Abstract
miRNAs are endogenous small RNA (sRNA) that play critical roles in plant development processes. Canna is an ornamental plant belonging to family Cannaceae. Here, we report for the first time the identification and differential expression of miRNAs in two contrasting flower color cultivars of Canna, Tropical sunrise and Red president. A total of 313 known miRNAs belonging to 78 miRNA families were identified from both the cultivars. Thirty one miRNAs (17 miRNA families) were specific to Tropical sunrise and 43 miRNAs (10 miRNA families) were specific to Red president. Thirty two and 18 putative new miRNAs were identified from Tropical sunrise and Red president, respectively. One hundred and nine miRNAs were differentially expressed in the two cultivars targeting 1343 genes. Among these, 16 miRNAs families targeting60 genes were involved in flower development related traits and five miRNA families targeting five genes were involved in phenyl propanoid and pigment metabolic processes. We further validated the expression analysis of a few miRNA and their target genes by qRT-PCR. Transcription factors were the major miRNA targets identified. Target validation of a few randomly selected miRNAs by RLM-RACE was performed but was successful with only miR162. These findings will help in understanding flower development processes, particularly the color development in Canna.
Introduction
The recent advancement in sequencing technologies has led to the discovery of a wide range of sRNAs from diverse group of organisms. This in turn has helped in understanding the diverse roles of sRNAs in gene regulation during growth and development of an organism. MicroRNAs (miRNAs) are a group of sRNAs, 21–24 nt in length, single stranded and non-coding in nature that are produced from RNA Polymerase II transcripts [1]. It is well established that miRNAs play a critical roles in post-transcriptional gene regulation and control many genes involved in various biological and metabolic processes [2, 3]. There are extensive studies to discover miRNAs and analyze their functions in model plant species, such as Arabidopsis and Rice [4]. With the advancement in next generation sequencing technologies, it has become possible to identify species-specific or lowly expressed miRNAs in non-model plants as well [5, 6].
Among other developmental processes, flower development is an important aspect in ornamental plants which largely determines the commercial value of the plants. miRNAs function throughout the flower development processes starting from the earliest floral induction to very late stage like floral organ development, floral organ polarity and defining floral boundaries etc.[7–17]. Some miRNAs regulate members of the florigen and integrator genes involved in flowering, and thus participate in complex genetic networks at floral transition phase [14]. Other miRNAs target and restrict the action of various genes that control different flower-related processes. Several miRNA families are evolutionarily conserved across species. There are at least nine such miRNA families which play critical roles in flower development. These include miR156, miR159, miR160, miR164, miR166/165, miR167, miR169, miR172, and miR319 [14]. These miRNAs mostly regulate flower development by targeting various transcription factors. For example, miR172 regulates floral organ identity and flowering time by translational repression or target cleavage of members of the APETELA2 (AP2) transcription factor genes [18–23]. miR159 is required for normal anther development which it controls through regulating the expression of genes that encode MYB transcription factors [7, 24]. miR156 targets squamosa promoter binding protein-like (SPL) transcription factor gene family to control the transition from the vegetative phase to the floral phase in Arabidopsis, rice, and maize [10, 25, 26].
Amongst other floral characteristics, color is an important trait for ornamental flowers. Flower color is mainly due to three classes of pigment: flavonoids, carotenoids and betalains [27]. Among them, a colored class of flavonoids, anthocyanin, confers a diverse range of color from orange to red and violet to blue. Carotenoids and betalains generally yield yellow color. The final color of a flower is determined by a combination of various factors: anthocyanin structures, the pH of the vacuole where anthocyanin localize, coexisting flavonoids (co-pigments) and metal ions etc.[27]. The flavonoids biosynthetic pathway genes including chalcone synthase, transcription factors regulating these genes and P450s relevant to flower color have also been well studied [28]. Besides others, SPL and R2R3-MYB transcription factors negatively regulate flavonoid biosynthesis [28, 29]. More recently the role of miRNAs in regulating these genes have been reported [5, 28]. Kim et al. (2012) in analyzing miRNAs from different rose cultivars observed enrichment of five miRNAs (miR171, miR166i, miR159e, miR845, and miR396e) in white cultivar of rose [5]. They suggested that these miRNAs may negatively regulate target genes to prevent accumulation of carotenoids or flavonoids resulting in white flowers. However, they could not validate any target genes and thus were not able to confirm the miRNA-directed cleavage of target genes involved in color determination in rose. But they validated miR156 and miR159 as the target of SPL and R2R3-MYB transcription factors, respectively [5]. Similarly, sequencing and degradome analysis of Apple miRNAs revealed large number of R2R3-MYB transcription factors primarily involved in anthocyanin biosynthesis were targets of miR858 [30]. It was further shown that a majority of those MYBs were co-targets of miR828 [31, 32]. These results indicate miRNAs play critical roles in regulation of color development in plant tissues.
However, in spite of rapid advancement in next generation sequencing technologies and our understanding on molecular mechanisms of flower development regulated by miRNAs in model as well as non-model plant species [5, 6], there are only a few studies on miRNAs from ornamental plants [5]. One of the reason may be due to the non availability of genomic resources from these group of plants. Because, it is necessary to predict and construct hairpin precursors of potential miRNAs by using neighbouring genomic sequences of the mapped sRNAs to distinguish high quality miRNAs from other sRNAs [33, 34]. Therefore, miRNA identification and deciphering their probable roles in flower and color development in ornamental flowers will greatly enhance our knowledge in the field.
Canna is a perennial flowering ornamental plant. It is reported to be originated in central and South America. There are 8–10 species of Canna which are widely used as ornamental plant [35]. It grows abundantly in the humid tropical and subtropical regions throughout the world. The flowers are born singly or in pairs and arranged into larger branched clusters at the tips of the flowering stems. Each flower appears to have five petals but these are actually modified stamens (staminodes) [36]. Since its first hybridization in 19th century, a large number of hybrids have evolved by crossing of different Canna species. These hybrids often grouped under the names of C. x generalis L.H. Bailey andC. x orchioides L.H. Baley [35]. There are over 1000 known cultivars ranging from less than 0.75 to 2.4 meter in height and variable in colors from creams, yellow, orange and red.
In this study, we report for the first time deep sequencing of miRNA from Canna cultivars, the only member of the family Cannaceae belonging to the order Zingiberales. To the best of our knowledge, except Musa acuminata no other plant genome sequence is known from this taxonomic order. Therefore, identification of miRNAs from this plant species is challenging. Our aim was to identify the conserved miRNAs and their differential expression in two contrasting flower color cultivars of Canna which might play important role in flower and color development. We also predicted a few novel miRNAs from both the cultivars. Overall,we identified 313 known and 50 putative novel miRNAs from both the cultivars. One hundred and nine miRNAs were differentially expressed between the two cultivars. The differentially expressed and the putative novel miRNAs may provide insight into the molecular mechanisms of flower color as well as other development processes in Canna.
Materials and Methods
Plant Samples
We have chosen two contrasting flower color Canna cultivars, the Tropical sunrise (TS) and Red president (RP) for deep sequencing of miRNA. The TS Canna have pale yellow flower with large glossy pointed dark green leaves, whereas the RP have large red flowers and lush green leaves “Fig 1”. The cultivars are maintained at the Institute's research field for various other research activities. Randomly selected first fully opened flowers from each of the two cultivars were collected from the field grown plants during mid of July, 2013. The flower tissues were immediately frozen in liquid nitrogen and brought to the laboratory for RNA isolation. Fresh tissues were also collected for phytochemical estimation.
