted peptides were ionized by electrospray (1.9 kV) followed by MS/MS analysis using collision induced dissociation on an LTQ Orbitrap Velos mass spectrometer (Thermo). MS data were acquired using the FTMS analyzer in profile mode at a resolution of 60,000 over a range of 375 to 1500 m/z. MS/MS data were acquired for the top 15 peaks from each MS scan using the ion trap analyzer in centroid mode and normal mass range with normalized collision energy of 35.0. Proteins were identified by database search using PEAKS Studio v7 [29]. Protein database contained yeast protein sequences downloaded from the UniProt database (release version 2013_04) concatenated with the list of common contaminants (keratins). Identification score thresholds were adjusted to yield 1% false positive of the peptide-to-spectrum matches. A score of 20 corresponded to a 1% peptide false discovery rate in most data sets. Post-translational modifications (PTMs) were determined using the PEAKS routine Spider. Modified spectra discussed in the text were required to have PTM-specific fragment ions to be higher that 5% of the base peak intensity and the identification score above 40.
NanoLC-LTQ-Orbitrap mass spectrometry. Identification of cross-linked peptides
To detect the cross-linked peptides, the LTQ-Orbitrap parameters were modified from the large-scale method: 1) the three most intense precursors were subjected to MS/MS analysis – each of the precursors was analyzed consecutively by HCD (higher-energy collision dissociation) detected in Orbitrap analyzer at a resolution of 7500, and CID detected in ion trap analyzer; 2) Each sample was run twice, the first time allowing all the charges, and the second time allowing only precursors of charge 4 and higher to be fragmented; 3) precursor selection intensity threshold was set to 5000; 4) for the 15N-labeled samples only the HCD fragmentation of the 10 most intense precursor ions at 7,500 resolution was used. Initial data refining and deconvolution of highly-charge fragments was performed by Peaks Studio 7 [29]. The refined spectra were exported as MGF files, followed by the searches by StavroX v2.0.6 and xComb (Supplemental Figure 4B) [31]. For the searches of 15N-labeled data-sets, the alphabet of amino-acids was expanded with their 15N-versions, and protein databases were modified accordingly to include both versions of Rim1. Different characters were used for the Rim1 lysine residues to account for the directionality of SDA cross-link (i.e. lysine-to-any). For the StavroX searches protein databases contained only Pif1 and Rim1 sequences. The database for the 15N-labeled analysis contained both 14N- and 15N-labeled Rim1 sequences. 15N-labeled data was also searched by PEAKS Studio 7, treating Rim1 peptides as variable modifications with either 14N or 15N-labels occurring at any position.
Identification of Pif1-interacting proteins using spectral counting
For the purpose of determining significant Pif1 interactors, we used the spectral counting approach [30,31] and G-test of independence [32] comparing spectral counts in the whole cell lysate, to the proteins in the TAP-tag pull-down [6]. Bonferroni correction for the multiple hypothesis testing was used and the proteins with the p-values below 5% significance threshold were called significant. When a protein was absent in one of the conditions, about 7-to-10 spectral counts (the exact number of counts differs between replicates and depends on the total number of protein IDs) in the other condition corresponded to the 5% significance threshold.
Protein-protein interaction network analysis
PPI data for human and yeast (discussed in the introduction section of this manuscript) were generated using Bioconductor cisPath package (R version 3, and cisPath version 1.4.6) and the PINA database [8], the iRefIndex database [9], and the STRING database [10].
Model of Pif1 structure
Pif1 structure was built using homology-modeling software SWISS-MODEL [32].
Protein structure visualization
To construct, manipulate, and visualize the model of Rim1-Pif1 interactions we used PyMOL (The PyMOL Molecular Graphics System, Version 1.6.0.0 Schrödinger, LLC).
