Abstract
Staphylococcus epidermidis is the most common cause of device-associated infections. It has been shown that active and passive immunization in an animal model against protein SesC significantly reduces S. epidermidis biofilm-associated infections. In order to elucidate its role, knock-out of sesC or isolation of S. epidermidis sesC-negative mutants were attempted, however, without success. As an alternative strategy, sesC was introduced into Staphylococcus aureus 8325–4 and its isogenic icaADBC and srtA mutants, into the clinical methicillin-sensitive S. aureus isolate MSSA4 and the MRSA S. aureus isolate BH1CC, which all lack sesC. Transformation of these strains with sesC i) changed the biofilm phenotype of strains 8325–4 and MSSA4 from PIA-dependent to proteinaceous even though PIA synthesis was not affected, ii) converted the non-biofilm-forming strain 8325–4 ica::tet to a proteinaceous biofilm-forming strain, iii) impaired PIA-dependent biofilm formation by 8325–4 srtA::tet, iv) had no impact on protein-mediated biofilm formation of BH1CC and v) increased in vivo catheter and organ colonization by strain 8325–4. Furthermore, treatment with anti-SesC antibodies significantly reduced in vitro biofilm formation and in vivo colonization by these transformants expressing sesC. These findings strongly suggest that SesC is involved in S. epidermidis attachment to and subsequent biofilm formation on a substrate.
Introduction
Of all coagulase-negative staphylococci, Staphylococcus epidermidis is the most common cause of infections associated with catheters and other indwelling medical devices [1, 2]. It is a permanent and ubiquitous colonizer of human skin, can easily contaminate the medical devices during insertion, and subsequently form a biofilm [2, 3]. The capacity to form a biofilm is considered as one of the major virulence factors of this bacterial species [4, 5].
Staphylococcal biofilms develop via a multifactorial process, which may differ between species and strains. Nevertheless, most of the factors involved are analogous in S. epidermidis and S. aureus and have a similar function in biofilm formation [1, 2, 3].
Up to now, based on extracellular matrix macromolecules constituting the biofilm, three mechanisms of biofilm formation in staphylococci are identified [6]. Production of polysaccharide intercellular adhesin [PIA, also called poly-N-acetylglucosamine (PNAG)] was the first and for a long time, the only mechanism of biofilm formation identified [7, 8]. Further studies showed the existence of other PIA- or ica-independent mechanisms in S. aureus and S. epidermidis. Based on in vivo and in vitro studies, the proteinaceous biofilm formation was identified. In this case, the cell-surface and cell-cell attachment is based on proteins [9, 10]. More recently, a third mechanism based on extracellular DNA (eDNA) constituting a cell-to-cell or cell-to-substratum connecting component was recognized. This eDNA originates from autolysis [11, 12].
It has been shown that staphylococcal surface proteins such as accumulation-associated protein (Aap), biofilm-associated proteins (Bap, and Bap homologue Bhp), extracellular matrix-binding protein (Embp), fibronectin- or fibrinogen-binding proteins (FnBPA, FnBPB and Fbe/SdrG), and the major autolysin (AtlE) mediate the formation of the network of multilayered cell clusters and filamentous proteins, and thus play an important role in the biofilm accumulation phase [7, 10, 13, 14]. In S. epidermidis and S. aureus, LPxTG motif-containing proteins which covalently link to the cell wall via sortase activity, are determinants in the pathogenesis of device-related infections [7].
Through unknown or not well-characterized mechanisms such as insertion and excision of the insertion sequence IS256 at specific hot spots of the S. epidermidis icaA and icaC genes, PIA/PNAG production, biofilm formation and biofilm phenotype may be phase variable, allowing to switch from PIA-dependent to proteinaceous phenotype [10, 15, 16]. In 2001, Knobloch et al. reported that NaCl affects biofilm formation through the activation of the σB operon, an important regulator of the ica operon, and thus can be used to distinguish ica-dependent from ica-independent biofilm formation [9, 17, 18]. By using dispersing agents such as sodium metaperiodate (SM), proteinase K (PK) and DNase I, the chemical composition of the biofilm extracellular polymeric substance can be determined and one can discriminate between PIA-dependent, proteinaceous and eDNA-based biofilms [9, 19].
So far, the roles of 5 S. epidermidis LPxTG proteins (Aap, Bhp, SdrF, SdrG, SesI) in the pathogenesis of S. epidermidis infections and biofilm formation have been studied [20, 21, 22]. We focused our research on the LPxTG motif-containing S. epidermidis surface protein SesC, a 676-amino acid (aa) protein with a predicted molecular mass of 75 kDa. The cytoplasmic precursor of SesC contains a 35-aa N-terminal signal peptide, required for Sec-dependent secretion and is cleaved off by the signal peptidase. The 37-aa C-terminal LPxTG-sorting signal is recognized by sortase, which will cleave the bond between the Thr and Gly and thereafter covalently link the 608-aa (68 kDa) remaining protein to the peptidoglycan layer. Using antibodies against the mature domain of the SesC protein, we were able to reduce S. epidermidis biofilm formation in vitro [23]. In addition, active and passive immunization against SesC could significantly reduce their biofilm formation on catheter fragments in animal models of subcutaneous and intravascular catheter infection [23]. However, the involvement and exact function of SesC in S. epidermidis biofilm formation have remained unknown, so far. In order to elucidate its role, knock-out of sesC or isolation of S. epidermidis sesC-negative mutants were attempted, however without success. Therefore, as an alternative strategy sesC was introduced into S. aureus strains and the effect of sesC expression in biofilm formation by these host strains was studied.
Materials and Methods
Bacterial strains, plasmids and media
Cloning experiments were performed in Escherichia coli DH5α competent cells (Invitrogen). E. coli DH5α transformants were grown in Lysogeny Broth (LB) or on LB agar at 37°C supplemented with ampicillin (100 μg/ml), as all plasmids used in this study (Table 1) contain an ampicillin resistance (bla) gene. All Staphylococcus strains (Table 1) were grown in brain heart infusion (BHI) medium or agar, and for biofilm formation assays also in BHI medium supplemented with 4% NaCl (BHI-NaCl) or 1% glucose (BHI-glucose). Bacterial CFU counting was done on Tryptone Soya agar (TSA, Oxoid) or blood agar plates (BD Biosciences). Whenever required, growth media were supplemented with appropriate antibiotics as follows: chloramphenicol at 10 μg/ml, erythromycin at 10 μg/ml and tetracycline at 5 μg/ml. Species identification and antibiograms for all clinical isolates were performed using a VITEK® 2 automated system (bioMérieux).