Fig 1. Two Canna cultivars having contrasting flower color (A) Tropical sunrise (B) Red president.
Quantification of Flavonoids and Carotenoids
Total flavonoids, carotenoids (carotene and xanthophylls) and anthocyanins were extracted from the staminodes of the two Canna cultivars. One gram of tissue was taken as starting material for extraction of the phytochemicals. Total flavonoids was extracted using AlCl3 method [37] and absorbance was measured at 440 nm using spectrophotometer (SpectraMax Plus 384, USA). The anthocyanins were extracted from staminodes in acidic methanol (1% HCl, w/v) for 48 hrs in dark with a ratio of 10 ml buffer to 1 gm sample following Mancinelli et al (1934) [38]. The absorbance of anthocyanin was measured at 530 nm. Carotenoids were extracted following the method of Chen and Yang (1992) [39]. The absorbance of carotene and xanthophylls was measured at 436 nm and 474 nm, respectively. All the absorbance measurements were taken with five technical and three biological replicates for each of the two cultivars.
sRNA libraries preparation and sequencing
Total RNA was extracted from staminodes tissue using Trizole-LS reagent (Invitrogen, USA) according to the manufacturer’s instructions. The integrity of RNA was checked by running on Agarose gel as well as using Agilent Bioanalyzer (Agilent Technologies, U.S.A). The RNA was run on 15% Tris-Borate-EDTA (TBE) urea denturating polyacrylamide gel and the 20 to 30 nt small RNA fraction was extracted and eluted. Next, the sRNA molecules were ligated to 5' adaptor and a 3' adaptor sequentially and then converted to cDNA by RT-PCR. A single cDNA library was prepared for each cultivar. The resulting libraries were sequenced at Center for Cellular and Molecular Platform (C-CAMP, Bangalore, India) using Illumina Hiseq platform. The raw data of both the libraries has been submitted to NCBI under accession no SRA310114.
Analysis of sRNAs
The raw reads from the two libraries were first cleaned by removing 5' and 3' adaptors and low quality reads (< 30 Q values)by using FASTAX-Toolkit. These clean reads were subjected to BLASTn search against Sanger RNA database (Rfam) (http://www.sanger.ac.uk/software/Rfam). The reads matching with other RNAs, including rRNA, tRNA, snRNA and snoRNA were excluded and remaining reads were designated as filtered reads. The filtered reads were submitted to the UEA sRNA toolkit-Plants version, miRProf pipeline (http://srna-workbench.cmp.uea.ac.uk/tools/analysis-tools/mirprof/) [40] at default parameters which aligned the sequences to miRBase 19.0 (http://www.miRBase.org) [41] and provided raw and normalized read counts of known miRNAs.
Since no other genomic information is available on Canna, we used Musa acuminata, taxonomically the nearest taxa to Canna with known genome sequence to predict miRNA precursors. Secondary structure was predicted using miRCat (http://srna-workbench.cmp.uea.ac.uk/tools/analysis-tools/mircat/) [40] at default parameters. The predicted secondary structure were analyzed manually by using following parameters 1) The miRNA and miRNA* are derived from opposite stem arms such that they form a duplex with two nucleotide 3' overhangs, 2) base-pairing between the miRNA and miRNA* is extensive such that there are typically four or fewer mismatches, 3) The frequency of asymmetric bulges is one or none and size of the bulges is no more than two nucleotide within miRNA/miRNA* duplex [42]. These miRNAs were further submitted to miRProf pipeline. The putative miRNAs that were mapped on previously reported miRNAs were designated as known and unmapped as putative novel miRNAs. The detailed pipeline of deep sequencing data analysis is shown in “S1 Fig”.
Determination of differentially expressed miRNAs
All the read counts were normalized to reads per million (RPM). If original miRNA expression in a library was zero, the normalized expression was adjusted to 0.001 according to a previous report [43]. Log2 fold change in miRNA expression were calculated between the two libraries only when read counts were more than 10 in any one of the libraries. A 2x2 contingency table was used to perform Pearson’s chi-square test of significance between the two libraries [44]. We considered expression to be significant when fold change was ≥ 1 and P value was ≤ 0.05 for a particular comparison.
Validation of known and putative novel miRNAs and their target genes by qRT-PCR
The expression level of known miRNAs were checked by using poly (A) method [45]. Five hundred nanograms of total RNA were poly (A) tailed and reverse transcribed using NCode miRNA first-strand cDNA synthesis kit (Invitrogen, USA) according to user’s manual. For novel miRNAs, cDNAs were synthesised from stem loop RT-PCR [46] by using SuperScript® III Reverse Transcriptase (Invitrogen, USA) for each of the four miRNAs following manufacturer's instructions. For the miRNA target identification, cDNAs were prepared by using GoScript™ Reverse Transcription System (Promega, USA). To check the expression level of the known and the novel miRNAs, the synthesized cDNAs were amplified using the miRNA specific forward primers and universal reverse primers [47]. To check the expression level of the target genes of the known miRNAs, cDNAs were amplified using gene specific primers. The primer sequences are listed in “S1 Table”. qRT-PCR was performed with DyNAmo Flash SYBR Green (Thermo) with cycling conditions: denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 20 s, annealing and extension together at 60°C for 60s. The amplification reaction was performed using ABI7300 real-time PCR system (Applied Biosystem). All the reactions were performed with two biological and three technical replicates. 5.8S and 18S rRNA were taken as an endogenous control for miRNAs and their target genes, respectively. The expression data of samples were normalized by using expression value of 5.8S and 18S rRNA and relative expression was calculated using 2−ΔΔCt method. The fold change values were determined using 2−ΔΔCt method [48].
Prediction of miRNA targets and GO analysis
The Musa acuminata transcripts were downloaded from the database (http://banana-genome.cirad.fr/content/download-dh-pahang) for target prediction. The psRNA target program (http://plantgrn.noble.org/psRNATarget/) was used for target prediction at default parameters. The GO terms of the miRNA targets were annotated according to their biological role, molecular function and cellular component by using the online GO term analysis tool (http://bioinfo.cau.edu.cn/agriGO/analysis.php).
Target validation by modified 5' RLM-RACE
To validate putative targets of a few selected miRNAs viz. miR156, miR159 and miR162 RLM 5′-RACE was carried out using First Choice RLM-RACE kit (Ambion,USA) following user's manual. Briefly, total RNA was isolated from flower tissue and adapter was ligated. The ligated RNA was used for cDNA synthesis using random hexamer. The PCR amplification was performed using the adapter specific outer primer and gene specific outer primers. Nested PCR amplification were performed using the adapter specific inner primer and gene specific inner primers “S1 Table”. The PCR product was separated on 2% agarose gel and distinct bands of the appropriate size of the miRNA target genes were eluted. The eluted PCR product was cloned in pGEM-T easy vector (Promega,USA) and at least ten individual clones were sequenced for each target.
