Abstract
Disruption in circadian gene expression, whether due to genetic variation or environmental factors (e.g., light at night, shiftwork), is associated with increased incidence of breast, prostate, gastrointestinal and hematologic cancers and gliomas. Circadian genes are highly expressed in the ovaries where they regulate ovulation; circadian disruption is associated with several ovarian cancer risk factors (e.g., endometriosis). However, no studies have examined variation in germline circadian genes as predictors of ovarian cancer risk and invasiveness. The goal of the current study was to examine single nucleotide polymorphisms (SNPs) in circadian genes BMAL1, CRY2, CSNK1E, NPAS2, PER3, REV1 and TIMELESS and downstream transcription factors KLF10 and SENP3 as predictors of risk of epithelial ovarian cancer (EOC) and histopathologic subtypes. The study included a test set of 3,761 EOC cases and 2,722 controls and a validation set of 44,308 samples including 18,174 (10,316 serous) cases and 26,134 controls from 43 studies participating in the Ovarian Cancer Association Consortium (OCAC). Analysis of genotype data from 36 genotyped SNPs and 4600 imputed SNPs indicated that the most significant association was rs117104877 in BMAL1 (OR = 0.79, 95% CI = 0.68–0.90, p = 5.59 × 10−4]. Functional analysis revealed a significant down regulation of BMAL1 expression following cMYC overexpression and increasing transformation in ovarian surface epithelial (OSE) cells as well as alternative splicing of BMAL1 exons in ovarian and granulosa cells. These results suggest that variation in circadian genes, and specifically BMAL1, may be associated with risk of ovarian cancer, likely through disruption of hormonal pathways.
Introduction
Almost every human cell contains an autonomous circadian clock that synchronizes gene transcription in a daily oscillation for many physiological processes allowing for adaptation to the 24 hour environmental day/night cycle. Circadian genes are known to regulate a variety of cellular processes including the cell cycle, apoptosis, and DNA damage repair [1]. Disruption in circadian gene expression, whether due to genetic variants or environmental factors (e.g., light at night, shiftwork), is associated with increased incidence and invasiveness of a variety of human cancers [2–5] such that in 2007 the International Agency for Research on Cancer classified shift work that involves circadian disruption as “a probable carcinogen” in humans [6]. Disruption of circadian rhythms is also associated with disturbances in menstrual function; female shift workers compared to non-shift workers are more likely to report menstrual irregularity and longer menstrual cycles [7]. Moreover, a recent study found that working nightshifts (i.e., 12:00–4:00 AM) was associated with an increased risk of serious and mucinous, invasive and borderline ovarian tumors in women who were 50 years of age and older [8]. Nevertheless, some studies have failed to find an association between shiftwork and cancer risk [9–11].
The molecular mechanism of the mammalian circadian rhythm is a transcriptional-translational-post-translational autoregulatory feedback loop [12]. The core of the loop consists of CLOCK and BMAL1 proteins, that form a dimer which binds to the E-box region in promoters of period (PER1, PER2, PER3) and cryptochrome (CRY1, CRY2) genes. Following transcription and translation, PER and CRY proteins form a complex with casein kinase 1 epsilon (CSNK1E) and translocate into the nucleus. Here they bind to BMAL1/CLOCK complex and inhibit their own transcription, which completes the basic auto regulatory loop. PER and CRY proteins are then tagged for proteasomal degradation via phosphorylation by CSNK1E and casein kinase 1 delta (CSNK1D) and subsequently by ubiquitination. This cycle lasts approximately 24 h. The BMAL1/CLOCK heterodimer also up regulates the transcription of Rev-erbα and Rora. Their protein products interact with ROR elements (RORE) in the promoter of BMAL1 gene, upregulating (RORα) or downregulating (REV-ERBα) its transcription [12,13].
Circadian rhythm genes in the hypothalamic suprachiasmatic nucleus (SCN) and reproductive tissues control the timing and length of the ovulatory cycle and pregnancy by their influence on hormones [14]. Estradiol, synthesized in the ovary in response to the stimulation by gonadotropins from the hypothalamic-pituitary-gonadal (HPG) axis, influences the expression of circadian rhythm genes, and in a complex loop-back mechanism the circadian rhythm proteins interfere with estradiol signaling [15]. Overexpression of CLOCK transcription factors may play a role in the pathogenesis of endometriosis [16], which is a risk factor for some subtypes of ovarian cancer [17–19]. Infertility is observed in knockout BMAL1, PER1, and PER2 mice [20–22]. These data are consistent with human studies indicating that genetic variation in BMAL1 is associated with increased rates of miscarriage [23]. Nulliparity is a well-established risk factor for ovarian cancer, although it is currently unclear whether this association is due to infertility or other biological factors (e.g., increased ovulation) [24–27].
Variation in circadian genes has been associated with cancer susceptibility and outcomes. CLOCK1, CRY1, CRY2, NPAS2, PER1, RORA and TIMELESS variants are associated with breast cancer risk [5,28–33], while polymorphisms in BMAL1, CLOCK1, CRY1, CRY2, CSNK1E, NPAS2, PER1, PER2, and PER3 are associated with prostate cancer risk [34–36]. CRY2 and NPAS2 variation is associated with risk of non-Hodgkin’s lymphoma [37,38] while polymorphisms in CLOCK1 are associated with colorectal cancer susceptibility [39]. PER1 and CLOCK1 variation is associated with glioma risk and outcome [40] and PER3 polymorphisms have been associated with hepatocellular carcinoma survival [41]. Interestingly, variation in many of these genes is also associated with dysregulation of circadian behaviors, including sleep and activity patterns [42,43], although data are conflicting [44,45]. To date, however, there are no published studies on the association of variation in circadian genes with ovarian cancer risk and invasiveness.
The goal of the current study was to examine variants in seven key circadian rhythm genes (BMAL1, CRY2, CSNK1E, NPAS2, PER3, REV1, TIMELESS) and two transcription factors (KLF10 and SENP3) activated by circadian rhythm gene expression as risk factors for epithelial ovarian cancer, histopathologic subtype, and invasiveness. SNPs were evaluated in a two-stage design: a discovery stage using two genome-wide association studies (GWAS) and a replication stage with approximately 44,000 cases and controls from 43 studies that comprise the Ovarian Cancer Association Consortium (OCAC).
Materials and Methods
Sample and procedure
The discovery set included 3,761 EOC cases and 2,722 controls in two ovarian cancer GWAS in North America and the United Kingdom (UK). Details of these studies have been previously published [46]. In brief, the North American study was comprised of four case-control studies genotyped using the Illumina 610-quad Beadchip Array™ (i.e., 1,814 cases and 1,867 controls) as well as a single case-control study genotyped on the Illumina 317K and 370K arrays (i.e., 133 cases and 142 controls). The UK study was comprised of four case-only studies genotyped on the Illumina 610-quad Beadchip Array™ and two common control sets genotyped on the Illumina 550K array (i.e., 1,814 cases and 713 controls). The North American and UK studies were analyzed separately and the results combined using fixed effects meta-analysis.
The replication sample consisted of 14,525 invasive EOC cases and 23,447 controls from 43 sites in the Ovarian Cancer Association Consortium (OCAC). An additional 1,747 participants with tumors of low malignant potential were also analyzed. The sample consisted of only participants with European ancestry due to small numbers belonging to other racial groups.
Gene and SNP selection
Seven essential circadian genes (BMAL1, CRY2, CSNK1E, NPAS2, PER3, REV1, TIMELESS) and two key transcription factor genes activated by circadian genes (KLF10, SENP3) were selected a priori for examination. On the Illumina 610quad, 241 tagSNPs in these genes were identified. The selection of SNPs for replication was informed by ranking of minimal p-values across four sets of results: 1) North American all histologies, 2) North American serous histology, 3) combined GWAS meta-analysis all histologies, and 4) combined GWAS meta-analysis serous histology. Of the 241 SNPs, 37 SNPs were significant in the GWAS discovery set.
Statistical analysis
Demographic and clinical characteristics of cases and controls were compared using t-tests for continuous variables and chi-square tests for categorical variables. Unconditional logistic regression, treating the number of minor alleles carried as an ordinal variable (i.e., log-additive model), was used to evaluate the association between each SNP and ovarian cancer risk. Per-allele log odds ratios (OR) and their 95% confidence intervals (CI) were estimated. Models were adjusted for study site and population substructure by including study-site indicators and the first five eigenvalues from principal components analysis. The number of principal components was based on the position of the inflexion of the principal components scree plot.
To maximize statistical power, the combined COGS dataset was used to perform SNP-specific analyses for all invasive EOC, the four main histological subtypes (serous, endometrioid, clear cell and mucinous), and tumors of low malignant potential (LMP). Odds ratios specific for each histological subtype were estimated by comparing cases of each subtype to all available controls as reference. Associations with a two-sided p value < 0.05 and a false discovery rate (FDR) q-value [47] < 0.10 were considered to be statistically significant.