Table 1. Staphylococcus strains and plasmids used in this study.
| Strains | Characteristic(s) | Reference |
|---|---|---|
| - S. epidermidis | ||
| 10b | Clinical isolate | [24] |
| - S. aureus | ||
| RN4220 | Restriction-negative derivative of 8325–4 | [25] |
| 8325–4 | NCTC8325 cured of prophages. 11-bp deletion in rsbU. | [9] |
| BH1CC | MRSA clinical isolate. Biofilm positive. SCCmec type II, MLST type 8, clonal complex 8. | [9] |
| MSSA4 | Clinical isolate | This study |
| 8325–4 ica::tet | icaADBC::tet; isogenic mutant of 8325–4 | [9] |
| 8325–4 srtA::tet | srtA::tet; isogenic mutant of 8325–4 | [26] |
| BH1CC ica::tet | icaADBC::tet; isogenic mutant of BH1CC | [9] |
| BH1CC srtA::tet | srtA::tet; isogenic mutant of BH1CC | [26] |
| Plasmids | ||
| pCN68 | E. coli-Staphylococcus shuttle vector | [27] |
| pSRsrtA5 | E. coli-Staphylococcus shuttle vector | [26] |
Cloning and expression of S. epidermidis sesC and sesK genes in S. aureus strains
The coding regions of S. epidermidis sesC (SE2232, Gene ID 1056520) and sesK (SE1501, Gene ID 1056680), were amplified using sesC- and sesK-specific primers (Table 2) containing additionally a SalI or SmaI restriction site for cloning purposes. Genomic DNA (gDNA) of biofilm-forming S. epidermidis strain 10b, a clinical isolate [24], was used as a template. The amplicons were ligated into SalI/SmaI-digested pCN68 E. coli—Staphylococcus shuttle vectors [27] yielding pCNsesC and pCNsesK. In this plasmid, PblaZ is the promoter. It is a highly active constitutive promoter; erythromycin was used as the selection marker. All recombinant plasmids were replicated in E. coli DH5α. Correctness of cloning was confirmed by restriction enzyme digestion, PCR, and nucleotide sequence analysis of the insert. Plasmids harvested from E. coli were first electroporated into the restriction-deficient S. aureus strain RN4220 and subsequently into other S. aureus strains. Presence and expression of sesC, sesK, sasF (Gene ID: 5775591), and icaA (Gene ID: 5776135) in transformed strains were evaluated using gel-based reverse transcription-PCR (RT-PCR) and Western blotting assays. Plasmid, gDNA and RNA isolation from bacterial strains and cDNA synthesis were performed as previously described [28].
Table 2. Primers used in this study.
| Name | Sequence (5’ → 3’) | RE site |
|---|---|---|
| sesCFs sesCF | ATGTCGACTTTATTAAAGGAGTATGTGTAAATG | SalI |
| sesCR | ATCCCGGGTGATGATGCCTATTACTATATATAA | SmaI |
| sesKF | ATGTCGACGACCTCTTAACTAATTATGTTATG | SalI |
| sesKR | ATCCCGGGTCTCGTTATTTTCACTCAAATATC | SmaI |
(Underlined sequences: restriction sites; bold sequence: start codon)
Biofilm formation assay
The amount of biofilm formed by the different strains was determined using a semi-quantitative adherence assay in 96-well polystyrene microtiter plates (BD Biosciences) as previously described [23, 26, 28]. Briefly, 20 μl of stock cultures were inoculated into 5 ml (selective) BHI medium and grown to the end-exponential growth phase in a shaking incubator at 37°C. Cultures were subsequently diluted to an OD600 of 0.005 (5.x106 CFU/ml) in fresh BHI medium whether or not supplemented with 4% NaCl or 1% glucose. 200 μl of the diluted cultures of bacteria were pipetted into sterile 96-well polystyrene microtiter plates and incubated overnight at 37°C without shaking.
After incubation, the wells were rinsed 3 times with phosphate-buffered saline (PBS) and dried afterwards. The adhered material was stained with 200 μl of a 1% (w/v) crystal violet (Sigma) solution for 10 min, and subsequently, the wells were washed 3 times with water and again dried. For quantification, 160 μl of 30% (v/v) acetic acid solution was added to each well to dissolve the crystal violet. The OD595 of the dissolved stain was measured in a multipurpose UV/VIS plate reader (VICTOR3 TM; PerkinElmer).
Biofilm treatment assays
The biofilm stability against sodium metaperiodate (SM), proteinase K (PK) or DNase I treatment was tested as described previously [29, 30, 31]. Briefly, 200 μl of an overnight grown culture diluted to an OD600 of 0.005 in BHI-glucose, were pipetted into sterile 96-well polystyrene microtiter plates and statically incubated overnight at 37°C. After 24 h incubation, the growth medium was replaced with 200 μl solution of SM (10 mM in 50 mM sodium acetate), of PK (Qiagen GmbH, 1 mg/ml in 100 mM NaCl, 20 mM Tris/HCl, pH 7.5) or of DNase I (Sigma, 2 mg/ml in 5 mM MgCl2) Subsequently, plates were incubated at 37°C for 2 h and the remaining biofilms were quantified as explained above.
To assess the effect of specific anti-SesC antibodies (αSesC-IgGs) produced as earlier described [23] on biofilm formation, 1x106 bacteria were in the first instance incubated with αSesC-IgGs (20 μg/ml bacterial suspension) for 2 h at 4°, and in a volume of 200 μl medium brought into a 96-well plate. Plates were incubated overnight at 37°C without shaking to allow bacterial growth and biofilm formation.
PIA quantification by PIA non-specific immunoblot assay
The relative amount of PIA present in a biofilm was determined as described [31], however with some modifications. Briefly, 1 ml of a diluted overnight culture of bacterial suspension (5x106 CFU/ml) in BHI-NaCl or BHI-glucose were pipetted in 24-well polystyrene microtiter plates (BD Biosciences) and next, plates were incubated overnight at 37°C. After incubation, spent medium was removed, 500 μl PBS was added into each well and the biofilm mass was removed from the surface via pipetting. Samples were transferred to 1.5 ml tubes which were next centrifuged for 3 min at 12000×g. Pellets and biofilm material were re-suspended in 0.5 M EDTA (pH 8) to an OD600 of 0.5 and PIA was extracted by boiling the samples for 5 min. After centrifugation at 18000×g, 250 μl of the supernatant was added to an Eppendorf tube with 25 μl PK solution (20 mg/ml). The mixture was incubated for 1 h at 60°C and afterwards PK was deactivated for 30 min at 80°C. Sample aliquots were applied to a nitrocellulose membrane, which was blocked with 5% (w/v) bovine serum albumin (BSA) in TTBS [Tris-buffered saline (100 mM Tris/HCl, 0.9% NaCl) with 0.05% Tween 20]. After washing the membrane 3 times in TTBS, it was incubated overnight at 4°C with wheat germ agglutinin-horseradish peroxidase conjugate (EY laboratories) in 1% (w/v) BSA-TTBS. After washing the membrane 3 times in TTBS, the presence of PIA was detected by the addition of Western blotting detection reagent (AmershamTM ECL, GE Healthcare), and visualized with a ChemiDoc™ XRS+ System (Bio-Rad).