Results
Capturing Pif1-Rim1 interaction using chemical cross-linking
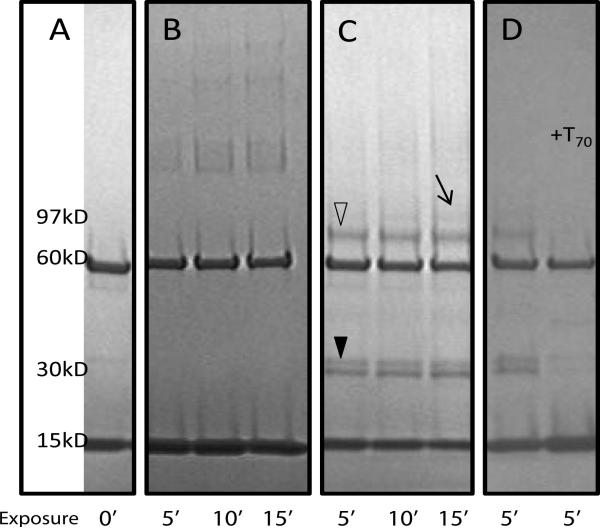
Amongst several different cross-linkers, we found that only formaldehyde and the short-length hetero-bifunctional cross-linker SDA were capable of capturing the interaction of the recombinant Rim1 and Pif1 (Supplemental Figure 1). Moreover, for a cross-link to be visible on the gel, Pif1 had to be stripped to its helicase domain, and Rim1 had to have a 6×His tag at the C-terminus. Mass spectrometric detection of peptides cross-linked by formaldehyde is difficult due to its broad specificity, poorly defined end products, thermal instability, and reversibility of the cross-link. SDA, on the other hand, forms well-defined end products and hence is more amenable to the CXMS analysis. The first step of SDA reaction involves modification of a reactive lysine with N-hydroxy-succinimide ester (NHS), and the second involves photo-activation which results in the non-specific cross-linking to any amino-acid in the vicinity of the reactive carbene radical. We found that the Rim1-Pif1 hetero-dimer is captured only if we use Rim1 first for the lysine modification in the NHS-reaction, followed by addition of Pif1 for the non-specific UV-cross-linking (Figure 1C). If SDA is reacted with Pif1 first, we see only Pif1 oligomers (Figure 1B). The Rim1-Rim1-Pif1 band (Figure 1C, arrow) contained 134 and 359 spectral counts derived from Rim1 and Pif1, respectively. The Rim1-Pif1 band (Figure 1C, open triangle) contained 181 and 573 spectral counts derived from Rim1 and Pif1, respectively. Adjusting for the protein constructs length (Rim1 – 127 amino acids, Pif1- 586 amino acids) results in the 1.72 Rim1:Pif1 ratio for the Rim1-Rim1-Pif1 band and in the 1.46 Rim1:Pif1 ratio for the Rim1-Pif1 band. The protein concentrations were determined by Bradford assay and the equivalent amount of proteins were loaded into each lane.
Figure 1. Rim1-Pif1 heterodimer detected with the SDA cross-linker.
Rim1 and Pif1 proteins were cross-linked with SDA using a 2-step procedure (see Methods). The reaction products were resolved on 5-15% gradient SDS-PAGE. Open triangle marks the position of Rim1-Pif1 heterodimer. Filled triangle marks the position of Rim1-Rim1 homodimer. Arrow points to the position of Pif1 bound by two molecules of Rim1. A, Rim1 and Pif1 without cross-linker. The Pif1 and Rim1 isoforms used in this work localize at 60 kD and 15 kD regions, respectively. B, lanes 1-3, Pif1 was reacted with SDA first, followed by addition of Rim1 and UV irradiations. C, lanes 1-3, Rim1 was reacted with SDA first, followed by the addition of Pif1 and UV irradiation. D, lane 1 – same reaction conditions as in C, lane 2 – ssDNA (T70) was added prior to cross-linking with SDA. UV exposure times at the second step of cross-linking are indicated, in minutes.
Pif1-Rim1 interaction is lost in the presence of ssDNA
Figure 1D shows that the Pif1-Rim1 heterodimer band is lost in the presence of ssDNA; performing the reaction in the presence of T70 abolishes the interaction. We used two different conditions: 1) Rim1 and Pif1 mixed together, with T70 added (shown); 2) Rim1 preincubated with T70 first. In both cases, there was no cross-link captured. This result indicates that the Rim1-DNA interaction is favored over its interaction with Pif1 and is possibly mediated by the same sites. This is consistent with our previously reported apparent dissociation constants for Rim1 with Pif1 compared to Rim1's interaction with ssDNA [6]. It is also possible that the conformation of the Rim1-Pif1 interaction changes upon binding to ssDNA, thereby reducing the crosslinking reaction. Interestingly, Figure 1D also shows that the amount of Rim1 dimer in the cross-linking reaction is reduced in the presence of ssDNA (indicated by closed triangle). The reduction in dimer is possibly due to inaccessibility of DNA-bound Rim1 to the cross-linking agent.
Identification of two different interaction sites
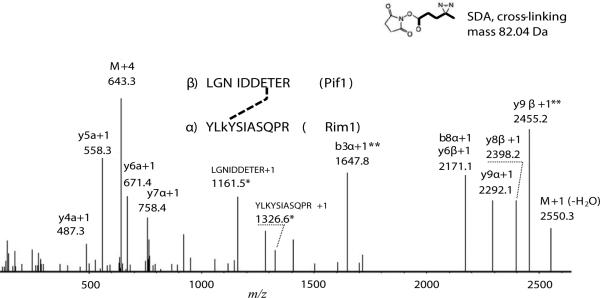
Figure 2 shows an HCD tandem mass spectrum of the SDA cross-linked peptide pair representative of the Rim1-Pif1 interaction, obtained from the analysis of Pif1-Rim1 heterodimer. The StavroX identification score was 109, higher that the 5% false positive rate score threshold, 68, determined by the decoy analysis (Supplemental Figure 4C). Fragments with masses corresponding to the fragmentation of the cross-links were verified manually (because StavroX does not account for the cross-link fragmentation) and are marked with ‘*’ on the Figure and provide additional confidence to the cross-link assignment. The original annotation by StavroX is shown in Supplemental Figure 4A. The observed retention time is consistent with the predicted hydrophobicity of the cross-linked pair. Across multiple experiments, the average retention time of the YLKSIASQPR peptide with the dead-end cross-link (SDA+H2O, Δmass of 100.05 Da) on the lysine was 19.7 min, while the average retention time of the unmodified LGNIDDETER peptide was 13.5 min. The cross-linked peptide pair eluted at 29.0 min, which is closed to the predicted retention time of 33 min calculated using the sequence specific retention time calculator [33]. The analysis suggests that Rim1's residue K29, within the YLKYSIASQPR peptide is cross-linked to one of the acidic residues within the LGNIDDETER peptide of Pif1.
