Abstract
Aim:
hERG potassium channels display miscellaneous interactions with diverse chemical scaffolds. In this study we assessed the hERG inhibition in a large compound library of diverse chemical entities and provided data for better understanding of the mechanisms underlying promiscuity of hERG inhibition.
Methods:
Approximately 300 000 compounds contained in Molecular Library Small Molecular Repository (MLSMR) library were tested. Compound profiling was conducted on hERG-CHO cells using the automated patch-clamp platform–IonWorks Quattro™.
Results:
The compound library was tested at 1 and 10 μmol/L. IC50 values were predicted using a modified 4-parameter logistic model. Inhibitor hits were binned into three groups based on their potency: high (IC50<1 μmol/L), intermediate (1 μmol/L< IC50<10 μmol/L), and low (IC50>10 μmol/L) with hit rates of 1.64%, 9.17% and 16.63%, respectively. Six physiochemical properties of each compound were acquired and calculated using ACD software to evaluate the correlation between hERG inhibition and the properties: hERG inhibition was positively correlative to the physiochemical properties ALogP, molecular weight and RTB, and negatively correlative to TPSA.
Conclusion:
Based on a large diverse compound collection, this study provides experimental evidence to understand the promiscuity of hERG inhibition. This study further demonstrates that hERG liability compounds tend to be more hydrophobic, high-molecular, flexible and polarizable.
Keywords: hERG, high-throughput screening, MLSMR library, automated electrophysiology, cardiotoxicity
Introduction
The acquired long QT syndrome is both a threat to public health and a major stumbling block for drug development1. It is most often caused through unintended blockade of the cardiac repolarizing potassium channel, IKr, encoded by the Human Ether-a-go-go related gene (hERG)2. Blockade of hERG channel was found to be associated with an increased duration of ventricular repolarization and prolongation of QT interval (long QT syndrome, or LQTS). hERG potassium channel displays promiscuous interactions with diverse chemical scaffolds3. Structurally and functionally unrelated drugs have been shown to block hERG channel, and some of these agents, including terfenadine, astemizole, droperidol and cisapride, etc, have been withdrawn from the market due to their potential to predispose individuals to sudden cardiac death. Due to the important implications in both drug discovery4 and clinical practice, evaluation of hERG liability has been required for any new chemical entities by regulatory agencies according to the ICH S7B guideline since 20055.
Manual patch clamp method is considered as gold standard for ion channel functional evaluation6. However the method is very labor-intensive with low throughput, and thus its application in drug discovery is limited. Ion flux assays with surrogate ions of Rubidium (Rb+) or Thallium (Tl+) have generated effective values for detecting potassium channel activity in high-throughput formats. However, ion flux assays are performed under less physiologically relevant conditions7,8 and may underestimate drug blocking potency8. Recent advance in automated electrophysiology technology has provided platforms such as IonWorks Quattro that can generate data with high information content and good quality at higher throughput9,10,11. This permits applications of automated electrophysiology in primary screening.
In addition to experimental approaches, to predict the hERG liability of lead candidates early in drug discovery, numerical in silico methods have been developed12,13,14,15,16,17,18. Since experimental data from a large collection of structurally diverse compounds were yet available, models was usually based on the existing data of limited number of hERG inhibitors generated using different methods. These models have displayed inconsistent prediction for hERG activity. It is probably due to lack of representation of the chemical space by the those inhibitors, or variations in detection methodologies to access hERG blockade. Development of in silico models may be benefited from the data of a large collection of diverse compounds generated by electrophysiology.
In 2005, NIH launched the decade-long of Molecular Libraries Program (MLPCN), offering the NIH-funded screen centers access to the large-scale screening capacity to identify small molecules that can be optimized as chemical probes by screening the molecular library small molecular repository (MLSMR)19. Based on the search in PubChem website (http://pubchem.ncbi.nlm.nih.gov), so far 553 primary screens for 486 protein targets have been completed, and the tested compound numbers varied from a few hundred up to approximately 300 000 compounds from the MLSMR compound collection. To prioritize the active compounds identified in other screenings based on their hERG liability and generate data to facilitate understanding of mechanisms underlying promiscuity regarding hERG inhibition, we conducted a screening of the (MLSMR) library of approximately 300 000 compounds at both 1 μmol/L and 10 μmol/L using automated electrophysiology. The hERG data are available in the website called hERGCentral (www.hergcentral.org), in which people can retrieve and analyze compound-hERG interaction based on their needs20. In additon, earlier we have uploaded part of the hERG screen data to PubChem website21 and reported some computational prediction analysis for hERG liability22. Here we reported the high-throughput screening of MLSMR compound targeting hERG potassium channels and effects of compound physiochemical properties on hERG inhibition. Such experimental data about hERG activity would provide further information to triage compounds in the MLSMR library in terms of their hERG liability and facilitate development of in silico models to better predict hERG blockade. In addition, our findings of the association of chemical properties with hERG potency verified the earlier findings by the large compound library23.
Materials and methods
Cell culture
Chinese hamster ovary (CHO) cells stably expressing hERG were grown in 50/50 DMEM/F-12 (Mediatech, Manassas, VA, USA) with 10% fetal bovine serum (Gemini Bio-products, West Sacramento, CA, USA), and 2 mmol/L L-glutamine (Invitrogen, Carlsbad, CA, USA) supplemented with G418 500 μg/mL.
Compound collection
Molecular Libraries Small Molecule Repository (MLSMR) library consisted of 318 950 compounds in 384-well plate format as single compound concentration at 5 mmol/L in DMSO (additional detail describing the library can be obtained from http://mlsmr.evotec.com). The compound quality evaluation and logistics have been handled by Evotec (US) Inc (Evotec, San Francisco, CA, USA) with purity of more than 90% determined by LCMS. The library was screened with the final concentrations of 1 μmol/L (DMSO 0.02%) and 10 μmol/L (DMSO 0.2%) in each well with dilutions of 1:5000 and 1:500, respectively. Control compound, Dofetilide (N-[4-(2-{[2-(4-methanesulfonamidophenoxy) ethyl] (methyl)amino}ethyl) phenyl]methanesulfonamide) (Fisher Scientific, Pittsburg, PA, USA) was dissolved in DMSO in a stock concentration of 20 mmol/L and diluted in the assay buffer at a final concentration of 1 μmol/L.
Automated patch-clamp assay
hERG activity was examined in the population-patch-clamp mode (PPC) of IonWorks Quattro™ (Molecular Devices, LLC, Sunnyvale, CA, USA), an automated patch clamp instrument. Compound effects were tested using dual compound additions at 1 μmol/L and 10 μmol/L. The CHO-K1 cells stably expressing hERG channels were freshly dislodged from flasks with Trypsin-EDTA (0.05%) for 4 min, spin down twice at 700×g, and suspended in the extracellular solution (in mmol/L) with 1.8×106 per mL: 137 NaCl, 4 KCl, 1 MgCl2, 1.8 CaCl2, 10 HEPES, and 10 Glucose, pH 7.4 adjusted with NaOH and dispensed into a 384-well population patch clamp (PPC) plate. The cell plating density was 6300 cells/well.