Detection of SesC
Western blot analysis of SesC was performed as described [32]. Cells of 10 ml overnight bacterial cultures grown at 37°C in BHI-glucose were harvested by centrifugation, resuspended within lysis buffer [10 mM Tris/HCl, 10 mM EDTA, 1 mM phenylmethanesulfonyl fluoride (PMSF), pH 7.5, 100 μg lysozyme/ml, 100 μg lysostaphin/ml] and incubated for 1 h at 37°C. Then, cells were broken by passing them three times through a French press at 69 MPa (SLM Aminco) followed by sonication (Branson 2510, 42 kHz) of samples on ice. Proteins were electrophoretically separated on a 12% sodium dodecyl sulphate polyacrylamide gel and subsequently transferred onto a PVDF membrane. The primary antibody (1:5000 dilution of rabbit anti-SesC polyclonal antibodies in TTBS) was added to the membranes, which were left overnight, followed by horseradish peroxidase-conjugated anti-rabbit IgGs as secondary antibodies [1:2500 dilution in TTBS+1% (w/v) skim milk] for 2 h. Finally, the presence of SesC was visualized using ECL Western blotting detection kit (GE Healthcare) in combination with a ChemiDoc™ XRS+ System (Bio-Rad).
Scanning electron microscopy
In vitro biofilm formation on cover Glasses (Ø 10 mm, Menzel GmbH) was visualized by scanning electron microscopy (SEM) as described [33]. Briefly, an overnight bacterial culture was diluted in BHI-glucose to an OD600 of 0.005, and 1 ml of the diluted culture was pipetted into the wells of sterile 24-well polystyrene microtiter plates, which each contained a glass disk. Plates were incubated overnight at 37°C without shaking, after which the disks were washed 3 times with PBS. Biofilms formed on the disks were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) by incubation for 2 h at room temperature. After fixation, disks were rinsed with 0.1 M sodium cacodylate buffer (pH 7.4) for 30 min with three changes. Thereafter, a post-fixation step was done with 1% osmiumtetroxide for 2 h at 4°C. Next, disks were rinsed with distilled water (2 times, 10 min) and then dehydrated in 10 min steps in a series of ascending ethanol baths (25%, 50%, 75%, 95% and 100%). Following a bath of hexamethyldisilazan, dehydrated air-dried samples were mounted on support stubs with C-stickers and silver glue, and sputter coated with platinum (Agar Scientific, Auto Sputter Coater). Finally, the samples were observed and images taken with a JSM7401F field emission scanning electron microscope (JEOL) in a high vacuum mode with a conventional Everhart-Thornley detector at 5kV accelerating voltage.
Investigation of the effect of SesC on the in vitro attachment to a catheter surface
In vitro bacterial attachment to the surface of a commercial polyurethane (PU) intravenous catheter (Arrow International) was examined as described [23, 34] with some modifications. Overnight cultures of S. aureus strains 8325–4 and 8325–4 (pCNsesC) were washed with saline (0.9 % NaCl) and diluted to an OD600 of 0.03 in saline. Seven mm catheter fragments were added to 2ml of bacterial suspension and the mixture was incubated at 37°C. After 2 h incubation, catheters were removed. After gentle rinsing with saline, catheters were placed in a tube containing 1 ml saline. Tubes were vortexed for 10 s, sonicated for 10 min at 40 kHz using a Branson water bath and again vortexed for 10 s. Thereafter, tube contents were 10-fold serially diluted and 50 μl aliquots of each dilution were plated on TSA plates using a spiral plater system (Spiral Plater Systems, Inc. Cincinnati, Ohio), and plates were incubated at 37°C overnight. Colonies were counted and the number of bacteria was defined as the mean of at least five quantitative cultures.
Jugular vein catheterized (JVC) mouse model
In order to investigate the involvement of SesC in catheter-related infections (CRIs) in vivo, we used a central venous catheter murine model [28, 35, 36] that reflects the clinical situation of catheter colonization by contaminated infusions. Briefly, 4-weeks-old Swiss-Webster mice (Taconic) were anesthetized with a single intraperitoneal (i.p.) injection of sodium pentobarbital (Nembutal, 40–60 mg/kg body weight) and placed on a heating pad to maintain the body temperature at 37°C. An anesthetized and surgically prepared animal was then placed in the dorsal recumbence under a dissecting microscope (Zeiss, Jena, Germany, 10x magnification). A small vertical incision was made using small scissors and the right jugular vein was identified, mobilized and exposed with blunt surgical dissection. A single lumen Intramedic polyethylene catheter (Becton Dickinson #427400; internal Ξ 0.28 mm, outer Ξ 0.61 mm, insertion length, 1 cm) was inserted into the right jugular vein via a small incision in the vein made with vein scissors and advanced into the superior vena cava. A ligature was then tied loosely around the catheter and the patency was verified. Once blood flow had been established, the catheter was anchored in place. Subsequently, a small midline skin incision was made between the scapulae. The catheter was subcutaneously tunneled by a straight surgical clamp and exteriorized through midline scapular incision. The incisions were then closed with stitches. The patency was tested and the catheter was flushed with 100 μl of saline, sealed with a plug and left in place throughout the experiment. Thereafter, mice were housed separately and monitored for recovery. 24h after surgery, the mice were inoculated via the catheter lumen with 100 μl of an S. aureus suspension (OD600 of 0.03 (3.107CFU/ml)). Then the catheter was flushed again with 100 μl saline so that bacteria entered the venous system of animals. At least every 12 h, animals were monitored for adverse events and all efforts were made to minimize animal suffering. All surgical procedures were performed under anesthesia with sodium pentobarbital (Nembutal) diluted in saline. After taking the blood samples at day 1 or 5 post-infection, animals were euthanized by CO2 inhalation and catheters and organs (spleen, liver, heart, vein and right kidney) were aseptically harvested from the animals. The organs were mechanically homogenized in saline and the portion of the catheter inserted into the vein (about 1 cm) was cut, gently washed and placed in a tube containing 1 ml saline. Tubes containing the catheter fragments were vortexed for 10 s, sonicated for 5 min at 40 kHz and again vortexed for 10 s. Serial dilutions of the organ homogenates and catheter fluid collections were cultured on blood agar plates using the spiral plating system, and plates were incubated at 37°C overnight. Colonies on all plates were counted and the number of bacteria was defined as the mean of at least 5 quantitative cultures. All in vivo experiments were repeated at least twice and conducted in compliance with the guidelines for animal experimentation. The Institutional Animal Care Commission and Ethical Committee of the KU Leuven approved all experimental protocols.
