Abstract
Norovirus infection is the most common cause of acute gastroenteritis in developed countries. Developing an assay based on a non-invasive biomarker for detecting incident norovirus infections could improve disease surveillance and epidemiological investigations. This project involved analysis of IgA and IgG norovirus-specific antibody responses in saliva samples from a Norwalk virus (Genogroup I, genotype 1 norovirus) challenge study involving infected and symptomatic, and non-infected asymptomatic individuals. Saliva was collected at the challenge, and two weeks and 40 days post-challenge. Samples were analyzed using the Luminex fluorometric and Meso Scale Discovery (MSD) electrochemiluminescence immunoassays. Recombinant P domains of Norwalk virus capsid protein, as well as similar recombinant proteins of two genogroup II noroviruses (VA387 and VA207) were used as antigens. Immunoconversions were defined as >4-fold increase in antibody responses to the norovirus antigens. Various sample pre-treatment options, buffers, saliva dilution ratios, and data adjustment approaches to control for sample-to-sample variability in saliva composition were compared using the Luminex assay. The results suggest that adjusting responses to the norovirus antigens for responses to the protein purification tag, glutathione-S-transferase (GST), significantly improved the odds of producing a correct immunoconversion test result. IgG-based tests were more accurate compared to IgA-based tests. At optimal conditions, both Luminex and MSD assays for Norwalk-specific IgG antibodies correctly identified all infected and non-infected individuals. There was no evidence of cross-reactivity of anti-Norwalk virus antibodies with genogroup II noroviruses. These results suggest that salivary antibody responses can be used for the detection of incident infections with Norwalk virus in prospective surveys.
Keywords: Salivary antibody, Volunteer challenge study, Norovirus, Immunoassay, Incident infection
1. Introduction
Noroviruses are a diverse group of single stranded RNA viruses which belong to the Caliciviridae family (Huang et al., 2005). These highly infectious viruses (Teunis et al., 2008) are the major cause of acute viral gastroenteritis in adults (Bon et al., 2005) and may be responsible for 10% to 20% of all endemic or non-outbreak cases of gastroenteritis (Marshall et al., 2003). Three genogroups (GI, GII, and GIV) have been detected in humans but two of them (GI and GII) are of particular importance to public health. Noroviruses usually cause mild, self-limited illness, with diarrhea and vomiting being the most common symptoms. However, illness can be severe in susceptible individuals, such as the elderly, young children, and hospitalized patients. It has been estimated that noroviruses cause up to 110,000 hospitalizations and nearly 1,000 deaths per year in the U.S. (Rockx et al., 2002; Lopman et al., 2011; Hall et al., 2012).
Each genogroup of noroviruses includes subgroups with many genetically distinct genotypes (Gallimore et al., 2004; Zheng et al., 2006). Most norovirus outbreaks are currently associated with person-to-person spread or contamination of food with genogroup II, genotype 4 (GII.4) noroviruses (Maunula and von Bonsdorff, 2005; Blanton et al., 2006). Waterborne norovirus outbreaks are also reported regularly (Kukkula et al., 1999; Parshionikar et al., 2003; Blackburn et al., 2004; Yoder et al., 2008). In addition, sporadic waterborne infections may be common as suggested by the presence of noroviruses in surface and ground water sources (Lodder et al., 2010; Lee et al., 2011; Lambertini et al., 2012). However, linking infections to contaminated drinking water and quantifying public health benefits of more efficient treatment of public water supplies remains a challenge (USEPA 2006). Prospective epidemiologic investigations involving the detection of incident infections would lead to a better understanding of the public health burden of waterborne norovirus infections and would provide data for assessing the benefits of specific measures to prevent these infections. Developing a non-invasive biomarker of norovirus infection would facilitate such investigations.
Because norovirus-specific antibodies typically increase after norovirus infection (Graham et al., 1994; Green et al., 2002; Lindesmith et al. 2003; Moe et al., 2004; Tsugawa et al., 2006; Tseng et al., 2007), an increase in these antibodies, or immunoconversion, can be used as a biomarker of infection. In general, norovirus-specific immunoglobulin (Ig) A antibodies in serum and saliva increase steeply within several days after infection and then start to decline within two weeks; while IgG responses typically peak between two and three weeks post-infection, and then gradually decline (Erdman et al., 1989; Monroe et al., 1993; Lindesmith et al., 2003 and 2005; Leon et al., 2008). Previous studies defined immunoconversion as at least a four-fold increase in norovirus-specific antibody response (Monroe et al., 1993; Moe et al., 2004). While the invasiveness of blood sampling may limit the application of serology in longitudinal community studies, salivary immunoassays relying on safe and non-invasive collection of oral fluid can enable large-scale and inexpensive population surveys (McKie et al., 2002; Morris-Cunnington et al., 2004a, b).
In a clinical setting, the preferred method for diagnosis of norovirus infections is detection of the viral nucleic acid in stool samples using reverse-transcription polymerase chain reaction (RT-PCR). While this method is valuable in investigating disease outbreaks, asymptomatic participants of community-based surveys are usually reluctant to donate fecal samples. Norovirus shedding in stool may also occur for a short period of time, which would necessitate frequent sampling in longitudinal surveys aimed at detecting incident infections in participants. Thus, a non-invasive sampling method which is well accepted by participants, such as saliva sampling, is likely to ensure a higher compliance rate and reduce the cost of data collection compared to stool and blood sampling.
Although non-invasive saliva sampling offers many advantages, greater temporal variability in saliva composition and concentration of immunoglobulins may confound the analysis of immunoconversions and cause false-negative and false-positive results. Two potential approaches to controlling for sample-to-sample variability are to (i) express results as a ratio of a norovirus-specific antibody response and a measure of total Ig in the sample or (ii) express results as a ratio of a norovirus-specific antibody response and an antibody response to an internal control, such as a recombinant protein purification tag.
This study aimed to optimize and evaluate a previously developed approach, which employs recombinant viral proteins (P particles) for the detection of incident infections (Griffin et al., 2011). It involved further development of specifications for the Luminex-based fluorescence suspension immunoassay (Luminex Corp., Austin, TX), which was used in the previous tests, and an initial demonstration of saliva antibody tests using another common immunoassay methodology, the Meso Scale Discovery (MSD; Rockville, MD) electrochemiluminescence (ECL) platform. The latter was conducted in order to expand the range of available options and facilitate a broader application of saliva testing in epidemiological investigations. Prospectively collected saliva samples from participants of a Norwalk virus (GI.1) challenge study were used to evaluate various assay conditions and data adjustment approaches to control for sample-to-sample variability in saliva composition and to assess potential cross-reactivity of Norwalk virus-specific antibodies with selected GII noroviruses, VA387 (GII.4) and VA207 (GII.9).
2. Methods
2.1. Saliva samples
This study used aliquots of prospectively-collected saliva samples from adult volunteers who participated in a randomized, double-blinded Norwalk virus challenge study (Leon et al., 2011). The objective of the parent study was to test the effectiveness of Norwalk virus inactivation in oysters by high hydrostatic pressure. All participants ingested either untreated or pressure-treated oysters seeded with Norwalk viruses. The protocols of the challenge study were approved by the Institutional Review Board (IRB) of Emory University. Twenty saliva samples from seven participants of the parent study were used in the current study. The participants were selected based on the availability of sufficient saliva samples collected at the day of challenge, and approximately 15 (± 1) and 40 (± 4) days post-challenge. Three of the seven volunteers developed gastrointestinal symptoms and excreted Norwalk virus in their feces (were infected). The remaining four volunteers did not excrete Norwalk virus and did not have symptoms (were not infected). Severity of symptoms or intensity of virus shedding was not used as selection criteria. Serum or salivary antibody responses had not been measured prior to the selection of these individuals.
Saliva samples were collected after stimulation with sour candy by spitting into sterile 5 mL tubes. Samples were centrifuged to remove debris and archived at −80°C. For this study, aliquots of saliva samples without individual identifying information were shipped in insulated containers with dry ice to laboratories of the U.S. Environmental Protection Agency (US EPA) for analysis.
2.2. Proteins and antibodies
To measure norovirus-specific antibody responses in saliva, norovirus recombinant P particles and GST control proteins were coupled to Luminex beads (Griffin et al., 2011) or coated on MSD plates as described in Table 1. Recombinant P particles of three noroviruses, Norwalk (GI.1), VA387 (GII.4), and VA207 (GII.9), were produced using an E. coli expression system and purified as described previously (Tan and Jiang, 2005; Tan et al., 2008). The purification procedure involved thrombin cleavage and removal of GST tags.
Table 1.
Protein coating conditions and antibodies used in confirmation tests.
| A. Luminex ass | |||
|---|---|---|---|
| Protein | Coupling concentration (µg per 500 µL) |
Confirmation tests | |
| Primary antibody | Secondary detection antibody | ||
| Specific antibody assay | |||
| Norwalk (GI.1) P particle | 5 | Guinea pig anti-GI and GII NoV (X. Jiang)a | Biotinylated donkey anti-guinea
pig (Jackson ImmunoResearch)d |
| VA387 (GII.4) P particle | 5 | Guinea pig anti-GI and GII NoV (X. Jiang)a | Biotinylated donkey anti-guinea
pig (Jackson ImmunoResearch)d |
| Guinea pig anti-GII.4 NoV VLP (A. Sutherland)a | Biotinylated donkey anti-guinea
pig (Jackson ImmunoResearch)d |
||
| Rabbit anti-GII.4 NoV VLP (A. Sutherland)a | Biotinylated donkey anti-rabbit
(Jackson ImmunoResearch)d |
||
| VA207 (GII.9) P particle | 5 | Guinea pig anti-GI and GII NoV (X. Jiang)a | Biotinylated donkey anti-guinea
pig (Jackson ImmunoResearch)d |
| Guinea pig anti-GII.4 NoV VLP (A. Sutherland)a | Biotinylated donkey anti-guinea
pig (Jackson ImmunoResearch)d |
||
| Rabbit anti-GII.4 NoV VLP (A. Sutherland)a | Biotinylated donkey anti-rabbit
(Jackson ImmunoResearch)d |
||
| GST | 5 | Rabbit anti-GST (Invitrogen)b | Biotinylated donkey anti-rabbit
(Jackson ImmunoResearch)d |
| Total antibody assay | |||
| Goat anti-human IgG | 25 | Human IgG (Jackson ImmunoResearch)c | Biotinylated anti-human IgG (KPL)e |
| Goat anti-human IgA | 25 | Human IgA (Jackson ImmunoResearch)c | Biotinylated anti-human IgA (KPL)e |
| B. MSD assay | |||
|---|---|---|---|
| Protein | Coating concentration (µg/mL) |
Confirmation tests | |
| Primary detection antibody | Secondary detection antibody | ||
| Specific antibody assay | |||
| Norwalk (GI.1) P particle | 4 | Mouse anti-GI.1 (Kim Labs)f | SULFO-TAG goat anti-mouse (MSD)i |
| VA387 (GII.4) P particle | 1 | Guinea pig anti-GII.4 NoV VLP (A. Sutherland)g | Biotinylated donkey anti-guinea
pig (Jackson ImmunoResearch) j |
| Rabbit anti-GII.4 NoV VLP (A. Sutherland) g | Biotinylated donkey anti-rabbit
(Jackson ImmunoResearch) j |
||
| Total antibody assay | |||
| Goat anti-human IgG | 1 | Human IgG (Jackson ImmunoResearch)h | Biotinylated anti-human IgG (KPL)j |
| Goat anti-human IgA | 1 | Human IgA (Jackson ImmunoResearch) h | Biotinylated anti-human IgA (KPL)j |
Norovirus (NoV)-specific antibodies were assayed at serial dilutions with four-fold increases from 1:1,000 to 1:128,000.