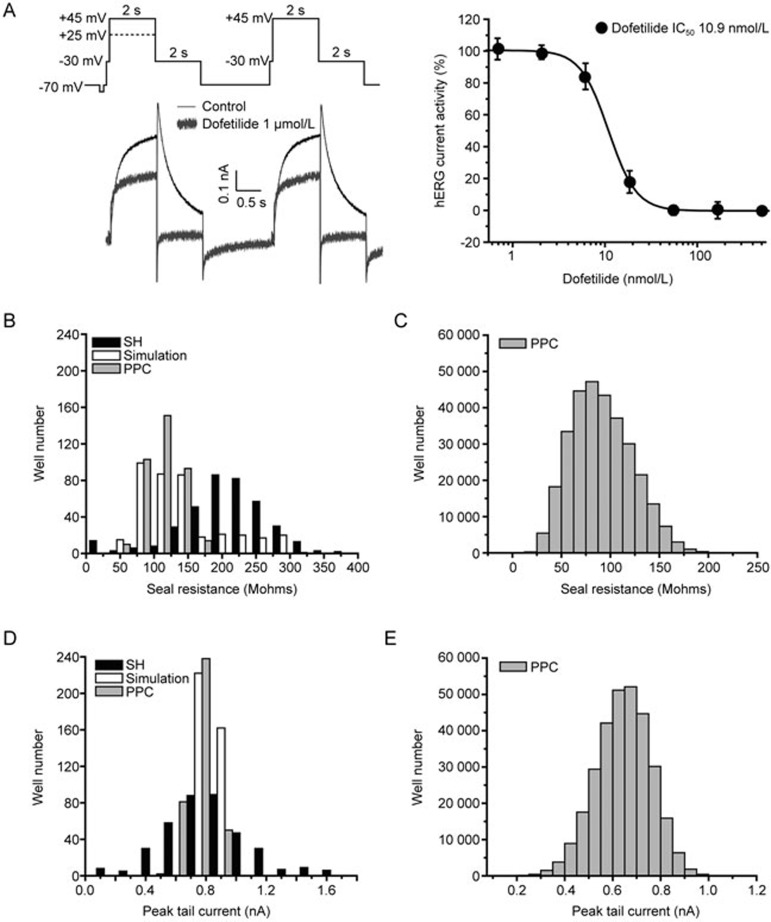
After dispensing, seal resistance of cells was measured for each well and cells were perforated by incubation with 50 μg/mL amphotericin B (Sigma, St Louis, MO, USA) in the internal solution composed of (in mmol/L): 40 KCl, 100 K-Gluconate, 1 MgCl2, 5 HEPES, 2 CaCl2, pH 7.2 adjusted with KOH. Activity of hERG was then measured with the recording protocol as shown in Figure 1A. hERG currents were evoked by two voltage pulses with a 3 s interval. The voltage pulse consisted of a 100 ms step to −30 mV, a conditioning pre-pulse (2 s duration, +25 mV or +45 mV) followed by a test pulse (2 s duration, −30 mV) from a holding potential at −70 mV. After a 3 s interval at −70 mV, a second pulse protocol was applied consisting of a 100 ms step to −30 mV, a pre-pulse (2 s duration, +45 mV) followed by a test pulse (2 s duration, −30 mV). Leak currents were linearly subtracted extrapolating the current elicited by a 100 ms step to −80 mV from a holding potential of −70 mV. Positive controls (1 μmol/L dofetilide) and negative controls [external buffer with 0.02% (first addition) and 0.2% (second addition) (v/v) DMSO] values were measured in each plate. Compound incubation duration was 3 min for both the first addition (1 μmol/L) and second addition (10 μmol/L). Z′ factor was calculated to evaluate data quality. Wells with a peak tail current amplitude bigger than 0.2 nA, a seal resistance more than 30 Mohms, and reduction of seal resistance less than 25% were included for data analysis.
Figure 1.
Seal resistance and current expression of hERG-CHO stable cell line measured using the IonWorks Quattro™ system. (A) Recording protocols and the typical hERG current traces in the absence and presence of 1 μmol/L Dofetilide. In the displayed protocol, the solid line represented the protocol used for majority of compounds while the dashed line presented a voltage pulse used in a small fraction of compounds of the screening. Dose-response curve for Dofetilide is displayed in the right panel. (B) Histograms of seal resistance recorded across 384 wells of single-hole (HT) (black color filled bars) and population patch clamp (PPC) (grey color filled bars) using hERG-CHO cells at a density 1.8×106 cells/mL. The bootstrap analysis was applied to the data from single-hole mode to simulate the distribution of seal resistance in the PPC mode (as shown in the unfilled bars). (C) The distribution of the seal resistance (PPC mode) for the screened library with the mean±SD values at 89.66±36.26 Mohms. (D) Histograms of peak tail current amplitude obtained from single-hole (black color filled bars) and PPC modes (grey color filled bars). Peak currents at the first test pulse were measured in each well. Similar to the seal resistance, the bootstrap analysis was performed to estimate the distribution of hERG tail current on PPC mode using experimental results from the single-hole mode (as shown in unfilled bars). (E) The distribution of the peak currents (PPC mode) for the screened library with the mean±SD values at 0.62±0.17 nA.
Peak amplitudes of tail currents and steady-state currents from the second pulse were measured before and after compound treatment. Compound effects were assessed by percentage changes of hERG peak tail currents and steady-state currents, which were calculated by dividing the difference between pre- and post-compound hERG currents by the respective pre-compound currents in the same well. No corrections for liquid junction potentials (estimated as −20 mV by comparing the hERG tail current reversal potential with the calculated Nernst potential for potassium ion) were applied. The current signal was sampled at 0.625 kHz.
Compound potency prediction for hERG inhibition
The 4-parameter logistic model (sigmoid dose response) is the commonly-used model to define IC50 value24,25. The formula of the model, which is derived from Hill equation, can be expressed as
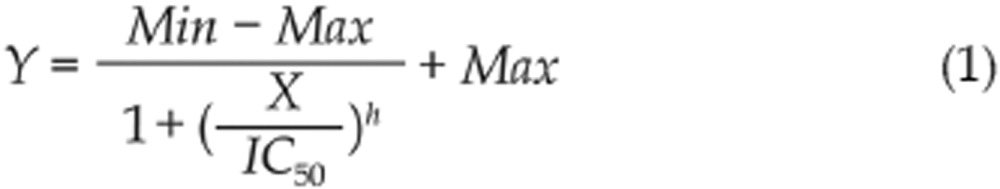 |
where Y is for the percent inhibition, X for the concentration, Min for the minimal percent inhibition, Max for the maximal percent inhibition and h for the Hill coefficient.
Compounds that inhibited hERG by more than the statistical threshold (3 SD) of the vehicle control at 10 μmol/L were considered as inhibitor hits and included for IC50 prediction. 1-point method was used to calculate the IC50 values at 1 μmol/L and 10 μmol/L, respectively, which was determined by the following formula (2) and (2.1), where the Hill coefficient was set to 1.
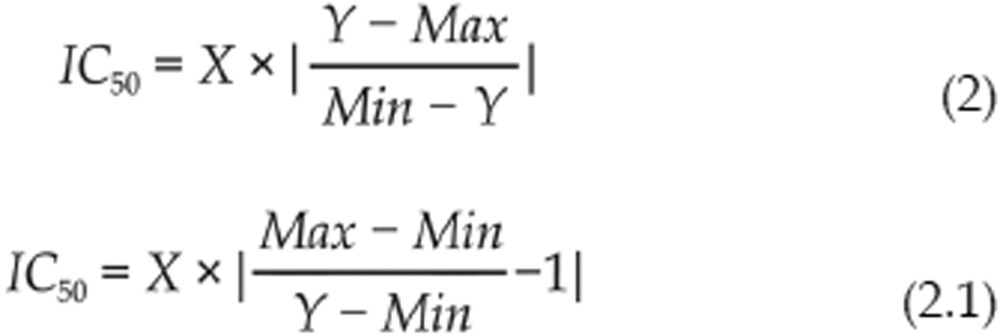 |
For this set of the data, Min is the mean of the 0% response and Max is the mean of 100% response from the control wells, which were used to define the 50% responses for all the compounds on the same plate.