Results
Flavonoids and Carotenoids contents of TS and RP flower tissues
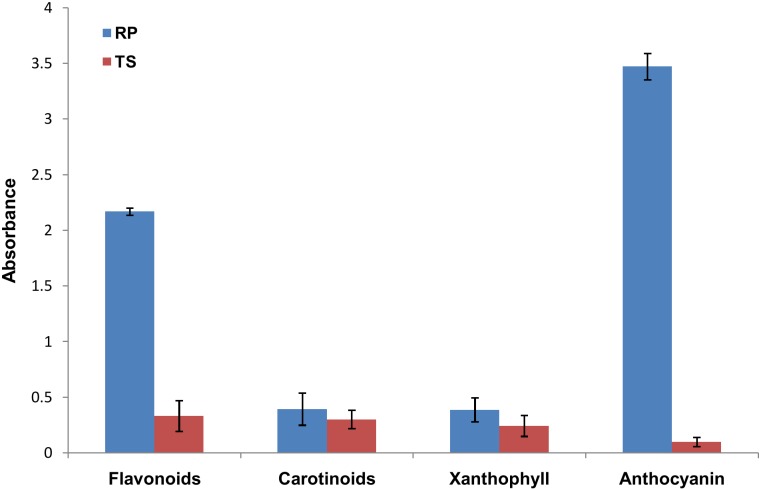
We estimated total flavonoids as well as anthocyanin and carotenoids from the flower tissue of the two cultivars. As shown in the “Fig 2”, the total flavonoids and anthocyanin content was much higher in RP than TS. On the other hand carotenoids and xanthophyll content of RP was very less as compared to the total flavonoids and anthocyanin. Carotenoids and xanthophyll content was less in TS than RP. These indicate flavonoids and anthocyanin are the major source of color pigment in Canna.
Fig 2. Flavonoids, Carotenoids, Xanthophyll and Anthocyanin content of TS and RP.
All the measurements were taken in three biological and five technical replicates. Error bars represent standard deviation of biological and technical replicates.
Sequencing of sRNA from Canna flower
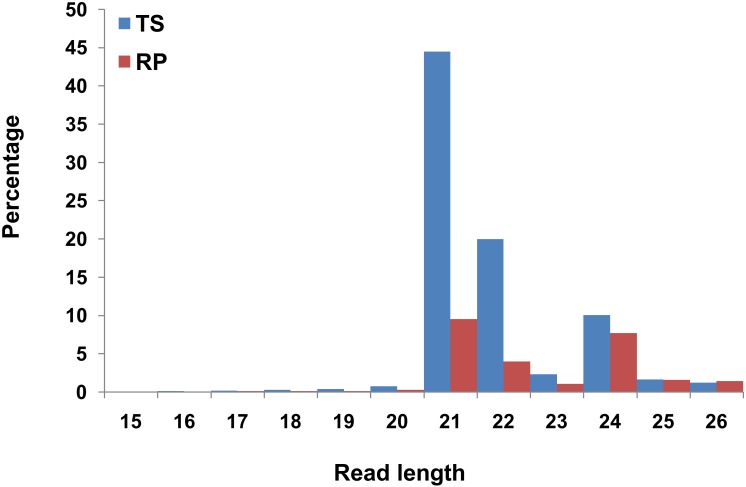
Two sRNA libraries from staminode tissuesof TS and RP were constructed. Sequencing of these sRNA libraries generated ~28.0 million and ~ 18.7 million absolute reads from TS and RP, respectively. The raw reads were processed to remove the adaptor sequences, low quality sequences and other small RNAs including tRNA, rRNA, snRNA and snoRNAs. Other small RNAs were filtered out by BLASTn against Rfam database. This resulted approximately 21.3 million and 10.89 million filtered reads from TS and RP, respectively “Table 1”. The size distribution of the raw reads is depicted in “Fig 3”. The majority of these RNA reads were of 21, 22 and 24 nt in length, whose production relied on DCL4, DCL2 and DCL3, respectively [49, 50]. Most abundant reads were of 21 nt length, the characteristic of canonical miRNAs. Filtered reads were mapped with miRBase19.0. 67.40% of the filtered reads of TS and 28.24% filtered reads of RP mapped with the miRBase 19.0 sequences “Table 1”. To identify the precursors of the known and the novel miRNAs, the filtered reads were further mapped to Musa acuminata genome (http://banana-genome.cirad.fr). This resulted in mapping of 42.9% reads of TS and 56.9% reads of RP to Musa acuminata genome "Table 1".
Table 1. Categorization and abundance of sRNA reads from TS and RP.
| absolute reads | unique reads | |||
|---|---|---|---|---|
| TS | RP | TS | RP | |
| Data Processing | ||||
| Total sequencing reads | 28040652 | 18475931 | 4594182 | 3324234 |
| rRNA | 6331713 | 6039143 | 3931 | 2773 |
| tRNA | 380941 | 1535094 | 9072 | 4964 |
| snoRNA | 7163 | 3384 | 2178 | 1303 |
| snRNA | 328 | 547 | 83 | 63 |
| Filtered reads | 21320507 | 10897763 | 4578918 | 3315131 |
| Reads mapped to miRBase19.0 | 14371676 | 3077840 | 3290 | 1703 |
| reads mapped to miRNA* | 114914 | 4523 | 535 | 165 |
| sRNA align to banana genome | 12029439 (42.9%) | 10512804 (56.9%) | ||
Fig 3. Length distribution of sRNAs in two libraries, TS and RP.
Two sRNA libraries were prepared from staminode tissue of two contrasting flower cultivars of Canna. Size distribution of the sRNA libraries show that the 21 nt size miRNAs were dominating ones in both the libraries.
Identification of known miRNAs in Canna
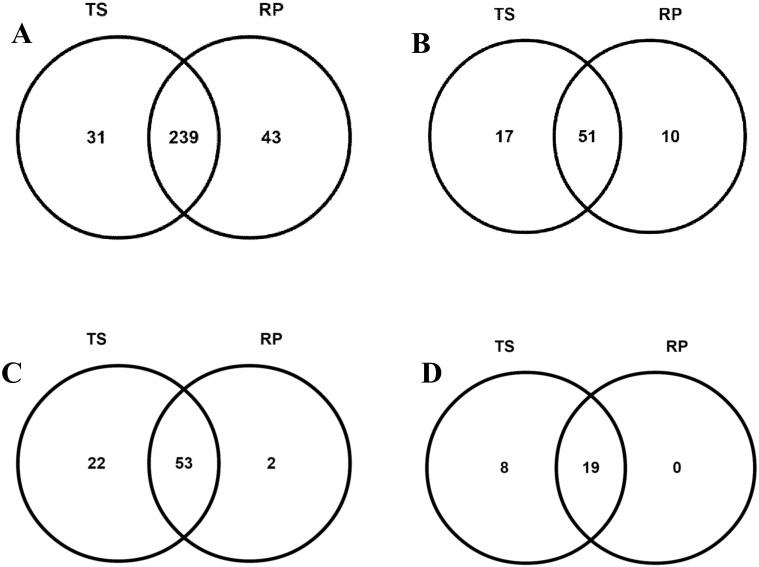
In order to identify the known miRNAs (both conserved and non-conserved), the filtered reads were mapped with miRBase 19.0. The small RNA sequences that matched with miRBase database were identified as the known miRNAs in Canna. A total of 313 miRNAs belonging to 78 miRNA families were identified from both the libraries. There were 271 miRNAs belonging to 68 miRNA families in TS and 282 miRNAs belonging to 61 miRNA families in RP “S2 Table”. Among these identified miRNAs, 239 miRNAs (51 families) were common in both TS and RP. Thirty one miRNAs (17 miRNA families) were specific to TS and 43 miRNAs (10 miRNA families) were specific to RP “Fig 4A and 4B”.
Fig 4. Venn diagrams of conserved and unique miRNAs between TS and RP.