Imputation analyses
These analyses were based on imputed genotypes from the four ovarian cancer GWAS studies (US GWAS, UK GWAS, COGS and Mayo clinic) with a total of 15,398 invasive EOC case subjects and 30,816 control subjects of white-European ancestry. Imputation of each dataset into the 1000 Genomes Project was performed using IMPUTE2 software [48]. We used the 1000 Genomes Project v3 as the reference with pre-phasing of the data using SHAPEIT [49]. SNP log-additive model meta-analysis was carried out for combining results across studies. Only imputed SNPs with r2 > 0.25 for each study were used in the analyses.
Functional analyses
An in vitro model of early-stage ovarian cancer has been previously described [45]. Briefly, Illumina HT12 gene expression microarrays were used to profile the transcriptome of 3D models of normal ovarian cells immortalized with TERT and overexpressing cMYC and a mutant KRAS or BRAF allele.
Results
Sample descriptives
All invasive cancers combined and the four main histological subtypes serous (n = 8,369), endometrioid (n = 2,067), clear cell (n = 1,024) and mucinous (n = 943) were analyzed. Sample characteristics are described in table 1. As expected, significant differences were observed between cases and controls on ovarian cancer risk factors including age, family history of ovarian cancer, age at menarche, body mass index (BMI), history of oral contraceptive use, endometriosis, and number of full term births (p values < 0.05). The proportion of serous histological subtype (57.6%) was higher than the other subtypes (14.2% endometrioid, 7.1% clear cell, 6.5% for mucinous, and 14.6% other).
Table 1.
Sample demographic and clinical characteristics (n= 37,972).
| Characteristics | Controls (n = 23,447) N (%) |
Invasive Cases (n = 14,525) N (%) |
p-value2 |
|---|---|---|---|
| Age (years) | |||
| Mean ± SD | 55.6 ± 11.9 | 58.1 ± 11.3 | <. 0001 |
| < 40 | 2027 (8.7) | 748 (5.2) | <. 0001 |
| 40–49 | 4771 (20.6) | 2544 (17.6) | |
| 50–59 | 7403 (31.9) | 4537 (31.3) | |
| 60–69 | 6098 (26.3) | 4324 (29.8) | |
| ≥ 70 | 2892 (12.5) | 2343 (16.2) | |
| Family history of ovarian cancer1 | |||
| No | 15425 (92.0) | 8634 (82.4) | <. 0001 |
| Yes | 1351 (8.0) | 1849 (17.6) | |
| Age at menarche (years) | |||
| Mean ± SD | 12.9 ± 1.7 | 12.8 ± 1.6 | 0.0314 |
| < 12 | 3128 (19.3) | 1856 (19.2) | 0.0772 |
| 12 | 3602 (22.2) | 2257 (23.4) | |
| 13 | 4357 (26.9) | 2621 (27.1) | |
| ≥ 14 | 5112 (31.6) | 2923 (30.3) | |
| Body mass inde × (kg/m2) | |||
| < 25 | 3834 (48.2) | 2528 (45.1) | 0.0006 |
| 25–29 | 2332 (29.3) | 1681 (30.0) | |
| ≥ 30 | 1797 (22.6) | 1396 (24.9) | |
| Oral contraceptive use | |||
| No | 6136 (37.5) | 4203 (43.7) | <. 0001 |
| Yes | 10230 (62.5) | 5419 (56.3) | |
| Histological subtypes | N/A | ||
| Serous | 8369 (57.6) | ||
| Endometroid | 2067 (14.2) | ||
| Clear Cell | 1024 (7.1) | ||
| Mucinous | 943 (6.5) | ||
| Others3 | 2122 (14.6) |
for the first degree relatives
t-test for a continuous variable and chi-square test for a categorical variable
Include mi × ed cell, other specified epithelial, undifferentiated, unknown (but known to be epithelial), nonepithelial, other or unknown if epithelial, or missing
Genotyped variants
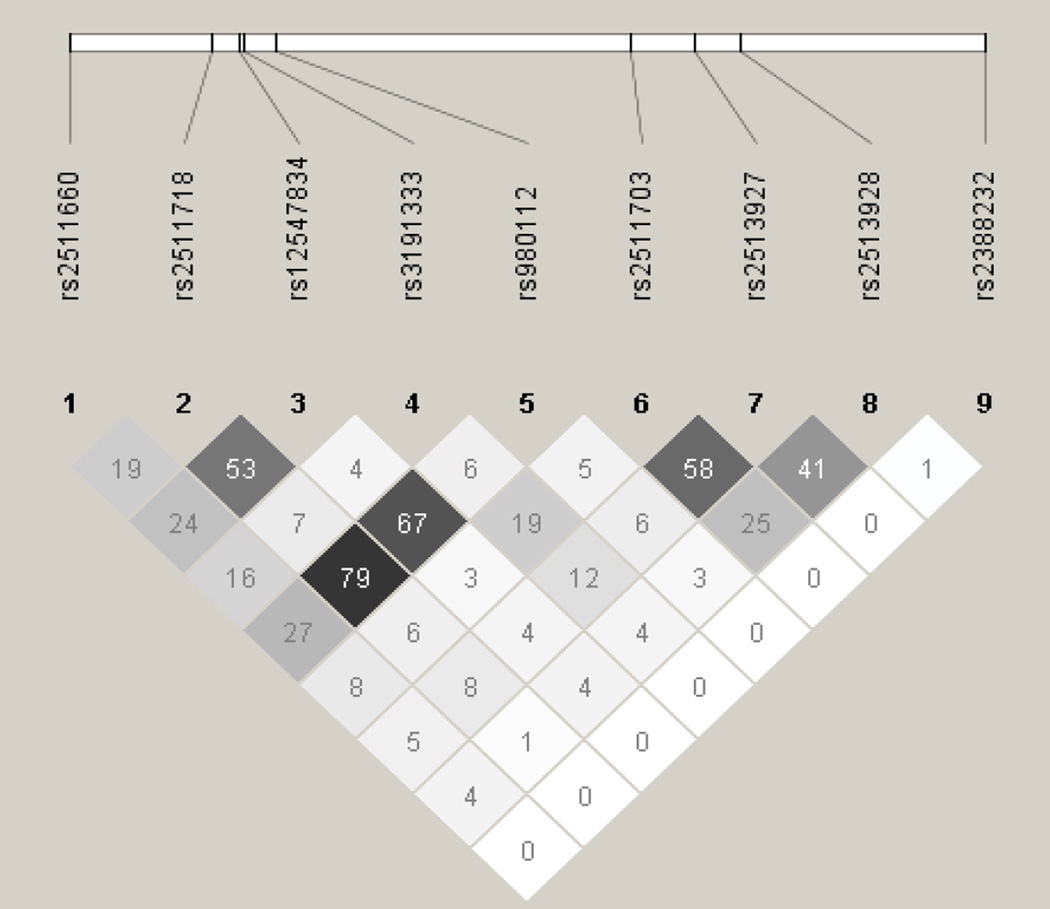
A total of 36 SNPs demonstrated p values < 0.05 in the screening stage and passed quality control. Of these, two in SENP3 (i.e., rs11656383, rs3499590) were rare variants (i.e., MAFs < 0.01) and were dropped from further analyses. Of the remaining 34 SNPs, 14 were associated with risk of overall EOC, histopathological subtype, and/or invasiveness (Table 2). Seven remained significant after applying the criterion of FDR < 0 .10. Specifically, one SNP was associated with risk of all invasive EOC, rs2513928 in KLF10 (OR = 0.95, 95% CI = 0.92–0.98, p = 1.75 × 10−3). Four SNPs in KLF10 were associated with risk of serous EOC (rs2513928: OR = 0.94, 95% CI = 0.91–0.98, p = 2.42 × 10−3; rs2511703: OR = 1.05, 95% CI = 1.02–1.09, p = 6.54 × 10−3; rs3191333: OR = 1.05, 95% CI = 1.02–1.10, p = 6.72 × 10−3; rs2513927: OR = 1.05, 95% CI = 1.01–1.09, p = 1.18 × 10−2). As shown in figure 1, linkage disequilibrium (LD) between the four significant SNPs in KLF10 was low to moderate. Risk of endometrioid EOC was associated with SENP3 rs6608 (OR = 1.13, 95% CI = 1.04–1.23, p = 4.43 ×10−3), CSNK1E rs135750 (OR = 1.13, 95% CI = 1.03–1.23, p = 7.09 × 10−3), REV1 rs3792152 (OR = 0.92, 95% CI = 0.86–0.98, p = 9.61 × 10−3), and BMAL1 rs10732458 (OR = 1.32, 95% CI = 1.07–1.63, p = 9.64 × 10−3). No SNPs were significantly associated with EOC invasiveness nor were any SNPs significantly associated with risk of mucinous or clear cell EOC after applying the criterion of FDR < 0.10.