Weighed 2-point method was used to integrate the two IC50 values from the 1-point method (1 μmol/L or 10 μmol/L) in a weighed sum. Adapted from the paper26, the weighed sum can be defined by the formula (3):
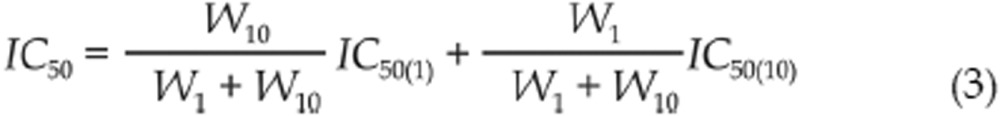 |
Wx (here W1 and W10) is the absolute distance between the compound response at concentration x μmol/L and the 50% response of control wells, expressed as Wx=|(percent inhibition at x μmol/L −middle value between Min and Max)|.
Data acquisition and analysis
Data from automated patch clamp assay were analyzed and exported in IonWorks 2.0.4.4 (Molecular Devices, LLC, Sunnyvale, CA, USA). Further analysis was performed in Matlab (Mathworks, MA, USA) and Excel (Microsoft, CA, USA). Graphic plots were created in Origin 7.0 (OriginLab Corporation, Northampton, MA, USA). To evaluate the quality of the screening, signal-to-noise ratio (S/N) and Z′ factor were calculated as reported27. The physiochemical properties for the MLSMR compounds were acquired and calculated by ACD software (ACD labs)28,29.
Statistical analysis
Statistical significance was determined by paired or unpaired Student t test (2-tail) for 2 groups as indicated (Excel, Microsoft, CA, USA). P<0.05 was considered statistically. Data were represented as mean±SEM.
Results
Assay optimization for hERG high-throughput screening
To develop an HTS assay for hERG using an automated patch clamp platform, IonWorks Quattro, two different hERG cell lines (CHO-K1 and HEK-293 as parental cells) were compared. These cells were tested using voltage protocol modified from a common two-pulse protocol for hERG (Figure 1A). hERG-CHO cell line was selected for HTS due to the relatively higher seal resistance than HEK-293 cells. To optimize cell density to achieve high success rate at lowest cell density, a series of cell densities (0.5–2.0×106 cells/mL) have been tested on IonWorks Quattro. 1.8×106 cells/mL (6300 cells/well) is the lowest cell density that ensures 95%–100% success rate. Typical hERG current traces were shown in the absence and presence of hERG inhibitor Dofetilide at 1 μmol/L. Averaged seal resistance and peak tail current amplitude are 201.70±49.10 Mohms and 0.76±0.29 nA in single hole mode (see Figure 1B and 1D). To predict performance of the cell line on PPC mode, the bootstrap analysis was carried out to simulate the distribution of seal resistance and peak tail current amplitude in the PPC mode (as shown in the grey color filled bars in Figure 1B and 1D) using the data collected from single-hole mode. Since up to 64 cells were tested in a single well on a PPC plate, 64 data points were randomly sampled from a set of 384 data points from a single-hole plate. The procedure was repeated for 384 times. The sampled 64 points would simulate for one well of the PPC mode recording. The simulated seal resistance was 103.70±53.33 Mohms, and peak tail current was 0.75±0.03 nA, which were confirmed in PPC plates with seal resistance of approximately 115.85±25.75 Mohms and current amplitude of 0.79±0.09 nA. Variation of seal resistance and peak tail current were smaller in PPC mode. In addition, success rate using PPC mode is higher. Therefore, PPC mode was applied through the entire screen. From the entire campaign of 1008 recording plates, seal resistance has an average of 89.66±36.26 Mohms, whereas averaged peak tail current amplitude is 0.62±0.17 nA (see Figure 1C and 1E).
Before the screening was started, eight well-characterized hERG inhibitors were validated using the optimized protocol. The IC50 values were consistent with literature reported values as shown in Supplementary Table S1. Through the screening, assay quality was assessed using known hERG inhibitor Dofetilide. The dashed line at +25 mV is the optional protocol used in part of the screening. Using the protocol shown in solid line, the half inhibition concentration (IC50) of tail current for Dofetilide is 10.9 nmol/L, consistent with the reported values30,31, suggesting reliable pharmacology evaluation.
Performance of the hERG high-throughput screening
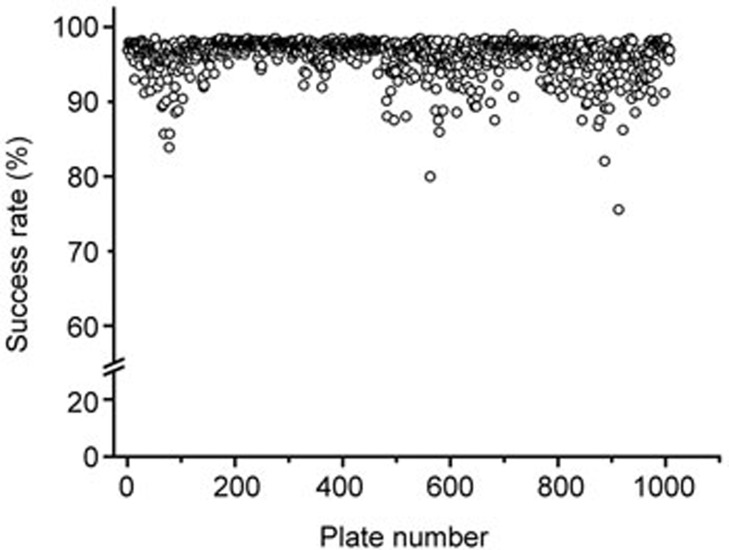
A high throughput screening was completed to profile the inhibitory effects on hERG for the MLSMR compounds. The compound application protocol included a sequential dual addition of 1 μmol/L and 10 μmol/L of testing compounds. The criteria for success were as follows: cells with a peak tail current amplitude bigger than 0.2 nA, a seal resistance more than 30 Mohms, and reduction of seal resistance after compound addition (first addition or second addition) lower than 25% were included for data analysis. The screened MLSMR library included 318 950 compounds and distributed in 1008 384-well compound plates. The averaged plate success rate was 96.08%±2.45% (Figure 2).
Figure 2.
Success rate of the hERG primary screening using automated patch clamp assay. Cells with peak tail current amplitude bigger than 0.2 nA, seal resistance more than 30 MOhms, and percentage reduction of seal resistance (caused by first or second compound addition) lower than 25% were included for data analysis. MLSMR library 318 950 compounds in 1008 384-well compounds plates were screened. The averaged success rate was 96.08%±2.45%.
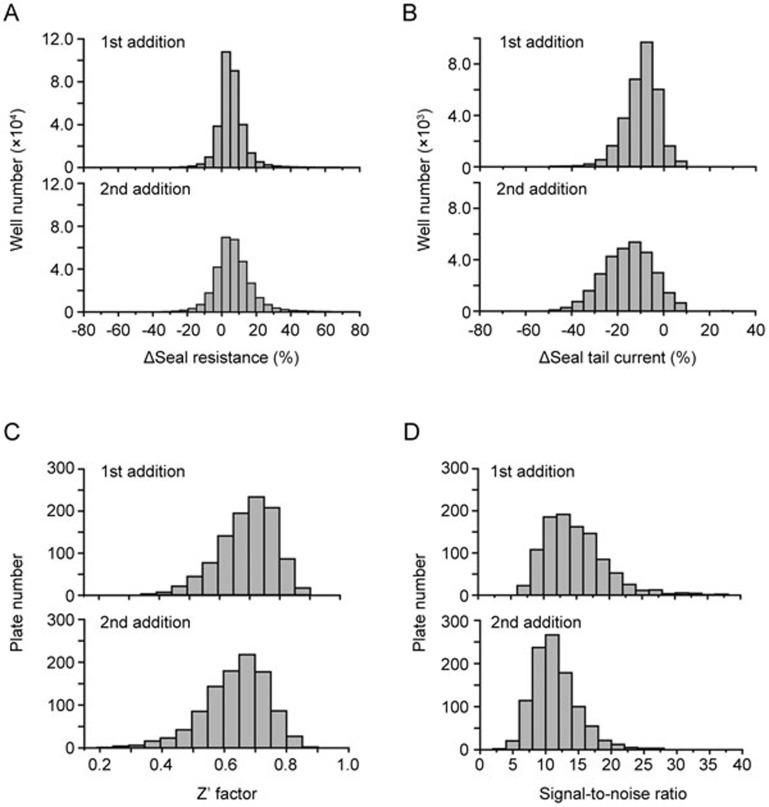
After the dual additions were implemented, the overall seal resistance compared to the pre-compound treatment was increased by 6.63%±9.13% (first addition) and 5.55%±12.56% (second addition), respectively (Figure 3A). To evaluate the current stability of hERG channel during the two additions, the percentage change of peak tail currents was calculated. The peak tail current in buffer control wells was decreased by 8.87%±7.54% (first addition) and 13.99%±11.84% (second addition), respectively (Figure 3B). The averaged Z′ factors were 0.70±0.09 (first addition) and 0.64±0.10 (second addition) (Figure 3C); the averaged signal-to-noise ratios were 14.72±4.60 (first addition) and 11.47±3.45 (second addition) (Figure 3D), suggesting a good assay quality for the high throughput screening.