(A) Total conserved and unique miRNAs between TS and RP, (B) Conserved and unique miRNA families between TS and RP, (C) Total conserved and unique miRNA* between TS and RP, (D) Conserved and unique miRNA* families between TS and RP.
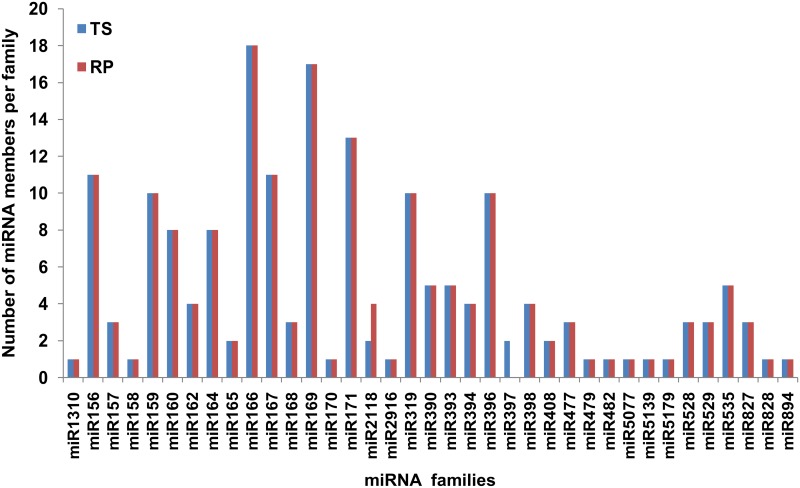
Among the known miRNAs, most of the conserved miRNA families had higher read abundance in both the libraries, e.g. miR166 had more than one hundred thousand reads. The non-conserved families had read abundance of one to four thousands “S2 Table”. Further, as reported earlier the conserved miRNA families had higher number of members per family as compared to the non-conserved miRNA [51]. For example, miRNA families miR156, miR159, miR166, miR167, miR169, miR171, miR319 and miR396 were highly conserved and exhibited higher number of members, whereas several known but non-conserved miRNAs familiesviz. miR408, miR477, miR479 and miR529 had one to three members per miRNAs family “Fig 5”.
Fig 5. Number of miRNAs members in each family identified from TS and RP by deep sequencing.
We further analyzed the miRNAs* and their families in both the libraries. A total of 75 and 55 miRNAs* belonging to 27 and 19 miRNA* families were identified in TS and RP, respectively “S3 Table”. Further analysis revealed that 53 miRNA* belonging to 19 miRNA* families were common in both the libraries. Twenty two miRNA* belong to eight miRNA* families were specific to TS, whereas only two miRNA* with no specific miRNA* families were found in RP “Fig 4C and 4D”. Interestingly, some miRNAs* have more read counts than their corresponding miRNAs “S4 Table”.
Identification and validation of novel miRNAs in Canna
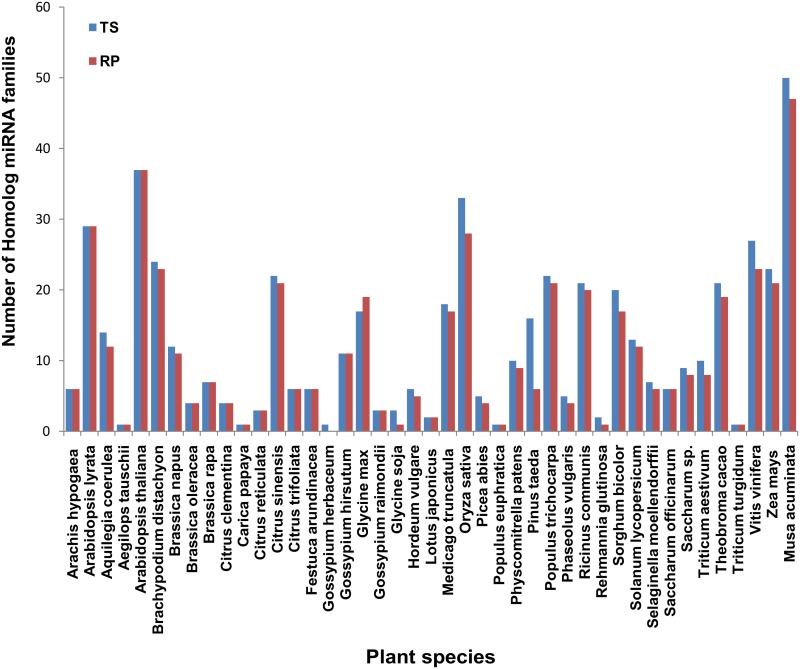
In order to identify the novel miRNAs, we mapped the raw reads with Musa acuminata genome [52]. Further, homology search of all the known miRNAs using miRBase 19.0 and reported Musa acuminata miRNAs [53] revealed that Canna miRNAs exhibited the highest number of homology with Musa, followed by Arabidopsis thaliana and Oryza sativa “Fig 6”. Therefore, we chose to map Canna sRNA reads with Musa acuminata. 42.90% and 56.90% reads of TS and RP, respectively mapped with Musa genome “Table 1”. Based on predicted secondary structures and the standard criteria of plant miRNAs [42], 205 and 160 miRNAs precursors and their corresponding mature miRNAs were predicted from TS and RP, respectively. These predicted miRNAs were again mapped with the plant miRNAs reported in miRBase 19.0. There were 32 and 18 miRNAs from TS and RP, respectively which did not map with known miRNAs and we predicted these miRNAs as putative novel miRNAs. Among these, 10 miRNAs were common in both TS and RP with three miRNAs showing their corresponding miRNA* sequences “S5 Table”, “S2 Fig”.Out of these putative novel miRNAs, four randomly selected miRNAs were further validated by stem loop RT-PCR “S3 Fig”.
Fig 6. The homology of identified miRNAs with other plant species.
Values on Y axis indicate the number of homological miRNA families between Canna and other plant species.
Expression pattern of known miRNAs in TS and RP
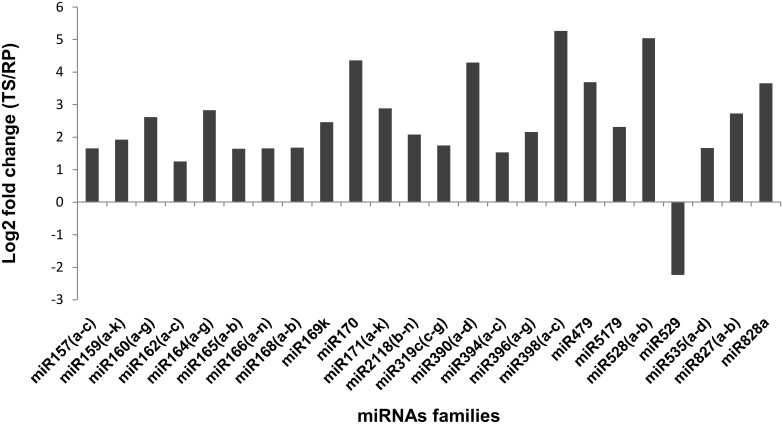
Differential expression of miRNAs was analyzed as described in materials and methods. Expression analysis showed 109 miRNAs belonging to 22 families were differentially expressed in the two cultivars “S6 Table”. Among the families, miR398 and miR528 expressed at five fold and miR170, miR390, miR479 and miR828 expressed at four fold higher in TS as compared to RP “Fig 7”. Expression of only miR529 was higher in RP as compared to TP (~ 2.2 fold). Further, miR397, miR397b and miR2118e, miR2118r expressed only in TS and RP, respectively “S6 Table”. These results indicate that ~ 34% miRNAs which were differentially expressed in two cultivars might play an important role in flower development.