Table 2.
Associations between Genotyped SNPs in Circadian Genes and EOC Incidence Overall, in Histological Subtypes, and Invasiveness.
| All invasive | Serous | Clear cell | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Chr | Min/Maj | MAF | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p |
| BMAL1 | rs1026071 | 11 | G/A | 0.30 | 0.98 (0.95–1.01) | 2.26 × 10–01 | 1.00 (0.96–1.04) | 9.38 × 10–01 | 0.88 (0.8–0.98) | 1.55 × 10–02 |
| BMAL1 | rs10732458 | 11 | A/G | 0.02 | 1.11 (0.99–1.23) | 6.91 × 10–02 | 1.10 (0.96–1.25) | 1.64 × 10–01 | 1.19 (0.88–1.6) | 2.52 × 10–01 |
| BMAL1 | rs10832027 | 11 | G/A | 0.33 | 0.98 (0.95–1.02) | 3.48 × 10–01 | 1.00 (0.96–1.04) | 9.79 × 10–01 | 0.92 (0.84–1.01) | 9.15 × 10–02 |
| BMAL1 | rs1562438 | 11 | A/G | 0.29 | 0.98 (0.95–1.02) | 3.07 × 10–01 | 1.00 (0.96–1.05) | 8.46 × 10–01 | 0.88 (0.80–0.97) | 1.35 × 10–02 |
| BMAL1 | rs16912751 | 11 | G/A | 0.05 | 0.98 (0.92–1.05) | 6.23 × 10–01 | 0.96 (0.88–1.04) | 3.42 × 10–01 | 1.13 (0.93–1.37) | 2.18 × 10–01 |
| BMAL1 | rs2896635 | 11 | T/A | 0.33 | 0.98 (0.95–1.02) | 3.14 × 10–01 | 1.00 (0.96–1.04) | 9.57 × 10–01 | 0.93 (0.84–1.02) | 1.17 × 10–01 |
| BMAL1 | rs3789327 | 11 | G/A | 0.48 | 1.01 (0.98–1.04) | 5.34 × 10–01 | 1.01 (0.97–1.04) | 7.88 × 10–01 | 1.04 (0.95–1.14) | 4.17 × 10–01 |
| BMAL1 | rs3816360 | 11 | A/G | 0.34 | 1.00 (0.96–1.03) | 7.75 × 10–01 | 1.02 (0.98–1.06) | 4.36 × 10–01 | 0.91 (0.82–1.00) | 4.31 × 10–02 |
| BMAL1 | rs4757151 | 11 | A/G | 0.47 | 1.00 (0.97–1.04) | 7.76 × 10–01 | 1.01 (0.98–1.05) | 5.46 × 10–01 | 0.97 (0.89–1.06) | 5.20 × 10–01 |
| BMAL1 | rs6486122 | 11 | G/A | 0.32 | 0.98 (0.95–1.02) | 2.83 × 10–01 | 1.00 (0.96–1.04) | 9.53 × 10–01 | 0.92 (0.83–1.01) | 8.10 × 10–02 |
| BMAL1 | rs7117836 | 11 | A/G | 0.02 | 1.10 (0.99–1.22) | 8.49 × 10–02 | 1.09 (0.96–1.24) | 1.65 × 10–01 | 1.19 (0.89–1.59) | 2.46 × 10–01 |
| BMAL1 | rs7947951 | 11 | A/G | 0.32 | 0.99 (0.95–1.02) | 3.60 × 10–01 | 1.00 (0.96–1.04) | 9.13 × 10–01 | 0.92 (0.84–1.01) | 9.30 × 10–02 |
| CRY2 | rs11038695 | 11 | A/G | 0.08 | 1.05 (0.99–1.11) | 1.11 × 10–01 | 1.03 (0.97–1.11) | 3.40 × 10–01 | 0.99 (0.84–1.17) | 9.25 × 10–01 |
| CSNK1E | rs135750 | 22 | G/C | 0.15 | 1.04 (1.00–1.09) | 6.14 × 10–02 | 1.03 (0.98–1.08) | 3.12 × 10–01 | 1.00 (0.89–1.13) | 9.73 × 10–01 |
| KLF10 | rs12547834 | 8 | G/A | 0.07 | 0.96 (0.90–1.02) | 1.43 × 10–01 | 0.94 (0.88–1.02) | 1.20 × 10–01 | 1.02 (0.85–1.21) | 8.49 × 10–01 |
| KLF10 | rs3191333 | 8 | A/G | 0.37 | 1.04 (1.01–1.07) | 2.42 × 10–02 | 1.05 (1.02–1.10) | 6.72 × 10–03 | 1.04 (0.95–1.14) | 3.95 × 10–01 |
| KLF10 | rs980112 | 8 | A/G | 0.10 | 0.97 (0.92–1.02) | 1.98 × 10–01 | 0.96 (0.90–1.03) | 2.42 × 10–01 | 1.06 (0.92–1.23) | 4.08 × 10–01 |
| KLF10 | rs2388232 | 8 | G/A | 0.27 | 1.01 (0.97–1.04) | 7.92 × 10–01 | 1.00 (0.96–1.04) | 9.22 × 10–01 | 1.11 (1.01–1.23) | 2.91 × 10–02 |
| KLF10 | rs2511703 | 8 | G/A | 0.43 | 1.04 (1.01–1.07) | 1.83 × 10–02 | 1.05 (1.02–1.09) | 6.54 × 10–03 | 1.00 (0.91–1.09) | 9.55 × 10–01 |
| KLF10 | rs2513927 | 8 | A/G | 0.49 | 1.04 (1.01–1.07) | 1.86 × 10–02 | 1.05 (1.01–1.09) | 1.18 × 10–02 | 1.00 (0.91–1.10) | 9.79 × 10–01 |
| KLF10 | rs2513928 | 8 | G/A | 0.46 | 0.95 (0.92–0.98) | 1.75 × 10–03 | 0.94 (0.91–0.98) | 2.42 × 10–03 | 0.94 (0.85–1.02) | 1.50 × 10–01 |
| KLF10 | rs2511660 | 8 | A/G | 0.22 | 0.97 (0.94–1.01) | 1.57 × 10–01 | 0.96 (0.92–1.00) | 6.95 × 10–02 | 0.99 (0.89–1.10) | 8.56 × 10–01 |
| KLF10 | rs2511718 | 8 | A/G | 0.12 | 0.98 (0.94–1.03) | 4.57 × 10–01 | 0.98 (0.92–1.04) | 4.47 × 10–01 | 1.06 (0.93–1.22) | 3.68 × 10–01 |
| NPAS2 | rs1053091 | 2 | A/G | 0.02 | 1.05 (0.93–1.19) | 4.14 × 10–01 | 1.10 (0.96–1.27) | 1.83 × 10–01 | 1.12 (0.79–1.59) | 5.17 × 10–01 |
| NPAS2 | rs13012930 | 2 | A/G | 0.17 | 0.96 (0.92–1.00) | 4.80 × 10–02 | 0.95 (0.91–1.00) | 4.11 × 10–02 | 0.98 (0.87–1.10) | 6.86 × 10–01 |
| NPAS2 | rs3768988 | 2 | G/A | 0.06 | 1.01 (0.95–1.07) | 8.18 × 10–01 | 1.02 (0.94–1.10) | 6.44 × 10–01 | 1.01 (0.84–1.22) | 9.09 × 10–01 |
| NPAS2 | rs7573323 | 2 | A/G | 0.03 | 0.97 (0.88–1.07) | 5.47 × 10–01 | 0.99 (0.88–1.11) | 8.61 × 10–01 | 0.87 (0.65–1.18) | 3.73 × 10–01 |
| PER3 | rs228644 | 1 | A/G | 0.40 | 1.00 (0.97–1.03) | 9.23 × 10–01 | 1.00 (0.96–1.03) | 8.38 × 10–01 | 0.97 (0.89–1.07) | 5.45 × 10–01 |
| PER3 | rs228682 | 1 | G/A | 0.40 | 1.00 (0.97–1.03) | 7.83 × 10–01 | 0.99 (0.96–1.03) | 7.32 × 10–01 | 0.97 (0.88–1.06) | 4.84 × 10–01 |
| PER3 | rs228698 | 1 | A/G | 0.04 | 1.00 (0.93–1.08) | 9.73 × 10–01 | 0.99 (0.90–1.08) | 7.67 × 10–01 | 0.90 (0.71–1.14) | 3.79 × 10–01 |
| PER3 | rs697693 | 1 | A/G | 0.19 | 0.99 (0.95–1.03) | 5.55 × 10–01 | 0.98 (0.94–1.03) | 5.02 × 10–01 | 1.07 (0.96–1.19) | 2.46 × 10–01 |
| REV1 | rs3792152 | 2 | A/G | 0.44 | 0.97 (0.94–1.00) | 6.47 × 10–02 | 0.97 (0.94–1.01) | 1.34 × 10–01 | 0.99 (0.90–1.08) | 7.96 × 10–01 |