Figure 2. Identification of Pif1-Rim1 interaction site using SDA cross-linking.
The charge-deconvoluted tandem MS/MS spectrum obtained using HCD fragmentation from the precursor at 643.078 m/z is shown. Mass deviation from the theoretical mass was 4.83 ppm. The precursor retention time was 29 min, which is close to the expected value, based on the hydrophobicity of the cross-linked peptides. Mass deviation of the fragments was within 20 ppm, on average. Possibility of one C13 atom was allowed in calculations of the mass deviation. The most intense ions and the ions indicative of cross-link fragmentation were annotated by StavroX, xComb, and manually. Fragments contacting cross-links are shown in bold. Fragments used to identify the cross-linked residue on Rim1 are marked with (**). For the details of StavroX and xComb annotations see Supplemental Figure 4. Stavro score was 109, with the 5% false discovery rate corresponding to the score above 68, according to the Stavro decoy analysis (Supplemental Figure 4, C). Greek letters α and β correspond to the cross-linked peptides from Rim1 and Pif1, respectively. Fragments related to the loss of the cross-link are marked with asterisk (*). Letter ‘M’ notes precursor-related ions
The second interaction site was identified by using 15N-labeled Rim1 and cross-linking was performed with a 1:1 15N:14N mixture. Using this method, the N-terminus of Rim1, the MDFSK peptide was shown to be cross-linked to Pif1. StavroX identified both 14N and 15N versions of this cross-linked peptide pair (with the scores of 143 and 163, respectively, and the 5% false discovery score threshold was 74), but no fragmentation ions were visible from the MDFSK portion. If one treats MDFSK as a post-translational modification, possible at any position, it is also easily identifiable by PEAKS (Figure 3). While in the 15N-labeling experiment we did not see the site determined in Figure 2, we note that it does not necessarily mean it is not there; it could simply mean that the peptide has poor ionization properties at the particular point in its LCMS profile.
Figure 3. 15N-labeling reveals the interaction of Pif1 with the N-terminus of Rim1.
14N and 15N-labeled Rim1 proteins were mixed with Pif1 and cross-linking using SDA was performed. De novo sequencing followed by matching to a sequence database using PEAKS Studio platform revealed Pif1 peptide cross-linked to the Rim1 N-terminal peptide, MDFSK. A, B, The N-terminal peptide of Rim1 has 6 nitrogen atoms, resulting in a visible isotopic doublet in MS spectra. The isotopic doublet shown is the sum of MS spectra over 2 min elution window. C, Sequence coverage map, annotated by PEAKS: MDFSK sequences does not contribute ions to the cross-linked spectra, and when treated as a non-specific modification results in multiple hits clustered around AIEDENE region, indicate by “#” (14N), and by “X” (15N). Oxidation of methionine is labeled with shaded “o”. D,E Two high scoring MSMS spectra are shown: 15N-labeled and 14N-labeled, as indicated. Masses of y20, y18, and y17[2+] fragments are indicated. F, The sequence of Pif1 isoform used in this work, with the peptide cross-linked to MDFSK highlighted.
K29A-Rim1 mutant reduces the amount of captured Rim1-Pif1 heterodimer
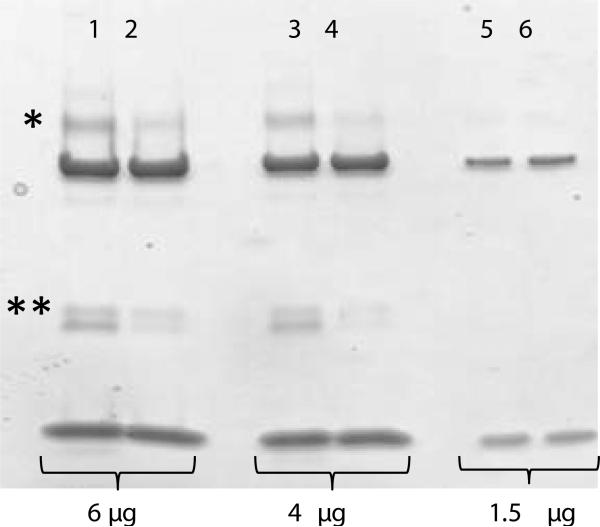
How important is the K29 residue for the Rim1-Pif1 interaction? Because the K29 site (Figure 2) was not confirmed by 15N labeling, and could potentially represent a spurious non-specific interaction, we decided to validate this interaction by mutating lysine to alanine at that position. The gel in the Figure 4 demonstrates that approximately half of the interaction was lost, judging by the intensity of the respective band. The mass-spectrometric analysis of the Rim1-Pif1 heterodimer region, as well as of the Rim1-Rim1 dimer band confirmed the K-to-A replacement. There were no cross-linked species identified from the Rim1-Pif1 hetero-dimer in this experiment. This result is consistent with two interaction sites between Pif1 and Rim1. A K29A mutation on Rim1 decreases but does not eliminate appearance of the cross-linked species. The product observed from cross-linking K29A Rim1 and Pif1 is likely due to the second interaction site on Rim1 at the N-terminus, which escaped the detection this time due to low quantity of the Rim1-Pif1 heterodimer.