Figure 3.
Data stability of the dual additions for seal resistance and hERG peak tail currents. The histograms of seal resistance and hERG currents are shown in (A) and (B). Seal resistance change (%) was determined by the formula 100 %*[(Rpost-1-Rpre)/Rpre] (first addition) and 100 %*[(Rpost-2-Rpre)/Rpre] (second addition) from the compound treated wells, which are represented as mean±SD values of 6.63±9.13 (%) and 5.55±12.56 (%), respectively. And the tail current change (%) was calculated by the formula 100%*[(Ipost-1-Ipre)/Ipre] (first addition) and 100%*[(Ipost-2-Ipre)/Ipre] (second addition) from the vehicle treated wells, which are represented as mean±SD values of −8.87%±7.54% and −13.99%±11.84%. The histogram distribution of Z′ factor and signal noise ratio for the first addition are shown in the upper panels of (C) and (D). The data exhibited mean±SD values of 0.70±0.09 (Z′ factor) and 14.72±4.60 (signal-to-noise ratio), respectively. For the second addition, the mean±SD values are 0.64±0.10 (Z′ factor) (lower panel of C) and 11.47±3.45 (signal-to-noise ratio) (lower panel of D), respectively.
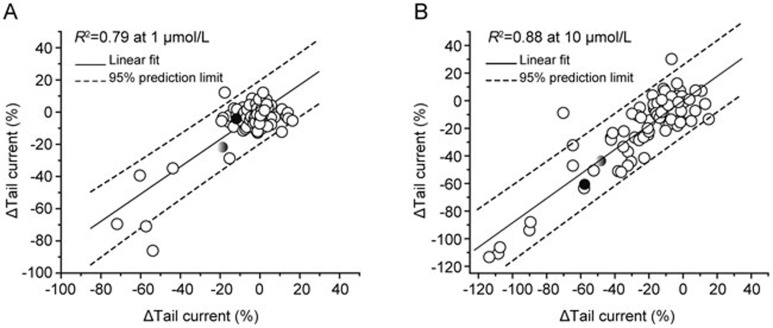
Coincidently, the MLSMR collection contains 85 pairs of compounds with identical chemical structures, including two known hERG inhibitors, Pyrilamine (grey filled circle) and Chlorpheniramine (black filled circle) (Figure 4). They were distributed in different plates and served as internal controls in this screening. The correlation efficient is 0.79 and 0.88 for the hERG activity change at 1 μmol/L (Figure 4A) and 10 μmol/L (Figure 4B), indicating a robust assay performance.
Figure 4.
Correlation between the same compounds but distributed in different plates. Within the MLSMR collection, we acquired data for 85 pairs of identical compounds but distributed in different plates, including two known hERG inhibitors, Pyrilamine (grey color filled circle) and Chlorpheniramine (black color filled circle). For the 85 pair of compounds, the correlation efficient is 0.79 and 0.88 for the peak tail current change at 1 μmol/L (A) and 10 μmol/L (B).
Prediction of hERG inhibitor potency
Concentrations producing 50% decrease in assay signal (IC50 values) are typically used to rank inhibitory compounds. IonWorks Quattro allows up to two additions. A benefit to have two concentrations (1 μmol/L and 10 μmol/L) over single concentration is to reduce false rates and provide data to estimate dose-response effects. For the IC50 prediction, the compound activity from single concentration would cause more negative predictions26, whereas the weighed 2-point method could fix the disadvantage of 1-point prediction. The IC50 values for the current screening were first calculated by 1-point32, and then by weighed 2-point methods26. Compared to the 1-point prediction method, weighed 2-point method helps to neutralize the unbalance from the one-concentration's disadvantage. The potency prediction model has been described in detail in Materials and methods. Using the 1-point and weighed 2-point prediction methods, we estimated IC50 values for the inhibitor hits.
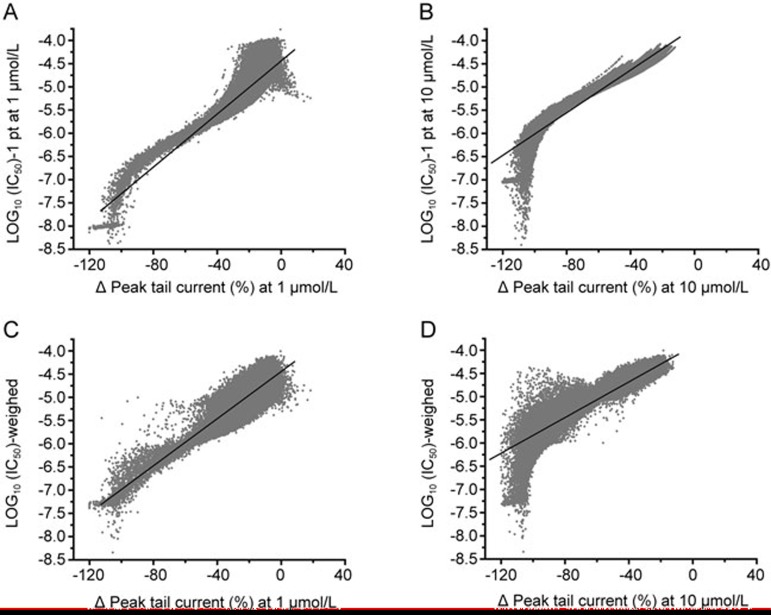
Relationship between percent inhibition and predicted Log10(IC50) (from 1-point and weighed 2-point) was plotted in Figure 5. Linear regression analysis was used to quantitatively assess the mathematical prediction for the IC50 values acquired from 1-point method. The R value is 0.90 and 0.95 for 1 μmol/L (Figure 5A) and 10 μmol/L, respectively (Figure 5B). The increased amount of variability is also observed in the predicted IC50 values for the concentration 1 μmol/L. Further the correlation between the percent inhibition and Log10(IC50) from the weighed IC50 list calculated by weighed method was generated. R value is 0.89 (at 1 μmol/L) (Figure 5C) and 0.91 (at 10 μmol/L) (Figure 5D). Lower R value at 1 μmol/L is because the integrated list includes the compounds that pass the hit selection criteria at 10 μmol/L but not at 1 μmol/L. The bump shown in Figure 5D is caused by the compounds that inhibited hERG at 10 μmol/L but inactive at 1 μmol/L. Therefore IC50 values of the hERG inhibitor hits from this set of data were binned into three classes, a high (IC50<1 μmol/L), an intermediate (1 μmol/L<IC50<10 μmol/L), and a low potential (IC50>10 μmol/L) for hERG channel inhibition26,32,33.
Figure 5.
Linear regression analysis to assess the mathematical prediction for the IC50. The R value is 0.90 and 0.95 respectively for 1 μmol/L (A) and 10 μmol/L (B) for the one-point method. Further the relationship between the percent inhibition and Log10 (IC50) from the weighed method was generated. R value is 0.89 (at 1 μmol/L) (C) and 0.91 (at 10 μmol/L) (D).