| SENP3 | rs6608 | 17 | A/G | 0.17 | 1.05 (1.00–1.09) | 3.35 × 10–02 | 1.04 (0.99–1.09) | 1.42 × 10–01 | 1.01 (0.90–1.14) | 8.81 × 10–01 |
| TIMELES | S rs7302060 | 12 | G/A | 0.41 | 0.99 (0.96–1.02) | 3.53 × 10–01 | 0.98 (0.94–1.01) | 2.09 × 10–01 | 0.97 (0.88–1.06) | 4.77 × 10–01 |
| Endometriod | Mucinous | LMP vs. controls | Invasive vs. LMP | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p |
| BMAL1 | rs1026071 | 0.98 (0.91–1.05) | 5.17 × 10–01 | 0.94 (0.85–1.05) | 2.63 × 10–01 | 0.99 (0.92–1.07) | 8.95 × 10–01 | 1.00 (0.92–1.08) | 9.17 × 10–01 |
| BMAL1 | rs10732458 | 1.32 (1.07–1.63) | 9.64 × 10–03 | 1.02 (0.72–1.44) | 9.12 × 10–01 | 0.77 (0.58–1.02) | 6.51 × 10–02 | 1.44 (1.09–1.92) | 1.17 × 10–02 |
| BMAL1 | rs10832027 | 0.99 (0.93–1.06) | 8.48 × 10–01 | 0.95 (0.86–1.05) | 2.75 × 10–01 | 1.00 (0.93–1.07) | 9.17 × 10–01 | 1.00 (0.92–1.07) | 9.04 × 10–01 |
| BMAL1 | rs1562438 | 0.97 (0.90–1.04) | 4.12 × 10–01 | 0.94 (0.85–1.05) | 2.74 × 10–01 | 1.00 (0.93–1.08) | 9.79 × 10–01 | 0.99 (0.92–1.07) | 8.80 × 10–01 |
| BMAL1 | rs16912751 | 0.90 (0.78–1.05) | 1.97 × 10–01 | 1.11 (0.91–1.36) | 2.94 × 10–01 | 0.88 (0.75–1.04) | 1.40 × 10–01 | 1.12 (0.95–1.33) | 1.73 × 10–01 |
| BMAL1 | rs2896635 | 0.99 (0.92–1.06) | 7.20 × 10–01 | 0.95 (0.86–1.05) | 3.04 × 10–01 | 1.00 (0.93–1.07) | 9.49 × 10–01 | 0.99 (0.92–1.07) | 8.21 × 10–01 |
| BMAL1 | rs3789327 | 1.00 (0.94–1.07) | 9.84 × 10–01 | 0.95 (0.86–1.04) | 2.53 × 10–01 | 1.01 (0.94–1.08) | 8.63 × 10–01 | 1.01 (0.94–1.08) | 8.01 × 10–01 |
| BMAL1 | rs3816360 | 0.99 (0.92–1.06) | 7.74 × 10–01 | 0.94 (0.85–1.04) | 2.52 × 10–01 | 1.02 (0.94–1.09) | 6.67 × 10–01 | 0.99 (0.92–1.06) | 7.53 × 10–01 |
| BMAL1 | rs4757151 | 0.99 (0.92–1.05) | 6.91 × 10–01 | 1.06 (0.97–1.17) | 1.97 × 10–01 | 0.98 (0.91–1.05) | 5.61 × 10–01 | 1.03 (0.96–1.11) | 3.73 × 10–01 |
| BMAL1 | rs6486122 | 0.99 (0.92–1.06) | 6.90 × 10–01 | 0.95 (0.86–1.05) | 3.12 × 10–01 | 0.99 (0.92–1.07) | 8.62 × 10–01 | 0.99 (0.92–1.07) | 8.83 × 10–01 |
| BMAL1 | rs7117836 | 1.24 (1.01–1.54) | 4.40 × 10–02 | 1.06 (0.76–1.48) | 7.36 × 10–01 | 0.76 (0.57–1.00) | 4.82 × 10–02 | 1.45 (1.09–1.92) | 9.81 × 10–03 |
| BMAL1 | rs7947951 | 0.99 (0.93–1.06) | 8.17 × 10–01 | 0.95 (0.86–1.05) | 2.94 × 10–01 | 1.00 (0.93–1.07) | 9.34 × 10–01 | 0.99 (0.92–1.07) | 8.78 × 10–01 |
| CRY2 | rs11038695 | 1.09 (0.97–1.22) | 1.48 × 10–01 | 0.97 (0.82–1.15) | 7.19 × 10–01 | 1.07 (0.94–1.21) | 2.88 × 10–01 | 0.98 (0.86–1.11) | 7.02 × 10–01 |
| CSNK1E | rs135750 | 1.13 (1.03–1.23) | 7.09 × 10–03 | 1.06 (0.93–1.20) | 3.90 × 10–01 | 1.02 (0.93–1.12) | 6.98 × 10–01 | 1.03 (0.93–1.13) | 6.10 × 10–01 |
| KLF10 | rs12547834 | 0.99 (0.87–1.13) | 8.75 × 10–01 | 0.86 (0.71–1.04) | 1.22 × 10–01 | 0.97 (0.84–1.11) | 6.56 × 10–01 | 0.99 (0.86–1.14) | 8.87 × 10–01 |
| KLF10 | rs3191333 | 1.03 (0.96–1.10) | 3.84 × 10–01 | 0.95 (0.86–1.05) | 3.01 × 10–01 | 0.99 (0.92–1.06) | 7.70 × 10–01 | 1.05 (0.97–1.13) | 2.21 × 10–01 |
| KLF10 | rs980112 | 0.95 (0.85–1.06) | 3.49 × 10–01 | 0.90 (0.76–1.05) | 1.85 × 10–01 | 0.99 (0.88–1.12) | 8.95 × 10–01 | 0.98 (0.87–1.11) | 7.73 × 10–01 |
| KLF10 | rs2388232 | 0.99 (0.92–1.06) | 7.81 × 10–01 | 0.98 (0.88–1.08) | 6.40 × 10–01 | 1.03 (0.96–1.12) | 3.86 × 10–01 | 0.97 (0.90–1.05) | 4.71 × 10–01 |
| KLF10 | rs2511703 | 1.05 (0.98–1.12) | 1.34 × 10–01 | 0.95 (0.87–1.05) | 3.17 × 10–01 | 0.98 (0.91–1.05) | 5.75 × 10–01 | 1.06 (0.98–1.13) | 1.32 × 10–01 |
| KLF10 | rs2513927 | 1.05 (0.99–1.13) | 1.11 × 10–01 | 0.94 (0.85–1.03) | 1.71 × 10–01 | 0.98 (0.92–1.05) | 5.94 × 10–01 | 1.06 (0.99–1.13) | 1.24 × 10–01 |
| KLF10 | rs2513928 | 0.95 (0.89–1.01) | 1.20 × 10–01 | 1.02 (0.93–1.12) | 6.88 × 10–01 | 0.96 (0.90–1.03) | 2.56 × 10–01 | 0.99 (0.92–1.06) | 6.95 × 10–01 |
| KLF10 | rs2511660 | 1.02 (0.94–1.10) | 6.80 × 10–01 | 0.96 (0.85–1.07) | 4.38 × 10–01 | 1.06 (0.98–1.15) | 1.43 × 10–01 | 0.92 (0.85–1.00) | 4.47 × 10–02 |
| KLF10 | rs2511718 | 0.96 (0.87–1.06) | 4.28 × 10–01 | 0.95 (0.82–1.09) | 4.52 × 10–01 | 1.01 (0.91–1.13) | 8.02 × 10–01 | 0.97 (0.87–1.09) | 6.41 × 10–01 |
| NPAS2 | rs1053091 | 0.84 (0.64–1.12) | 2.45 × 10–01 | 1.02 (0.71–1.47) | 9.00 × 10–01 | 1.10 (0.85–1.44) | 4.69 × 10–01 | 0.93 (0.71–1.22) | 5.88 × 10–01 |
| NPAS2 | rs13012930 | 1.02 (0.93–1.11) | 7.23 × 10–01 | 0.91 (0.80–1.03) | 1.31 × 10–01 | 1.04 (0.95–1.14) | 3.73 × 10–01 | 0.92 (0.84–1.01) | 9.47 × 10–02 |
| NPAS2 | rs3768988 | 0.93 (0.81–1.07) | 3.02 × 10–01 | 1.01 (0.84–1.22) | 8.90 × 10–01 | 1.09 (0.95–1.26) | 2.06 × 10–01 | 0.93 (0.80–1.07) | 2.83 × 10–01 |
| NPAS2 | rs7573323 | 0.92 (0.74–1.13) | 4.13 × 10–01 | 0.83 (0.60–1.16) | 2.78 × 10–01 | 0.83 (0.66–1.05) | 1.12 × 10–01 | 1.18 (0.93–1.49) | 1.76 × 10–01 |
| PER3 | rs228644 | 0.97 (0.91–1.04) | 3.76 × 10–01 | 1.07 (0.97–1.17) | 1.82 × 10–01 | 0.99 (0.92–1.06) | 6.91 × 10–01 | 1.02 (0.95–1.09) | 6.69 × 10–01 |
| PER3 | rs228682 | 0.97 (0.91–1.04) | 3.51 × 10–01 | 1.07 (0.97–1.17) | 1.90 × 10–01 | 0.98 (0.92–1.06) | 6.37 × 10–01 | 1.02 (0.95–1.09) | 6.65 × 10–01 |
