Abstract
Recent studies on copy number variation (CNV) have suggested that an increasing burden of CNVs is associated with susceptibility or resistance to disease. A large number of genes or genomic loci contribute to complex diseases such as autism. Thus, total genomic copy number burden, as an accumulation of copy number change, is a meaningful measure of genomic instability to identify the association between global genetic effects and phenotypes of interest. However, no systematic annotation pipeline has been developed to interpret biological meaning based on the accumulation of copy number change across the genome associated with a phenotype of interest. In this study, we develop a comprehensive and systematic pipeline for annotating copy number variants into genes/genomic regions and subsequently pathways and other gene groups using Biofilter – a bioinformatics tool that aggregates over a dozen publicly available databases of prior biological knowledge. Next we conduct enrichment tests of biologically defined groupings of CNVs including genes, pathways, Gene Ontology, or protein families. We applied the proposed pipeline to a CNV dataset from the Marshfield Clinic Personalized Medicine Research Project (PMRP) in a quantitative trait phenotype derived from the electronic health record – total cholesterol. We identified several significant pathways such as toll-like receptor signaling pathway and hepatitis C pathway, gene ontologies (GOs) of nucleoside triphosphatase activity (NTPase) and response to virus, and protein families such as cell morphogenesis that are associated with the total cholesterol phenotype based on CNV profiles (permutation p-value < 0.01). Based on the copy number burden analysis, it follows that the more and larger the copy number changes, the more likely that one or more target genes that influence disease risk and phenotypic severity will be affected. Thus, our study suggests the proposed enrichment pipeline could improve the interpretability of copy number burden analysis where hundreds of loci or genes contribute toward disease susceptibility via biological knowledge groups such as pathways. This CNV annotation pipeline with Biofilter can be used for CNV data from any genotyping or sequencing platform and to explore CNV enrichment for any traits or phenotypes. Biofilter continues to be a powerful bioinformatics tool for annotating, filtering, and constructing biologically informed models for association analysis – now including copy number variants.
Keywords: Copy number burden, functional annotation, electronic medical record, precision medicine
1. Introduction
Precision medicine, an emerging approach for prevention and treatment strategies that takes into account individual variability in genes, lifestyle, and environment for each person, has become one of the main research interests of biomedical science [1]. Recently, a precision medicine initiative was announced as a new research initiative that plans to boost progress toward a new era of personalized medicine [1]. Thus, collecting and utilizing patients’ rich information through electronic health records (EHRs) is one of the most important keys in precision medicine in order to tailor disease prevention and effective treatment strategies. First, precision medicine will need to be tested in many pilot studies to guide clinical practice.
The electronic MEdical Record and GEnomics (eMERGE) is a national network organized and funded by the National Human Genome Research Institute (NHGRI) that combines DNA repositories linked with electronic medical record (EMR) systems for performing large scale, high-throughput genetic association studies [2]. Many genome-wide association studies (GWAS) have been performed for multiple phenotypes generated from the eMERGE network [3,4]. In addition, a phenome-wide association study (PheWAS) approach has been used to query genotype-phenotype associations between targeted single-nucleotide polymorphisms (SNPs) and multiple phenotypes and to detect pleiotropy [5]. Despite many efforts to investigate genotype-phenotype associations, genetic studies to date have still only identified a small fraction of the heritability of complex traits [6]. Many alternative approaches to improve the ‘missing heritability’ problem have been proposed such as investigating gene-gene interactions associated with phenotypes or a systems genomics approach [7,8]. In addition, an alternative explanation for the ‘missing heritability’ could be copy number variations (CNVs) [9].