Selection and Ranking of hERG inhibitor hits
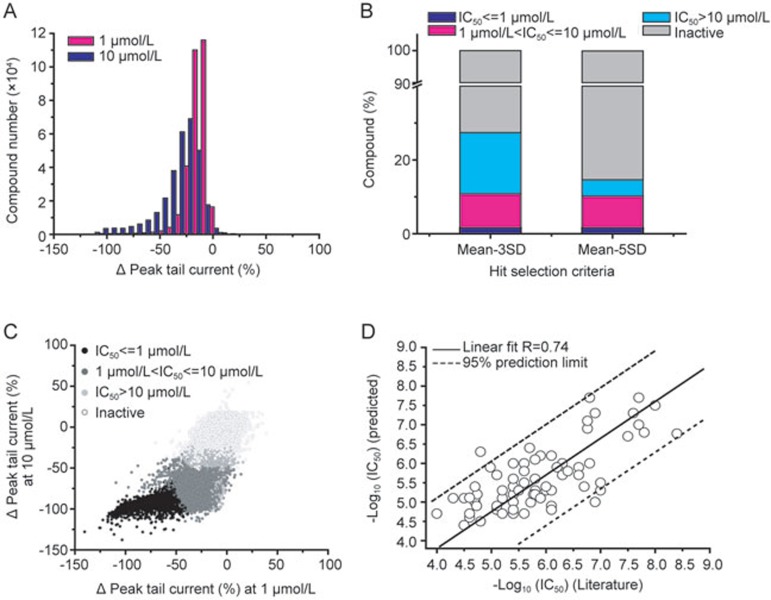
Compound effects on the hERG peak tail current at 1 μmol/L and 10 μmol/L were plotted in histograms. In comparison to 1 μmol/L, the dose-dependent shift of tail current change was observed at 10 μmol/L (Figure 6A). And more compounds displayed higher percentage (>50%) of inhibition at 10 μmol/L than at 1 μmol/L. The scatter plot of tail current change at 1 μmol/L and 10 μmol/L is shown in Figure 6C, which is color-coded by the aforementioned potency classes. As shown in Figure 6C, most of defined hERG inhibitors showed dose-dependent inhibition on hERG channel activity from 1 μmol/L to 10 μmol/L. Based on the potency prediction methods, the tested MLSMR library compounds were classified into 4 classes: IC50<1 μmol/L, 1 μmol/L<IC50<10 μmol/L, IC50>10 μmol/L and inactive (Figure 6B and 6C). If hit rates were determined by the hit selection criteria of 3 SD or 5 SD of vehicle controls, the hit rates at 10 μmol/L were 27.44% and 14.71% for the 3 SD and 5 SD criteria, respectively. If the hits were classified based on potency, using the criterion of “mean of Control-3 SD at 10 μmol/L”, the inhibitor hit rates are 1.64%, 9.17% and 16.63% for IC50<1 μmol/L, 1 μmol/L<IC50<10 μmol/L and IC50>10 μmol/L, respectively; if using the criterion of “mean of Control-5 SD at 10 μmol/L”, the hit rates are 1.64%, 8.63% and 4.44% for the 3 potency ranges, respectively. Clearly, the stringency of the hit selection criteria mainly affects the less potent compounds. Furthermore, the potency-based data provide more information than the single-concentration one.
Figure 6.
Summary of MLSMR compound library on hERG channels. (A) Distribution of compound effects on peak tail current. Black bars are for tail current percentage change at 1 μmol/L and red bars are for 10 μmol/L. (B) Effects of inhibitor select criteria on hit rates. hERG inhibitors were ranked based on their IC50s calculated using weighed 1-point method based on the two hit selection criteria (Mean of Control-3SD or Mean of Control-5SD) at 10 μmol/L. Black box: IC50<=1 μmol/L; grey box: 1 μmol/L<IC50<=10 μmol/L; light grey box: IC50>10 μmol/L; white box: inactive. (C) Correlation of compound effects at 1 μmol/L and 10 μmol/L. The scatter plot of tail current change for each tested compound at 1 μmol/L (horizontal axis) and 10 μmol/L (Vertical axis), is color-coded by the potency classes. Black filled circles indicate inhibitors with IC50<=1 μmol/L; grey filled circles indicate inhibitors with IC50 between 1 μmol/L and 10 μmol/L; light grey filled circles indicate inhibitors with IC50>10 μmol/L with the cutoff at mean-3SD of vehicle control at 10 μmol/L; open circles indicate hERG activators and inactive compounds. (D) Correlation between the predicted and literature reported IC50 for 74 known hERG inhibitors.
74 reported hERG inhibitors were found in the screened MLSMR library. The reported IC50 values of these inhibitors obtained using patch clamp were acquired from literatures. The detailed information was shown in Supplementary Table S2. The 74 compounds were used as internal controls to evaluate the quality of the high-throughput screening and the potency prediction methods. The correlation was described as a plot against the predicted IC50 values by weighed one-point method from the automated electrophysiological primary screening. Linear regression for IC50 values is shown in Figure 6D. The predicted IC50 values correlated well with the reported values with coefficient R=0.74.
Promiscuity of hERG inhibition affected by physiochemical properties
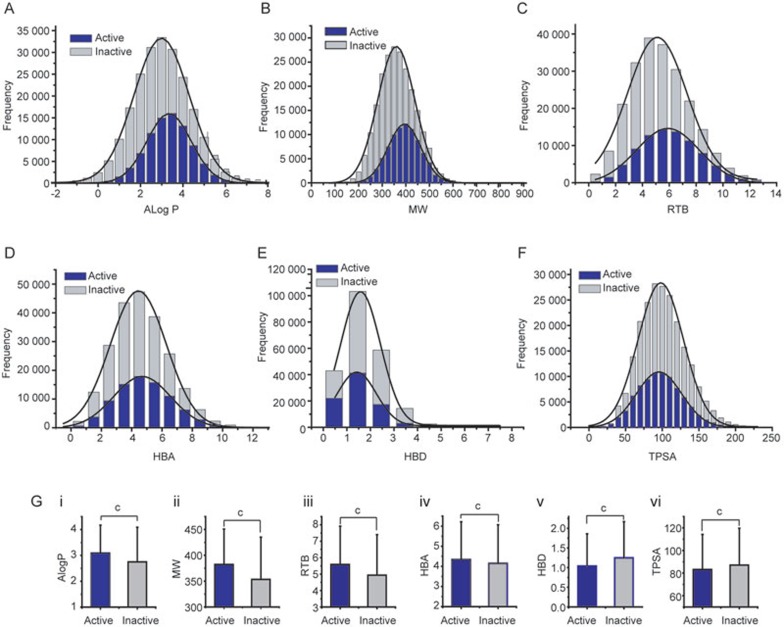
Multiple in silico models have been developed to predict the hERG liability of lead candidates by academic and industrial groups34. Studies implicated that some general structural properties seemed to be linked to the hERG inhibition, including the octanol-water partition coefficient (ALogP), molecular weight (MW), rotatable bond number (RTB), hydrogen bond acceptor number (HBA), hydrogen bond donor number (HBD) and topological polar surface area (TPSA)23. The correlation of the compound physiochemical properties and hERG inhibition were quantitatively analyzed. The distributions of these six parameters for hERG inhibitors (“Active”) and non-inhibitors (“Inactive”) at 10 μmol/L are shown in Figure 7. In comparison to inactive compounds, the distributions of ALogP, MW, RTB of hERG active compounds were clearly right-shifted, while the distributions of HBA, HBD and TPSA were not shifted (Figure 7A-7F). To compare the difference of the mean values of active and inactive classes, student's t-test was used. The P-value is <<0, <<0, <<0, 2.44E−139, <<0, 8.36E−195 for ALogP (Active 3.09±1.08 (mean±SD) and Inactive 2.74±1.34), MW (Active 382.32±68.58 and Inactive 352.76±81.72), RTB (Active 5.59±2.33 and Inactive 4.94±2.46), HBA (Active 4.34±1.88 and Inactive 4.15±1.92), HBD (Active 1.04±0.82 and Inactive 1.25±0.92) and TPSA (Active 83.20±31.00 and Inactive 87.09±32.65), indicating that the distributions of hERG actives and inactives can be significantly distinguished from each other by these parameters (Figure 7G).