Figure 4. K29A-Rim1 mutant shows reduced cross-linking with Pif1.
Lanes 1,3,5, Wt-Rim1 is cross-linked with the helicase domain of Pif1 using SDA (conditions are the same as in Figure 1D). Lanes 2,4,6, K29A-Rim1 mutant is cross-linked to helicase domain of Pif1. The positions of the cross-linked Rim1-Pif1 pair and of the cross-linked Rim1 homo-dimer are indicated by single and double asterisk, respectively. The amount of total protein loaded into each lane is indicated below the gel image.
K29A mutant shows reduced capacity for ssDNA binding
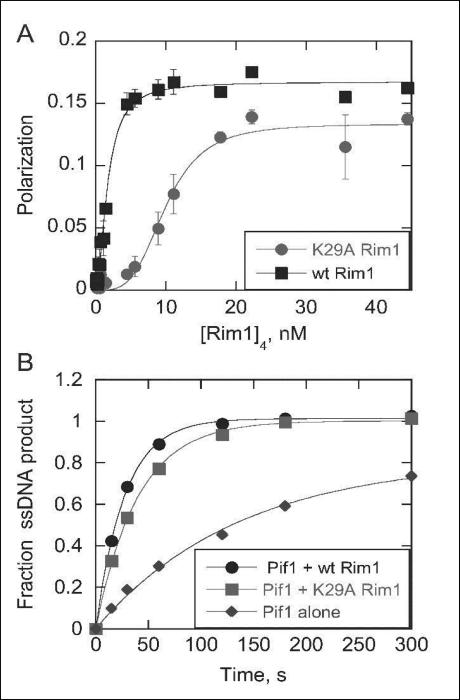
Figure 5A demonstrates that the K-to-A substitution of the 29th residue of Rim1 results in diminished ssDNA binding, with K0.5 increasing from 1.7 to 10 nM. The cooperativity in binding is also increased in the K29A Rim1 mutant. The Hill coefficient increases from 1.7 for wtRim1 binding to ssDNA to 3.7 for K29A Rim1 binding. This could be due to the decreased affinity for ssDNA with the K29A mutant making the affinity of Rim1 for itself stronger, relative to ssDNA, leading to the appearance of increased cooperativity in binding.
Figure 5.
A, Binding of wt Rim1 and K29A Rim1 to 3’-FT70 was measured by polarization. Data were fit with the Hill equation to obtain a K0.5 of 1.7 nM For wt Rim1 and 10.0 nM for K29A Rim1. Binding of K29A Rim1 was highly cooperative, with a Hill coefficient of 3.7. The Hill coefficient for wt Rim1 binding was 1.7. B, Effect of Rim1 on Pif1 catalyzed DNA unwinding. Unwinding of a 30bp duplex by Pif1 was measured in the presence and absence of Rim1. Data was fit to a single exponential to obtain unwinding rate constants of 0.036 s−1, 0.025 s−1, and 0.0073 s−1 for unwinding by Pif1 + wt Rim1, Pif1 + K29A Rim1, and Pif1 alone, respectively.
K29A Rim1 stimulated Pif1 helicase activity
Previously, we established that Rim1 stimulates the Pif1 helicase activity [6]. Figure 5B shows that the K29A Rim1 is still almost as effective as wild type Rim1 in the stimulation of the Pif1. This result is in contrast to the reduction of ssDNA binding of the K29A Rim1 and indicates that the two activities, ssDNA binding and stimulation of Pif1 helicase activity, are independent. This is consistent with crosslinking results that indicate that the Rim1-Pif1 interaction competes with the Rim1-ssDNA interaction (Figure 1D). The specific mechanism by which Rim1 stimulates Pif1 helicase activity remains to be determined.
Mitochondrial proteome
We are interested in the interaction of Rim1 and Pif1, which are both known mitochondrial proteins. We therefore, sought to obtain a reference mitochondrial proteome. We isolated mitochondria from S. cerevisiae cells grown on YPG media, and performed a proteomics survey using the standard proteomics method (SDS-PAGE, in-gel digestion, followed by LTQ-Orbitrap mass spectrometry). The list of proteins identified with PEAKS Spider at 1% false discovery rate for the peptide-to-spectrum matches is presented in Supplemental Table 1. Notably; Rim1 was identified with 168 and 246 spectra in replicates 1 and 2. Certain peptides were clearly more visible than others. In the case of recombinant Rim1, as expected, modification with a 6×His tag at the Rim1 C-terminus dramatically increases the C-terminus visibility in the MS (e.g. Supplemental Figures 2,3). At the same time, Pif1 protein was not found in this analysis.