Disease-associated rare/common CNVs have been identified through multiple studies [10,11]. However, one conclusion from the extensive CNV association studies is that there are hundreds or thousands of genes or genomic regions that contribute to disease susceptibility for certain disorders such as autism. Thus, total genomic copy number burden, as an accumulation of copy number change, is a meaningful measure of genomic change that may contribute to phenotypes that are associated with many genes/regions. Previously, we found that autism is associated with increased levels of copy number burden [12]. However, one of the current limitations of this approach is that it is difficult to interpret biological meaning based on the accumulation of copy number change genome-wide. Is it the amount of copy number change that is important or is it which genes/pathways the copy number change occurs that is important? In this study, we develop a comprehensive and systematic pipeline for annotating copy number variants into genes/genomic regions and subsequently pathways and other gene groups using Biofilter – a bioinformatics tool that aggregates over a dozen publicly available databases of prior biological knowledge [13]. Next we conduct enrichment tests of biologically defined groupings of CNVs including pathways, Gene Ontology (GO), or protein families. We applied the proposed pipeline to a CNV data set in a cholesterol phenotype from the Marshfield Clinic, a study site of the eMERGE network. We identified several significant pathways, GOs, and protein families that are associated with the cholesterol phenotype based on CNV profiles. The results discussed herein demonstrate the utility of the proposed pipeline as a novel method for annotating the results of CNV burden analysis underlying complex traits such as cholesterol.
2. Methods
2.1. Data
Median total cholesterol as a phenotype for this study was extracted from the EHR from the Marshfield Personalized Medicine Research Project (PMRP) [14]. Table 1 shows the descriptive statistics of the data set. High-density SNP genotyping was performed on DNA samples at the Center for Inherited Disease Research (CIDR) using the Illumina 660W-Quad. After quality controls (QC), 3,399 samples with available median total cholesterol phenotype from the Marshfield PMRP were selected for the present study. DNA samples from this site were genotyped using the Illumina 660W-Quad array as previously described [15]. QC is described in further detail in the CNV Burden Analysis section.
Table 1.
Descriptive statistics on Marshfield Median Total Cholesterol data set. Total number of samples after QC is presented.
| Phenotype | Sex |
Birthdate Year* (Mean±SD) | Total | |
|---|---|---|---|---|
| Male | Female | |||
| Median Total Cholesterol | 1,428 | 1,971 | 3.1779±1.1772 | 3,399 |
Birthdate Year denotes decade of birth where 1=1910, 2=1920, 3=1930, 4=1940, 5=1950, and 6=1960.
2.2. CNV Burden Analysis
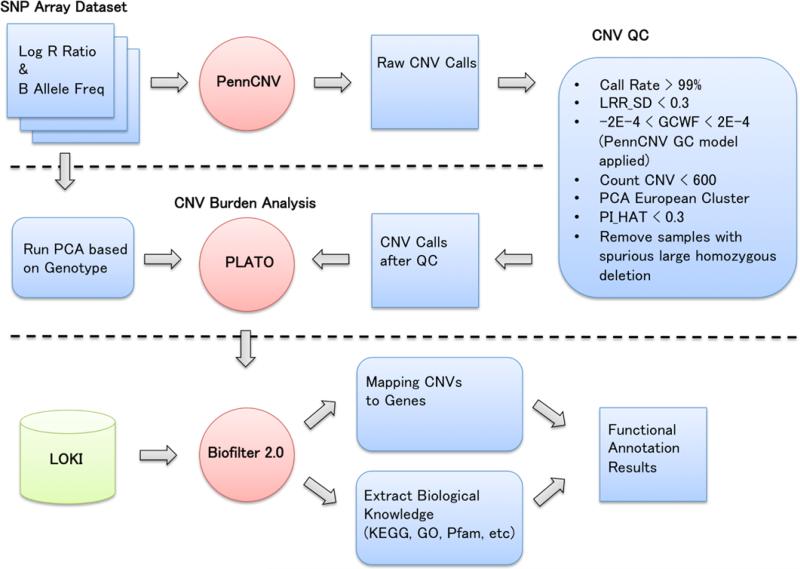
Figure 1 shows the illustration of the entire pipeline. In order to detect CNV, log R ratio and B Allele Frequency values were extracted from the Illumina 660W-Quad array. The PennCNV software, based on a hidden Markov model, was used for calling CNVs [16]. First, individual CNV calls were generated as raw CNV calls and then several QC steps were performed. CNVs that had a high success rate of attempted SNPs, a low standard deviation of normalized intensity, and low genomic wave artifacts passed QC thresholds. All samples had genetically inferred European ancestry and any genotypic duplicates were removed. In addition, samples with spurious large homozygous deletions were removed. After QC, 3,399 samples were analyzed for the CNV burden analysis. Linear regression models using PLATO software [17] were fit to the data to evaluate the associations between CNV burden, i.e. accumulation of duplication or deletion in each individual, or collectively, as total base pairs of altered copy number (i.e. total CNV burden), and the median total cholesterol phenotype. Analyses were adjusted for potential confounders, including age (decade of birth), sex, and the first three principal components of ancestry that were generated from the PCA analysis based on SNP data set.