Study of the involvement of SesC in virulence and biofilm formation
Nine mice were divided into 3 groups of 3 mice. Overnight culture cells of S. aureus strains 8325–4, 8325–4 (pCN) and 8325–4 (pCNsesC) grown to the late exponential/early stationary growth phase in (selective) BHI medium and the cells were pelleted, re-suspended and diluted to an OD600 of 0.03 (circa 3x107 CFU/ml) in 0.9% NaCl with the suitable antibiotics. After the 24-h recovery period, animals in one group were inoculated through the catheter lumen with 100 μl of one of the 3 prepared bacterial suspensions. Five days post-infection, the infection rate and the bacterial load on the implanted catheter were determined and compared between the 3 groups.
The effect of rabbit polyclonal αSesC-IgGs on infection rates and biofilm formation
Bacterial suspensions of S. aureus strains 8325–4 and 8325–4 (pCNsesC) were freshly prepared and diluted till an OD600 ~ 0.03. The diluted suspensions were incubated for 2 h at 4°C without IgGs, with αSesC-IgGs (80 μg/ml) or pre-immune IgGs (80 μg/ml). Fifteen mice divided into 5 groups of 3 mice groups received 100 μl of inoculum: i) 8325–4 without any IgG; ii) 8325–4 pre-incubated with αSesC-IgGs (80μg/ml); iii) 8325–4 (pCNsesC); iv) 8325–4 (pCNsesC) pre-incubated with pre-immune IgGs (80μg/ml); v) 8325–4 (pCNsesC) pre-incubated with αSesC-IgGs (80μg/ml). Since we expected similar impact for pre-immune IgG and αSesC-IgGs on 8325–4, we considered just αSesC-IgGs. Each inoculum was administered to each mouse through the lumen of the implanted catheter.
Statistical analysis
Analyses of data were pooled from at least two independent experiments, and were performed using GraphPad prism 6 software. The data from in vitro and in vivo experiments involving wild-type, mutant and transformed strains were subjected to a one-way analysis of variance (1-way ANOVA) to find significant differences. A P value of <0.05 was considered a significant difference. Data for the transformants carrying mock plasmids are not always shown as they did not show any significant difference with wild-type conditions (data are available upon request).
Results
Heterologous expression of sesC in S. aureus switches the biofilm phenotype from PIA-dependent to proteinaceous
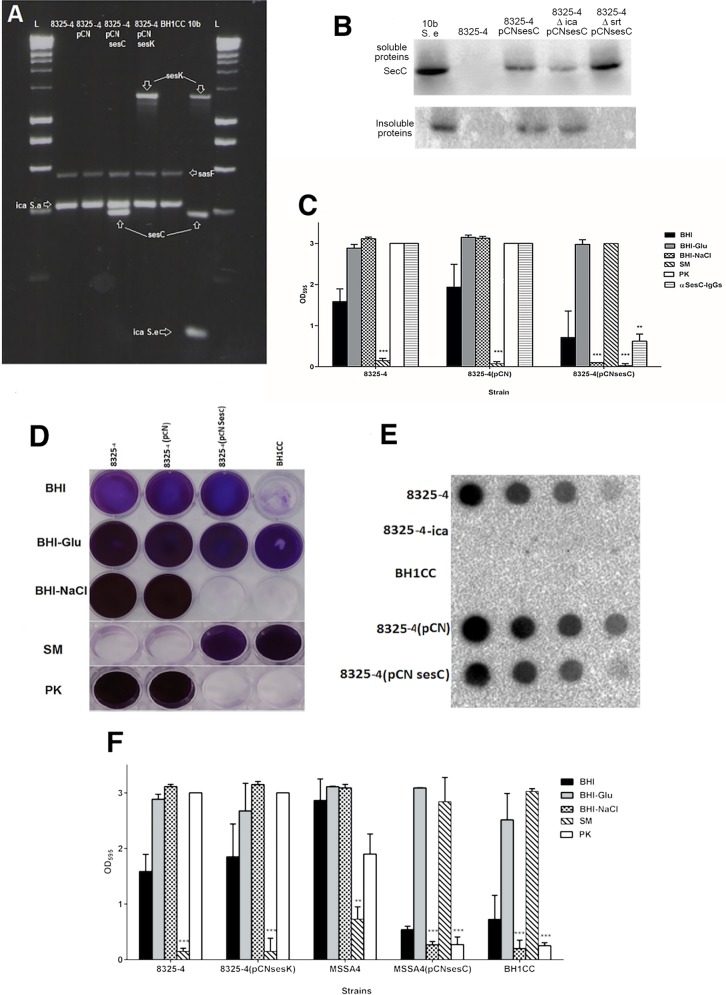
The S. epidermidis sesC gene was cloned into a pCN plasmid resulting in pCNsesC. The recombinant plasmid was introduced into the laboratory S. aureus strain 8325–4, which makes a PIA-type biofilm, and into the hospital-associated MRSA strain BH1CC, which makes a eDNA and proteinaceous biofilm (AtlE/FnBP-dependent) [14, 19, 29]. The expression of sesC and the presence of the corresponding protein were confirmed by gel-based RT-PCR assay and Western blot assay (Fig 1A and 1B). Western blot confirmed the expression of SesC in the transformant strains. Heterologous expression of sesC had no effect on the BH1CC biofilm phenotype (data not shown), but inhibited biofilm formation by 8325–4 transformants cultivated in BHI-NaCl (Fig 1C and 1D). Furthermore, 8325–4 (pCNsesC) biofilms grown in BHI-glucose were dispersed with proteinase K (PK) but not sodium metaperiodate (SM), which did disperse wild-type 8325–4 biofilms (Fig 1C and 1D). This is consistent with a biofilm phenotypic switch from PNAG- to protein-mediated in these transformants. Nevertheless, quantification of PIA showed no changes in the rate of PIA production in 8325–4 (pCNsesC) in comparison to the wild-type strain (Fig 1E).
Fig 1. Effect of transformation of S. aureus strains with sesC or sesK on the biofilm formation.