Affinity purification of Pif1-interacting proteins from yeast grown on glycerol media
Previous experiments that yielded Rim1 as a strong Pif1 interactor were performed with cells grown on sucrose, and using a lower resolution mass spectrometry platform (MALDI-TOF) [6]. Here, we repeat those experiments with yeast grown on glycerol as a sole carbon source (thereby enforcing functional mitochondria). Additionally, we are using a more sensitive and more accurate mass-spectrometry platform (LTQ-Orbitrap-Velos). Also, in [6], we detected Rim1 with only one peptide that was used for quantification via the i-Dirt method [26] Our goals here were 1) to increase Rim1 sequence coverage, 2) to map Rim1's PTMs, and also 3) to capture other potential Pif1 interactors.
We performed the pull-down using two biological replicates, compared the proteins in the pull-down to the proteins in the cytosol, and calculated percentage enrichment using adjusted spectral counts. Table 2 shows the most significant Pif1 interactors, which include Rim1.
Table 2.
The most significant Pif1 interactors as determined by the TAP-Pif1 pull down and LTQ-Orbitrap mass spectrometry.
| accession a | descriptionb | Replicate 1 | Replicate 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S | I | F | P-val | S | I | F | P-val | ||
| P01870 | RABIT Ig gamma chain C region | 4 | 4247 | 3639 | 0 | 3 | 4333 | 5006 | 0 |
| P07271 | DNA repair and recombination protein PIF1 | 0.1 | 1595 | 54663 | 0 | 0.1 | 755 | 26168 | 0 |
| P07259 | Protein URA2 | 17 | 133 | 27 | 5.1E-67 | 14 | 76 | 19 | 5.1E-36 |
| P35732 | RNA polymerase II degradation factor 1 | 44 | 53 | 4 | 8.4E-12 | 12 | 50 | 14 | 2.0E-22 |
| P07342 | Acetolactate synthase catalytic subunit, mitochondrial | 0.1 | 58 | 1988 | 3.8E-39 | 0.1 | 25 | 866 | 9.6E-18 |
| P53297 | PAB1-binding protein 1 | 0.1 | 54 | 1851 | 1.5E-36 | 0.1 | 25 | 866 | 9.6E-18 |
| P08566 | Pentafunctional AROM polypeptide | 4 | 47 | 40 | 1.3E-26 | 4 | 30 | 26 | 2.4E-16 |
| P32445 | Single-stranded DNA-binding protein RIM1 | 5 | 58 | 40 | 2.2E-32 | 5 | 29 | 20 | 5.8E-15 |
| P25367 | [PIN+] prion protein RNQ1 | 3 | 61 | 70 | 1.9E-36 | 3 | 26 | 30 | 9.1E-15 |
| Q04119 | Endopolyphosphatase | 0.1 | 10 | 343 | 8.5E-08 | 0.1 | 17 | 589 | 1.8E-12 |
| P02407 | 40S ribosomal protein S17-A | 19 | 30 | 5 | 6.8E-09 | 23 | 38 | 6 | 2.4E-11 |
| P0CX41 | 60S ribosomal protein L23-A | 28 | 56 | 7 | 7.3E-18 | 33 | 44 | 5 | 4.7E-11 |
| P20606 | Small COPII coat GTPase SAR1 | 15 | 20 | 5 | 1.1E-05 | 7 | 21 | 10 | 3.4E-09 |
| P12612 | T-complex protein 1 subunit alpha | 1 | 25 | 86 | 3.6E-16 | 0.1 | 11 | 381 | 1.7E-08 |
| P07244 | Bifunctional purine biosynthetic protein ADE5,7 | 10 | 33 | 11 | 5.1E-14 | 6 | 15 | 9 | 1.8E-06 |
| P28007 | H/ACA ribonucleoprotein complex subunit 1 | 1 | 9 | 31 | 5.1E-06 | 1 | 9 | 31 | 4.7E-06 |
| P47079 | T-complex protein 1 subunit theta | 4 | 35 | 30 | 3.0E-19 | 4 | 12 | 10 | 7.9E-06 |
| P53123 | Ribosomal RNA methyltransferase MRM2 | 0.1 | 7 | 240 | 8.5E-06 | 0.1 | 7 | 243 | 7.9E-06 |
| P53305 | Mitochondrial 37S ribosomal protein S27 | 1 | 6 | 21 | 3.8E-04 | 0.1 | 6 | 208 | 3.8E-05 |
| P00958 | Methionine--tRNA ligase, cytoplasmic | 0.1 | 19 | 651 | 1.0E-13 | 1 | 7 | 24 | 8.6E-05 |
| P25626 | 54S ribosomal protein IMG1 | 3 | 9 | 10 | 1.2E-04 | 0.1 | 5 | 173 | 1.8E-04 |
Columns named S and I show spectral counts in the whole-cell lysate and the pull-down, respectively. Column named F shows fold enrichment as calculated using adjusted spectral counts (30), (32). P-values were determined by G-test of independence for each replicate, as in (32). Mitochondrial proteins are highlighted in bold. Bait related proteins are shown in italics.
protein accession and description are as in Swiss-Prot database (www.uniprot.org). Only proteins that had P-values below 0.001, fold enrichment above 4, and spectral counts in the pull down above 5, in each of the replicates are shown. The fractional spectral count of 0.1 indicates absence of a protein.
protein accession and description are as in Swiss-Prot database (www.uniprot.org). Only proteins that had P-values below 0.001, fold enrichment above 4, and spectral counts in the pull down above 5, in each of the replicates are shown. The fractional spectral count of 0.1 indicates absence of a protein.