Fig. 1.
Illustration of the pipeline for functional annotation based on the results of the CNV burden analyses. PennCNV is used for calling CNVs, then copy number burden analysis is performed using CNV calls after QC. A new function of Biofilter 2.0 provides functional annotation results based on copy number burden.
2.3. Biofilter 2.0
Biofilter 2.0 is a software tool that provides a convenient single interface for high-throughput annotation, filtering of genetic data via accessing multiple publicly available human genetic data sources, and constructing biologically informed models for association analysis [13]. This software uses a build-in database called the Library of Knowledge Integration (LOKI), which contains a number of public data resources. LOKI includes not only information about the genomic locations of SNPs and genes, but also information about biological networks, connections, and/or pathways to be used for determining relationships between genes. For more information, see: http://ritchielab.psu.edu/software/.
A new function was added in Biofilter 2.0 for CNV analyses. CNV data, which are specified by a chromosome and base pair range from any genotyping or sequencing platform, can be mapped to genes (Fig 1). These CNV regions can be mapped to genes based on percent of overlap of the genes with the CNV region or based on the number of base pairs overlapped. In addition, biological knowledge such as pathway, GO, or Pfam along with list of its gene members can be extracted using Biofilter 2.0 for the functional annotation calculation based on the results of CNV burden analyses (Fig 1). For the current study, 281 Kyoto Encyclopedia of Gene and Genomes (KEGG) pathways, 1,454 GOs, and 2,908 Pfams were used.
2.4. Functional Annotation based on CNV Profiles
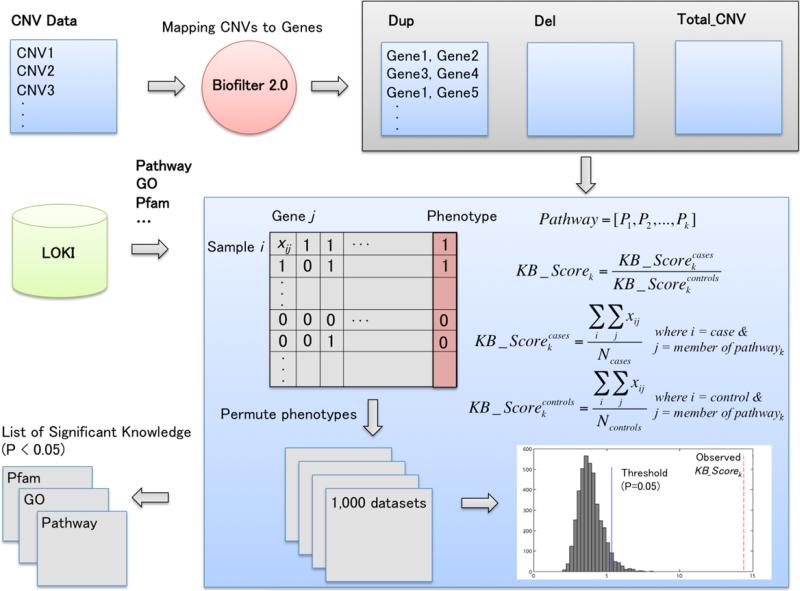
Figure 2 describes the overview of the functional annotation calculation based on CNV profiles. After the CNV data set was mapped to genes using Biofilter 2.0, functional enrichment tests can be used for the functional annotation. However, an over-representation analysis (ORA) approach, which is one of the most common methods for the pathway analysis, is not appropriate for annotating the results of CNV burden analyses since it does not consider the frequency information of rare and/or common CNV across samples. Thus, we propose a new functional annotation method based on the results of CNV burden analyses in order to capture the frequency information of rare and common CNV. Knowledgebase score (KB_Score) was calculated via aggregating not only frequency of genes within a specific pathway but also frequency of those genes across samples (Fig 2). KB_Scores can be obtained from cases and controls, respectively, and then each KB_Score should be normalized by dividing by the number of cases/controls. Next, a final KB_Score can be achieved as a ratio of each KB_Score, which is generated from the cases/controls, respectively. After calculating KB_Score per pathway, we randomly permuted the phenotype 1,000 times to generate random data sets, that is, the phenotypes are randomly associated with the CNV profiles. To assess whether annotated biological knowledge (observed KB_Score) is more significant than expected by chance, any observed KB_Score higher than the 950th highest KB_Score in the permuted data set was recorded in a final list of significant biological knowledge (Fig 2). Even though the proposed method is suitable for case/control phenotypes, continuous phenotypes can be dichotomized based on quartiles as described more below.