Fig 7. Differentially expressed miRNA families between TS and RP.
Validation of miRNA expression by qRT-PCR
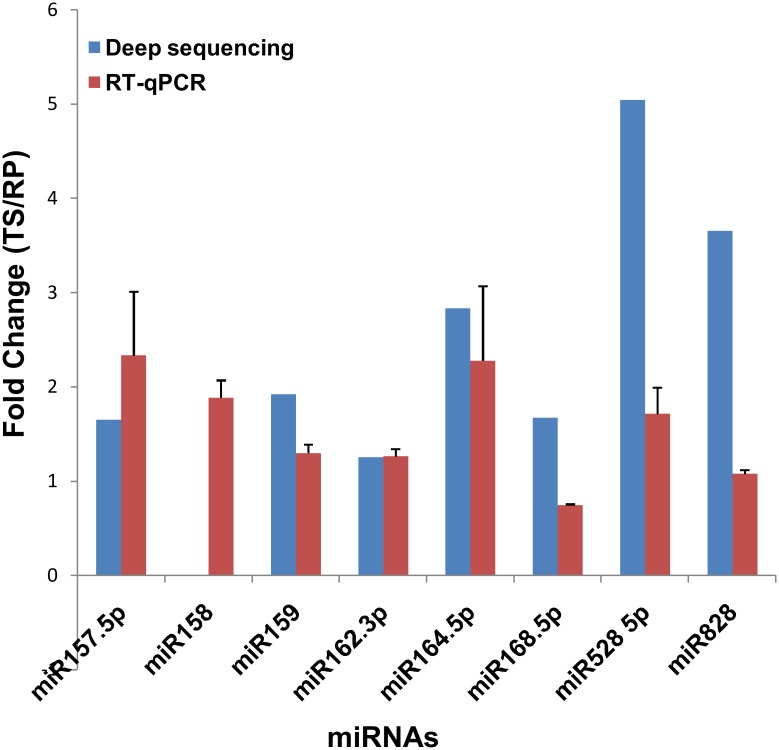
Out of 22 differentially expressed miRNA families as indicated in deep sequencing, eight families were randomly chosen for validation of expression pattern using qRT-PCR. As shown in “Fig 8”, seven miRNA families exhibited similar expression level in both deep sequencing and qRT-PCR analysis.
Fig 8. Comparison of the miRNA expression levels determined by deep sequencing and qRT-PCR.
Blue and red colors indicate the fold change obtained by deep sequencing and qRT-PCR, respectively. The error bars indicate the standard deviation obtained from biological and technical replicates.
Identification and enrichment analysis of miRNAs target genes
The biological role of miRNAs is better understood by the functions of their target genes. It is also reported that the conserved miRNAs may regulate the conserved targets [42]. Therefore, with the aim of better understanding the biological role of the conserved and the novel miRNAs, we searched for putative target genes by using a plant sRNA target analysis tool, psRNATarget [54] at default parameters. The details of the target gene ID, nature of inhibition, unpaired energy are shown in “S7 Table”. A total of 1343 targets were identified for 109 significantly differentially expressed miRNAs. Transcription factors were the major miRNA targets identified. Among these transcription factors, squamosa promoter binding proteins like (SPL), MYB-domain transcription factors (MYB), ARF-auxin responsive factors (ARF), NAC-domain transcription factors (NAC), CUC-cup shaped cotyledon (CUC), homeodomain leucine zipper transcription factors (HD-ZIP) and TCP domain transcription factors (TCP) were the important ones “S7 Table”. Other miRNA targets were growth regulating factors, laccase, transporter activity, ATPase domain containing protein, kinase proteins and chaperone proteins etc. The targets for the putative novel miRNAs were mainly transporter genes, amino transferses, heat shock proteins and protein kinase “S8 Table”.
Experimental validation of known miRNA targets
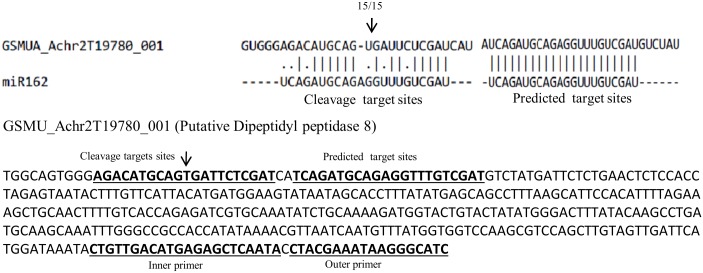
Cleavage site of predicted target of conserved miRNAs in Canna were validated by using a modified form of 5' RLM-RACE. Though we selected a few miRNAs for target validation, however we were successful in validating the target of only miR162. As reported earlier, miR162 had two putative targets viz. dipeptidyl peptidase 8 [55] and endoribonuclease dicer homolog 1 [56]with expected E value less than two. The latter target was experimentally validated in earlier report [56]. However, in Canna only Dipeptidyl peptidase 8 was identified as the target of miR162. The Dipeptidyl peptidase 8 has two adjacent target sites for miR162that forms miRNA/target duplex. Out of these two sites, one was perfect complementary to miR162 and other one had one nucleotide indel and five nuclotides mismatch in miRNA/target duplex region. In RLM-RACE experiment, only the latter traget was identified “Fig 9”. While this type of miRNA target site was not very common for most of the validated plant miRNA targets but has been observed in another case [57].
Fig 9. Detection of cleavage site through RLM-RACE.
5' RLM RACE was used to map the cleavage sites. The partial mRNA sequence from the target genes were aligned with the miRNA. The arrow indicates the cleavage site, and the number above the arrow denotes the frequency of the sequenced clones.
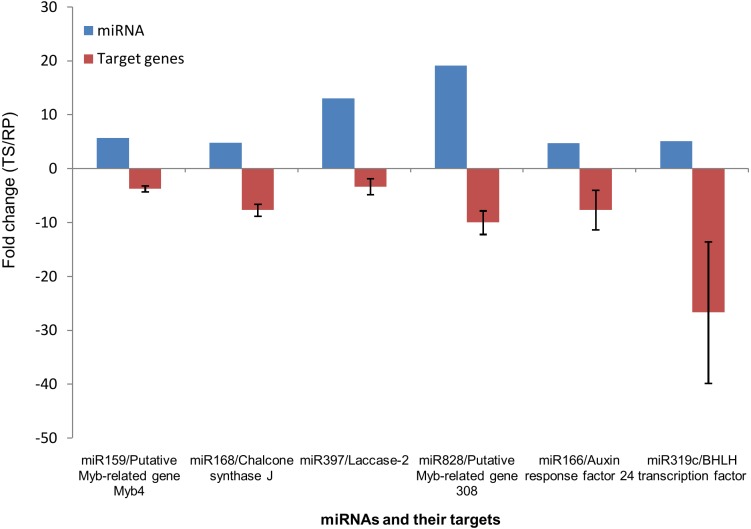
Further, we analyzed the expression level of the predicted miRNA target genes which were involved in pigment and flower development processes by qRT-PCR. In TS library, the expression of MYB related genes, chalcone synthase and laccase which are involved in pigment biosynthesis were down regulated, where as the expression of miRNAs viz. miR159, miR828, miR168 and miR397 which target these genes were up regulated. While, miR159 and miR828 known to target different MYB related genes [32], miR397 is known to target laccase genes [58]. On the other hand, in RP, the expression level of these genes and the miRNAs targeting these genes were up and down regulated, respectively “Fig 10”. Further, on the basis of miRNA expression data, target analysis and validation experiment by qRT-PCR, we predict miR168, which targets ARGONAUTE1 (AGO1) [59] may also target chalcone synthase gene in Canna flowers. These results indicate the involvement of these miRNAs in regulation of pigment related genes in Canna flowers.