Discussion
Mass spectrometric detection of the products of the short-length broad specificity cross-linking reaction
There have been very few reports describing detailed characterization of cross-linked peptides that also include site assignment, obtained using a short non-specific cross-linker, such as SDA (non-specific at the second step). In our work we demonstrated the possibility of analyzing such peptides without resorting to any enrichment or other cross-link specific detection parameters, other than high charge filter. We observe that cross-linked species typically have LC-MS intensities 10 to 100 fold lower than the base peak. While low, this is still well within the dynamic range of modern mass spectrometers. We also have attempted ETD, without success – as the signal-to-noise ratio was inadequate for identification of cross-linked species. To further advance the method to the large scale, it could be beneficial to perform the trypsin digestion in the presence of 18O-labeled water, which leads to incorporation of 4 18O-atoms into the cross-linked species, as opposed to just two, in the case of linear peptides. It is important to have good precursor intensity and to account for any asparagine to aspartate conversion, while using this method. In addition, 18O labelling often suffers from potentially incomplete labeling and back-exchange from 18O to 16O, which will generate a mix of 18O and 16O peptide C-terminal. This will further complicate the data analysis and lower intensity of the cross-linked peptides. When attempted, we were not able to detect 18O-atom incorporation into the Rim1-Pif1 cross-links, because the precursor intensities were low (not shown).
Bioinformatics pipe-line for the cross-link identification
Data refinement proved to be important for the cross-link detection – specifically deconvolution of the initial peak list, so that fragments in MS/MS are recalculated to the charge 1+ state. This is a default feature of PEAKS Software Suite 7; we used the spectra refined by PEAKS for all other downstream analyses. Deconvolution enhances confidence in the site identification dramatically, compared to the non-deconvoluted spectra, as does using the quality filter.
Rim1-Pif1 interaction was not captured with chemo-selective cross-linkers DSS and EDC
We did not observe the Rim1-Pif1 heterodimer using DSS cross-linker, which cross-links primary amino groups (Supplemental Figure 1, lane 18). There are two possibilities why this was the case: 1) there is no suitable pair of primary amino groups at the appropriate distance in the Rim1-Pif1 interaction interface; or 2) DSS interferes with the interaction. The reason for the absence of the Rim1-Pif1 cross-linked product in the case of reaction with the the amino-to-carboxy cross-linker EDC (Supplemental Figure 1, lanes 19,20) is less obvious. Indeed, according to the analysis of the SDA cross-linked product, cross-linking occurs between the 3-rd lysine residue of Rim1's peptide YLKYSIASQPR and the 7-th glutamate in the Pif’1 peptide LGNIDDETER (Figure 2, Supplemental Figure 4). Therefore, there are amino- and carboxy- groups at a short distance that should be available for the EDC reaction. However, the possibility, that the cross-linker itself disrupts the interaction remains – it is possible the o-Acylisourea modification, which occurs at the first step of the EDC reaction destabilizes the Rim1-Pif1 heterodimer.
The two sites of Rim1-Pif1 interactions
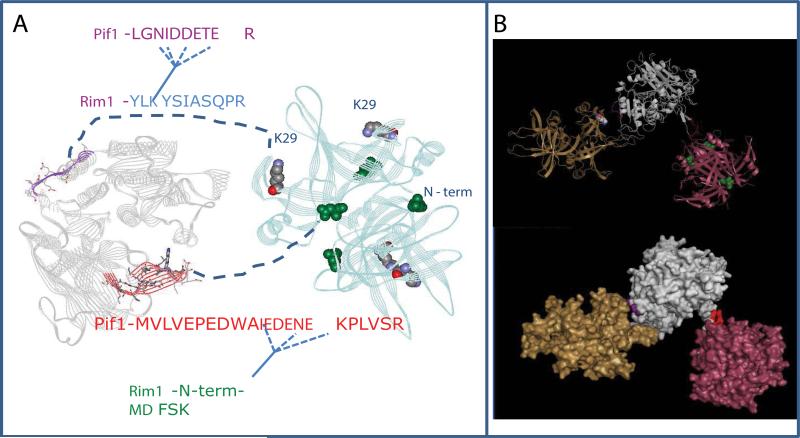
Figure 6 shows the model of the Rim1-Pif1 interaction. The Pif1 structural model is based on the homologus RecD2 helicase, and Rim1 is an unpublished crystal structure (K. Varughese, personal communication). The two cross-linking sites mark the interface of the interaction, one of which is composed of a β-hairpin in the SH3 domain of Pif1 which fits into a cleft between the two Rim1 dimers to interact with the N-terminus of Rim1. The two sites on Pif1 are too far apart to interact with just one Rim1 tetramer, and most likely involve interaction with the two Rim1 tetramers. It is not currently clear from the obtained data, whether the Rim1-Pif1 complex has two different forms, fulfilling the two interaction sites either with the Rim1:Pif1 stoichiometry of 4:1, or the interaction with the two tetramers occurs at the same time – and the actual stoichiometry of Rim1:Pif1 complex is 8:1. The second site of interaction is mediated through K29 in the OB-fold domain of Rim1 and an acidic patch on domain 2A (the second RecA-like domain) of Pif1. The OB domain also interacts with DNA, and the competition between DNA and protein binding is clear (Figure 1D).