Fig. 2.
Overview of the functional annotation calculation based on CNV profiles. After the CNV data set was mapped to genes using Biofilter 2.0, functional enrichment tests can be used to identify significantly enriched biological knowledge such pathway, GO or Pfam. KB, knowledgebase.
3. Results and Discussion
3.1. The Results of CNV Burden Analysis
We assessed the significance of the association between CNV burden variables (duplication, deletion, and total CNV burden) and median total cholesterol using linear regression. Through the CNV burden analysis, duplication and total CNV burden were significantly associated with cholesterol phenotype, P = 0.0023, P = 0.0099, respectively. Thus, duplication regions and total CNV regions were mapped to genes using Biofilter 2.0. Since functional annotation results were similar between different overlap criteria between CNV regions and genes (data not shown), CNV data was mapped to genes based on 1bp overlap criteria for further analysis. From 3,399 samples 7,150 distinct genes and 9,587 distinct genes were mapped based on duplication and total CNV, respectively.
3.2. Significant Pathways, GOs, and Pfams Associated with Cholesterol
Since the proposed method for annotating the results of CNV burden analyses is appropriate for the case/control phenotype, the median total cholesterol, a continuous phenotype, was dichotomized in three different ways based on quartiles: (1) 4th quartile (cases) vs. 3rd, 2nd, 1st quartiles (controls); (2) 4th, 3rd quartiles (cases) vs. 2nd, 1st quartiles (controls); (3) 4th, 3rd, 2nd quartiles (cases) vs. 1st quartile (controls). We compared the annotation results between different dichotomized phenotypes (Table 2). Since total number of significant biological knowledge between dichotomized phenotypes was not too different and there were many distinct biological knowledge that were shared between at least two dichotomized phenotypes, we chose the first way of dichotomized phenotype, top quartile as cases vs. three bottom quartiles as controls, for further analysis. This approach is also commonly used in many epidemiology studies in order to calculate odds ratio for the continuous phenotype [18].
Table 2.
Comparison of total number of significant knowledge features based on different dichotomized cholesterol phenotypes. Each element is the number of significant knowledge features from the functional annotation calculation (P<0.05).
| Knowledge Type | CNV Type | Dichotomizing Cholesterol Phenotype |
Shared Significant Knowledge* | ||
|---|---|---|---|---|---|
| 4th Quartile vs. 3rd, 2nd, 1st Quartiles | 4th, 3rd Quartiles vs. 2nd, 1st Quartiles | 4th, 3rd, 2nd Quartiles vs. 1st Quartile | |||
| Pathway | Dup | 32 | 38 | 36 | 28 |
| Total CNV | 20 | 10 | 21 | 13 | |
| GO | Dup | 39 | 62 | 62 | 43 |
| Total CNV | 50 | 38 | 58 | 26 | |
| Pfam | Dup | 43 | 46 | 35 | 22 |
| Total CNV | 63 | 57 | 68 | 20 | |
Shared significant knowledge denotes the number of distinct knowledge that appears in at least more than two dichotomized phenotypes.
Through the proposed functional annotation method, significant pathways, GOs, and Pfams were obtained based on the selected dichotomized phenotype. Table 3 shows the results of pathway knowledge for duplication and total CNV burden. We restricted the significance threshold (permutation P-value <0.01) to remove marginally significant results. Based on a stricter threshold, 6 pathways were found from duplication burden and 2 pathways were selected from total CNV burden as pathways associated with the cholesterol phenotype (Table 3). Similarly, significant GOs and Pfams were also found (Table 4 and Table 5).
Table 3.