Anti-GST antibody was assayed at two-fold dilutions from 0.25 to 0.003906 µg/mL.
Human IgG and IgA were assayed at 10-fold serial dilutions from 100 to 0.001 ng/mL.
Biotinylated secondary detection antibodies were prepared at 8 µg/mL and used with 10 µg/mL of the reporter, streptavidin, R-phycoerythrin conjugate (SAPE; Invitrogen, Carlsbad, CA).
Biotinylated anti-human IgG and IgA secondary antibodies were prepared at 4 µg/mL and used with 8 µg/mL of SAPE.
Mouse anti-GI.1 monoclonal antibody was assayed at 10-fold concentrations from 0.1 ng/mL to 10 µg/mL.
Guinea pig and rabbit anti-GII.4 sera were assayed at 10-fold dilutions from 1:1,000 to 1:100,000,000 (responses were still detectable at 1:10,000,000 dilution).
Human IgG and IgA were assayed at 10-fold serial dilutions from 1,000 to 0.001 ng/mL.
SULFO-TAG labeled goat anti-mouse secondary detection antibodies were prepared at 1 µg/mL.
Biotinylated secondary detection antibodies were prepared at 1 µg/mL and used with 0.5 µg/mL of the reporter, SULFO-TAG labeled streptavidin (MSD).
For quantification of total IgA and IgG in saliva, goat anti-human IgAα and IgGγ (both from KPL, Gaithersburg, MD) were coupled to Luminex microbeads and coated on MSD plates (Table 1).
2.3. Luminex assay
The Luminex immunoassay is a suspension sandwich assay that utilizes sets of 5.6 µm microspheres with unique internal fluorescent properties. Each antigen is covalently coupled to a specific set of microspheres. Different sets of microspheres coupled to various antigens are simultaneously incubated with the sample in a multiplex format. A dual laser Luminex analyzer is used to determine the type of each microsphere in the multiplex and to quantify the fluorescence intensity of the reporter signal.
2.3.1. Microsphere coupling
Norovirus P particles, GST, or isotype-specific anti-human capture antibodies (for total IgA and IgG tests) were covalently coupled to Luminex carboxylated microspheres as described previously (Griffin et al., 2011). Specifically, 50 mM 2-(N-morpholino) ethanesulfonic acid (MES), pH 5.0 was used as coupling buffer. Proteins were added to the coupling suspension as described in Table 1A. All coupled microspheres were stored in PBS, 0.1% bovine serum albumin (BSA), 0.05% sodium azide, 0.02% Tween 20, pH 7.4 at 4 °C until use.
Confirmation of coupling norovirus antigens and GST to Luminex beads was performed as described previously (Griffin et al., 2011) and defined in Table 1A. To demonstrate successful coupling of anti-human IgA and IgG antibodies to Luminex beads, purified human IgA and IgG were assayed as indicated in Table 1A.
2.3.2. Saliva testing
In previous studies which detected salivary antibodies to measles, mumps and rotavirus, heat treatment and centrifugation of saliva were used for inactivation and clarification of samples (Friedman, 1982; Friedman et al., 1989 and 1993). To examine the effects of heat pre-treatment of saliva on the Luminex norovirus assay results, aliquots of saliva samples were incubated in a water bath at 56°C for 60 min and then centrifuged at 13,000×g for 10 min at 20°C. Supernatant was carefully removed and used to prepare dilutions for analysis with the Luminex immunoassay. Untreated and heat-treated saliva samples were analyzed for specific responses to three noroviruses and to GST in two quadruplex Luminex assays for IgA and IgG responses.
Saliva samples were diluted in PBS-1% BSA standard Luminex assay buffer or in PBS-0.025% Tween 20–0.5% polyvinyl alcohol-0.8% polyvinyl pyrrolidone buffer (hereafter called PVX buffer) to investigate the effects of dilution buffer on the immunoassay performance. Previous studies indicated that pre-incubation of serum samples in PVX buffer reduced nonspecific reactivity of antibodies in Luminex assays (Waterboer et al., 2006; van Gageldonk et al., 2011).
Saliva samples were assayed in Luminex microplate wells at 1:4 or 1:32 final dilutions as described in Table 2. A total of five different saliva processing and assay specification options were used in both IgA and IgG Luminex tests (Table 2). Due to insufficient volumes of some saliva samples, not all possible combinations of buffers, dilutions and heat pre-treatment options were tested. For example, as tests at 1:32 dilution in PVX buffer and 1:4 dilution in PBS-BSA buffer demonstrated that heat pre-treatment of saliva samples had a detrimental effect on assay’s results, additional tests comparing untreated and heat-treated samples at 1:4 dilution in PVX and at 1:32 dilution in PBS-BSA were not conducted.
Table 2.
Sensitivity, specificity and overall proportion of correct results of norovirus immunoconversion tests under various assay conditions and data adjustment approaches, Luminex assay data (%).
| A. IgA responses | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No adjustment | Ratio to total IgA | Ratio to GST IgA | Proporti on of correct results for Norwalk Virus |
Overall proporti on of correct resultse |
||||||||||
| Assay conditio ns |
Sensitivi ty NVa |
Specifici ty NVb |
Specifici ty VA207c |
Specifici ty VA387d |
Sensitivi ty NVa |
Specifici ty NVb |
Specifici ty VA207c |
Specifici ty VA387d |
Sensitivi ty NVa |
Specifici ty NVb |
Specifici ty VA207c |
Specifici ty VA387d |
||
| PBS- BSA, no heat, 1:4 |
33 | 75 | 86 | 86 | 33 | 100 | 86 | 100 | 67 | 100 | 100 | 100 | 71 | 86 |
| PBS- BSA, heat, 1:4 |
33 | 75 | 57 | 86 | 33 | 50 | 71 | 71 | 67 | 100 | 100 | 100 | 62 | 75 |
| PVX, no heat, 1:4 |
33 | 75 | 86 | 86 | 33 | 75 | 71 | 100 | 33 | 100 | 100 | 100 | 62 | 81 |
| PVX, no heat, 1:32 |
33 | 100 | 100 | 100 | 33 | 100 | 100 | 100 | 33 | 100 | 100 | 100 | 71 | 90 |
| PVX, heat, 1:32 |
33 | 25 | 43 | 57 | 33 | 75 | 86 | 71 | 67 | 100 | 100 | 86 | 57 | 68 |
| Average | 33 | 70 | 74 | 83 | 33 | 80 | 83 | 89 | 53 | 100 | 100 | 97 | 65 | 80 |
| Proporti on of correct resultse |
70 | 77 | 92 | |||||||||||
| B. IgG responses | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No adjustment | Ratio to total IgG | Ratio to GST IgG | Proporti on of correct results for Norwalk virus |
Overall proporti on of correct resultse |
||||||||||
| Assay conditio ns |
Sensitivi ty NVa |
Specifici ty NVb |
Specifici ty VA207c |
Specifici ty VA387d |
Sensitivi ty NVa |
Specifici ty NVb |
Specifici ty VA207c |
Specifici ty VA387d |
Sensitivi ty NVa |
Specifici ty NVb |
Specifici ty VA207c |
Specifici ty VA387d |
||
| PBS- BSA, no heat, 1:4 |
100 | 75 | 86 | 86 | 100 | 100 | 86 | 86 | 100 | 100 | 100 | 100 | 95 | 92 |
| PBS- BSA, heat, 1:4 |
100 | 50 | 71 | 71 | 100 | 100 | 86 | 86 | 100 | 100 | 100 | 71 | 90 | 84 |
| PVX, no heat, 1:4 |
100 | 75 | 86 | 86 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 95 | 95 |
| PVX, no heat, 1:32 |
100 | 100 | 100 | 100 | 67 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 95 | 98 |
| PVX, heat, 1:32 |
100 | 25 | 57 | 57 | 67 | 75 | 71 | 86 | 67 | 100 | 100 | 71 | 71 | 73 |
| Average | 100 | 65 | 80 | 80 | 87 | 95 | 89 | 91 | 93 | 100 | 100 | 89 | 90 | 89 |
| Proporti on of correct resultse |
80 | 90 | 95 | |||||||||||
Value in each cell is the sensitivity of Norwalk virus (NV) immunoconversion tests (percent of true positive results or proportion of the 3 infected individuals who immunoconverted to Norwalk virus).
Value in each cell is the specificity of Norwalk virus immunoconversion tests (percent of true negative results or proportion of the 4 non-infected individuals who did not immunoconvert to Norwalk virus).
Value in each cell is the specificity of immunoconversion tests for the VA207 norovirus (percent of true negative results or proportion of the 7 study participants who did not immunoconvert to the VA207 norovirus).
Value in each cell is the specificity of immunoconversion tests for the VA387 norovirus (percent of true negative results or proportion of the 7 study participants who did not immunoconvert to the VA387 norovirus).
Value in each cell is the overall proportion of correct results among all immunoconversion tests conducted.