| PER3 | rs228698 | 1.04 (0.89–1.23) | 6.04 × 10–01 | 1.08 (0.86–1.36) | 4.89 × 10–01 | 0.96 (0.81–1.15) | 6.66 × 10–01 | 1.01 (0.85–1.21) | 8.76 × 10–01 |
| PER3 | rs697693 | 0.99 (0.91–1.08) | 8.61 × 10–01 | 0.92 (0.81–1.04) | 1.67 × 10–01 | 1.04 (0.95–1.13) | 3.81 × 10–01 | 0.96 (0.88–1.05) | 3.45 × 10–01 |
| REV1 | rs3792152 | 0.92 (0.86–0.98) | 9.61 × 10–03 | 0.99 (0.90–1.09) | 8.32 × 10–01 | 0.98 (0.91–1.05) | 4.87 × 10–01 | 1.01 (0.94–1.09) | 7.65 × 10–01 |
| SENP3 | rs6608 | 1.13 (1.04–1.23) | 4.43 × 10–03 | 1.00 (0.88–1.14) | 9.90 × 10–01 | 1.01 (0.92–1.10) | 9.00 × 10–01 | 1.04 (0.94–1.14) | 4.79 × 10–01 |
| TIMELESS | rs7302060 | 1.01 (0.95–1.08) | 7.22 × 10–01 | 0.97 (0.88–1.07) | 5.10 × 10–01 | 0.93 (0.87–1.00) | 4.86 × 10–02 | 1.06 (0.99–1.14) | 1.09 × 10–01 |
SNP: Single Nucleotide Polymorphism, Chr: Chromosome, Min/Maj: Minor and Major Allele, MAF: Minor Allele Frequency, LMP: Low Malignant Potential, OR: Odds Ratio
Note: odds ratio is calculated based on per-minor allele, bolded SNPs indicate an association of p < 0.05 with overall EOC or histologic subtype.
Figure 1.
Linkage Disequilibrium (r2) among Single Nucleotide Polymorphisms in KLF10.
Imputed variants
A total of 4600 imputed SNPs in the nine genes of interest (BMAL1, CRY2, CSNK1E, NPAS2, PER3, REV1, TIMELESS, KLF10, SENP3) were then examined for association with all invasive EOC. A total of 304 SNPs across all nine genes met criteria for statistical significance (p < 0.05). Top hits in each gene with good imputation quality [r2 > 0.8] are shown in table 3. Across all genes, the most significant imputed SNP was rs117104877 in BMAL1 (OR = 0.79, 95% CI = 0.68–0.90, p = 5.59 × 10−4).
Table 3.
Associations between the Top Imputed SNP in Each Gene with Good Imputation Quality (r2 > 0.8) and EOC Incidence Overall.
| Gene | SNP | Min/Maj | MAF | OR (95% CI) | p |
|---|---|---|---|---|---|
| BMAL1 | rs117104877 | G/A | 0.017 | 0.79 (0.68–0.90) | 5.59 × 10–4 |
| CRY2 | rs10838527 | G/A | 0.082 | 1.05 (0.99–1.11) | 7.66 × 10–2 |
| CSNK1E | rs111427515 | G/T | 0.008 | 1.25 (1.06–1.47) | 6.60 × 10–3 |
| KLF10 | rs2511699 | A/G | 0.461 | 0.96 (0.93–0.99) | 4.13 × 10–3 |
| NPAS2 | rs732375 | T/A | 0.134 | 1.07 (1.02–1.11) | 3.76 × 10–3 |
| PER3 | rs228640 | A/G | 0.297 | 1.04 (1.01–1.07) | 1.24 × 10–2 |
| REV1 | rs3792146 | T/C | 0.547 | 1.03 (1–1.06) | 2.71 × 10–2 |
| SENP3 | rs143094271 | A/G | 0.023 | 0.86 (0.77–0.95) | 4.01 × 10–3 |
| TIMELESS | rs2638286 | C/T | 0.030 | 1.05 (0.96–1.15) | 2.56 × 10–1 |
SNP: Single Nucleotide Polymorphism, Min/Maj: Minor and Major Allele, MAF: Minor Allele Frequency, OR: Odds Ratio
Note: odds ratio is calculated based on per-minor allele
Evaluating the functional role of BMAL1 in ovarian cancer
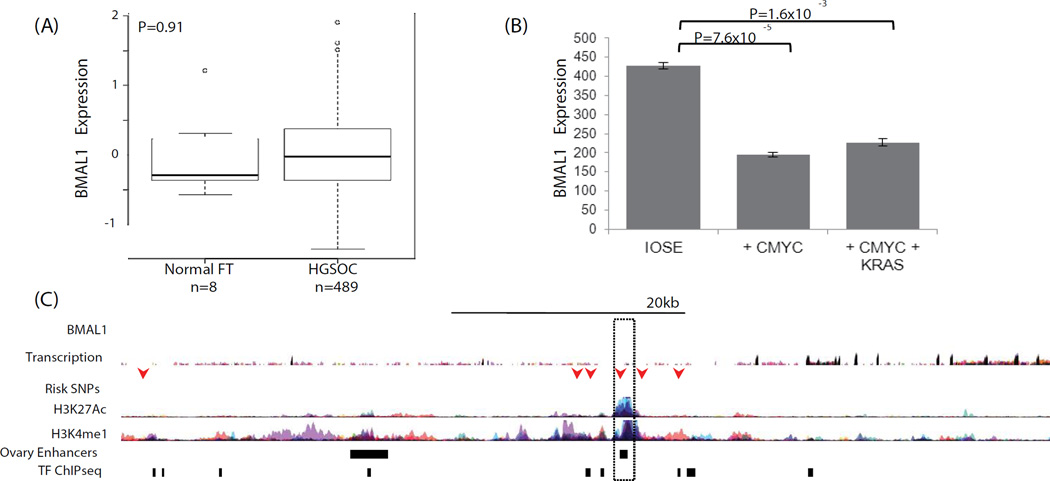
The role of BMAL1 in ovarian cancer was examined using in silico analysis of existing biological datasets in ovarian normal and tumor tissues and an in vitro cell biology model of early stage ovarian cancer development. We evaluated gene expression in normal fallopian tubes (n = 8) compared to high-grade serous ovarian carcinomas (HGSOCs, n = 489) using data from The Cancer Genome Atlas (TCGA), but there was no evidence that BMAL1 was differentially regulated in EOCs as compared to normal tissue (Figure 2).
Figure 2.
(A) BMAL1 is not differentially expressed in TCGA expression data for 8 normal fallopian tubes and 489 high-grade serous EOCs; however, in an early stage model of ovarian cancer, (B) BMAL1 is downregulated in partially transformed ovarian epithelial cells overexpressing cMYC. BMAL1 downregulation is cMYC dependent, and not enhanced by the expression of a mutant KRAS allele. (C) 6 SNPs at the BMAL1 locus coincide with marks of active regulatory elements (H3K27Ac and H3K4me1) or transcription factor binding sites (TF ChiPseq) (arrows). One SNP, rs2896635 coincides with a commonly used enhancer that is active in ovarian stromal tissue (dashed box), and which targets the BMAL1 gene. ENCODE data and data from [44].
BMAL1 expression was further investigated in an early stage transformation model of EOC based on overexpression of CMYC in the ovarian surface epithelium (OSE) [50]. BMAL1 was significantly down regulated in this model, but down regulation was not enhanced by expression of a mutant KRAS allele (Figure 2b). Risk associated SNPs were located within intronic regions of BMAL1 (Figure 2c) and clustered around a commonly described enhancer, suggesting that risk SNPs may influence enhancer activity. Rs2896635 in particular coincides with an enhancer used in many cell types, including an enhancer that is active in ovarian stromal cells that targets the BMAL1 gene [51]. This suggests that non-cell autonomous signaling pathways may be involved in risk at this locus.