Using a semi-quantitative microtiter plate assay the level of biofilm formation in different media, the phenotype of biofilms and effect of αSesC-IgG antibodies on biofilm formation of different strains were identified. Biofilm formation in medium supplemented with NaCl or the effect of dispersal agents were used to discriminate the phenotype of biofilms. (A) Expression of sesC, sesK, sasF and icaA in cDNA of transformed strains was evaluated using the gel-based reverse transcription-PCR assay. (B) Detection of SesC production in S. epidermidis 10b, 8325–4, 8325–4 (pCNsesC) and 8325–4 ica::tet (pCNsesC) 8325–4 srt::tet (pCNsesC)) in soluble and insoluble fractions via Western blot assay. (C) The effect of S. aureus 8325–4 transformation with sesC. Introduction of a plasmid carrying sesC but not the mock plasmid changed the biofilm formation of transformants in BHI-NaCl and also the effects of PK and SM were opposite. (D) Microtiter plate assay demonstrating the effect of the inducers of biofilm formation (glucose and NaCl) and dispersal agents on biofilm formation of strains (E) quantification of PIA production using PIA non-specific immunoblot assay for biofilm in BHI-Glu. (F) Effect of transformation with sesK on the biofilm formation of strain 8325–4 and also the effect of sesC on the biofilm formation of strain MSSA4 in comparison with MRSA strain BH1CC. (SM: sodium metaperiodate, PK: proteinase K; error bars mean standard deviation)
In order to confirm the relation between SesC production and a phenotypic switch of biofilm growth, another ica-positive, PIA-dependent biofilm-forming S. aureus strain, the clinical isolate MSSA4, was transformed with pCNsesC. As observed in 8325–4, the biofilm phenotype of MSSA4 switched from PIA-dependent to proteinaceous following introduction of pCNsesC (Fig 1F).
Moreover, the effect of αSesC-IgGs on biofilm formation by strains 8325–4 and 8325–4 (pCNsesC) grown overnight in BHI-glucose was investigated. αSesC-IgGs had no effect on 8325–4 or 8325–4 carrying the empty plasmid, but inhibited biofilm formation by 8325–4 (pCNsesC) up to 80% (Fig 1C and 1D). Treatment with DNaseI did not have any significant impact on the biofilms (data not shown).
Scanning electron microscopy (SEM) images from 8325–4, 8325–4 with mock plasmid, 8325–4 (pCNsesC) and BH1CC biofilms, respectively, showed morphological differences (Fig 2). In BHI-glucose, strain 8325–4 formed porous and less condensed biofilms whereas 8325–4 (pCNsesC) formed more condensed and smoother biofilms with a glue-like matrix as seen in BH1CC biofilms. 8325–4 ica::tet did not form biofilm whereas 8325–4 ica::tet (pCNsesC) presented as a biofilm forming strain (Fig 2). To evaluate whether the phenomenon of biofilm phenotypic switching is due to the specific function of SesC or to high-level constitutive production of any LPxTG surface protein, the sesC gene in pCNsesC was replaced with sesK that encodes another LPxTG protein in S. epidermidis. Unlike sesC, which is present in all S. epidermidis strains, sesK is only present in circa 10% of S. epidermidis isolates [28]. Additionally, it was previously shown that anti-SesC antibodies could reduce S. epidermidis biofilm formation, whereas anti-SesK antibodies had no effect [28]. Transformation with pCNsesK had no impact on the biofilm phenotype of 8325–4 (Fig 1F), showing that the effect of heterologous expression of sesC in PIA-producing S. aureus may be specific.
Fig 2. SEM images of biofilms formed by 8325–4, its sesC-expressing transformants and BH1CC as controls.
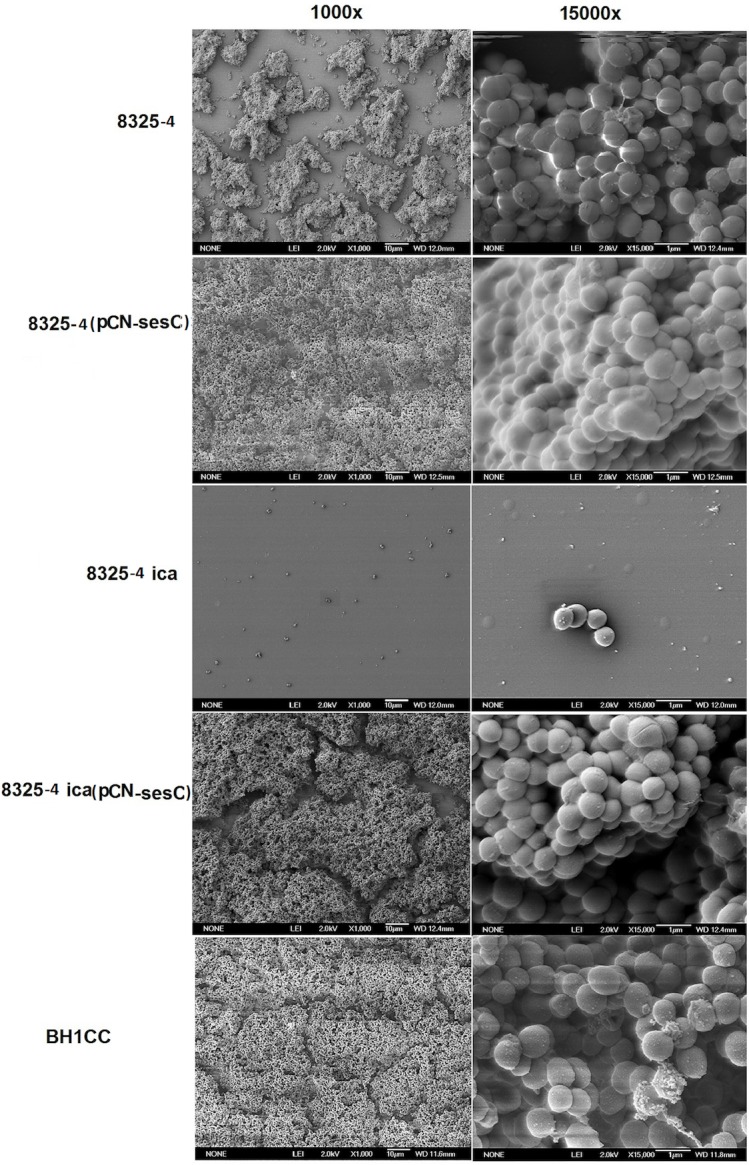
Biofilm growth on glass disks was allowed during overnight incubation at 37°C in BHI supplemented with 1% glucose. The next day, samples were fixed and sputter coated with platinum. The images show bacteria attached on the surface of disks at 1000x, 15000x magnification.