Figure 6. Model of Pif1-Rim1 interaction.
A, Summary of the protein-protein interactions defined in cross-linking experiments. The model of Pif1 helicase domain is on the left. The X-ray structure of Rim1 tetramer is on the right. The two interaction sites identified in this work are highlighted. The X-ray structure has N-terminal aspartate instead of methionine. We propose the model of Rim1-Pif1 interaction, where 1) the beta-hairpin of Pif1, highlighted in red fits into the cleft between two Rim1 dimers; and 2) the stretch of acidic residues within Pif1's peptide LGNIDDETER is likely to be in the vicinity of K29 residue of Rim1. B, The possible spatial arrangement of Rim1-Pif1 interactions. It is unlikely that the two interaction sites are realized at the same time within the (Rim1)4:Pif1 heterodimer. Instead, the most consistent with the current structural information is the model, where two Rim1 tetramers participate in the interaction with Pif1. Pif1 helicase domain is shown in grey, Rim1 tetramers are shown in yellow and pink.
Our data provide insight into the possible mechanism of activation of Pif1 helicase by Rim1. Clear disruption of the interaction between Pif1 and Rim1 by DNA indicates that a DNA-bound Rim1 is less likely to interact with Pif1. It may, therefore, serve as guide to direct Pif1 to the appropriate position. Interestingly, the N-terminus of Rim1 is also involved in interaction between different molecules of Rim1. The 15N-labeling experiment identified an 14N-15N mixed species corresponding to the C-terminus of Rim1 cross-linking to the N-terminus of Rim1 during the analysis of the Rim1 dimer band (Supplemental Figures 2,3). In this experiment, prior to the addition of Pif1, we mixed 15N and 14N-labeled Rim1 tetramers. The discovery of mixed 14N-15N cross-linked species can be explained either by interaction between two different tetramers or dissociation and reformation of the new tetramers with mixed subunits. We think that the direct interaction between two tetramers is more likely for the following two reasons: 1) Rim1 tetramer itself is very stable, not prone to dissociation; 2) Only C-terminus-to-N-terminus 14N-15N species were observed, with all others being either 14N-14N, or 15N-15N only. It is therefore a possibility that Pif1-Rim1 interaction at the Rim1's N-terminus happens after two Rim1 tetramers dissociate. The exact sequence of events and dynamics of Rim1-Pif1-DNA interaction demands further investigation.
Biological Context of Rim1 and Pif1 interaction
Our mitochondrial proteome analysis (Supplemental Table 1) shows that Rim1 is a relatively abundant protein, ranking within the first 100 of the most abundant proteins identified, while Pif1 was not detected. The absence of Pif1 in the mitochondrial proteome is consistent with its low abundance under normal growth conditions. It is known that Pif1 becomes important during the conditions of mitochondrial DNA damage (5), and we plan to explore the Rim1-Pif1 interaction in this context in the future studies.
Conclusions
Using a combination of proteomics methods and chemical cross-linking with the short-length, hetero-bifunctional cross-linker SDA, we showed that Rim1 and Pif1 interact directly. We further characterized the Pif1-Rim1 heterodimer and identified two different sites of cross-link incorporation. One site involves one of the putative RecA-like domains of Pif1 and OB-fold domain of Rim1, and the other site involves a β-hairpin in the SH3 domain of Pif1 and Rim1's N-terminus. We were able to validate both interactions by the site-directed mutagenesis and by 15N-labeling. The presented workflow will further aid identification of protein-protein interactions using non-specific cross-linkers.
Supplementary Material
Acknowledgements
The work presented in this paper is supported by NIH Grant R01 GM098922 (to Kevin Raney), and by a UAMS Cancer Pilot Grant (to Boris Zybaylov). The UAMS Proteomics core is supported by the Arkansas IDeA Network for Biomedical Research Excellence (P20GM103429), the UA Center for Protein Structure and Function (P30GM103450), and the UAMS Center for Microbial Pathogenesis and Host Inflammatory Responses (P20GM103625).
Footnotes
Data Availability
All the raw mass-spectrometry data, R scripts, StavroX and PEAKS identification results are available by request from the authors.