To test for norovirus-specific IgA responses, biotinylated goat anti-human IgAα detection antibody (Jackson ImmunoResearch) was utilized at 30 µg/mL; the IgG tests used biotinylated donkey anti-human IgG Fc-specific detection antibody (Jackson ImmunoResearch), also at 30 µg/mL. Both were followed by incubation with the reporter, streptavidin, R-phycoerythrin conjugate (SAPE; Invitrogen, Carlsbad, CA) at 10 µg/mL as described previously (Griffin et al., 2011).
Saliva samples were tested for total IgA and IgG using a separate duplex assay involving two sets of Luminex beads coupled to goat anti-human IgAα and IgGγ capture antibodies. Samples were diluted in PBS-1% BSA and assayed with Luminex beads at 1:5,000 final dilution. Purified human IgA and IgG were assayed on the same plate at serial dilutions as standards (for details, see Griffin et al., 2011). Immunoglobulins were detected using a mixture of biotinylated goat anti-human IgA and anti-human IgG secondary detection antibodies (KPL) at 4 µg/mL each with a final incubation with SAPE at 8 µg/mL to obtain reporter signal.
2.3.3. Luminex analysis
MultiScreen BV 1.2 µm filter microplates (Millipore, Billerica, MA) pre-washed with 200 µL of wash buffer (PBS-0.05% Tween 20) were used in all coupling confirmation and salivary antibody tests. Microspheres were sonicated and diluted to a concentration of 100 microspheres of each type per µL in assay buffer (PBS-1% BSA or PVX). Then, 50 µL of suspension (5,000 beads of each type) and 50 µL of sample (primary antibody or diluted saliva) were added to wells. Plates were incubated for 1 h at room temperature in the dark on a microplate shaker at 500 rpm, washed 3 times, and beads resuspended in 50 µL of assay buffer. Next, 50 µL of biotinylated detection antibody in assay buffer was added to wells, followed by incubation for 30 min at room temperature in the dark on a plate shaker, and three wash and aspiration cycles. Beads were resuspended in 50 µL of assay buffer and then 50 µL of SAPE in assay buffer was added to wells. After incubation for 30 min at 500 rpm, and three wash and aspiration cycles, beads resuspended in 100 µL of assay buffer were analyzed using a Luminex-100 instrument at default settings. Median fluorescence intensity (MFI) of the reporter signal was used in data analysis.
2.4. MSD assay
The MSD electrochemiluminescence (ECL) technology uses proprietary microplates with carbon electrodes integrated in the bottom of each well. Capture antibodies or microbial antigens are attached to the electrode by passive absorption. The assay uses ruthenium tris(bipyridine) as an electrochemiluminescent label. The label, which can be conjugated either directly to the secondary antibody or to streptavidin, emits light at 620 nm when electrochemically stimulated. The mechanism of ECL production has been described in detail elsewhere (Deaver, 1995). Microplates are analyzed using an MSD instrument, which passes an electrical current through plate-associated electrodes and quantifies light emitted from each well using custom designed optics and photodetectors. Previous studies reported that MSD assays have a wide dynamic range and low background (Sourial et al. 2009, Thompson et al. 2012).
MSD assay tests employed a subset of 16 remaining samples from six individuals (three infected and three non-infected). All samples were tested for IgA and IgG antibody responses to Norwalk virus P particles and for IgG responses to VA387 P particles, as well as for total IgA and IgG antibodies. All tests were conducted without heat pre-treatment of saliva samples, at 1:4 final sample dilution using a standard MSD assay buffer. Limited quantities of VA207 P particles prohibited salivary antibody testing on the MSD platform. In addition, testing for GST, which would have required re-analysis of samples on a separate microplate, was not conducted due to a shortage of saliva.
2.4.1. MSD plate coating
This study used standard MSD 96 well Multi-Assay® single spot microplates (MSD catalog number L15XA) with a single antigen coated on the bottom of each well. Plates were coated with norovirus proteins or, for the total Ig test, with isotype-specific anti-human antibodies using a manufacturer-recommended procedure adapted to these proteins. Specifically, norovirus P particles and anti-human capture antibodies were diluted in the standard coating buffer (PBS at pH 7.4) and 30 µL of solution applied to each microplate well. Plates were gently tapped to ensure that the coating solution completely submerged the electrodes, covered using adhesive seals, and incubated at room temperature overnight. Following incubation, 150 µL of 3% MSD Blocker A (BSA) in PBS was added to each well, and plates were sealed and incubated for 1 h at room temperature with shaking at approximately 1,000 rpm. Plates were then washed four times in MSD Wash Buffer using an automatic plate washer (BioTrak II).
Monoplex MSD assays were optimized for analysis of salivary antibody responses to Norwalk and VA387 P-particles, and for total salivary antibodies. For each coating protein, coating concentrations of 0.5, 1, 2 and 4 µg/mL were compared. Norovirus-specific primary antibodies were analyzed at serial dilutions to confirm coating of MSD plates with Norwalk and VA387 P-particles (Table 1B). Optimal concentrations of antigens were determined by comparing dose-response curves as described previously for the Luminex assay (Griffin et al., 2011).
2.4.2. Saliva testing
Saliva samples were diluted 1:4 in 1% PBS solution of MSD Blocker A without heat pre-treatment. To assess potential non-specific binding to microplate wells, all samples from infected individuals were also assayed on blank wells which were blocked with BSA, but not coated with viral protein. Biotinylated secondary detection antibodies identical to those used with the Luminex specific antibody assay were utilized at 1 µg/mL with 0.5 µg/mL MSD SULFO-TAG labeled streptavidin.
To analyze total salivary Ig, samples were diluted 1:10,000 (for IgA tests) or 1:20,000 (for IgG tests) in 1% MSD Blocker A in PBS (the dilutions were selected based on preliminary tests so that the results were in the middle of the assay linearity range). Purified human IgA and IgG were assayed on the same plate at serial dilutions from 0.001 ng/mL to 1 µg/mL. Biotinylated secondary anti-human IgA and anti-human IgG detection antibodies were identical to those used in the Luminex total antibody assay. The detection antibodies were assayed at 1 µg/mL with 0.5 µg/mL SULFO-TAG labeled streptavidin.
2.4.3. MSD analysis
25 µL samples (primary detection antibody or diluted saliva) were applied to dry wells of an antigen-coated MSD microtiter plate. Plates were sealed and incubated at room temperature for 1 h with shaking, then washed four times using MSD Wash Buffer. Biotinylated detection antibodies and SULFO-TAG labeled streptavidin were diluted in 1% Blocker A in PBS immediately prior to analysis. The optimal concentrations of detection antibodies and labeled streptavidin (see 2.4.1 and 2.4.2; Table 1B) were determined using separate assay optimization tests in which secondary antibodies were assayed at concentrations from 0.25 to 2.0 µg/mL with two-fold increases and streptavidin at 0.25 and 0.5 µg/mL (data not shown). The mixture was added to plates at 25 µL per well. Plates were again sealed and incubated at room temperature for 1 h with shaking. After washing plates four times in MSD Wash Buffer and tapping dry, 150 µL of MSD Read Buffer T was added to each well, and plates were analyzed on a MSD Sector imager model 2400.
2.5. Statistical analysis of assay data
Statistical data analysis was conducted using Excel and SAS version 9.2. The objectives of the analysis were (i) to compare various conditions (dilutions, pre-treatment options and assay buffers) in the Luminex assay; and (ii) to compare various approaches to adjusting the assay data in order to control for sample-to-sample variability in saliva composition.
Geometric mean values of reporter signals from samples analyzed in duplicate were used in statistical analysis. Assay data for each individual were dichotomized as presence or absence of an immunoconversion, which was defined as at least four-fold increase in virus-specific antibody response from the baseline (day of Norwalk virus challenge) in at least one follow-up sample. IgA and IgG immunoconversions were analyzed separately.
The analysis of Luminex data for immunoconversions was conducted using crude responses from specific antibody assays, as well as two different data adjustment approaches. In the first adjustment approach, norovirus-specific IgA or IgG antibody responses were divided by the corresponding (geometric mean) responses from the separate immunoassay which measured total IgA and IgG. In the second adjustment approach, specific responses to norovirus proteins were divided by responses to the GST protein purification tag measured in the same quadruplex immunoassay (e.g. Norwalk-specific IgG response divided by IgG response to GST in the same microplate well). The resulting norovirus-to-total Ig or norovirus-to-GST ratios for each sample were then used in analysis of immunoconversions.
Sensitivity was defined as the ratio of true positive results (the number of infected individuals who immunoconverted) to the total number of infected individuals (in this study, three individuals). Specificity of the Norwalk virus assay was defined as the ratio of true negative results in the analysis of immunoconversions to Norwalk virus assay to the total number of non-infected individuals (in this study, four individuals). In addition, the specificity of GII norovirus tests was defined as the ratio of all true negative results for each of the GII noroviruses (VA387 and VA207) to the total number of tests conducted (in this study, seven individuals).
Sensitivity values for Norwalk virus immunoconversion tests, and specificity values for Norwalk and both GII tests were calculated for all combinations of Luminex assay conditions (five options) and data adjustment approaches (three options), separately for IgA and IgG tests. Average sensitivity and specificity (for Norwalk and GII tests) values were then calculated for each data adjustment approach, for IgA and IgG tests (Table 2). Overall proportions of correct results for Norwalk virus and GII norovirus tests were calculated for each assay condition and each data adjustment approach as ratios of the total number of correct responses to the total number of tests conducted.
Finally, statistical tests were conducted to compare results produced using different data adjustment approaches. For this analysis, data from each immunoconversion test were dichotomized as correct (true positive or true negative) or incorrect (false positive or false negative). Datasets for IgA and IgG tests included 315 observations each (seven individuals by three viruses by five assay conditions by three data adjustment approaches). Data were analyzed using mixed effects logistic regression models with random intercept (SAS procedure NLMIXED). Regression analysis was conducted separately for IgA and IgG, and also using a pooled dataset including results for both antibody isotypes. For assessing the effect of data adjustment approaches, the assay conditions were modeled as a random effect and data adjustment options as a fixed effect (Table 3). For assessing the effects of assay conditions, model specifications were changed with assay conditions modeled as a fixed effect and data adjustment options as a random effect (see section 3.1).
Table 3.