Expression of sesC promotes biofilm production by an ica mutant of 8325–4
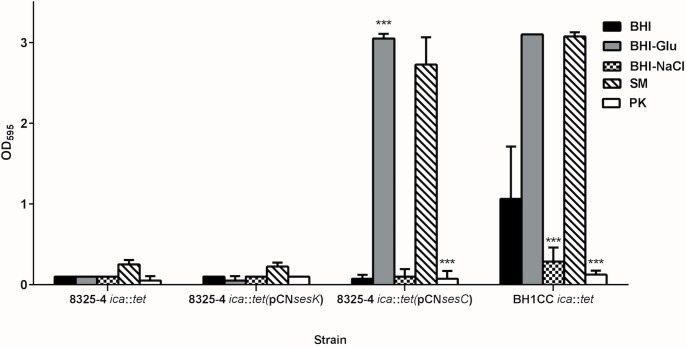
Previously published data revealed that deletion of the ica operon encoding PIA biosynthesis impaired PIA-dependent biofilm production by 8325–4 but had no impact on the biofilm formation by MRSA strain BH1CC which expresses an AtlE/FnBP-mediated biofilm phenotype [14]. Transformation of 8325–4 ica::tet with sesC—in contrast to sesK—restored the biofilm formation to approximately wild-type levels in BHI-glucose (Fig 3). Furthermore, SEM analysis showed that the morphology of 8325–4 ica::tet (pCNsesC) biofilms was similar to 8325–4 (pCNsesC) biofilm (data not shown). Interestingly, when grown in medium supplemented with NaCl, which induces a PIA-type biofilm, 8325–4 ica::tet (pCNsesC) was unable to produce biofilm while in BHI-glucose biofilm growth of 8325–4 ica::tet (pCNsesC) occurred. In addition, these biofilms were dispersed by PK, indicating a proteinaceous biofilm phenotype for this transformed mutant (Fig 3).
Fig 3. Effect of transformation with sesC and sesK on biofilm formation by non-biofilm-forming 8325 ica::tet mutant.
Transformation of this non-biofilm-forming mutant with sesC restored its biofilm formation and converted it to protein-mediated biofilm-forming strain that cannot form biofilm in the presence of NaCl and is dispersed by PK but not SM. Transformation with sesK had no effect. Mutation of the ica in PIA-independent biofilm-forming BH1CC strain had no effect on its biofilm formation. (SM: sodium metaperiodate, PK: proteinase K; error bars mean standard deviation)
Surface expression of SesC is involved in biofilm formation
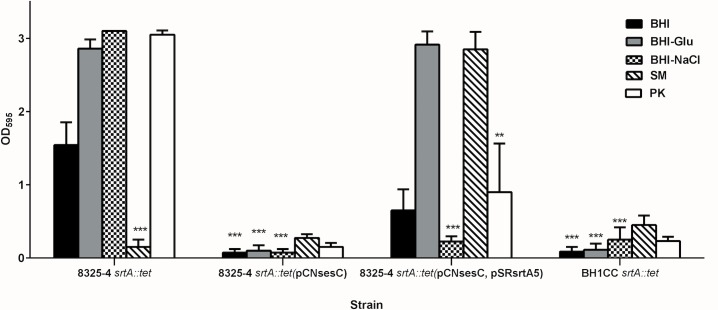
LPxTG proteins are known to be anchored to the bacterial peptidoglycan by sortases [37, 38, 39]. Deletion of srtA inhibits LPxTG protein-dependent biofilm formation. In 2008, O'Neill et al. reported that deletion of srtA in BH1CC impairs its biofilm forming activity, while biofilm formation of 8325–4 srtA::tet was not affected [26]. Introduction of pCNsesC in the latter strain, however, completely impaired biofilm production, further indicating the dominant role of SesC over PIA-type biofilm production (Fig 4). Unlike 8325–4, in the absence of sortase the 8325–4 srtA::tet (pCNsesC) strain was unable to form biofilm. The presence of SesC in the soluble and insoluble proteins fractions of the transformant strain was investigated and showed that in the 8325–4 srtA::tet (pCNsesC) SesC is only expressed in the soluble fraction (Fig 1B). This strongly suggests that presence of srt is necessary for sorting SesC to the cell wall.
Fig 4. Effect of transformation with sesC on biofilm formation by the biofilm-forming isogenic srtA mutants of 8325–4.
Effect of srtA mutation on biofilm formation of PIA-dependent biofilm-forming strain 8325–4, (Atl/FnBP)-mediated biofilm-forming strain BH1CC, the sesC-tranformed 8325–4 srtA::tet strain and complementation of 8325–4 srtA::tet (pCNsesC) with srtA, was evaluated using the quantitative microtiter plate assay. (SM: sodium metaperiodate, PK: proteinase K; error bars mean standard deviation)
Complementation of 8325–4 srtA:: tet (pCNsesC) with the plasmid pSRsrtA5 carrying the S. aureus srtA gene [26] restored biofilm formation (Fig 4). The biofilm of this complemented strain was only induced in BHI-glucose 1%, not in BHI-NaCl, and dispersed only with Proteinase K and not with sodium metaperiodate.
Heterologous expression of sesC increases colonization of polyurethane intravenous catheters in vitro and in vivo
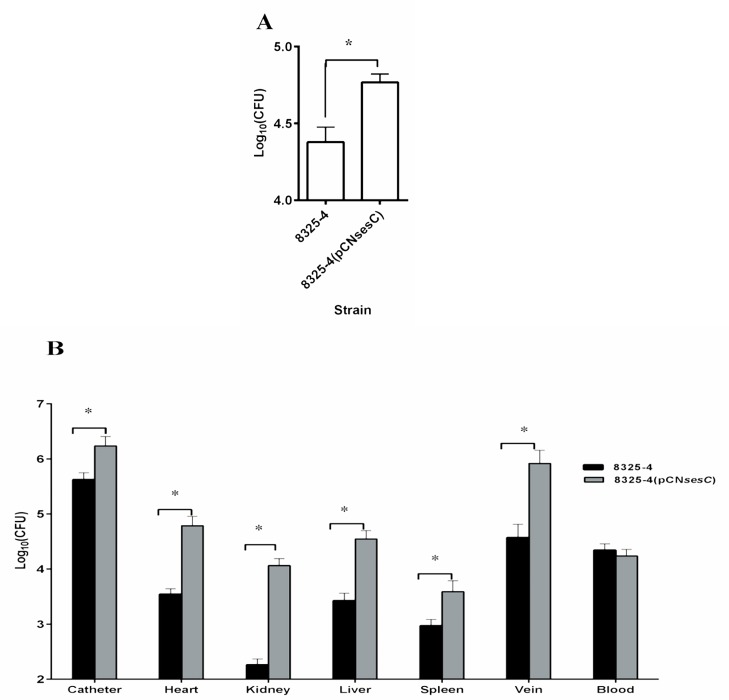
Transformation of S. aureus 8325–4 with sesC significantly increased its attachment to polyurethane intravenous catheters in vitro (P<0.01; 1-way ANOVA) (Fig 5A). Using a jugular vein catheterized (JVC) mouse model, the number of bacteria recovered from the catheter implanted in animals and afterwards infected with 8325–4 (pCNsesC) was also significantly higher than after administration of 8325–4 (P<0.05; 1-way ANOVA) (Fig 5B). Interestingly, 8325–4 (pCNsesC) did not only show an increased catheter colonization ability compared to the non-transformed strain, but also the overall infection rate was raised. The number of 8325–4 versus 8325–4 (pCNsesC) cells in blood were similar, but the number of 8325–4 (pCNsesC) cells recovered from organs such as spleen, liver, heart, vein and kidney were significantly increased compared to 8325–4 (10- to 100-fold, P<0.05; 1-way ANOVA) (Fig 5B). These results indicate that SesC is a colonization factor that may promote S. epidermidis catheter colonization, which in turn is the first step in biofilm formation and the establishment of a chronic infection.