References
- 1.Chung WH. To peep into Pif1 helicase: multifaceted all the way from genome stability to repair-associated DNA synthesis. J Microbiol. 2014;52:89–98. doi: 10.1007/s12275-014-3524-3. [DOI] [PubMed] [Google Scholar]
- 2.Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol Pharm Bull. 2007;30:1685–1692. doi: 10.1248/bpb.30.1685. [DOI] [PubMed] [Google Scholar]
- 3.Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimada K, Gasser SM. DNA replication: Pif1 pulls the plug on stalled replication forks. Curr Biol. 2012;22:R404–405. doi: 10.1016/j.cub.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Cheng X, Qin Y, Ivessa AS. Loss of mitochondrial DNA under genotoxic stress conditions in the absence of the yeast DNA helicase Pif1p occurs independently of the DNA helicase Rrm3p. Mol Genet Genomics. 2009;281:635–645. doi: 10.1007/s00438-009-0438-6. [DOI] [PubMed] [Google Scholar]
- 6.Ramanagoudr-Bhojappa R, Blair LP, Tackett AJ, Raney KD. Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 2013;41:1029–1046. doi: 10.1093/nar/gks1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merz S, Westerman B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009;10:R95. doi: 10.1186/gb-2009-10-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowley MJ, Pinese M, Kassahn KS, Waddell N, Pearson JV, et al. PINA v2.0: mining interactome modules. Nucleic Acids Res. 2012;40:D862–D865. doi: 10.1093/nar/gkr967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paramelle D, Miralles G, Subra G, Martinez J. Chemical cross-linkers for protein structure studies by mass spectrometry. Proteomics. 2013;13:438–456. doi: 10.1002/pmic.201200305. [DOI] [PubMed] [Google Scholar]
- 12.Zybailov BL, Glazko GV, Jaiswal M, Raney KD1. Large Scale Chemical Cross-linking Mass Spectrometry Perspectives. J Proteomics Bioinform. 2013;6:001. doi: 10.4172/jpb.S2-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaarur N, Xu X, Lestienne P, Meriin AB, McComb M, et al. RuvbL1 and RuvbL2 enhance aggresome formation and disaggregate amyloid fibrils. EMBO J. 2015;34:2363–2382. doi: 10.15252/embj.201591245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capelo JL, Carreira RJ, Fernandes L, Lodeiro C, Santos HM, et al. Latest developments in sample treatment for 18O-isotopic labeling for proteomics mass spectrometry-based approaches: a critical review. Talanta. 2010;80:1476–1486. doi: 10.1016/j.talanta.2009.04.053. [DOI] [PubMed] [Google Scholar]
- 15.Ye X, Luke B, Andresson T, Blonder J. 18O stable isotope labeling in MS-based proteomics. Brief Funct Genomic Proteomic. 2009;8:136–144. doi: 10.1093/bfgp/eln055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chowdhury SM, Du X, Tolic N, Wu S, Moore RJ, et al. Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 2009;81:5524–5532. doi: 10.1021/ac900853k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo J, Fishburn J, Hahn S, Ranish J. An integrated chemical cross-linking and mass spectrometry approach to study protein complex architecture and function. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clifford-Nunn B, Showalter HD, Andrews PC. Quaternary diamines as mass spectrometry cleavable crosslinkers for protein interactions. J Am Soc Mass Spectrom. 2012;23:201–212. doi: 10.1007/s13361-011-0288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritzsche R, Ihling CH, Götze M, Sinz A. Optimizing the enrichment of cross-linked products for mass spectrometric protein analysis. Rapid Commun Mass Spectrom. 2012;26:653–658. doi: 10.1002/rcm.6150. [DOI] [PubMed] [Google Scholar]
- 20.Leitner A, Reischl R, Walzthoeni T, Herzog F, Bohn S, et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Götze M, Pettelkau J, Schaks S, Bosse K, Ihling CH, et al. StavroX--a software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom. 2012;23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 22.Gomes AF, Gozzo FC. Chemical cross-linking with a diazirine photoactivatable cross-linker investigated by MALDI- and ESI-MS/MS. J Mass Spectrom. 2010;45:892–899. doi: 10.1002/jms.1776. [DOI] [PubMed] [Google Scholar]
- 23.Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods. 2005;2:261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 24.Kölbel K, Ihling CH, Sinz A. Analysis of peptide secondary structures by photoactivatable amino acid analogues. Angew Chem Int Ed Engl. 2012;51:12602–12605. doi: 10.1002/anie.201205308. [DOI] [PubMed] [Google Scholar]
- 25.Gregg C, Kyryakov P, Titorenko VI. Purification of mitochondria from yeast cells. J Vis Exp. 2009 doi: 10.3791/1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, Rout MP, et al. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005;4:1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- 27.Tackett AJ, Morris PD, Dennis R, Goodwin TE, Raney KD. Unwinding of unnatural substrates by a DNA helicase. Biochemistry. 2001;40:543–548. doi: 10.1021/bi002122+. [DOI] [PubMed] [Google Scholar]
- 28.Tyler RC, Sreenath HK, Singh S, Aceti DJ, Bingman CA, et al. Auto-induction medium for the production of [U-15N]- and [U-13C, U-15N]-labeled proteins for NMR screening and structure determination. Protein Expr Purif. 2005;40:268–278. doi: 10.1016/j.pep.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 29.Ma B, Zhang K, Hendrie C, Liang C, Li M, et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 30.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 31.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 32.Zybailov B, Friso G, Kim J, Rudella A, Rodriguez VR, et al. Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol. Cell Proteomics. 2009;8:1789–1810. doi: 10.1074/mcp.M900104-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.