Discussion
Circadian genes appear to play an important role in regulating reproductive cycles, including ovulation, the length of the estrous cycle, and maintenance of pregnancy. The current study examined variation in nine key genes involved in circadian rhythm regulation or their transcription (BMAL1, CRY2, CSNK1E, KLF10, NPAS2, PER3, REV1, SENP3, TIMELESS) as predictors of epithelial ovarian cancer risk, histopathologic subtype, and invasiveness. We found that 14 of the 34 genotyped SNPs in the discovery set were associated with risk of overall EOC, histopathological subtype, and/or invasiveness at p < 0.05. Seven remained significant after applying the criterion of FDR < 0.10. Specifically, risk of overall and serous EOC was associated with variants in KLF10 while risk of endometrioid EOC was associated with variants in SENP3, CSNK1E, REV1, and BMAL1. Of 4600 imputed variants in the nine genes of interest, 304 were found to be associated with overall EOC risk at p <. 05. Significant variants were found in all nine genes with the most significant located in BMAL1. Additional functional analyses of BMAL1 indicated that it was down regulated as a consequence of overexpressing cMYC in the OSE, although differential regulation was not observed in HGSOCs compared to normal fallopian tube tissue. Taken together, these results suggest that circadian rhythm genes may play a role in the development of EOC, particularly the genes KLF10 and BMAL1.
While previous research has implicated circadian genes in the development of several types of human cancer, the current study is the first to our knowledge to examine relationships with risk of ovarian cancer. Findings regarding the Krüppel-like factor 10 (KLF10) gene are consistent with a sizable body of experimental data indicating that KLF10 acts to inhibit cellular proliferation and induce apoptosis in a variety of cell types via regulation of transforming growth factor beta (TGFβ) and in turn SMAD [52–58]. KLF10 is a circadian transcriptional regulator that links the molecular clock to energy metabolism [59]. KLF10 displays robust BMAL1-dependent circadian expression; the KLF10 promoter recruits BMAL1 and is transactivated by the CLOCK/BMAL1 dimer through a conserved E-box response element. To our knowledge the role of KLF10 in human ovarian cancer has not been investigated, although estrogen is known to increase KLF10 gene transcription [60,61]. KLF10 expression is reduced in breast tumors relative to normal tissue and is inversely correlated with stage of disease [62,63]. The KLF10-TGFβ-SMAD pathway has been implicated in the development of several other human cancers including those of the prostate, pancreas, kidney, lymphoma, and brain [53,64–67].
Our findings regarding BMAL1 are interesting in light of data suggesting that this gene may regulate the p53 tumor suppressor pathway. Specifically, silencing of BMAL1 gene expression prevents cell cycle arrest upon p53 activation in human fibroblast cells [68] and mouse colon and fibroblast cells [69]. These data are consistent with research suggesting that BMAL1 is transcriptionally silenced via hypermethylation in hematologic malignancies; reintroduction of BMAL1 causes growth inhibition, while BMAL1 depletion by RNA interference increases tumor growth [70]. The BMAL1 protein also has been shown to bind to the promoter region of VEGF where it regulates transcription and promotes angiogenesis [71].
Evidence suggests that, controlling for stage, histological subtype, and grade, low BMAL1 and CRY1 expression together significantly predict lower overall survival in ovarian cancer patients [72]. Previous research also suggests significantly lower BMAL1 and CRY1 expression in EOC cells compared to normal ovarian tissue [72]. The current study demonstrated downregulation of BMAL1 when cMYC was overexpressed in an early stage ovarian cancer transformation model, resulting in increasing ovarian epithelial cell transformation. Nevertheless, we did not observe differential regulation of BMAL1 when comparing EOC cells to normal fallopian tube tissue. Our findings suggest that down regulation of BMAL1 may be an early event in ovarian carcinogenesis and that BMAL1 is a novel cMYC target. SNPs statistically significant in the current study lie within intronic sequences of the BMAL1 gene and mechanisms by which they impact BMAL1 expression have yet to be elucidated. Nevertheless, our data suggest that this risk locus may modulate ovarian cancer risk by altering the ovarian stromal microenvironment, for example by influencing the character of ovarian fibroblasts or granulosa cells, both of which express BMAL1. In conclusion, our results highlight the significance of circadian rhythm gene variation in EOC susceptibility and suggest an early role for the BMAL1 gene in EOC pathogenesis.
Acknowledgments
Individual acknowledgements by study
We thank all the individuals who took part in this study and all the researchers, clinicians and technical and administrative staff who have made possible the many studies contributing to this work. In particular, we thank: D. Bowtell, A. deFazio, D. Gertig, A. Green, P. Parsons, N. Hayward, P. Webb and D. Whiteman (AUS); G. Peuteman, T. Van Brussel and D. Smeets (BEL); the staff of the genotyping unit, S LaBoissiere and F Robidoux (Genome Quebec); U. Eilber and T. Koehler (GER); L. Gacucova (HMO); P. Schurmann, F. Kramer, W. Zheng, T. W. Park, Simon, K. Beer- Grondke and D. Schmidt (HJO); S. Windebank, C. Hilker and J. Vollenweider (MAY); the state cancer registries of AL, AZ, AR, CA, CO, CT, DE, FL, GA, HI, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WYL (NHS); L. Paddock, M. King, L. Rodriguez-Rodriguez, A. Samoila, and Y. Bensman (NJO); M. Sherman, A. Hutchinson,N. Szeszenia--- Dabrowska, B. Peplonska, W. Zatonski, A. Soni, P. Chao and M. Stagner (POL); C. Luccarini,P. Harrington the SEARCH team and ECRIC (SEA); I. Jacobs, M. Widschwendter, E. Wozniak, N. Balogun, A. Ryan and J. Ford (UKO); Carole Pye (UKR); A. Amin Al Olama, K. Michilaidou, K. Kuchenbaker (COGS).
Main funding
The scientific development and funding for this project were funded by the following: NIH R01 CA-1491429 (Phelan PI); the US National Cancer Institute (R01-CA076016); the COGS project is funded through a European Commission’s Seventh Framework Program grant (agreement number 223175 HEALTH F2 2009–223175); the Genetic Associations and Mechanisms in Oncology (GAME-ON): a NCI Cancer Post-GWAS Initiative (U19-CA148112); the Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith (PPD/RPCI.07).
Investigator-specific funding
K.L is supported by a K99/R00 grant from the National Cancer Institute (Grant number 1K99CA184415-01). G.C.-T. is supported by the National Health and Medical Research Council; B.K. is supported by the American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124.; L.E.K. is supported by a Canadian Institute of Health Research New Investigator Award (MSH-87734). AWL is supported by NIEHS T32 training grant (T32ES013678).
Funding of included studies
Funding of the constituent studies was provided by the California Cancer Research Program (00-01389V-20170, N01-CN25403, 2II0200); the Canadian Institutes of Health Research (MOP-86727); Cancer Australia; Cancer Council Victoria; Cancer Council Queensland; Cancer Council New South Wales; Cancer Council South Australia; Cancer Council Tasmania; Cancer Foundation of Western Australia; the Cancer Institute of New Jersey; Cancer Research UK (C490/A6187, C490/A10119, C490/A10124); the Danish Cancer Society (94-222-52); the ELAN Program of the University of Erlangen-Nuremberg; the Eve Appeal; the Helsinki University Central Hospital Research Fund; Helse Vest; the Norwegian Cancer Society; the Norwegian Research Council; the Ovarian Cancer Research Fund; Nationaal Kankerplan of Belgium; Grant-in-Aid for the Third Term Comprehensive 10-Year Strategy For Cancer Control from the Ministry of Health Labour and Welfare of Japan; the L & S Milken Foundation; the Polish Ministry of Science and Higher Education (4 PO5C 028 14, 2 PO5A 068 27); the Roswell Park Cancer Institute Alliance Foundation; the US National Cancer Institute (K07-CA095666, K07-CA143047,K22-CA138563, N01-CN55424, N01-PC67001, N01-PC067010, N01-PC035137, P01-CA017054, P01-CA087696, P30-CA072720, P50-CA105009, P50-CA136393, R01-CA014089, R01-CA016056, R01-CA017054, R01-CA049449, R01-CA050385, R01-CA054419, R01-CA058598, R01-CA058860, R01-CA061107, R01-CA061132, R01-CA067262, R01-CA071766, R01-CA074850, R01-CA080742, R01-CA080978, R01-CA083918, R01-CA087538, R01-CA092044, R01-095023, R01-CA122443, R01-CA112523, R01-CA114343, R01-CA126841, R01-CA136924, R03-CA113148, R03-CA115195, U01-CA069417, U01-CA071966 and Intramural research funds); the US Army Medical Research and Material Command (DAMD17-01-1-0729, DAMD17-02-1-0666, DAMD17-02-1-0669, W81XWH-07-0449, W81XWH-10-1-02802); the US Public Health Service (PSA-042205); The National Health and Medical Research Council of Australia (199600 and 400281); the German Federal Ministry of Education and Research of Germany Programme of Clinical Biomedical Research (01GB 9401); the State of Baden-Wurttemberg through Medical Faculty of the University of Ulm (P.685); the Minnesota Ovarian Cancer Alliance; the Mayo Foundation; the Fred C. and Katherine B. Andersen Foundation; the Lon V. Smith Foundation (LVS-39420); the Oak Foundation; the OHSU Foundation; the Mermaid I project; the Rudolf-Bartling Foundation; the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Imperial College London, University College Hospital “Womens Health Theme” and the Royal Marsden Hospital; Work Safe BC 14.