Fig 10. Correlation of miRNAs and their target genes expression in TS and RP.
The target genes of selected miRNAs were analyzed by qRT-PCR. The expression level of these target genes (red) were compared with the expression level of corresponding miRNAs (blue) estimated from sequencing data. Error bars indicate mean ± standard deviation of three biological and three technical replicates.
Functional annotation of differentially expressed miRNAs
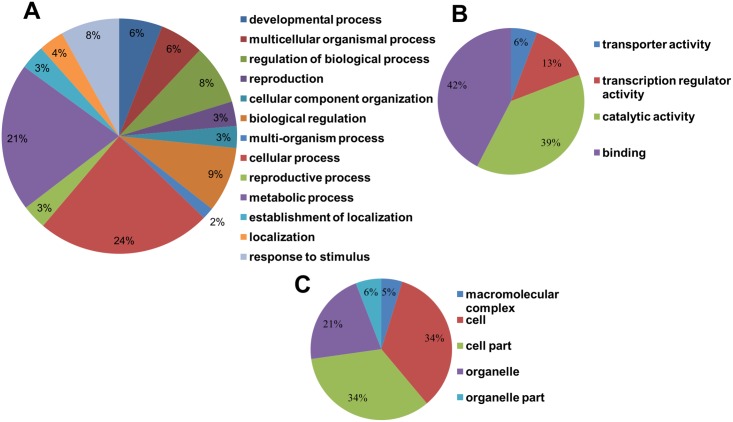
For better understanding the role of identified miRNA targets, we performed AgriGO analysis,a promising method for uncovering the miRNA-gene regulatory network. A total of 1343 identified targets of 109 differentially expressed miRNAs gave 633 GO terms. These were further categorized in Biological process (398 terms), Cellular component (84 terms) and Molecular function (151 terms). Under biological processes, major target genes were involved in cellular process (24%), metabolic process (21%), biological regulation (9%) and developmental process (6%) “Fig 11A”. In molecular component major target genes were involved in binding processes (42%), catalytic activity (39%) and transcription regulator activity (13%) “Fig 11B”. The cellular component included major three targets which were involved in cell part (34%), cell (34%) and organelle function and developments (21%) “Fig 11C”. We were interested to see the major targets involved in flower development related traits. Our analysis revealed that 16 miRNAs which targeted 60 genes were involved in flower development related traits such as fertilization, floral whorl development, fruit development and sexual reproduction “S9 Table”. Since the cultivars were contrasting in terms of flower colors, we further analyzed the targets for secondary metabolite pathways including pigment biosynthesis. This revealed five miRNAs families which targets five genes involved in phenyl propanoid metabolic process and pigment metabolic process “S9 Table”. Expression of all these miRNAs that are related to flower development and pigment biosynthesis were higher in TS than RP, except for miR529 “Fig 7”.
Fig 11. Gene ontology enrichment analysis of the predicted targets of Canna miRNAs.
Categorization of miRNA-targets genes were performed according to the(A) Biological Processes, (B) Molecular function and (C) Cellular component.
Discussion
Canna is one of the important ornamental flowering plant because of its attractive and vibrant flower colors. Yet,it's use in cut flower trade is limited because of its very short shelf life. The petals drupe within a few hours of plugging. However, it has great potential to be used as cut flower if flower related traits including shelf life can be enhanced. In spite of having great values, there is no report on molecular studies especially related to flower development in Canna. Understanding the molecular mechanisms in shaping flower ornamental quality in Canna will provide great value in breeding of different Canna cultivars.
A large number of miRNAs have been reported from different plant species [59]. Most of these studies are based on plant species whose genome is well characterized [60–62]. More recently focus has been made to identify new miRNAs from plant species with unknown genome sequence [5, 6]. miRNAs are known to regulate almost all developmental processes in organisms. Identification of new miRNAs and analysis of differential expression of miRNAs in two contrasting cultivars will provide further insight into their role in plant developmental processes. A total of 313 miRNAs (78 miRNA families) and corresponding 130 miRNA* (56 miRNA* families) were identified from both the cultivars. Due to lack of genome sequences or ESTs of Canna, miRNA precursors were predicted based on sRNAs mapped with Musa acuminata genome, which is taxonomically nearest taxa having known genome sequence. The mapping of sRNA to cross species is possible as miRNAs are highly conserved between species.This also maximizes the screening of novel miRNAs in plants for which genome is not available [5, 12, 63]. This approach has generally been followed for miRNA profiling in organisms with unknown genome sequences [5, 63]. However, this may under/ over-estimate the actual numbers of miRNAs due to evolutionarily loss/gain of genomic region and expression pattern, especially when one compares with taxonomically distant clad, as in the present study. Nevertheless, this approach has provided fair miRNA profiling in cross-species studies [5, 63, 64]. Identification of large numbers of miRNAs in Canna based on mapping of very distant taxa suggests that Canna shares many conserved miRNAs between ornamental and fruit bearing plants. Similarly,a very high degree of conservation of miRNAs was also observed between Rosaceae and Strawberry [5]. We further detected a large numbers of miRNA* familes indicating existence of authentic miRNAs. The abundance of miRNA* is usually much lower than that of their corresponding miRNAs. This is because of rapid degradation of miRNA* after their complementary miRNA sequences are selected from the miRNA/miRNA* duplex and loaded into the AGO protein [65, 66]. However, we observed a few miRNAs* sequences whose read counts were more than their corresponding miRNAs. Similar observation wasalso reported in other study [67]. Further, in both the libraries the reads of conserved miRNAs were larger as compared to non-conserved miRNAs. For example, miR166 have more than one hundred thousand reads. It is reported that conserved miRNAs generally have high reads abundance than non conserved miRNAs [63].
Discovery of novel miRNAs from species with unknown genomic information is more challenging. However, based on mapping with nearest genome sequence and subsequent prediction of secondary structures using the standard criteria of plant miRNAs [42], we identified 32 and 18 miRNAs as putative novel miRNAs in TS and RP, respectively. Predicted target genes for these miRNAs were related to transporter gene, heat shock protein etc. Further characterization and functional validation of these miRNAs and their target genes may provide insight into the Canna specific flower development process. Because of very contrasting flower color and differences in some other morphological traits between the two cultivars, we hypothesized that there would be high degree of differential expression of a range of miRNAs. As expected, we observed as many as 109 miRNAs which were differentially expressed in the two cultivars. Amongst 109 differentially expressed miRNAs, expression of only miR 529was higher in RP cultivar where as all other miRNAs expressed at 1–5 fold higher in TS as compared to RP cultivar.