Mixed effect logistic regression analysis of the effects of data adjustment approach on the odds of producing a correct immunoconversion test result in the multiplex Luminex assay. The results are presented as: point estimate of odds ratios (95% confidence intervals), p value.
| Data adjustment approach |
IgA results | IgG results | Pooled IgA and IgG results |
|---|---|---|---|
| No adjustment | Reference | Reference | Reference |
| Ratio to total Ig | 1.4 (0.6, 3.4), p = 0.4 | 2.4 (0.6, 10.7), p = 0.2 | 1.7 (0.8, 3.9), p = 0.17 |
| Ratio to GST | 5.2 (1.8, 14.9), p = 0.005 | 5.5 (1.1, 29.1), p = 0.04 | 5.2 (2.1, 13.0), p = 0.001 |
3. Results
3.1. Luminex assay
3.1.1 Comparison of data adjustment approaches to control for sample-to-sample variability in saliva composition
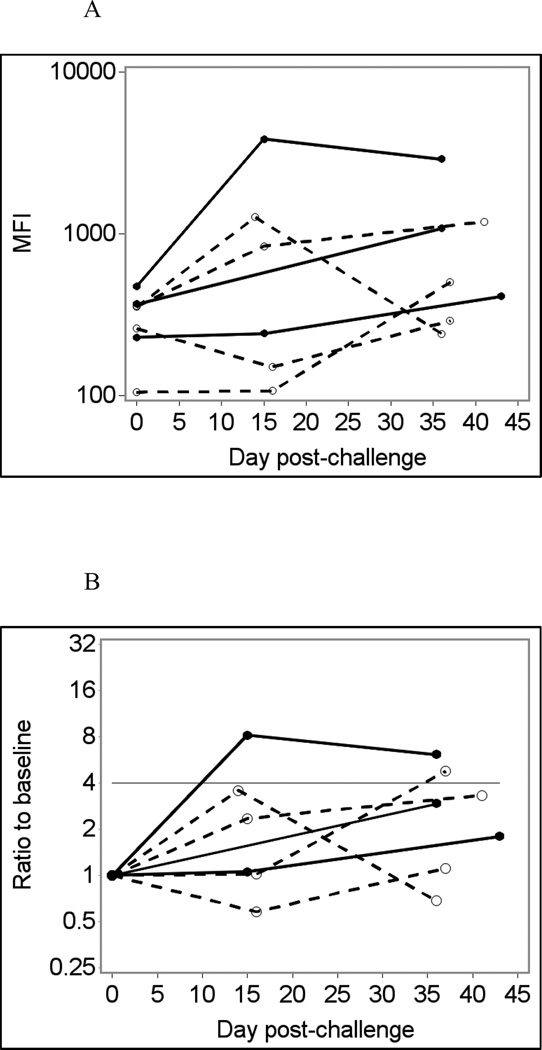
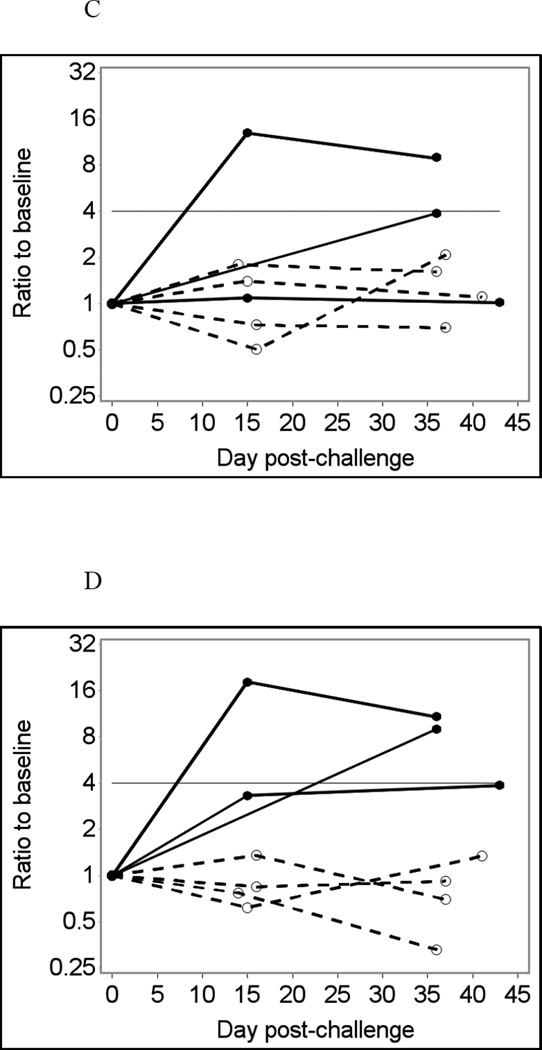
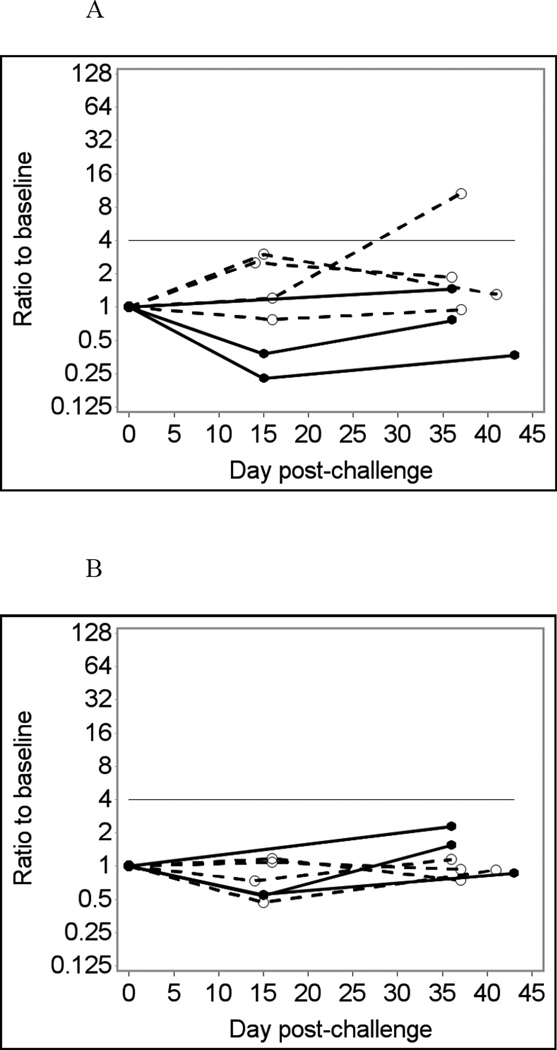
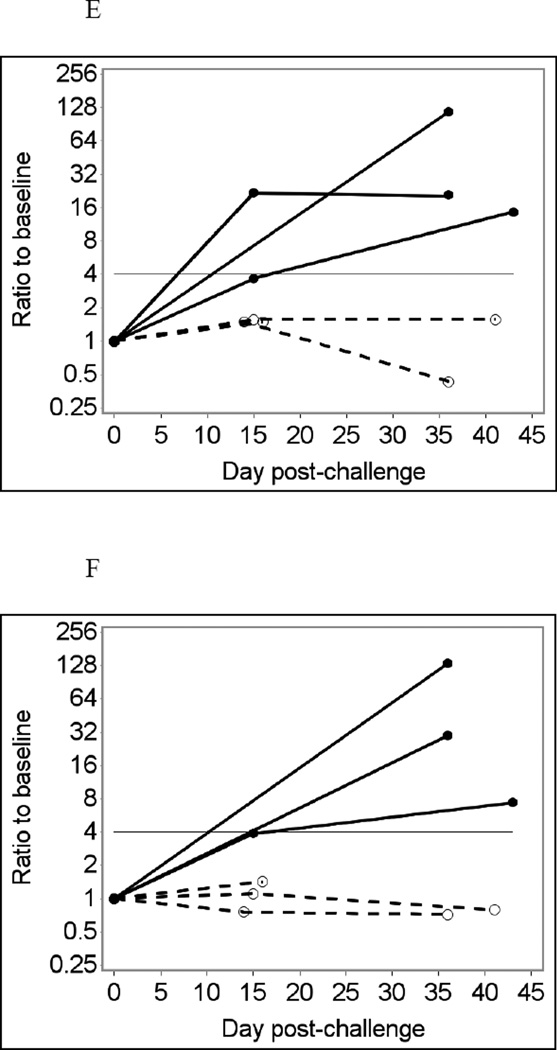
Fig. 1 shows temporal changes in IgA (A – D) and IgG (E – H) responses to the Norwalk virus P particles in infected and uninfected individuals using saliva samples diluted four-fold in PBS-BSA without heat treatment as an example. Using crude (unadjusted) responses to Norwalk virus P particles to detect immunoconversions caused two false-negative and one false-positive result in the IgA assay (Fig. 1A and B), and one false-positive result in the IgG assay (Fig. 1E and F). Adjusting for total Ig improves the assay performance, eliminating false-positive results in both IgA and IgG assays (Fig. 1 C and G). Adjusting for GST (Fig 1D and H) further improves the assay performance with only one false-negative result in the IgA assay (which is just short of being positive). It is also evident that adjusting for GST improves the overall differentiation of results from infected and non-infected individuals.
Fig. 1.
Norwalk virus-specific salivary antibody responses in seven participants of a Norwalk virus challenge study using the Luminex immunoassay with saliva diluted 1:4 in PBS-BSA buffer without heat pre-treatment.
Solid lines represent antibody responses in Norwalk virus-infected individuals; dashed lines represent responses in uninfected individuals. More than four-fold increase in antibody response (above the horizontal line) is considered an immunoconversion.
(A) Crude IgA responses to Norwalk virus in MFI units;
(B) Fold increases from baseline in Norwalk-specific IgA responses;
(C) Fold increases from baseline in Norwalk-specific IgA adjusted for total IgA;
(D) Fold increases from baseline in Norwalk-specific IgA adjusted for GST IgA;
(E) Crude IgG responses to Norwalk virus in MFI units;
(F) Fold increases from baseline in Norwalk-specific IgG responses;
(G) Fold increases from baseline in Norwalk-specific IgG adjusted for total IgG;
(H) Fold increases from baseline in Norwalk-specific IgG adjusted for GST IgG.