The list of significant pathways. Significant pathways associated with cholesterol were selected based on CNV burden data set (P<0.01). Continuous cholesterol phenotype was dichotomized at the 75th percentile in order to perform the proposed functional annotation pipeline, comparing CNVs in the top quartile (‘high’) with those in the bottom 3 quartiles (‘low’).
| CNV Type | Significant Pathways | Permutation P-value |
|---|---|---|
| Dup | Hepatitis C | 0.001998 |
| Toll-like receptor signaling pathway | 0.001998 | |
| Cytokine-cytokine receptor interaction | 0.006993 | |
| Shigellosis | 0.006993 | |
| RIG-I-like receptor signaling pathway | 0.007992 | |
| Influenza A | 0.00999 | |
| Total CNV | Toll-like receptor signaling pathway | 0.001 |
| Renal cell carcinoma | 0.002997 | |
Table 4.
The list of significant protein families. Significant protein families associated with cholesterol were selected based on CNV burden data set (P<0.01). Continuous cholesterol phenotype was dichotomized at the 75th percentile in order to perform the proposed functional annotation pipeline comparing CNVs in the top quartile (‘high’) with those in the bottom 3 quartiles (‘low’).
| CNV Type | Significant Pfams | Permutation P-value |
|---|---|---|
| Dup | Cell morphogenesis central region | 0.001998 |
| Cell morphogenesis C-terminal | 0.001998 | |
| Cell morphogenesis N-terminal | 0.001998 | |
| RAVE protein 1 C terminal | 0.00999 | |
| Zinc-binding domain | 0.00999 | |
| Total CNV | Poly (ADP-ribose) glycohydrolase (PARG) | 0.001 |
| Adenylate and Guanylate cyclase catalytic domain | 0.001 | |
| Thrombospondin type 1 domain | 0.000999 | |
| ADAM-TS Spacer 1 | 0.000999 | |
| Cell morphogenesis central region | 0.001998 | |
| Cell morphogenesis C-terminal | 0.001998 | |
| Cell morphogenesis N-terminal | 0.001998 | |
| Reprolysin (M12B) family zinc metalloprotease | 0.008991 | |
| Zinc binding domain | 0.00999 | |
Table 5.
The list of significant GOs. Significant GOs associated with cholesterol were selected based on CNV burden data set (P<0.01). Continuous cholesterol phenotype was dichotomized at the 75th percentile in order to perform the proposed functional annotation pipeline comparing CNVs in the top quartile (‘high’) with those in the bottom 3 quartiles (‘low’).
| CNV Type | Significant GOs | Permutation P-value |
|---|---|---|
| Dup | Nucleoside triphosphatase activity | 0.001 |
| GTPase activity | 0.001 | |
| Pyrophosphatase activity | 0.001 | |
| Cellular defense response | 0.000999 | |
| Hydrolase activity, acting on acid anhydrides | 0.000999 | |
| Caspase regulator activity | 0.005994 | |
| Histone acetyltransferase activity | 0.006993 | |
| Response to virus | 0.007992 | |
| Microtubule organizing center organization and biogenesis | 0.008991 | |
| Centrosome organization and biogenesis | 0.008991 | |
| Nuclear envelope | 0.00999 | |
| Total CNV | Sensory organ development | 0.001 |
| Nicotinic acetylcholine-gated receptor-channel complex | 0.000999 | |
| Nicotinic acetylcholine-activated cation-selective channel activity | 0.000999 | |
| Double stranded DNA binding | 0.001998 | |
| Cyclase activity | 0.002997 | |
| Phosphorus-oxygen lyase activity | 0.002997 | |
| Secondary metabolic process | 0.005994 | |
| Learning and/or memory | 0.006993 | |
| Serotonin receptor activity | 0.007992 | |
| Microvillus | 0.008991 | |
| Amino acid transmembrane transporter activity | 0.008991 | |
3.3. Biological interpretation
Previously, many studies have reported that hypercholesterolemia or lower cholesterol levels are associated with CNV [19]. In addition, hyperlipidemia is associated with many other diseases such as myocardial infarction. For example, one study found several CNVs to have a link to myocardial infarction and hyperlipidemia [20]. Most of these studies were focused on specific CNVs or genes within CNV regions. However, we found that CNV burden is associated with cholesterol level. This is the first study to identify the association between the cholesterol quantitative trait and CNV burden in the literature. This suggests that cholesterol levels may also be associated with global genetic effects of many genes/regions.