Since we were interested to explore flower related traits and their regulation, we focused on major targets of miRNAs involved in this process. After assessing the GO terms by enrichment analysis we found most of the conserved miRNAs were involved in development of various flower parts. Functional annotation showed that16 miRNAs which targeted 60 genes were involved in flower related traits. Most of these predicted targets that are involved in Canna flower development have already been confirmed in model plants [14]. For example, it is reported that miR164 defines the flower boundary by targeting no apical meristem protein [13, 68]. The floral boundary, which also refers to the overall size of the flower was smaller in TS than RP. This is consistent with expression of miR164 which was higher in TS as compared to RP (further validated by qRT-PCR).
Flower color is one of the important characteristic of any flowering ornamental plant. Therefore, it is important to understand the color development in individual plant species for better utilization of genotypes by breeders. Flower colors are determined by accumulation of secondary metabolites such as flavonoids, carotenoids, and betalains [27, 69]. The biochemical estimation showed that flavonoids were the major source of color development in Canna. The low level of carotenoids in TS as compared to RP, but comparable amount of carotenoids in both the cultivars indicated carotenoids may not be the major source of pigments in floral color development of Canna. A previous study also suggested higher flavonoids content in red flower variety of Canna than yellow one [70]. We examined the miRNAs and their targets which were involved in the color development pathways. Five miRNAs were identified which were differentially expressed between the two cultivars. Amongst these, miR159 and miR828 target different types of R2R3-MYB transcription factors. These transcription factors form an (MYB-bHLH-WDR) initiation complex and are involved in the early steps of flavonoid biosynthesis pathway [71]. Thus miR159 and miR828 plays important role in regulation of flavonoid biosynthesis pathway. The expression of these miRNAs was 2–3 folds higher in TS cultivar as compared to RP cultivar. The higher level of expression in TS might down regulate the early steps of flavonoid biosynthesis pathway and hence inhibit pigment biosynthesis. Similarly, expression of miR168 which target chalcone synthase (CHS) was also higher in TS as compared to RP. Chalcone synthase (CHS) is involved in the first step of flavonoid pathway. On the other hand miR535 and miR397 target the intermediate enzymes, coumarate CoA ligase and laccase which are intermediate enzymes in flavonoid pathway. All these miRNAs were expressed at higher level in TS as compared to RP. The validation of some of these target gene expression by qRT-PCR further reconfirmed our findings about their role in flower color develpment in Canna. However, flower color development is known to be regulated by combinatorial mechanisms and may not be an act of straight regulation by single or a few miRNAs and their targets. Therefore, it is difficult to elucidate the molecular mechanism of miRNA-directed regulation of color determination in Canna. More particularly when no other genomic resources are available from the species. Further, target validation of a few selected miRNAs including those involved in pigment biosynthesis genes largely failed in our experiment. We validated target of miR162 that encodes an enzyme dipeptidyl peptidase 8. This target carries two miRNA target sites, one with a high level of complementarity and the other one is a duplex with six nucleotides mismatches. The identified new target demonstrates that miRNA/target duplex need not to be a strictly complementary to their target site. Though this type of unconventional traget sites is uncommon but was reported in ealier study [57]. Nevertheless, miRNA profiling of these two contrasting Canna cultivars will certainly provide potential clue about Canna flower color development.
Conclusions
Understanding the mechanism of processes associated with flower development in different flowering plants is an important aspect of miRNA regulated processes. Here, we reported differential expression of miRNAs in two cultivars of Canna. Our analysis showed that there were 31 and 43 miRNAs which were specifically expressed in TS and RP cultivars. These miRNAs might play a crucial role in cultivar specific flower development processes. The integrated analysis of miRNAs and their targets suggested that differential expression of miRNAs in two cultivars might be responsible in regulation of morphological as well as pigment biosynthesis pathways. The putative new miRNAs might provide further clue in gene regulation of flower development processes in ornamental plants, particularly in Canna.
Supporting Information
(PDF)
(PDF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors acknowledge University Grant Commission, New Delhi, for fellowship to AMT and PM. The work was supported by Council for Scientific and Industrial Research (CSIR), New Delhi granted project,OLP-088.
Data Availability
All relevant data are within the paper and its Supporting Information files. miRNA sequences have been deposited in NCBI under accession no. SRA310114.
Funding Statement
This work was supported by the Council of Scientific and Industrial Research, New Delhi, India. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. cell. 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 2.Mangrauthia SK, Agarwal S, Sailaja B, Madhav MS, Voleti S. MicroRNAs and their role in salt stress response in plants Salt Stress in Plants: Springer; 2013. p. 15–46. [Google Scholar]
- 3.Sun G. MicroRNAs and their diverse functions in plants. Plant molecular biology. 2012;80(1):17–36. 10.1007/s11103-011-9817-6 [DOI] [PubMed] [Google Scholar]
- 4.Yang T, Xue L, An L. Functional diversity of miRNA in plants. Plant Science. 2007;172(3):423–32. [Google Scholar]
- 5.Kim J, Park JH, Lim CJ, Lim JY, Ryu J-Y, Lee B-W, et al. Small RNA and transcriptome deep sequencing proffers insight into floral gene regulation in Rosa cultivars. BMC genomics. 2012;13(1):657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakhotia N, Joshi G, Bhardwaj AR, Katiyar-Agarwal S, Agarwal M, Jagannath A, et al. Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing. BMC plant biology. 2014;14(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131(14):3357–65. [DOI] [PubMed] [Google Scholar]
- 8.Amasino R. Seasonal and developmental timing of flowering. The Plant Journal. 2010;61(6):1001–13. 10.1111/j.1365-313X.2010.04148.x [DOI] [PubMed] [Google Scholar]
- 9.Cartolano M, Castillo R, Efremova N, Kuckenberg M, Zethof J, Gerats T, et al. A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nature genetics. 2007;39(7):901–5. [DOI] [PubMed] [Google Scholar]
- 10.Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. The Plant Journal. 2007;49(4):683–93. [DOI] [PubMed] [Google Scholar]
- 11.Irish VF. The flowering of Arabidopsis flower development. The Plant Journal. 2010;61(6):1014–28. 10.1111/j.1365-313X.2009.04065.x [DOI] [PubMed] [Google Scholar]
- 12.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. [DOI] [PubMed] [Google Scholar]
- 13.Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131(17):4311–22. [DOI] [PubMed] [Google Scholar]
- 14.Luo Y, Guo Z, Li L. Evolutionary conservation of microRNA regulatory programs in plant flower development. Developmental biology. 2013;380(2):133–44. 10.1016/j.ydbio.2013.05.009 [DOI] [PubMed] [Google Scholar]
- 15.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant molecular biology. 2008;67(1–2):183–95. 10.1007/s11103-008-9310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srikanth A, Schmid M. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences. 2011;68(12):2013–37. 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. The Plant Cell Online. 2003;15(7):1563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. The Plant cell. 2003;15(11):2730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glazińska P, Zienkiewicz A, Wojciechowski W, Kopcewicz J. The putative miR172 target gene InAPETALA2-like is involved in the photoperiodic flower induction of Ipomoea nil. Journal of plant physiology. 2009;166(16):1801–13. 10.1016/j.jplph.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 21.Jung J-H, Park C-M. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta. 2007;225(6):1327–38. [DOI] [PubMed] [Google Scholar]