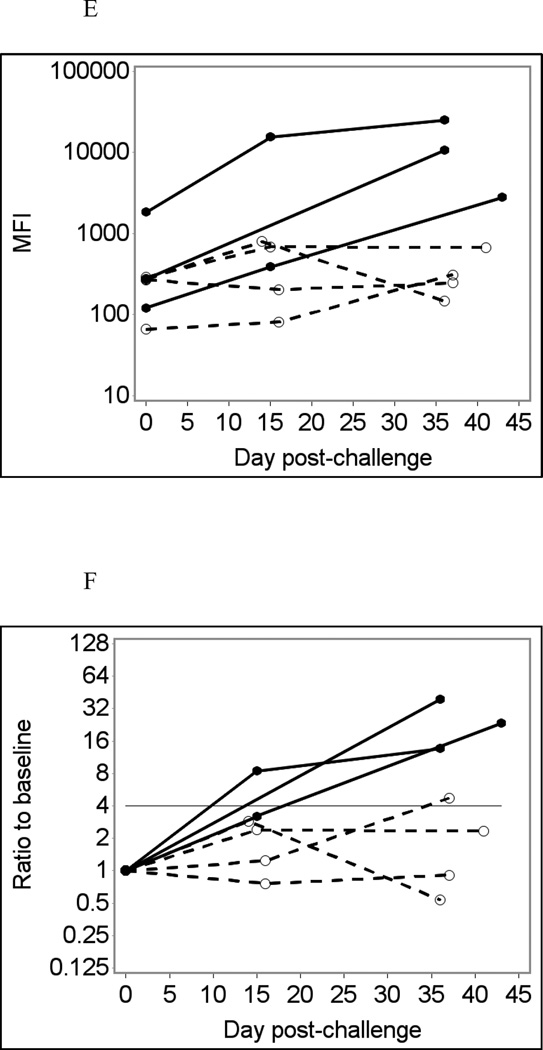
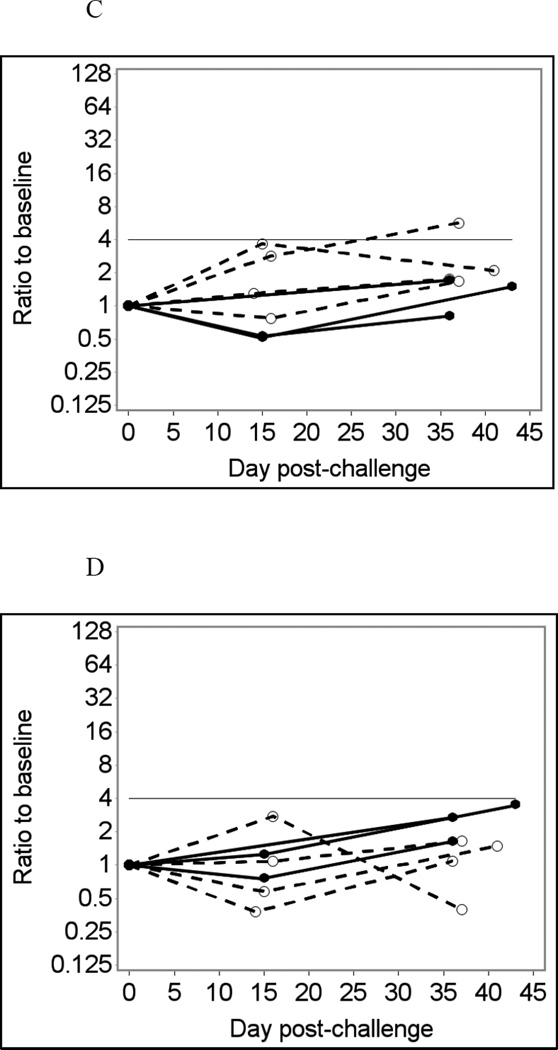
Fig. 2 shows selected data on temporal changes in IgG responses to GII noroviruses, VA207 (A – B) and VA387 (C – D). Using unadjusted assay data (Fig. 2A and C) produced two false positive results in different individuals who were not infected with Norwalk: one in the VA207 assay and another in the VA387 assay. Adjusting for GST eliminated these false-positive results (Fig. 2B and D). None of the three Norwalk virus-infected individuals had an immunoconversion to GII noroviruses in these tests although one of these individuals had a 3.52-fold increase in VA387-specific IgG response, nearing the immunoconversion threshold.
Fig. 2.
GII norovirus-specific salivary IgG responses in seven participants of a Norwalk virus challenge study using the Luminex immunoassay with saliva diluted 1:4 in PBS-BSA buffer without heat pre-treatment.
Solid lines represent antibody responses in Norwalk virus-infected individuals; dashed lines represent responses in uninfected individuals. More than four-fold increase in antibody response (above the horizontal line) is considered an immunoconversion.
(A) Fold increases from baseline in VA207-specific IgG responses;
(B) Fold increases from baseline in VA207-specific IgG adjusted for GST IgG;
(C) Fold increases from baseline in VA387-specific IgG responses;
(D) Fold increases from baseline in VA387-specific IgG adjusted for GST IgG.
Table 2 summarizes the results of all Luminex tests expressed as sensitivity and specificity of the Norwalk virus immunoassay, and specificity of the immunoassays for two GII noroviruses. Preliminary analysis (not shown) demonstrated that all false-positive immunoconversions in GII assays occurred in individuals who did not develop Norwalk infection after challenge and that antibody responses to VA387 and VA207 antigens were rather similar. It is important to note that the proportions of correct results in Table 2 are calculated for the purposes of comparing assay conditions; they do not necessarily reflect the proportions of correct results that may be observed in a population-based survey.
Adjusting for GST responses consistently produced the highest proportions of correct immunoconversion results in both IgA and IgG assays and for all assay conditions. Statistical analysis using mixed effect logistic regression models showed that adjusting for GST significantly improved the odds of obtaining a correct result compared to using unadjusted data (Table 3). Adjusting for total Ig produced smaller statistically insignificant improvements compared to using unadjusted data (Table 3).
3.1.2 Comparison of assay conditions
Heat pre-treatment of saliva samples consistently reduced sensitivity and specificity compared to no pre-treatment in both assay buffers, both dilutions, and all data adjustment approaches tested (Table 2). Furthermore, mixed effect logistic regression analysis with assay condition as a fixed effect and data adjustment approach as a random effect suggested that in the pooled IgA and IgG data set, the odds ratio of correct assignment in experiments with heat treatment compared to no treatment was 0.25 (0.12, 0.52), p = 0.004. When analyzed separately for IgA and IgG, the effects of heat treatment were consistently detrimental but short of being significant (data not shown).
Comparing the results of tests at 1:4 and 1:32 final dilutions (PVX buffer, no heat treatment) shows that the highest overall proportions of correct assignments for IgA and IgG tests were achieved at 1:32 sample dilution (Table 2). This effect was not statistically significant with p = 0.14 for the combined IgA and IgG data. It also has to be noted that using a higher dilution predictably resulted in an improved specificity at the expense of a reduced sensitivity (one false negative result in the IgG assay). Due to the larger number of specificity tests, the improvement in specificity has more influence on the overall proportion of correct assignments than the decline in sensitivity.
Results of tests conducted using PBS-BSA and PVX buffers (no heat treatment, 1:4 final dilution) were similar and showed no significant effect (not shown). While using PVX buffer did not provide clear advantages compared to the PBS-BSA buffer, it created problems with bead acquisition in the Luminex device (where multiple microscopic beads are analyzed one-by-one after coming through a special channel) due to the higher viscosity of this buffer and formation of foam in microplate wells, especially at the 1:4 dilution of saliva.
3.1.3 Comparison of IgA and IgG assays
Fig. 1 shows that Norwalk-specific salivary IgA responses were weaker than IgG responses in infected individuals (Fig. 1A and E). Similarly, much weaker fold increases in Norwalk-specific IgA responses compared to the corresponding IgG responses were observed in infected individuals. For GST-adjusted results, maximum fold-increases in IgA responses in infected individuals were 3.9 (false-negative result), 9.0 and 18.2 while respective maximum ratios to baseline for IgG responses were 55.0, 61.2 and 27.8. The maximum ratios to baseline in non-infected individuals varied from 0.77 to 1.35 in the Norwalk-specific IgA assay and from 0.85 to 1.66 in the IgG assay. Summary of results presented in Table 2 also shows that IgG tests had consistently higher sensitivity than IgA, while specificity of IgA and IgG tests were rather similar.