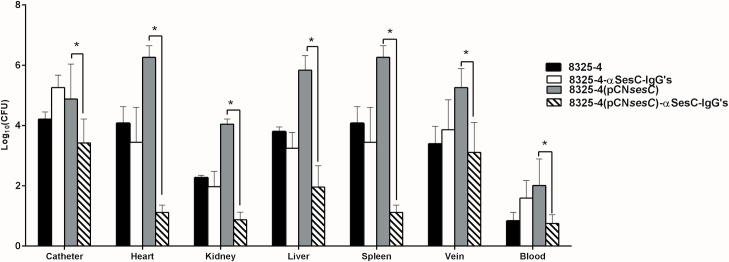
Fig 5. Effect of transformation with sesC on in vitro and in vivo catheter colonization and in vivo organ colonization.
(A) Seven mm catheter fragments were inoculated with 8325–4 or its sesC-expressing transformant. After 2 h incubation at 37°C, catheters were rinsed and adherent bacteria were detached by sonication and the numbers of bacteria recovered from each catheter fragments were quantified by quantitative cultures. (B) In vivo, catheterized animals were inoculated through the catheter lumen with 8325–4 strain or its 8325–4 (pCNsesC) transformant. At day 5 post-infection, the numbers of bacteria attached to the catheters or recovered from organs were quantified by quantitative cultures. The sesC-expressing transformant 8325–4, (pCNsesC) has a higher rate of colonization of different organs up to 100-fold compare to its parental strain. *: P<0.05
Antibodies against SesC have therapeutic benefit in an 8325–4 (pCNsesC)-induced catheter-related infection
The rate of catheter and organ colonization significantly decreased (100–100.000 fold; P<0.01–0.001; 1-way ANOVA) in the JVC mouse model group inoculated with 8325–4 (pCNsesC), which were pre-incubated with αSesC-IgGs versus untreated 8325–4 (pCNsesC) (Fig 6). Pre-immune IgGs had no significant effect on the catheter and organ colonization by 8325–4 or its sesC-expressing transformant (data not shown).
Fig 6. Effect of αSesC-IgGs on catheter colonization and infection rate by 8325–4 and its sesC-expressing transformant.
Catheterized animals were inoculated with bacteria pre-incubated with αSesC-IgGs or pre-immune IgGs for 2 h at 4°C. The next day, animals were sacrificed and the bacteria were recovered from catheter and organs and quantified by quantitative cultures. Pre-incubation with αSesC-IgGs significantly reduced the rate of catheter and organ colonization by 8325–4 (pCNsesC) from 10 to 10000-fold. *p<0.05
Discussion
Surface proteins have been shown to play important roles in S. epidermidis and S. aureus biofilm formation, especially by MRSE and MRSA in device-related infections [10, 14, 18, 40]. We previously reported that rabbit polyclonal antibodies directed against the extracellular domain of S. epidermidis LPXTG surface protein SesC (or αSesC-IgGs) can significantly inhibit S. epidermidis biofilm formation in vitro and in vivo in a rat model of subcutaneous catheter-related infection (CRI) as well as in a mouse model of jugular vein CRI [23, 28]. It has also been demonstrated that active immunization with the recombinantly produced extracellular domain of SesC decreased S. epidermidis biofilm formation in a rat model of subcutaneous CRI [28]. Data obtained in this study are consistent with previous observations and demonstrate that SesC plays an important role in biofilm formation even in another genetic background.
We have tried, but were unable to knockout sesC. It may be that knockout of sesC is associated with a lethal phenotype in these bacteria. On the other hand, using antisense RNAs to knock down sesC in S. epidermidis and subsequently applying gel-based reverse transcription-PCR (RT-PCR) assay and Western blot analysis, we could not see any changes in sesC expression in the transformed strain compared with the parental strain.
PCR screening to determine the incidence of sesC in 300 clinical isolates of Staphylococcus spp. including 175 S. epidermidis, 33 isolates of methicillin-resistant S. aureus (MRSA), 50 isolates of methicillin-sensitive S. aureus (MSSA) and 42 strains belonging to various non-epidermidis CoNS species, revealed that sesC is present in all S. epidermidis strains, but not in other staphylococci, indicating that it is potentially a conserved, S. epidermidis-specific gene.
Despite the lack of S. epidermidis sesC mutants, the unraveling of the function of SesC in biofilm formation was attempted by introduction of sesC into S. aureus strains that formed different types of biofilm (PIA-dependent, PIA-independent or non-biofilm-forming). Using gel-based RT-PCR assay and Western blot analysis, we confirmed sesC expression and the production of the corresponding protein in these transformed strains. Our findings revealed that transformation with sesC had no impact on the strains, which display proteinaceous biofilm phenotypes, while it caused a switch from PIA-dependent biofilm-formation to a proteinaceous-type biofilm in MSSA strains 8325–4 and MSSA4. As this biofilm phenotype switching was not associated with transformation of 8325–4 with sesK, it can be concluded that the switch is not necessarily associated with high expression level of any LPxTG surface protein or with the genetic background of the transformed strain, but is specifically caused by SesC. Results obtained also show that presence of SesC is dominant over the presence of PIA and impairs the role of PIA in cell-cell interaction.
To further confirm the direct involvement of SesC in biofilm formation, we transformed the non-biofilm-forming, isogenic ica mutant of S. aureus strain 8325–4 (being 8325–4 ica::tet) with sesC. Transformation with sesC converted this non-biofilm-forming mutant to a proteinaceous biofilm-forming strain similar to its parental strain transformed with sesC. These data suggest a direct role for SesC in biofilm formation, because even in the absence of PIA we have the same effect of SesC on biofilm formation.