- 22.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Developmental cell. 2005;8(4):517–27. [DOI] [PubMed] [Google Scholar]
- 23.Varkonyi-Gasic E, Lough RH, Moss SM, Wu R, Hellens RP. Kiwifruit floral gene APETALA2 is alternatively spliced and accumulates in aberrant indeterminate flowers in the absence of miR172. Plant molecular biology. 2012;78(4–5):417–29. 10.1007/s11103-012-9877-2 [DOI] [PubMed] [Google Scholar]
- 24.Tsuji H, Aya K, Ueguchi‐Tanaka M, Shimada Y, Nakazono M, Watanabe R, et al. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. The Plant Journal. 2006;47(3):427–44. [DOI] [PubMed] [Google Scholar]
- 25.Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature genetics. 2010;42(6):541–4. 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- 26.Miura K, Ikeda M, Matsubara A, Song X-J, Ito M, Asano K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature genetics. 2010;42(6):545–9. 10.1038/ng.592 [DOI] [PubMed] [Google Scholar]
- 27.Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. The Plant Journal. 2008;54(4):733–49. 10.1111/j.1365-313X.2008.03447.x [DOI] [PubMed] [Google Scholar]
- 28.Gou J-Y, Felippes FF, Liu C-J, Weigel D, Wang J-W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. The Plant cell. 2011;23(4):1512–22. 10.1105/tpc.111.084525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends in plant science. 2010;15(10):573–81. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 30.Xia R, Zhu H, An Y, Beers EP, Liu Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 2012;13(6):R47 10.1186/gb-2012-13-6-r47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, et al. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nature communications. 2014;5. [DOI] [PubMed] [Google Scholar]
- 32.Li C, Lu S. Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC genomics. 2014;15(1):277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nature biotechnology. 2008;26(4):407–15. 10.1038/nbt1394 [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Li L. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics. 2011;27(18):2614–5. 10.1093/bioinformatics/btr430 [DOI] [PubMed] [Google Scholar]
- 35.Patra B, Acharya L, Mukherjee AK, Panda MK, Panda M. Molecular characterization of ten cultivars of Canna lilies (Canna Linn.) using PCR based molecular markers (RAPDs and ISSRs). Int J Integr Biol. 2008;2:129–37. [Google Scholar]
- 36.Glinos E, Cocucci A. Pollination biology of Canna indica (Cannaceae) with particular reference to the functional morphology of the style. Plant systematics and evolution. 2011;291(1–2):49–58. [Google Scholar]
- 37.Mishra T, Das AP, Sen A. Phytochemical Screening and ln-vitro Antioxidant Profiling of Solvent Fractions of Canna edulis Ker Gawler. Free Radicals and Antioxidants. 2012;2(1):13–20. [Google Scholar]
- 38.Mancinelli AL, Schwartz OM. The Photoregulation of Anthocyanin Synthesis IX. The photosensitivity of the response in dark and light-grown tomato seedlings. Plant and cell physiology. 1984;25(1):93–105. [Google Scholar]
- 39.Chen B, Yang S. An improved analytical method for the determination of carotenes and xanthophylls in dried plant materials and mixed feeds. Food chemistry. 1992;44(1):61–6. [Google Scholar]
- 40.Moxon S, Schwach F, Dalmay T, MacLean D, Studholme DJ, Moulton V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008;24(19):2252–3. 10.1093/bioinformatics/btn428 [DOI] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic acids research. 2008;36(suppl 1):D154–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, et al. Criteria for annotation of plant MicroRNAs. The Plant cell. 2008;20(12):3186–90. 10.1105/tpc.108.064311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2005;25(17):2537–45. [DOI] [PubMed] [Google Scholar]
- 44.May P, Liao W, Wu Y, Shuai B, McCombie WR, Zhang MQ, et al. The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nature communications. 2013;4. [DOI] [PubMed] [Google Scholar]
- 45.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50(4):244–9. 10.1016/j.ymeth.2010.01.026 [DOI] [PubMed] [Google Scholar]
- 46.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant methods. 2007;3(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen D, Suhrkamp I, Wang Y, Liu S, Menkhaus J, Verreet JA, et al. Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytologist. 2014;204(3):577–94. 10.1111/nph.12934 [DOI] [PubMed] [Google Scholar]
- 48.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature protocols. 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 49.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS biology. 2004;2(5):e104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Current Biology. 2005;15(16):1494–500. [DOI] [PubMed] [Google Scholar]
- 51.Körbes AP, Machado RD, Guzman F, Almerão MP, de Oliveira LFV, Loss-Morais G, et al. Identifying conserved and novel microRNAs in developing seeds of Brassica napus using deep sequencing. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Hont A, Denoeud F, Aury J-M, Baurens F-C, Carreel F, Garsmeur O, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488(7410):213–7. 10.1038/nature11241 [DOI] [PubMed] [Google Scholar]
- 53.Montes RA, de Fatima Rosas-Cardenas F, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G, et al. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nature communications. 2014;5:3722 10.1038/ncomms4722 . [DOI] [PubMed] [Google Scholar]
- 54.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic acids research. 2011;39(suppl 2):W155–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee WS, Gudimella R, Wong GR, Tammi MT, Khalid N, Harikrishna JA. Transcripts and MicroRNAs Responding to Salt Stress in Musa acuminata Colla (AAA Group) cv. Berangan Roots. PLoS One. 2015;10(5):e0127526 10.1371/journal.pone.0127526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Current Biology. 2003;13(9):784–9. [DOI] [PubMed] [Google Scholar]
- 57.Brousse C, Liu Q, Beauclair L, Deremetz A, Axtell MJ, Bouché N. A non-canonical plant microRNA target site. Nucleic acids research. 2014;42(8):5270–9. 10.1093/nar/gku157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu S, Li Q, Wei H, Chang M-J, Tunlaya-Anukit S, Kim H, et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proceedings of the National Academy of Sciences. 2013;110(26):10848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Yu J, Li D, Zhang Z, Liu F, Zhou X, et al. PMRD: plant microRNA database. Nucleic acids research. 2010;38(suppl 1):D806–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PloS one. 2007;2(2):e219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(34):12753–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Montes RAC, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G, Marsch-Martínez N, et al. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nature communications. 2014;5. [DOI] [PubMed] [Google Scholar]
- 63.Dong M, Yang D, Lang Q, Zhou W, Xu S, Xu T. Microarray and Degradome Sequencing Reveal MicroRNA Differential Expression Profiles and Their Targets in Pinellia pedatisecta. PloS one. 2013;8(9):e75978 10.1371/journal.pone.0075978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bocs S, editor The Banana Genome Hub. Plant and Animal Genome XXII Conference; 2014: Plant and Animal Genome.
- 65.Chen X. Small RNAs and their roles in plant development. Annual Review of Cell and Developmental. 2009;25:21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–87. 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- 67.Devers EA, Branscheid A, May P, Krajinski F. Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant physiology. 2011;156(4):1990–2010. 10.1104/pp.111.172627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B. CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development. 2004;131(4):915–22. [DOI] [PubMed] [Google Scholar]
- 69.Nishihara M, Nakatsuka T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnology letters. 2011;33(3):433–41. 10.1007/s10529-010-0461-z [DOI] [PubMed] [Google Scholar]
- 70.Vankar PS, Srivastava J. Comparative study of total phenol, flavonoid contents and antioxidant activity in Canna indica and Hibiscus rosa sinensis: Prospective natural food dyes. International journal of food engineering. 2008;4(3). [Google Scholar]
- 71.Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris NN, Walker AR, et al. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant physiology. 2009;151(3):1513–30. 10.1104/pp.109.142059 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. miRNA sequences have been deposited in NCBI under accession no. SRA310114.