Figure 7.
Effect of compound physiochemical properties on hERG inhibitors and non-hERG inhibitors. (A) Octanol-water partition coefficient (ALogP); (B) Molecular weight (MW); (C) Rotatable bond number (RTB); (D) Hydrogen bond acceptor number (HBA); (E) Hydrogen bond donor number (HBD); (F) Topological polar surface area (TPSA); (G) Bar chart for the comparison between hERG active and inactive compounds. cP<0.01, student t-test. “Active” represents hERG inhibitors passing the statistical threshold (mean-3 SD) of vehicle control at 10 μmol/L; “Inactive” is for those compounds not passing the statistical threshold (mean-3 SD) of vehicle control at 10 μmol/L.
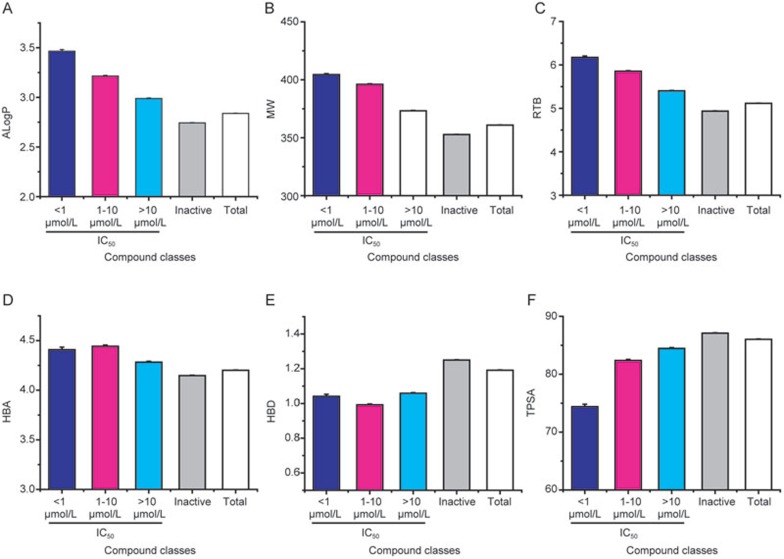
In addition, the most potent hERG inhibitors tend to be positively correlative to ALogP, MW, RTB, and negatively correlative to TPSA as shown in Figure 8. However hydrogen bond parameters (HBA and HBD) didn't show significant difference between hERG inhibitors with differential potency but did between the active and inactive classes (Figure 7G). Furthermore the linear correlation between hERG inhibition at 10 μmol/L and the six parameters were made, a relatively better correlation was generated for ALogP (r=−0.16), MW (−0.22) and RTB (r=−0.13) compared with the other three, HBA (r=−0.05), HBD (r=+0.04) and TPSA (r=+0.06). Our large set of hERG data from HTS primary screening further confirmed and demonstrated that compounds with hERG library tend to be more hydrophobic, high-molecular, flexible and polarizable.
Figure 8.
Compound physiochemical properties affected potency of hERG inhibitors. (A) Octanol-water partition coefficient (ALogP); (B) Molecular weight (MW); (C) Rotatable bond number (RTB); (D) Hydrogen bond acceptor number (HBA); (E) Hydrogen bond donor number (HBD); (F) Topological polar surface area (TPSA). Dark blue bar: IC50<1 μmol/L; pink bar: 1 μmol/L<IC50<10 μmol/L; light blue bar: IC50>10 μmol/L; grey bar: hERG inactive compounds; white bar: hERG inactive compounds.
Discussion
hERG channel has become one of the primary anti-targets for cardiac safety profiling in drug development and compound profiling against hERG has been a mandatory procedure required by FDA because undesired blockade causes potentially serious side effects. There are considerable interests in developing computational models to predict hERG inhibition. However those models have not yet produced satisfactory predictions given limited compounds and differential data quality caused by variations in detection methodologies. Several high-throughput screening approaches have been established for identification of hERG modulators, including ion flux assay (Rb+) and fluorescence-based assay (Tl+), etc8,35,36,37,38. Although these assays could identify hERG modulators effectively and showed better correlation with electrophysiological assays, the interferences from the parental cells may interact with compounds, and thus cause higher false-positive or false-negative hit rate7. In addition, these assays are lack of voltage control of membrane potential and provide less information content compared with voltage-clamping. In contrast, automated patch clamp assay offers the optimal properties suited for high throughput screening of ion channels.
To better understand the promiscuity mechanisms of hERG inhibition by a large collection of structurally diverse compounds, we completed the HTS of the MLSMR compound collection (approximately 300 000 compounds) using automated electrophysiological platform–IonWorks Quattro™. We summarized the assay as follows. First, automated electrophysiological assay directly measures hERG channel currents with controlled membrane potential, in which channels are activated at more physiological conditions avoiding using high concentration of K+. Second, automated patch clamp generates data with good quality at high throughput level. The HTS assays were completed with high success rate within and across plates. Third, automated electrophysiological assay undergoes less interference from the parental cells. Among the random-distributed 74 reported hERG inhibitors within the library, the predicted IC50 values produced by IonWorks Quattro correlate well with the reported IC50 values, suggesting low false-positive and false-negative rate. Finally, the automated electrophysiological assay uses dual controls with the tested cell itself as pre-compound control to evaluate compound effect and vehicle controls to monitor current stability in parallel, thus increasing reliability of data. Run-down issue is often encountered for channel recording in manual patch. However, the current reduction level in the current hERG assay is in the acceptable range with current decrease by 9% (first addition) and 14% (second addition), respectively.
In addition, for the automated electrophysiology-based large scale screening, we used the dual-compound-addition mode. This also increased the challenge for the cells compared to single addition. Cells with robust expression and higher seal resistance are the key for the success of hERG screening. In the campaign, we have learnt that some factors are very important for assay quality of automated electrophysiological screening, including 1) cell lines with high expression of target proteins; 2) good cell culture conditions; 3) optimal cell-dissociation conditions; 4) rapid cell handling process after dissociation; 5) optimal cell seeding density. Those factors can also be applied to other automated electrophysiological platforms. After a series of optimizations based on aforementioned factors, our screening has yielded data with good recording stability, and achieved averaged success rate of 96%. The data consistency among those pairs of identical compounds randomly distributed in the compound library further confirmed good performance of the assay, suggesting the reliable assay.
A promiscuous target exhibits interactions with miscellaneous ligands39. hERG, a representative of promiscuous targets, attracts a wide range of structurally diverse compounds to block the channel due to the relative large inner cavity compared to the other members of Kv channel family36,40,41. In the current screening, hERG channels were inhibited by substantially diverse compounds in the MLSMR collection with a hit rate of 27% at 10 μmol/L when 3-SD criterion was used for hERG inhibitor hit selection. The hit rate is much more than those of other ion channel targets we have screened (usually the hit rates are within the range 0.5%–1.5% for other Kv channels) for the same diverse compound library, consistent with the promiscuity.