In order to better understand possible mechanisms of the association between the cholesterol phenotype and CNV burden, the proposed functional annotation test was performed based on CNV profiles. Six pathways, hepatitis C, toll-like receptor signaling pathway, cytokine-cytokine receptor interaction, shigellosis, RIG-I-like receptor signaling pathway, and influenza A were found in the annotation results based on duplication burden. In particular, toll-like receptor (TLR) signaling pathway is a well-known pathway that acts an important role in atherosclerosis [21]. A prior study found that TLR4 can directly interfere with cholesterol metabolism in macrophages, which suggests that TLR4 could affect disease pathology [21]. The results from a second study revealed that Hepatitis C virus entry, in cooperation with CD81 and scavenger receptor B type I, is also partially dependent on membrane cholesterol [22]. In addition, TLR signaling pathways and renal cell carcinoma were obtained based on total CNV burden. In a recent study of patients who underwent surgery for renal cell carcinoma, preoperative serum cholesterol was implicated as an independent factor for prognosis. Lower cholesterol levels were found to be associated with advanced disease and worse survival, which may be due to cholesterol's increased storage in tumour cells and role in new membrane biosynthesis [23]. For Pfam, 5 and 9 protein families were found based on duplication and total copy number burden, respectively. Interestingly, many cell morphogenesis-related protein families were found, in line with findings that cholesterol is important for proper cell morphogenesis due to its role in maintaining membrane order [24]. Among many significant GOs, nucleoside triphosphatase activity (NTPase) was found to be associated with cholesterol. Nuclear membrane cholesterol both modulates NTPase activity and can alter activity when oxidized [25]. Furthermore, similarly to the aforementioned Hepatitis C virus, cholesterol is important for membrane fusion during virus infection into host cells, as the enrichment of cholesterol helps to maintain membrane fluidity in the cell [26]. Taken together these results demonstrate the utility of the proposed pipeline for annotating the results of CNV burden analysis underlying complex traits such as total cholesterol phenotype.
4. Conclusions
In this study, we developed a systematic pipeline for annotating copy number variants into genes/genomic regions and subsequently pathways and other biological knowledge using Biofilter 2.0. In addition, a new method that takes into account the frequency information of genes in rare/common CNVs was proposed and led to the finding of many biologically relevant pathways, GOs, and protein families associated with cholesterol. Based on the copy number burden analysis, it follows that with larger copy number changes and a greater accumulation of copy number changes, it is more likely that genes known to influence disease risk and phenotypic severity will be affected. Thus, our study suggests the proposed pipeline could improve the interpretability of copy number burden analysis where hundreds of loci or genes contribute toward disease susceptibility via biological knowledge groups such as pathways. This CNV annotation pipeline with Biofilter can be used for CNV data from any genotyping or sequencing platform and to explore CNV enrichment for any traits or phenotypes. Biofilter is open source and freely available at http://ritchielab.psu.edu/software. Biofilter continues to be a powerful bioinformatics tool for annotation, filtering, and constructing biologically informed models for association analysis – now including copy number variants.
As demonstrated by this and other studies, CNV burden analysis is a new powerful method to investigate the association between accumulated genetic effects and many traits or phenotypes. In particular, the development of an appropriate annotation pipeline for CNV burden analysis will be valuable to better understand possible mechanisms associated with phenotypes in the context of accumulated effect of rare/common CNVs. As more well-designed genetic and phenotypic data are generated based on EHR for better precision medicine, CNV burden analysis continues to demonstrate the strengths along with the proposed annotation pipeline.
Acknowledgments
This work was funded by NHGRI grant U01 HG006389, NHLBI grant U01 HL065962, and CTSI: UL1 RR033184-01. This work is also supported by a grant with the Pennsylvania Department of Health using Tobacco CURE Funds.