References
- 1.Greene MW. Circadian rhythms and tumor growth. Cancer Lett. 2012;318:115–123. doi: 10.1016/j.canlet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Bratsun DA, Merkuriev DV, Zakharov AP, Pismen LM. Multiscale modeling of tumor growth induced by circadian rhythm disruption in epithelial tissue. J Biol Phys. 2015 doi: 10.1007/s10867-015-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knutsson A, Alfredsson L, Karlsson B, Akerstedt T, Fransson EI, et al. Breast cancer among shift workers: results of the WOLF longitudinal cohort study. Scand J Work Environ Health. 2013;39:170–177. doi: 10.5271/sjweh.3323. [DOI] [PubMed] [Google Scholar]
- 4.Hansen J, Lassen CF. Nested case-control study of night shift work and breast cancer risk among women in the Danish military. Occup Environ Med. 2012;69:551–556. doi: 10.1136/oemed-2011-100240. [DOI] [PubMed] [Google Scholar]
- 5.Wang F, Yeung KL, Chan WC, Kwok CC, Leung SL, et al. A meta-analysis on dose-response relationship between night shift work and the risk of breast cancer. Ann Oncol. 2013;24:2724–2732. doi: 10.1093/annonc/mdt283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Straif K, Baan R, Grosse Y, Secretan B, El Ghissassi F, et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. 2007;8:1065–1066. doi: 10.1016/S1470-2045(07)70373-X. [DOI] [PubMed] [Google Scholar]
- 7.Baker FC, Driver HS. Circadian rhythms, sleep, and the menstrual cycle. Sleep Med. 2007;8:613–622. doi: 10.1016/j.sleep.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Bhatti P, Cushing-Haugen KL, Wicklund KG, Doherty JA, Rossing MA. Nightshift work and risk of ovarian cancer. Occup Environ Med. 2013;70:231–237. doi: 10.1136/oemed-2012-101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamdar BB, Tergas AI, Mateen FJ, Bhayani NH, Oh J. Night-shift work and risk of breast cancer: a systematic review and meta-analysis. Breast Cancer Res Treat. 2013;138:291–301. doi: 10.1007/s10549-013-2433-1. [DOI] [PubMed] [Google Scholar]
- 10.Ijaz S, Verbeek J, Seidler A, Lindbohm ML, Ojajärvi A, et al. Night-shift work and breast cancer--a systematic review and meta-analysis. Scand J Work Environ Health. 2013;39:431–447. doi: 10.5271/sjweh.3371. [DOI] [PubMed] [Google Scholar]
- 11.Poole EM, Schernhammer ES, Tworoger SS. Rotating night shift work and risk of ovarian cancer. Cancer epidemiology, biomarkers & prevention. 2011;20:934–938. doi: 10.1158/1055-9965.EPI-11-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng X, Sehgal A. Speed control: cogs and gears that drive the circadian clock. Trends Neurosci. 2012;35:574–585. doi: 10.1016/j.tins.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon I, Choe HK, Son GH, Kim K. Mammalian molecular clocks. Exp Neurobiol. 2011;20:18–28. doi: 10.5607/en.2011.20.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boden MJ, Varcoe TJ, Kennaway DJ. Circadian regulation of reproduction: from gamete to offspring. Prog Biophys Mol Biol. 2013;113:387–397. doi: 10.1016/j.pbiomolbio.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 15.de la Iglesia HO, Schwartz WJ. Minireview: timely ovulation: circadian regulation of the female hypothalamo-pituitary-gonadal axis. Endocrinology. 2006;147:1148–1153. doi: 10.1210/en.2005-1311. [DOI] [PubMed] [Google Scholar]
- 16.Khan MA, Sengupta J, Mittal S, Ghosh D. Genome-wide expressions in autologous eutopic and ectopic endometrium of fertile women with endometriosis. Reprod Biol Endocrinol. 2012;10:84. doi: 10.1186/1477-7827-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merritt MA, De Pari M, Vitonis AF, Titus LJ, Cramer DW, et al. Reproductive characteristics in relation to ovarian cancer risk by histologic pathways. Hum Reprod. 2013;28:1406–1417. doi: 10.1093/humrep/des466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart LM, Holman CD, Aboagye-Sarfo P, Finn JC, Preen DB, et al. In vitro fertilization, endometriosis, nulliparity and ovarian cancer risk. Gynecol Oncol. 2013;128:260–264. doi: 10.1016/j.ygyno.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Matalliotakis IM, Cakmak H, Krasonikolakis GD, Dermitzaki D, Fragouli Y, et al. Endometriosis related to family history of malignancies in the Yale series. Surg Oncol. 2010;19:33–37. doi: 10.1016/j.suronc.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Miller BH, Olson SL, Turek FW, Levine JE, Horton TH, et al. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr Biol. 2004;14:1367–1373. doi: 10.1016/j.cub.2004.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez JD, Hansen A, Ord T, Bebas P, Chappell PE, et al. The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J Biol Rhythms. 2008;23:26–36. doi: 10.1177/0748730407311254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilorz V, Steinlechner S. Low reproductive success in Per1 and Per2 mutant mouse females due to accelerated ageing? Reproduction. 2008;135:559–568. doi: 10.1530/REP-07-0434. [DOI] [PubMed] [Google Scholar]
- 23.Kovanen L, Saarikoski ST, Aromaa A, Lönnqvist J, Partonen T. ARNTL (BMAL1) and NPAS2 gene variants contribute to fertility and seasonality. PLoS One. 2010;5:e10007. doi: 10.1371/journal.pone.0010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braem MG, Onland-Moret NC, van den Brandt PA, Goldbohm RA, Peeters PH, et al. Reproductive and hormonal factors in association with ovarian cancer in the Netherlands cohort study. Am J Epidemiol. 2010;172:1181–1189. doi: 10.1093/aje/kwq264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piek JM, Kenemans P, Zweemer RP, van Diest PJ, Verheijen RH. Ovarian carcinogenesis, an alternative theory. Gynecol Oncol. 2007;107:355. doi: 10.1016/j.ygyno.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Tworoger SS, Fairfield KM, Colditz GA, Rosner BA, Hankinson SE. Association of oral contraceptive use, other contraceptive methods, and infertility with ovarian cancer risk. Am J Epidemiol. 2007;166:894–901. doi: 10.1093/aje/kwm157. [DOI] [PubMed] [Google Scholar]
- 27.Brinton LA, Westhoff CL, Scoccia B, Lamb EJ, Althuis MD, et al. Causes of infertility as predictors of subsequent cancer risk. Epidemiology. 2005;16:500–507. doi: 10.1097/01.ede.0000164812.02181.d5. [DOI] [PubMed] [Google Scholar]
- 28.Dai H, Zhang L, Cao M, Song F, Zheng H, et al. The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast Cancer Res Treat. 2011;127:531–540. doi: 10.1007/s10549-010-1231-2. [DOI] [PubMed] [Google Scholar]
- 29.Fu A, Leaderer D, Zheng T, Hoffman AE, Stevens RG, et al. Genetic and epigenetic associations of circadian gene TIMELESS and breast cancer risk. Mol Carcinog. 2012;51:923–929. doi: 10.1002/mc.20862. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman AE, Zheng T, Yi CH, Stevens RG, Ba Y, et al. The core circadian gene Cryptochrome 2 influences breast cancer risk, possibly by mediating hormone signaling. Cancer Prev Res (Phila) 2010;3:539–548. doi: 10.1158/1940-6207.CAPR-09-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi C, Mu L, de la Longrais IA, Sochirca O, Arisio R, et al. The circadian gene NPAS2 is a novel prognostic biomarker for breast cancer. Breast Cancer Res Treat. 2010;120:663–669. doi: 10.1007/s10549-009-0484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y, Stevens RG, Leaderer D, Hoffman A, Holford T, et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res Treat. 2008;107:421–425. doi: 10.1007/s10549-007-9565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truong T, Liquet B, Menegaux F, Plancoulaine S, Laurent-Puig P, et al. Breast cancer risk, nightwork, and circadian clock gene polymorphisms. Endocr Relat Cancer. 2014;21:629–638. doi: 10.1530/ERC-14-0121. [DOI] [PubMed] [Google Scholar]