3.2. MSD assay results
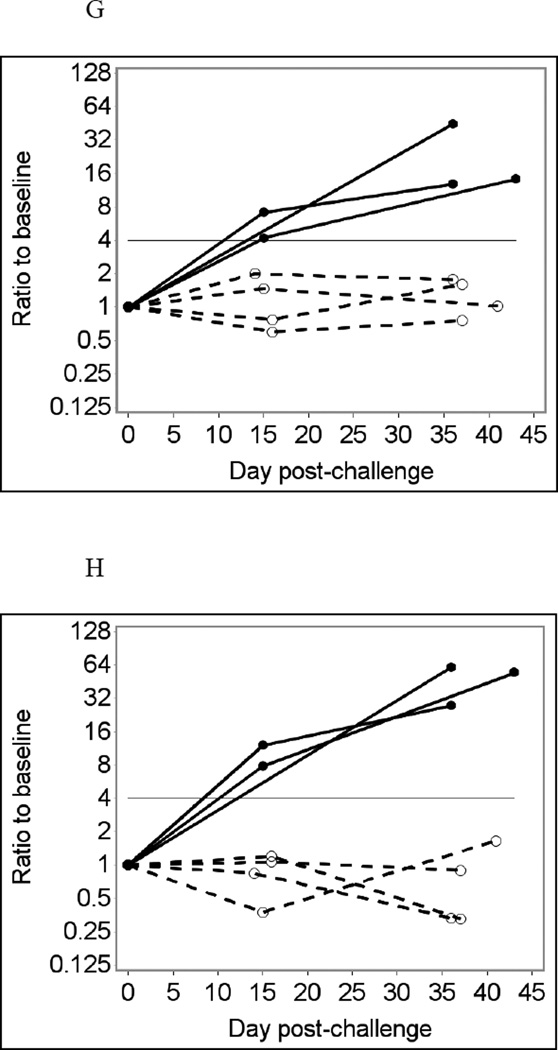
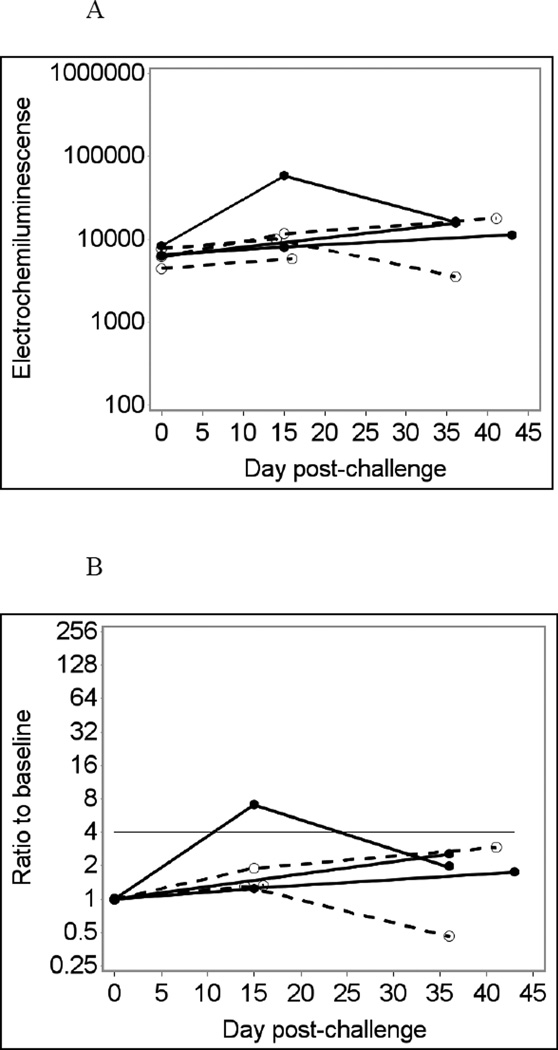
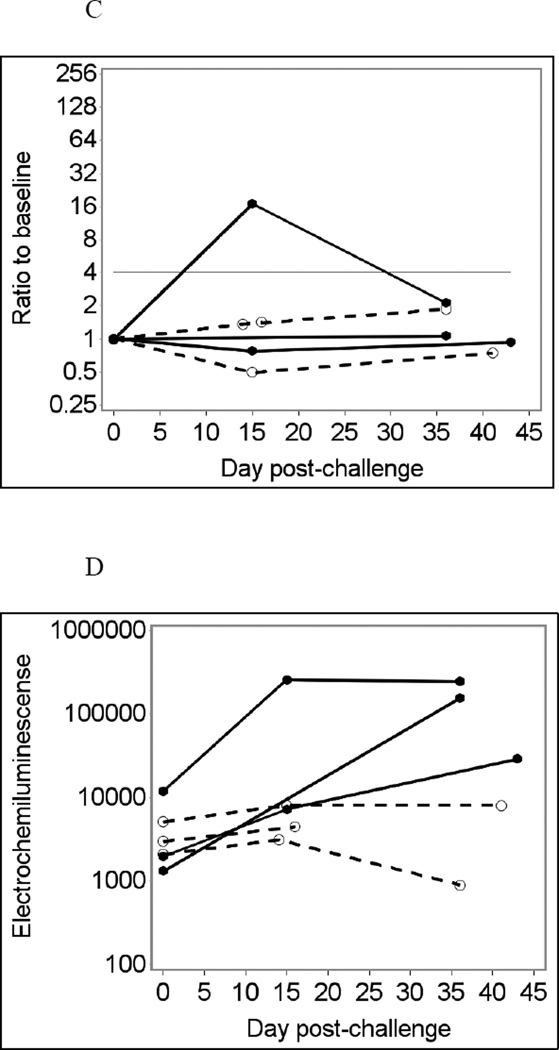
Sixteen leftover saliva samples from six individuals were tested using the MSD assay at 1:4 final dilution without heat treatment, in duplicate (Fig. 3A and D). For Norwalk-specific IgA responses, the MSD assay produced one true positive and two false-negative immunoconversion results (33% sensitivity), while its specificity was 100% (Fig. 3B). The MSD assay for Norwalk virus produced 100% specificity and 100% sensitivity for IgG responses (Fig. 3E). The MSD assay for IgG responses to VA387 norovirus also produced 100% specificity (data not shown). Antibody responses to VA207 norovirus and to GST were not measured in the MSD assay (see section 2.4). Adjusting for total Ig did not produce noticeable improvement in the results of MSD tests (Fig. 3C and F). No immunoconversions were observed with saliva samples assayed in BSA-coated MSD wells, which were used as controls (data not shown). Comparing the results of MSD and Luminex analyses at similar conditions (no heat pre-treatment, saliva diluted 1:4) showed very similar temporal patterns of antibody responses in the same individuals (not shown) confirming that the two assays produce largely consistent results.
Fig. 3.
Norwalk virus-specific salivary IgG responses in six participants of a Norwalk virus challenge study using the MSD immunoassay with saliva diluted 1:4 in PBS-BSA buffer without heat pre-treatment.
Solid lines represent antibody responses in Norwalk virus-infected individuals; dashed lines represent responses in uninfected individuals. More than four-fold increase in antibody response (above the horizontal line) is considered an immunoconversion.
(A) Crude IgA responses to Norwalk virus in ECL units;
(B) Fold increases from baseline in Norwalk-specific IgA responses;
(C) Fold increases from baseline in Norwalk-specific IgA adjusted for total IgA;
(D) Crude IgG responses to Norwalk virus in ECL units;
(E) Fold increases from baseline in Norwalk-specific IgG responses;
(F) Fold increases from baseline in Norwalk-specific IgG adjusted for total IgG.
4. Discussion
In this study, salivary immunoassays based on the Luminex and MSD platforms were used to successfully detect incident Norwalk virus (GI.1) infections in participants of a Norwalk virus challenge study and to differentiate infected and non-infected individuals. This study demonstrated that dividing each norovirus-specific antibody response by response to GST in the same sample significantly improves the sensitivity and specificity of the Norwalk virus immunoconversion test based on the multiplex Luminex assay data and the specificity of the immunoconversion tests for both GII noroviruses compared to using unadjusted Luminex data. Adjusting anti-norovirus responses for the total immunoglobulin level of the same isotype produced weaker, statistically not significant improvements in assay performance compared to using unadjusted data.
Although the method of P particle production involves cleaving and removing the GST purification tag (Tan and Jiang, 2005; Tan et al., 2008), separate tests using anti-GST antibodies (not shown) demonstrated reactivity with all three types of norovirus proteins, suggesting that GST removal was incomplete. In these experiments, however, GST served as an internal control for overall non-specific binding of salivary antibodies which varied from sample to sample substantially due to intrinsic characteristics of saliva. These may include total concentration of Ig, which were measured in this survey, as well as other factors that were not explored. While GST responses were analyzed in the same wells with norovirus responses, total IgA and IgG levels were measured using a separate duplex Luminex assay, which could be a source of additional random variation and a factor partially explaining the improved sensitivity and specificity of immunoconversions tests with GST adjustment. While Luminex beads are blocked with BSA as part of the standard bead coupling procedure, non-specific responses to GST-coupled beads were always stronger than responses to beads coated with BSA only (not shown), which made GST a preferable internal control producing more quantifiable responses.
Converting crude MFI and ECL values for total antibody tests into Ig concentrations by standard curve analysis (for method description, see Griffin et al., 2011) and adjusting for concentrations of total antibodies did not improve Norwalk virus assay performance compared to simply adjusting for MFI or ECL values for total antibodies. This was expected because saliva dilutions for the total antibody assay (5,000x to 20,000x) were selected based on the results of preliminary tests of saliva at serial dilutions so that the results are in the middle of the assay linearity ranges.
Previously conducted experiments used heat pre-treated saliva samples to detect salivary antibodies to noroviruses (Lindesmith et al., 2003), and to measles, mumps, and rotavirus (Friedman, 1982; Friedman et al., 1989 and 1993). This study demonstrated that heat pre-treatment and centrifugation of saliva samples had a detrimental impact on the performance of the Luminex immunoassay for Norwalk virus resulting in a lower proportion of correct results. It can be speculated that centrifugation after heat treatment caused antibodies to be trapped in the pellet and therefore not retained in the supernatant and used in subsequent Luminex tests (see section 2.3.2).
The PVX buffer is known to reduce non-specific binding of serum antibodies in the Luminex assays (Waterboer et al., 2006; van Gageldonk et al., 2011). It has been suggested that human sera contains broadly reactive (heterophile) antibodies that directly bind to Luminex beads, resulting in a higher nonspecific background (Waterboer et al., 2006; van Gageldonk et al., 2011). It is recommended to preincubate serum in PVX buffer to significantly reduce this nonspecific background. It has also been proposed that nonspecific binding is likely caused by changes in antigen structure during Luminex bead coupling (van Gageldonk et al., 2011). Van Gageldonk et al. (2011) found that use of PVX buffer only caused a reduction in background with certain antigens (i.e. diphtheria toxoid), but not others (i.e. tetanus and pertussis antigens). In the tests with norovirus antigens in this study, using PVX and PBS-BSA buffers produced very similar results. It may be that there are fewer heterophile antibodies in saliva compared to serum, and thus less nonspecific binding regarding these broadly reactive antibodies. Additionally, the norovirus P particles may be less susceptible to conformational changes during bead coupling, which also allows for reduced nonspecific binding. Furthermore, the more viscous PVX buffer interfered with the Luminex fluidics system at a low dilution of saliva in this study. Therefore, using the standard Luminex PBS-BSA buffer is selected as the default option.
Due to the viscosity of saliva samples, at least 1:4 final dilution in the well is required in the Luminex assay. Lower saliva dilutions can clog filter-bottom microplates which results in a high proportion of unsuccessful Luminex tests as well as spurious results due to increased nonspecific binding (not shown). It may be advisable to re-analyze samples at a higher dilution when it is more important to achieve a high specificity than high sensitivity. For example, high specificity can be crucial in a population based survey in non-outbreak settings where only a small proportion of participants may experience norovirus infections.
Norwalk-specific IgG response was a more accurate biomarker of incident infections than IgA response. All three infected individuals had strong IgG responses, which were very clearly distinguishable from responses in uninfected individuals. In contrast, one of the three infected individuals had a very weak IgA response to Norwalk virus and did not exceed the immunoconversion threshold. IgA responses increase after infection sooner than IgG and peak approximately two weeks after infection (Erdman et al., 1989; Moe et al., 2004). IgA tests can be used along with IgG if time interval between infection and follow-up sample collection is relatively short.