References
- 1.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Namjou B, Keddache M, Marsolo K, Wagner M, Lingren T, et al. EMR-linked GWAS study: investigation of variation landscape of loci for body mass index in children. Front Genet. 2013;4:268. doi: 10.3389/fgene.2013.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, et al. Robust replication of genotype phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype phenotype interactions. Nat Rev Genet. 2015;16:85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- 8.Hall MA, Verma SS, Wallace J, Lucas A, Berg RL, et al. Biology-Driven Gene-Gene Interaction Analysis of Age-Related Cataract in the eMERGE Network. Genet Epidemiol. 2015;39:376–384. doi: 10.1002/gepi.21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prakash SK, LeMaire SA, Guo DC, Russell L, Regalado ES, et al. Rare copy number variants disrupt genes regulating vascular smooth muscle cell adhesion and contractility in sporadic thoracic aortic aneurysms and dissections. Am J Hum Genet. 2010;87:743–756. doi: 10.1016/j.ajhg.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly JJ, Glessner JT, Almoguera B, Crosslin DR, Jarvik GP, et al. Copy number variation analysis in the context of electronic medical records and large-scale genomics consortium efforts. Front Genet. 2014;5:51. doi: 10.3389/fgene.2014.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girirajan S, Johnson RL, Tassone F, Balciuniene J, Katiyar N, et al. Global increases in both common and rare copy number load associated with autism. Hum Mol Genet. 2013;22:2870–2880. doi: 10.1093/hmg/ddt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pendergrass SA, Frase A, Wallace J, Wolfe D, Katiyar N, et al. Genomic analyses with biofilter 2.0: knowledge driven filtering, annotation, and model development. BioData Min. 2013;6:25. doi: 10.1186/1756-0381-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peissig PL, Rasmussen LV, Berg RL, Linneman JG, McCarty CA, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic hea lth records. J Am Med Inform Assoc. 2012;19:225–234. doi: 10.1136/amiajnl-2011-000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, et al. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet. 2011 doi: 10.1002/0471142905.hg0119s68. Chapter 1: Unit1 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Li M, Hadley D, Liu R, Glessner J, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, et al. Finding unique filter sets in PLATO: a precursor to efficient interaction analysis in GWAS data. Pac Symp Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]
- 18.Volk HE, Lurmann F, Penfold B, Hertz-Picciotto I, McConnell R. Traffic-related air pollution, particulate matter, and autism. JAMA Psychiatry. 2013;70:71–77. doi: 10.1001/jamapsychiatry.2013.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollex RL, Hegele RA. Genomic copy number variation and its potential role in lipoprotein and metabolic phenotypes. Curr Opin Lipidol. 2007;18:174–180. doi: 10.1097/MOL.0b013e32802e6c12. [DOI] [PubMed] [Google Scholar]
- 20.Shia WC, Ku TH, Tsao YM, Hsia CH, Chang YM, et al. Genetic copy number variants in myocardial infarction patients with hyperlipidemia. BMC Genomics 12 Suppl. 2011;3:S23. doi: 10.1186/1471-2164-12-S3-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtiss LK, Tobias PS. Emerging role of Toll-like receptors in atherosclerosis. J Lipid Res. 2009;50(Suppl):S340–345. doi: 10.1194/jlr.R800056-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapadia SB, Barth H, Baumert T, McKeating JA, Chisari FV. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J Virol. 2007;81:374–383. doi: 10.1128/JVI.01134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Martino M, Leitner CV, Seemann C, Hofbauer SL, Lucca I, et al. Preoperative serum cholesterol is an independent prognostic factor for patients with renal cell carcinoma (RCC). BJU Int. 2015;115:397–404. doi: 10.1111/bju.12767. [DOI] [PubMed] [Google Scholar]
- 24.Arita Y, Nishimura S, Ishitsuka R, Kishimoto T, Ikenouchi J, et al. Targeting cholesterol in a liquid-disordered environment by theonellamides modulates cell membrane order and cell shape. Chem Biol. 2015;22:604–610. doi: 10.1016/j.chembiol.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Ramjiawan B, Czubryt MP, Massaeli H, Gilchrist JS, Pierce GN. Oxidation of nuclear membrane cholesterol inhibits nucleoside triphosphatase activity. Free Radic Biol Med. 1997;23:556–562. doi: 10.1016/s0891-5849(97)00003-8. [DOI] [PubMed] [Google Scholar]
- 26.Tanner LB, Lee B. The greasy response to virus infections. Cell Host Microbe. 2013;13:375–377. doi: 10.1016/j.chom.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]