- 34.Markt SC, Valdimarsdottir UA, Shui IM, Sigurdardottir LG, Rider JR, et al. Circadian clock genes and risk of fatal prostate cancer. Cancer Causes Control. 2015;26:25–33. doi: 10.1007/s10552-014-0478-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y, Stevens RG, Hoffman AE, Fitzgerald LM, Kwon EM, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. 2009;69:9315–9322. doi: 10.1158/0008-5472.CAN-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu LW, Zhu Y, Yu K, Zheng T, Yu H, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2008;11:342–348. doi: 10.1038/sj.pcan.4501024. [DOI] [PubMed] [Google Scholar]
- 37.Hoffman AE, Zheng T, Stevens RG, Ba Y, Zhang Y, et al. Clock-cancer connection in non-Hodgkin’s lymphoma: a genetic association study and pathway analysis of the circadian gene cryptochrome 2. Cancer Res. 2009;69:3605–3613. doi: 10.1158/0008-5472.CAN-08-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Leaderer D, Guss C, Brown HN, Zhang Y, et al. Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin’s lymphoma. Int J Cancer. 2007;120:432–435. doi: 10.1002/ijc.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karantanos T, Theodoropoulos G, Gazouli M, Vaiopoulou A, Karantanou C, et al. Association of the clock genes polymorphisms with colorectal cancer susceptibility. J Surg Oncol. 2013;108:563–567. doi: 10.1002/jso.23434. [DOI] [PubMed] [Google Scholar]
- 40.Madden MH, Anic GM, Thompson RC, Nabors LB, Olson JJ, et al. Circadian pathway genes in relation to glioma risk and outcome. Cancer Causes Control. 2014;25:25–32. doi: 10.1007/s10552-013-0305-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao B, Lu J, Yin J, Liu H, Guo X, et al. A functional polymorphism in PER3 gene is associated with prognosis in hepatocellular carcinoma. Liver Int. 2012;32:1451–1459. doi: 10.1111/j.1478-3231.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- 42.Evans DS, Parimi N, Nievergelt CM, Blackwell T, Redline S, et al. Common genetic variants in ARNTL and NPAS2 and at chromosome 12p13 are associated with objectively measured sleep traits in the elderly. Sleep. 2013;36:431–446. doi: 10.5665/sleep.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim AS, Chang AM, Shulman JM, Raj T, Chibnik LB, et al. A common polymorphism near PER1 and the timing of human behavioral rhythms. Ann Neurol. 2012;72:324–334. doi: 10.1002/ana.23636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choub A, Mancuso M, Coppedè F, LoGerfo A, Orsucci D, et al. Clock T3111C and Per2 C111G SNPs do not influence circadian rhythmicity in healthy Italian population. Neurol Sci. 2011;32:89–93. doi: 10.1007/s10072-010-0415-1. [DOI] [PubMed] [Google Scholar]
- 45.Barclay NL, Eley TC, Mill J, Wong CC, Zavos HM, et al. Sleep quality and diurnal preference in a sample of young adults: associations with 5HTTLPR, PER3, and CLOCK 3111. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:681–690. doi: 10.1002/ajmg.b.31210. [DOI] [PubMed] [Google Scholar]
- 46.Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, et al. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet. 2013;45:362–370. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storey JD. A direct approach to false discovery rates. J Roy Statist Soc Ser B. 2002;64:479–98. [Google Scholar]
- 48.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrenson K, Sproul D, Grun B, Notaridou M, Benjamin E, et al. Modelling genetic and clinical heterogeneity in epithelial ovarian cancers. Carcinogenesis. 2011;32:1540–1549. doi: 10.1093/carcin/bgr140. [DOI] [PubMed] [Google Scholar]
- 51.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hefferan TE, Reinholz GG, Rickard DJ, Johnsen SA, Waters KM, et al. Overexpression of a nuclear protein, TIEG, mimics transforming growth factor-beta action in human osteoblast cells. J Biol Chem. 2000;275:20255–20259. doi: 10.1074/jbc.C000135200. [DOI] [PubMed] [Google Scholar]
- 53.Tachibana I, Imoto M, Adjei PN, Gores GJ, Subramaniam M, et al. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J Clin Invest. 1997;99:2365–2374. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem. 1998;273:25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- 55.Ribeiro A, Bronk SF, Roberts PJ, Urrutia R, Gores GJ. The transforming growth factor beta(1)-inducible transcription factor TIEG1, mediates apoptosis through oxidative stress. Hepatology. 1999;30:1490–1497. doi: 10.1002/hep.510300620. [DOI] [PubMed] [Google Scholar]
- 56.Jin W, Di G, Li J, Chen Y, Li W, et al. TIEG1 induces apoptosis through mitochondrial apoptotic pathway and promotes apoptosis induced by homoharringtonine and velcade. FEBS Lett. 2007;581:3826–3832. doi: 10.1016/j.febslet.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 57.Cook T, Urrutia R. TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am J Physiol Gastrointest Liver Physiol. 2000;278:G513–G521. doi: 10.1152/ajpgi.2000.278.4.G513. [DOI] [PubMed] [Google Scholar]
- 58.Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene. 2002;21:5783–5790. doi: 10.1038/sj.onc.1205681. [DOI] [PubMed] [Google Scholar]
- 59.Guillaumond F, Gréchez-Cassiau A, Subramaniam M, Brangolo S, Peteri-Brünback B, et al. Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol Cell Biol. 2010;30:3059–3070. doi: 10.1128/MCB.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leclerc N, Luppen CA, Ho VV, Nagpal S, Hacia JG, et al. Gene expression profiling of glucocorticoid-inhibited osteoblasts. J Mol Endocrinol. 2004;33:175–193. doi: 10.1677/jme.0.0330175. [DOI] [PubMed] [Google Scholar]
- 61.Hofbauer LC, Hicok KC, Khosla S. Effects of gonadal and adrenal androgens in a novel androgen-responsive human osteoblastic cell line. J Cell Biochem. 1998;71:96–108. [PubMed] [Google Scholar]
- 62.Subramaniam M, Hefferan TE, Tau K, Peus D, Pittelkow M, et al. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J Cell Biochem. 1998;68:226–236. [PubMed] [Google Scholar]
- 63.Reinholz MM, An MW, Johnsen SA, Subramaniam M, Suman VJ, et al. Differential gene expression of TGF beta inducible early gene (TIEG), Smad7, Smad2 and Bard1 in normal and malignant breast tissue. Breast Cancer Res Treat. 2004;86:75–88. doi: 10.1023/B:BREA.0000032926.74216.7d. [DOI] [PubMed] [Google Scholar]
- 64.Eid MA, Kumar MV, Iczkowski KA, Bostwick DG, Tindall DJ. Expression of early growth response genes in human prostate cancer. Cancer Res. 1998;58:2461–2468. [PubMed] [Google Scholar]
- 65.Barna G, Sebestyén A, Chinopoulos CC, Nagy K, Mihalik R, et al. TGF beta 1 kills lymphoma cells using mitochondrial apoptotic pathway with the help of caspase-8. Anticancer Res. 2002;22:3867–3872. [PubMed] [Google Scholar]
- 66.Zohrabian VM, Nandu H, Gulati N, Khitrov G, Zhao C, et al. Gene expression profiling of metastatic brain cancer. Oncol Rep. 2007;18:321–328. [PubMed] [Google Scholar]
- 67.Ivanov SV, Ivanova AV, Salnikow K, Timofeeva O, Subramaniam M, et al. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem Biophys Res Commun. 2008;370:536–540. doi: 10.1016/j.bbrc.2008.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mullenders J, Fabius AW, Madiredjo M, Bernards R, Beijersbergen RL. A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway. PLoS One. 2009;4:e4798. doi: 10.1371/journal.pone.0004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeng ZL, Wu MW, Sun J, Sun YL, Cai YC, et al. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J Biochem. 2010;148:319–326. doi: 10.1093/jb/mvq069. [DOI] [PubMed] [Google Scholar]
- 70.Taniguchi H, Fernández AF, Setién F, Ropero S, Ballestar E, et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 2009;69:8447–8454. doi: 10.1158/0008-5472.CAN-09-0551. [DOI] [PubMed] [Google Scholar]
- 71.Jensen LD, Cao Z, Nakamura M, Yang Y, Bräutigam L, et al. Opposing effects of circadian clock genes bmal1 and period2 in regulation of VEGF-dependent angiogenesis in developing zebrafish. Cell Rep. 2012;2:231–241. doi: 10.1016/j.celrep.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 72.Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira J, Higashimoto M, et al. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand. 2008;87:1060–1070. doi: 10.1080/00016340802348286. [DOI] [PubMed] [Google Scholar]