This salivary immunoassay-based method is intended for detecting incident infections in longitudinal studies. At least two saliva samples collected from the same individual on different dates are needed in order to detect an immunoconversion (at least four-fold increase in antibody response in the second sample compared to the first sample). This assay is not intended to assess the proportions of “positive” or “negative” antibody responses to Norwalk virus in cross- sectional settings. For a transient infection, like norovirus, a strong antibody response can be a marker of a relatively recent infection or an ongoing infection. Estimating an incidence rate of infection from cross-sectional survey data would be problematic because the magnitude of antibody response and rate of its temporal decline after infection can vary substantially among individuals.
This study did not detect cross-reactivity of salivary antibodies against Norwalk virus with two GII noroviruses, VA387 (GII.4) and VA207 (GII.9). These results are consistent with previous serological studies that demonstrated genogroup-specific antibody responses with a lower level of antibody cross-reactivity between genogroups than within genogroups (Parker et al., 2005; Hansman et al., 2006; LoBue et al., 2006; Okame et al., 2007; Lindesmith et al., 2010). It remains to be demonstrated that this Norwalk antigen-based immunoassay can detect infections with other genogroup I noroviruses.
An assay that can differentiate infections with GI and GII noroviruses will be particularly useful in epidemiological studies of waterborne norovirus infections as GI noroviruses are more likely to be transmitted through drinking water. It has been shown that from 31% to 85% of waterborne norovirus outbreaks are caused solely by GI noroviruses (Maunula et al., 2005; Lysen et al., 2009; Matthews at el., 2012).
The Luminex and MSD assays using the same antigens produced very similar results demonstrating that the salivary antibody approach can be implemented on various platforms and expanding options available for further development and application in epidemiological research. The selection of the assay platform for further applications of this method would largely depend on the availability of equipment in the laboratory. Although both Luminex and MSD platforms provide multiplexing capabilities, this study employed multiplex Luminex assays and monoplex MSD assays. Multiplex MSD assays require special plates with multiple antigens coated on predefined spots in each well. Coating multi-spot MSD wells with antigens can only be performed by the assay manufacturer under a custom order. While developing and using a multiplex MSD assay was impracticable in this study due to the small number of samples involved, it may be a cost effective approach in investigations involving larger sets of samples.
This study used spit samples collected in the presence of survey staff after stimulation with sour candy. The composition of oral fluid collected using other methods can be different. For example, oral fluid samples collected using Oracol™ samplers can have higher concentrations of IgG than spit samples (Griffin et al., 2011). In the present survey, mean concentrations of total IgA and total IgG were 16 µg/mL and 7 µg/mL respectively compared to 21 µg/mL and 50 µg/mL in Oracol oral fluid samples reported by Griffin et al. (2011). Sample-to-sample variability in the composition of saliva or oral fluid may also be greater in population based surveys when participants are allowed to collect samples on their own without supervision of the survey staff. These factors may have an effect on assay sensitivity and specificity.
The small sample size limits the interpretation of the results to a proof of concept. Small sample sizes are common in experiments in which human volunteers are intentionally infected with pathogens. For example, a similarly small sample size was recently used in a volunteer study which demonstrated infectivity of a protozoan parasite Cryptosporidium meleagridis in healthy adults (N = 5, Chappell et al., 2011). Previous norovirus challenge studies with small sample sizes provided useful information about immunobiology of GII.2 norovirus infection (N = 15; Lindesmith et al., 2005), length and amount of Norwalk virus shedding (N = 16; Atmar et al., 2008), and infectivity of Norwalk virus and its persistence in water (N = 13; Seitz et al., 2011).
In this study, Luminex and MSD salivary IgG assays produced 100% sensitivity and 100% specificity in detecting incident Norwalk virus (GI.1) infections at optimal conditions. These tests also produce 100% negative results when using GII norovirus antigens. Further assay validation using saliva samples from individuals infected with genogroup II noroviruses is necessary for assessing sensitivity of these tests.
A larger study involving more individuals with known infection status would be essential for producing more accurate estimates of assay sensitivity and specificity (which are bound to be less than 100%). It should also be noted that this study only used samples from individuals who were not infected or who were infected and had typical norovirus infection symptoms. Patterns of antibody responses in infected asymptomatic individuals may potentially be different. In a community-based survey of norovirus infections, the salivary immunoassay method should always be used in combination with symptom diaries to differentiate symptomatic and asymptomatic infections. Positive and negative predictive values of the method will need to be assessed for different scenarios, such as outbreak investigations vs. community survey in non-outbreak settings. Ensuring a high specificity of the immunoconversion test is especially important for non-outbreak situations when most survey participants do not become infected during a follow-up period.
This study demonstrated that the use of salivary antibodies in conjunction with recombinant Norwalk virus P particles may enable inexpensive and non-invasive surveillance of incident Norwalk virus infections in prospective epidemiological studies. Further verification of assay performance in a larger population of individuals with known infection status would be necessary to provide better estimates of assay performance (specificity, sensitivity, predictive values), characterize the uncertainty of these estimates, and to evaluate potential cross-reactivity of anti-Norwalk virus antibodies with other noroviruses. Further method development should also aim at incorporating antigens of other noroviruses to enable the detection of all norovirus infections and, potentially, differentiation between genogroup I and genogroup II norovirus infections in longitudinal survey settings.
Highlights.
A Norwalk virus immunoassay was tested using saliva from a human challenge study
An accurate indicator of infection was a four-fold increase in anti-Norwalk IgG
Anti-Norwalk virus antibodies did not cross-react with genogroup II noroviruses
Luminex and MSD immunoassay platforms provided similar results
This salivary immunoassay may enable low cost longitudinal community-based surveys
Acknowledgements
The authors are thankful to Jefferson Inmon for his laboratory assistance and to Jennifer Cashdollar and Elizabeth Hedrick (US EPA) for critical review of the draft manuscript. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of US EPA. Mention of trade names, products, or services does not convey, and should not be interpreted as conveying official US EPA approval, endorsement or recommendation. This work was supported in part (to Juan S. Leon) by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number K01AI087724. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None of the authors has any conflict of interest.
References
- Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 2008;14:1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn BG, Craun GF, Yoder JS, Hill V, Calderon RL, Chen N, Lee SH, Levy DA, Beach MJ Centers for Disease Control and Prevention (CDC) Surveillance for waterborne-disease outbreaks associated with drinking water--United States, 2001–2002. MMWR Surveill. Summ. 2004;53:23–45. [PubMed] [Google Scholar]
- Blanton LH, Adams SM, Beard RS, Wei G, Bulens SN, Widdowson MA, Glass RI, Monroe SS. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000–2004. J. Infect. Dis. 2006;193:413–421. doi: 10.1086/499315. [DOI] [PubMed] [Google Scholar]
- Bon F, Ambert-Balay K, Giraudon H, Kaplon J, Le Guyader S, Pammepuy M, Gallay A, Vaillant V, de Valk H, Chikhi-Brachet R, Flahaut A, Pothier P, Kohli E. Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J. Clin. Microbiol. 2005;43:4659–4664. doi: 10.1128/JCM.43.9.4659-4664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell CL, Okhuysen PC, Langer-Curry RC, Akiyoshi DE, Widmer G, Tzipori S. Cryptosporidium meleagridis: infectivity in healthy adult volunteers. Am. J. Trop. Med. Hyg. 2011;85:238–242. doi: 10.4269/ajtmh.2011.10-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaver DR. A new non-isotopic detection system for immunoassays. Nature. 1995;377:758–760. doi: 10.1038/377758a0. [DOI] [PubMed] [Google Scholar]
- Erdman DD, Gary GW, Anderson LJ. Serum immunoglobulin A response to Norwalk virus infection. J. Clin. Microbiol. 1989;27:1417–1418. doi: 10.1128/jcm.27.6.1417-1418.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman MG. Radioimmunoassay for the detection of virus-specific IgA antibodies in saliva. J. Immunol. Methods. 1982;54:203–211. doi: 10.1016/0022-1759(82)90061-8. [DOI] [PubMed] [Google Scholar]
- Friedman MG, Phillip M, Dagan R. Virus-specific IgA in serum, saliva, and tears of children with measles. Clin. Exp. Immunol. 1989;75:58–63. [PMC free article] [PubMed] [Google Scholar]
- Friedman MG, Segal B, Zedaka R, Sarov B, Margalith M, Bishop R, Dagan R. Serum and salivary responses to oral tetravalent reassortant rotavirus vaccine in newborns. Clin. Exp. Immunol. 1993;92:194–199. doi: 10.1111/j.1365-2249.1993.tb03379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore CI, Green J, Lewis D, Richards AF, Lopman BA, Hale AD, Eglin R, Gray JJ, Brown DW. Diversity of noroviruses co-circulating in the north of England from 1998 to 2001. J. Clin. Microbiol. 2004;42:1396–1401. doi: 10.1128/JCM.42.4.1396-1401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DY, Jiang X, Tanaka T, Opekun AR, Madore HP, Estes MK. Norwalk virus infection of volunteers: new insights based on improved assays. J. Infect. Dis. 1994;170:34–43. doi: 10.1093/infdis/170.1.34. [DOI] [PubMed] [Google Scholar]