The compound library was screened at 1 μmol/L and 10 μmol/L, concentrations commonly used in screenings, allowing assessment of dose-dependent effects. The prediction power based on 1 or 2 concentrations varies with the chosen concentrations and suffers from increasing variability at IC50 values far away from the chosen concentrations. Therefore the weighed method is more accurate for those IC50 values between 1 μmol/L and 10 μmol/L. hERG inhibitors were binned into three potency categories, a high (IC50<1 μmol/L, 1.64%), an intermediate (1 μmol/L<IC50<10 μmol/L, 9.17%), and a low potential (IC50>10 μmol/L, 16.63%). The high hit rate (totally 27%) demonstrated the promiscuous properties of hERG channels. Usually the favorable lead compounds are in the sub-micromolar range of potency or at least less than 10 μmol/L. According to S7B guideline, it would be assumed serious proarrhythmic risk if any candidate drug significantly blocked hERG channel function. Thus almost all the hERG inhibitors (∼27%) would be abandoned in the earlier stage during lead developing stages to avoid the cardiac safety issue. It brought the drug research community a lot of challenge to reduce or avoid the hERG-related toxicity. With more human data available, we have gradually realized that not every hERG liability drug is proarrhythmic, such as verapamil and ranolazine27. They blocked hERG channels, but not associated with Torsade de Pointes (TdP), which may be caused by the potent blockade of L-type calcium channels (verapamil) or sodium channels (ranolazine). S7B strategy has been gradually questioned as an inaccurate surrogate for TdP risk in the last decade. Thus the comprehensive in vitro proarrhythmia assay (CiPA), a combo-cardiac safety assay, has been proposed to evaluate proarrhythmic risk for candidate drugs8. Other six human cardiac ion channels [INa (INaF, INaL), Ito, ICaL, IKs and IK1] present in ventricular myocardium are suggested to join hERG as first CiPA assays8. To comply with this, authors would suggest that the hERG liability compounds should be further tested in other cardiac ion channel panels to predict their potential proarrythmic potential.
Based on the current screening of 300 000 molecules on hERG channels, the six physiochemical properties, commonly used in drug-like profiling, are significantly different between hERG inhibitors and non-hERG inhibitors. These properties are able to significantly separate hERG actives from inactives. As three of these properties increase, including compound lipophilicity (manifested as ALogP), molecular weight (MW), and rotatable bond number (RTB), the potency of hERG inhibition increases, whereas topological polar surface area (TPSA) increase, the potency of hERG inhibition decreases. The findings from the current study are in line with those computational models34,40,42. Among these parameters, HBA and HBD values of hERG inhibitors were significantly different from those of non-inhibitors. However there was no significant difference between hERG inhibitors with differential potency. Even though they showed poorer correlation with hERG activity, the linear relationship indicated that the properties affected the hERG activity of the compounds. The poorer the correlation is, the weaker the capability of discriminating hERG inhibitors would be. The rank order of these properties relating to hERG inhibition was ALogP>MW>RTB>TPSA>HBA>HBD. Among these parameters, lipophilicity (ALogP), flexibility (RTB), topological polar surface area (TPSA) have been successfully used to predict hERG blockage in some prediction models as reviewed in the paper by Raschi et al34. HBD and HBA were usually used for prediction combined with other parameters including lipophilicity. Therefore based on the analysis, an integrated prediction model may be considered for lead compound optimization to avoid the off-target effect on hERG channels.
In summary, we first reported a high throughput screening on hERG channels of a large-scale diverse compound library using automated electrophysiology. The screening was completed with good quality. Furthermore an improved potency prediction method was developed based on the dual concentrations of data and the inhibitor hits were binned into differential potency ranges. The high hit rate (∼27%) demonstrated the promiscuity property of hERG channels by a diverse range of compound structures. The data would provide the information for other users of the MLSMR compound collection to triage compounds in the early stage20. Analysis of hERG data further demonstrated that hERG liability compounds tend to be more hydrophobic, high-molecular, flexible and polarizable. However, none of current available models integrates those properties to predict the unfavorable hERG blockage. The study thus suggests a possibility that the combination of more physiochemical properties would render higher confidence for prediction of hERG inhibition. In the meanwhile, we should realize that hERG liability compounds are carrying the potential to be proarrhythmic but not necessarily with TdP risk.
Author contribution
Hai-bo YU, Bei-yan ZOU and Min LI designed research; Hai-bo YU, Bei-yan ZOU performed research, analyzed data and wrote the paper; Xiao-liang WANG and Min LI contributed new reagents or analytic tools.
Acknowledgments
This work was supported by the grant from the National Institutes of Health of USA (U54 MH084691) (to Min LI), and National Natural Science Foundation of China (Grant 81470163 and 81503056) (to Hai-bo YU).
Footnotes
Supplementary Tables are available at Acta Pharmacologica Sinica's website.
Supplementary Information
The well-characterized hERG inhibitors were validated by automated patch clamp-IonWorks Quattro™.
Reported hERG inhibitors were validated by automated patch clamp-IonWorks Quattro™
References
- 1Fermini B, Fossa AA. The impact of drug-induced QT interval prolongation on drug discovery and development. Nat Rev Drug Discov 2003; 2: 439–47. [DOI] [PubMed] [Google Scholar]
- 2Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: hERG encodes the IKr potassium channel. Cell 1995; 81: 299–307. [DOI] [PubMed] [Google Scholar]
- 3Zhang KP, Yang BF, Li BX. Translational toxicology and rescue strategies of the hERG channel dysfunction: biochemical and molecular mechanistic aspects. Acta Pharmacol Sin 2014; 35: 1473–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Babcock JJ, Li M. hERG channel function: beyond long QT. Acta Pharmacol Sin 2013; 34: 329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5ICH S7B. Guideline on safety pharmacology studies for assessing the potential for delayed ventricular repolarization (QT interval prolongation) by human pharmaceuticals 2005. [PubMed]
- 6Witchel HJ, Milnes JT, Mitcheson JS, Hancox JC. Troubleshooting problems with in vitro screening of drugs for QT interval prolongation using HERG K+ channels expressed in mammalian cell lines and Xenopus oocytes. J Pharmacol Toxicol Methods 2002; 48: 65–80. [DOI] [PubMed] [Google Scholar]
- 7Zou B, Yu H, Babcock JJ, Chanda P, Bader JS, McManus OB, et al. Profiling diverse compounds by flux- and electrophysiology-based primary screens for inhibition of human Ether-a-go-go related gene potassium channels. Assay Drug Dev Technol 2010; 8: 743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Schmalhofer WA, Swensen AM, Thomas BS, Felix JP, Haedo RJ, Solly K, et al. A pharmacologically validated, high-capacity, functional thallium flux assay for the human Ether-a-go-go related gene potassium channel. Assay Drug Dev Technol 2010; 8: 714–26. [DOI] [PubMed] [Google Scholar]
- 9Mattmann ME, Yu H, Lin Z, Xu K, Huang X, Long S, et al. Identification of (R)-N-(4-(4-methoxyphenyl)thiazol-2-yl)-1-tosylpiperidine-2-carboxamide, ML277, as a novel, potent and selective Kv7.1 (KCNQ1) potassium channel activator. Bioorg Med Chem Lett 2012; 22: 5936–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Cheung YY, Yu H, Xu K, Zou B, Wu M, McManus OB, et al. Discovery of a series of 2-phenyl-N-(2-(pyrrolidin-1-yl)phenyl)acetamides as novel molecular switches that modulate modes of Kv7.2 (KCNQ2) channel pharmacology: identification of (S)-2-phenyl-N-(2-(pyrrolidin-1-yl)phenyl)butanamide (ML252) as a potent, brain penetrant Kv7.2 channel inhibitor. J Med Chem 2012; 55: 6975–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Yu H, Wu M, Townsend SD, Zou B, Long S, Daniels JS, et al. Discovery, synthesis, and structure activity relationship of a series of N-Aryl- bicyclo[2.2.1]heptane-2-carboxamides: Characterization of ML213 as a novel KCNQ2 and KCNQ4 potassium channel opener. ACS Chem Neurosci 2011; 2: 572–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Song M, Clark M. Development and evaluation of an in silico model for hERG binding. J Chem Inf Model 2006; 46: 392–400. [DOI] [PubMed] [Google Scholar]
- 13Doddareddy MR, Klaasse EC, Shagufta, Ijzerman AP, Bender A. Prospective validation of a comprehensive in silico hERG model and its applications to commercial compound and drug databases. ChemMedChem 2010; 5: 716–29. [DOI] [PubMed] [Google Scholar]
- 14Buturak B, Durdagi S, Noskov SY, Ildeniz AT. Designing of multi-targeted molecules using combination of molecular screening and in silico drug cardiotoxicity prediction approaches. J Mol Graph Model 2014; 50: 16–34. [DOI] [PubMed] [Google Scholar]
- 15Bidault Y. A flexible approach for optimising in silico ADME/Tox characterisation of lead candidates. Expert Opin Drug Metab Toxicol 2006; 2: 157–68. [DOI] [PubMed] [Google Scholar]
- 16Beresford AP, Segall M, Tarbit MH. In silico prediction of ADME properties: are we making progress? Curr Opin Drug Discov Devel 2004; 7: 36–42. [PubMed] [Google Scholar]
- 17Beattie KA, Luscombe C, Williams G, Munoz-Muriedas J, Gavaghan DJ, Cui Y, et al. Evaluation of an in silico cardiac safety assay: using ion channel screening data to predict QT interval changes in the rabbit ventricular wedge. J Pharmacol Toxicol Methods 2013; 68: 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Liu LL, Lu J, Lu Y, Zheng MY, Luo XM, Zhu WL, et al. Novel Bayesian classification models for predicting compounds blocking hERG potassium channels. Acta Pharmacol Sin 2014; 35: 1093–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Schreiber SL, Kotz JD, Li M, Aube J, Austin CP, Reed JC, et al. Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell 2015; 161: 1252–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Du F, Yu H, Zou B, Babcock J, Long S, Li M. hERGCentral: a large database to store, retrieve, and analyze compound-human Ether-a-go-go related gene channel interactions to facilitate cardiotoxicity assessment in drug development. Assay Drug Dev Technol 2011; 9: 580–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21https://pubchem.ncbi.nlm.nih.gov/bioassay/493156.
- 22Du F, Babcock JJ, Yu H, Zou B, Li M. Global analysis reveals families of chemical motifs enriched for hERG inhibitors. PLoS One 2015; 10: e0118324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Wang S, Li Y, Wang J, Chen L, Zhang L, Yu H, et al. ADMET evaluation in drug discovery. 12. Development of binary classification models for prediction of hERG potassium channel blockage. Mol Pharm 2012; 9: 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Weiss JN. The Hill equation revisited: uses and misuses. FASEB J 1997; 11: 835–41. [PubMed] [Google Scholar]
- 25Gadagkar SR, Call GB. Computational tools for fitting the Hill equation to dose-response curves. J Pharmacol Toxicol Methods 2015; 71: 68–76. [DOI] [PubMed] [Google Scholar]
- 26Krippendorff BF, Lienau P, Reichel A, Huisinga W. Optimizing classification of drug-drug interaction potential for CYP450 isoenzyme inhibition assays in early drug discovery. J Biomol Screen 2007; 12: 92–9. [DOI] [PubMed] [Google Scholar]
- 27Bowlby MR, Peri R, Zhang H, Dunlop J. hERG (KCNH2 or Kv11.1) K+ channels: screening for cardiac arrhythmia risk. Curr Drug Metab 2008; 9: 965–70. [DOI] [PubMed] [Google Scholar]
- 28Hernandez-Covarrubias C, Vilchis-Reyes MA, Yepez-Mulia L, Sanchez-Diaz R, Navarrete-Vazquez G, Hernandez-Campos A, et al. Exploring the interplay of physicochemical properties, membrane permeability and giardicidal activity of some benzimidazole derivatives. Eur J Med Chem 2012; 52: 193–204. [DOI] [PubMed] [Google Scholar]
- 29Manchester J, Walkup G, Rivin O, You Z. Evaluation of pKa estimation methods on 211 druglike compounds. J Chem Inf Model 2010; 50: 565–71. [DOI] [PubMed] [Google Scholar]
- 30Mo ZL, Faxel T, Yang YS, Gallavan R, Messing D, Bahinski A. Effect of compound plate composition on measurement of hERG current IC50 using PatchXpress. J Pharmacol Toxicol Methods 2009; 60: 39–44. [DOI] [PubMed] [Google Scholar]
- 31Pearlstein RA, Vaz RJ, Kang J, Chen XL, Preobrazhenskaya M, Shchekotikhin AE, et al. Characterization of hERG potassium channel inhibition using CoMSiA 3D QSAR and homology modeling approaches. Bioorg Med Chem Lett 2003; 13: 1829–35. [DOI] [PubMed] [Google Scholar]
- 32Gao F, Johnson DL, Ekins S, Janiszewski J, Kelly KG, Meyer RD, et al. Optimizing higher throughput methods to assess drug-drug interactions for CYP1A2, CYP2C9, CYP2C19, CYP2D6, rCYP2D6, and CYP3A4 in vitro using a single point IC50. J Biomol Screen 2002; 7: 373–82. [DOI] [PubMed] [Google Scholar]
- 33Sebaugh JL. Guidelines for accurate EC50/IC50 estimation. Pharm Stat 2011; 10: 128–34. [DOI] [PubMed] [Google Scholar]
- 34Raschi E, Ceccarini L, De Ponti F, Recanatini M. hERG-related drug toxicity and models for predicting hERG liability and QT prolongation. Expert Opin Drug Metab Toxicol 2009; 5: 1005–21. [DOI] [PubMed] [Google Scholar]
- 35Titus SA, Beacham D, Shahane SA, Southall N, Xia M, Huang R, et al. A new homogeneous high-throughput screening assay for profiling compound activity on the human ether-a-go-go-related gene channel. Anal Biochem 2009; 394: 30–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Ding M, Stjernborg L, Albertson N. Application of cryopreserved cells to hERG screening using a non-radioactive Rb+ efflux assay. Assay Drug Dev Technol 2006; 4: 83–8. [DOI] [PubMed] [Google Scholar]
- 37Cheng CS, Alderman D, Kwash J, Dessaint J, Patel R, Lescoe MK, et al. A high-throughput HERG potassium channel function assay: an old assay with a new look. Drug Dev Ind Pharm 2002; 28: 177–91. [DOI] [PubMed] [Google Scholar]
- 38Rezazadeh S, Hesketh JC, Fedida D. Rb+ flux through hERG channels affects the potency of channel blocking drugs: correlation with data obtained using a high-throughput Rb+ efflux assay. J Biomol Screen 2004; 9: 588–97. [DOI] [PubMed] [Google Scholar]
- 39Tang W, Kang J, Wu X, Rampe D, Wang L, Shen H, et al. Development and evaluation of high throughput functional assay methods for hERG potassium channel. J Biomol Screen 2001; 6: 325–31. [DOI] [PubMed] [Google Scholar]
- 40Bridgland-Taylor MH, Hargreaves AC, Easter A, Orme A, Henthorn DC, Ding M, et al. Optimisation and validation of a medium-throughput electrophysiology-based hERG assay using IonWorks HT. J Pharmacol Toxicol Methods 2006; 54: 189–99. [DOI] [PubMed] [Google Scholar]
- 41Murphy SM, Palmer M, Poole MF, Padegimas L, Hunady K, Danzig J, et al. Evaluation of functional and binding assays in cells expressing either recombinant or endogenous hERG channel. J Pharmacol Toxicol Methods 2006; 54: 42–55. [DOI] [PubMed] [Google Scholar]
- 42Aptula AO, Cronin MT. Prediction of hERG K+ blocking potency: application of structural knowledge. SAR QSAR Environ Res 2004; 15: 399–411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The well-characterized hERG inhibitors were validated by automated patch clamp-IonWorks Quattro™.
Reported hERG inhibitors were validated by automated patch clamp-IonWorks Quattro™