- Green KY, Belliot G, Taylor JL, Valdesuso J, Lew JF, Kapikia AZ, Lin FC. A predominant role for Norwalk-like viruses as agents of epidemic gastroenteritis in Maryland nursing homes for the elderly. J. Infect. Dis. 2002;185:133–146. doi: 10.1086/338365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin SM, Chen IM, Fout GS, Wade TJ, Egorov AI. Development of a multiplex microsphere immunoassay for the quantitation of salivary antibody responses to selected waterborne pathogens. J. Immunol. Methods. 2011;364:83–93. doi: 10.1016/j.jim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Hall AJ, Curns AT, McDonald LC, Parashar UD, Lopman BA. The roles of Clostridium difficile and norovirus among gastroenteritis-associated deaths in the United States, 1999–2007. Clin. Infect. Dis. 2012;55:216–223. doi: 10.1093/cid/cis386. [DOI] [PubMed] [Google Scholar]
- Hansman GS, Natori K, Shirato-Horikoshi H, Ogawa S, Oka T, Katayama K, Tanaka T, Miyoshi T, Sakae K, Kobayashi S, Shinohara M, Uchida K, Sakurai N, Shinozaki K, Okada M, Seto Y, Kamata K, Nagata N, Tanaka K, Miyamura T, Takeda N. Genetic and antigenic diversity among noroviruses. J. Gen. Virol. 2006;87:909–919. doi: 10.1099/vir.0.81532-0. [DOI] [PubMed] [Google Scholar]
- Huang P, Farkas T, Zhong W, Tan M, Thornton S, Morrow AL, Jiang X. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 2005;79:6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkula M, Maunula L, Silvennoinen E, von Bonsdorff CH. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J. Infect. Dis. 1999;180:1771–1776. doi: 10.1086/315145. [DOI] [PubMed] [Google Scholar]
- Lambertini E, Borchardt MA, Kieke BA, Jr, Spencer SK, Loge FJ. Risk of viral acute gastrointestinal illness from nondisinfected drinking water distribution systems. Environ. Sci. Technol. 2012;46:9299–9307. doi: 10.1021/es3015925. [DOI] [PubMed] [Google Scholar]
- Lee SG, Jheong WH, Suh CI, Kim SH, Lee JB, Jeong YS, Ko G, Jang KL, Lee GC, Paik SY. Nationwide groundwater surveillance of noroviruses in South Korea, 2008. Appl. Environ. Microbiol. 2011;77:1466–1474. doi: 10.1128/AEM.01996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon JS, Souza M, Wang Q, Smith ER, Saif LJ, Moe CL. Immunology of norovirus infection. In: Vajdy M, editor. Immunity against Mucosal Pathogens. Springer Science & Business Media, B.V; 2008. pp. 219–262. [Google Scholar]
- Leon JS, Kingsley DH, Montes JS, Richards GP, Lyon GM, Abdulhafid GM, Sietz SR, Fenandez ML, Teunis PF, Flick GJ, Moe CL. Randomized, double-blinded clinical trial for human norovirus inactivation in oysters by high hydrostatic pressure processing. Appl. Environ. Microbiol. 2011;77:5476–5482. doi: 10.1128/AEM.02801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith LC, Donaldson E, Leon J, Moe CL, Frelinger JA, Johnston RE, Weber DJ, Baric RS. Heterotypic humoral and cellular immune responses following Norwalk virus infection. J Virol. 2010;84:1800–1815. doi: 10.1128/JVI.02179-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L, Moe C, Lependu J, Frelinger JA, Treanor J, Baric RS. Cellular and humoral immunity following Snow Mountain virus challenge. J. Virol. 2005;79:2900–2909. doi: 10.1128/JVI.79.5.2900-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, Stewart P, LePendu J, Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- LoBue AD, Lindesmith L, Yount B, Harrington PR, Thompson JM, Johnston RE, Moe CL, Baric RS. Multivalent norovirus vaccines induce strong mucosal and systemic blocking antibodies against multiple strains. Vaccine. 2006;24:5220–5234. doi: 10.1016/j.vaccine.2006.03.080. [DOI] [PubMed] [Google Scholar]
- Lodder WJ, van den Berg HH, Rutjes SA, de Roda Husman AM. Presence of enteric viruses in source waters for drinking water production in The Netherlands. Appl. Environ. Microbiol. 2010;76:5965–5971. doi: 10.1128/AEM.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopman BA, Hall AJ, Curns AT, Parashar UD. Increasing rates of gastroenteritis hospital discharges in U.S. adults and the contribution of norovirus, 1996–2007. Clin. Infect. Dis. 2011;52:466–474. doi: 10.1093/cid/ciq163. [DOI] [PubMed] [Google Scholar]
- Lysén M, Thorhagen M, Brytting M, Hjertqvist M, Andersson Y, Hedlund KO. Genetic diversity among food-borne and waterborne norovirus strains causing outbreaks in Sweden. J. Clin. Microbiol. 2009;47:2411–2418. doi: 10.1128/JCM.02168-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JE, Dickey BW, Miller RD, Felzer JR, Dawson BP, Lee AS, Rocks JJ, Kiel J, Montes JS, Moe CL, Eisenberg JN, Leon JS. The epidemiology of published norovirus outbreaks: a review of risk factors associated with attack rate and genogroup. Epidemiol. Infect. 2012;140:1161–1172. doi: 10.1017/S0950268812000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JA, Hellard ME, Sinclair MI, Fairley CK, Cox BJ, Catton MG, Kelly H, Wright PJ. Incidence and characteristics of endemic Norwalk-like virus-associated gastroenteritis. J. Med. Virol. 2003;69:568–578. doi: 10.1002/jmv.10346. [DOI] [PubMed] [Google Scholar]
- Maunula L, Miettinen IT, von Bonsdorff CH. Norovirus outbreaks from drinking water. Emerg. Infect. Dis. 2005;11:1716–1721. doi: 10.3201/eid1111.050487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunula L, von Bonsdorff CH. Norovirus genotypes causing gastroenteritis outbreaks in Finland 1998–2002. J. Clin. Virol. 2005;34:186–194. doi: 10.1016/j.jcv.2005.03.004. [DOI] [PubMed] [Google Scholar]
- McKie A, Vyse A, Maple C. Novel methods for the detection of microbial antibodies in oral fluid. Lancet Infect. Dis. 2002;2:18. doi: 10.1016/s1473-3099(01)00169-4. [DOI] [PubMed] [Google Scholar]
- Moe CL, Sair A, Lindesmith L, Estes MK, Jaykus LA. Diagnosis of Norwalk virus infection by indirect enzyme immunoassay detection of salivary antibodies to recombinant norwalk virus antigen. Clin. Diagn. Lab. Immunol. 2004;11:1028–1034. doi: 10.1128/CDLI.11.6.1028-1034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe SS, Stine SE, Jiang X, Estes MK, Glass RI. Detection of antibody to recombinant Norwalk virus antigen in specimens from outbreaks of gastroenteritis. J. Clin. Microbiol. 1993;31:2866–2872. doi: 10.1128/jcm.31.11.2866-2872.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Cunnington MC, Edmunds WJ, Miller E, Brown DW. A novel method of oral fluid collection to monitor immunity to common viral infections. Epidemiol. Infect. 2004a;132:35–42. doi: 10.1017/s0950268803001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Cunnington MC, Edmunds WJ, Miller E, Brown DW. A population-based seroprevalence study of hepatitis A virus using oral fluid in England and Wales. Am. J. Epidemiol. 2004b;159:786–794. doi: 10.1093/aje/kwh107. [DOI] [PubMed] [Google Scholar]
- Okame M, Shiota T, Hansman G, Takagi M, Yagyu F, Takanashi S, Phan TG, Shimizu Y, Kohno H, Okitsu S, Ushijima H. Anti-norovirus polyclonal antibody and its potential for development of an antigen-ELISA. J. Med. Virol. 2007;79:1180–1186. doi: 10.1002/jmv.20906. [DOI] [PubMed] [Google Scholar]
- Parker TD, Kitamoto N, Tanaka T, Hutson AM, Estes MK. Identification of Genogroup I and Genogroup II broadly reactive epitopes on the norovirus capsid. J. Virol. 2005;79:7402–7409. doi: 10.1128/JVI.79.12.7402-7409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parshionikar SU, Willian-True S, Fout GS, Robbins DE, Seys SA, Cassady JD, Harris R. Waterborne outbreak of gastroenteritis associated with a norovirus. Appl. Environ. Microbiol. 2003;69:5263–5268. doi: 10.1128/AEM.69.9.5263-5268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B, De Wit M, Vennema H, Vinje J, De Bruin E, Van Duynhoven Y, Koopmans M. Natural history of human calicivirus infection: a prospective cohort study. Clin. Infect. Dis. 2002;35:246–253. doi: 10.1086/341408. [DOI] [PubMed] [Google Scholar]
- Seitz SR, Leon JL, Schwab KJ, Lyon GM, Dowd M, McDaniels M, Abdulhafid G, Fernandez ML, Lindesmith LC, Baric RS, Moe CL. Norovirus infectivity in humans and persistence in water. Appl, Environ. Microbiol. 2011;77:6884–6888. doi: 10.1128/AEM.05806-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourial S, Marcusson-Stahl M, Cederbrant K. Meso Scale Discovery and Luminex Comparative Analysis of Calbindin D28K. J. Biomed. Biotech. 2009 doi: 10.1155/2009/187426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Fang P, Chachiyo T, Xia M, Huang P, Fang Z, Jiang W, Jiang X. Noroviral P particle: structure, function and applications in virus-host interaction. Virol. 2008;382:115–123. doi: 10.1016/j.virol.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Jiang X. The P domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J. Virol. 2005;79:14017–14030. doi: 10.1128/JVI.79.22.14017-14030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunis PF, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. Norwalk virus: how infectious is it? J. Med. Virol. 2008;80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- Thompson DK, Huffman KM, Kraus WE, Kraus VB. Critical Appraisal of Four IL-6 Immunoassays. PLOS One. 2012;7:e30659. doi: 10.1371/journal.pone.0030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng FC, Leon JS, MacCormack JN, Maillard JM, Moe CL. Molecular epidemiology of norovirus gastroenteritis outbreaks in North Carolina, United States: 1995–2000. J. Med. Virol. 2007;79:84–91. doi: 10.1002/jmv.20729. [DOI] [PubMed] [Google Scholar]
- Tsugawa T, Numata-Kinoshita K, Honma S, Nakata S, Tatsumi M, Sakai Y, Natori K, Takeda N, Kobayashi S, Tsutsumi H. Virological, serological, and clinical features of an outbreak of acute gastroenteritis due to recombinant genogroup II norovirus in an infant home. J. Clin. Microbiol. 2006;44:177–182. doi: 10.1128/JCM.44.1.177-182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA. GWR 40 CFR Parts 9, 141, and 142 National Primary Drinking Water Regulations: Ground Water Rule; Final Rule. Federal Register / 2006;71(216) [Google Scholar]
- van Gageldonk PGM, von Hunolstein C, van der Klis FRM, Berbers GAM. Improved specificity of a multiplex immunoassay for quantitation of anti-diphtheria toxin antibodies with the use of diphtheria toxoid. Clin. Vaccine Immunol. 2011;18:1183–1186. doi: 10.1128/CVI.05081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterboer T, Sehr P, Pawlita M. Suppression of non-specific binding in serological Luminex assays. J. Immunol. Methods. 2006;309:200–204. doi: 10.1016/j.jim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Yoder J, Roberts V, Craun GF, Hill V, Hicks LA, Alexander NT, Radke V, Calderon RL, Hlavsa MC, Beach MJ, Roy SL Centers for Disease Control and Prevention (CDC) Surveillance for waterborne disease and outbreaks associated with drinking water and water not intended for drinking--United States, 2005–2006. MMWR Surveill. Summ. 2008;57:39–62. [PubMed] [Google Scholar]
- Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]