To answer the question whether SesC affects PIA biofilm formation in mutated srtA strain, S. aureus 8325–4 srtA::tet was transformed with sesC. Deletion of srtA has no impact on biofilm formation of PIA-dependent biofilm-forming strain 8325–4, but impairs the normal display of LPxTG surface proteins, including SesC. After the introduction of sesC, we observed that this biofilm-forming strain converted to a non-biofilm-forming strain. To confirm the localization of SesC in 8325–4 srtA::tet (pCNsesC) strain, we checked the presence of SesC in the soluble and insoluble protein fraction. Since SesC is a cell wall-anchored LPXTG protein, basically we expected to find it in the insoluble fraction. However, SesC was observed only in the soluble part. These data suggest that the presence of sortase is necessary to sort SesC to the cell wall [41].
Complementation of the non-biofilm-forming sesC-expressing mutant with srtA by means of transformation with pSRsrtA5 converted it to a proteinaceous biofilm-forming strain. These data confirm that transformation with sesC is sufficient to switch the mechanism of biofilm formation to a proteinaceous type biofilm on condition that SesC is sorted to its place on the surface, attached to the peptidoglycan layer. Our data show that SesC expression does not have any negative effect on PIA production. There is some evidence that may explain this phenomenon. Previous groups reported that the generation of PIA is not sufficient to form biofilm [42]. Vergara-Irigaray et al. showed that the clinical MRSA strain 132 is able to alternate between a proteinaceous and a polysaccharidic biofilm matrix, depending on environmental conditions, and strain S115 generates PIA but is a non-biofilm-forming strain. This might be because of the existence of a defect in the export of PIA by IcaC or IcaB [42]. Similarly to the effect of FnAB on biofilm phenotype in S. aureus strain S132 and BH1CC, there is the possibility that here, the extracellular location of SesC changes the architecture of the cell wall to the extent that PIA can no longer link the cells. This hypothesis is consistent with the observed effect of biofilm dispersal agents on sesC- and ica-positive strains. The observation that high level expression of another surface protein (SesK) did not have a similar effect may suggest some form of interaction between SesC and PIA that is absent for SesK. Different expression levels of SesC and SesK in spite of using the same expression vectors may offer another explanation.
Expression of SasG, a surface protein in S. aureus, can similarly as SesC switch the biofilm of PIA-dependent strains SH1000 and 8325–4 to protein-mediated biofilm [39] SasG like SesC is a fibrinogen-binding protein, and SasG masked binding to fibrinogen mediated by both ClfB and the FnBPs. Biofilm formation by SasG is also likely to be protease dependent, because the broad spectrum protease inhibitor a2-macroglobulin inhibited the biofilm formation process of the strain SH1000 transformed with sasG [39]. By looking at the protein sequences alignment of SesC and SasG, we realized they have 26% identity.
Similar to the presented observations, a recent report illustrated the impact of introducing the methicillin resistant gene mecA into the PNAG-producing MSSA strain 8325–4 [19]. This generated a heterogeneously oxacillin resistant (HeR) strain, from which a homogeneous, high-level resistant (HoR) derivative was isolated following exposure to oxacillin. Transcription of icaADBC and production of PNAG were impaired in the 8325–4 HoR derivative, which instead produced a proteinaceous biofilm that was significantly inhibited by antibodies against the mecA-encoded penicillin binding protein 2a (PBP2a). HoR derivatives of 8325–4 icaADBC::tet, 8325–4 fnbAB::tet, 8325–4 atl::cat and 8325–4 srtA::tet exhibited a similar biofilm phenotype as 8325–4 HoR [19, 43].
SEM images confirm the presence of a morphologically different biofilm for 8325–4 in comparison to its transformant harboring pCNsesC. These findings are also supported by the effect of αSesC-IgGs on established biofilms of SesC-producing transformants and also in vitro catheter colonization data that suggest a role for SesC in attachment to the catheter surfaces.
The obtained in vivo data are consistent with our previous findings in suggesting an important role for SesC in infection. Transformation with sesC increased the organ infection rate up to 100-fold. This can be partially explained by the fact that fibrinogen is one of the components of the extracellular matrix [44]. Our previous report showed that transformation of S. aureus strain RN4220 with sesC increased the fibrinogen-binding ability of transformants, suggesting SesC as a potential Fg-binding MSCRAMM [23]. But, our experiments also show enhanced adherence of the transformant strains to an uncoated catheter in vitro, suggesting that the adherence effects are not entirely or solely mediated by binding to host factors.
Reduction of in vivo catheter and organ colonization by SesC-producing S. aureus strains in the presence of αSesC-IgGs indicate the specificity of the antibody, surface expression of SesC, and involvement of SesC in catheter and organ colonization.
Although we have to be cautious in extrapolating conclusions based on data obtained in S. aureus to S. epidermidis, we conclude that SesC is a virulence factor associated with the early stages in S. epidermidis biofilm formation, such as adhesion and colonization possibly favoring chronic, persistent infections on indwelling biomaterials. The biofilm formation versatility and flexibility of S. epidermidis may be due in part to the presence of SesC and similar factors that help S. epidermidis to adapt to changing environmental conditions.
In future studies, SesC can be considered as a valuable vaccine target against S. epidermidis infections.
Data Availability
All relevant data are within the paper.
Funding Statement
Laleh Khodaparast received no specific funding for this work.
References
- 1.O’Gara JP, Humphreys H. Staphylococcus epidermidis biofilms : importance and implications. Society. 2001;50: 582–587. [DOI] [PubMed] [Google Scholar]
- 2.Otto M. Staphylococcus epidermidis-the “accidental” pathogen. Nat Rev Microbiol. 2009;7: 555–567. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uçkay I, Pittet D, Vaudaux P, Sax H, Lew D, Waldvogel F. Foreign body infections due to Staphylococcus epidermidis. Ann Med. 2009;41: 109–119. 10.1080/07853890802337045 [DOI] [PubMed] [Google Scholar]
- 4.Raad I, Alrahwan a, Rolston K. Staphylococcus epidermidis: emerging resistance and need for alternative agents. Clin Infect Dis. 1998;26: 1182–1187. 10.1086/520285 [DOI] [PubMed] [Google Scholar]
- 5.Götz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43: 1367–1378. 10.1046/j.1365-2958.2002.02827.x [DOI] [PubMed] [Google Scholar]
- 6.Rawlinson LAB, O’Gara JP, Jones DS, Brayden DJ. Resistance of Staphylococcus aureus to the cationic antimicrobial agent poly(2-(dimethylamino ethyl)methacrylate) (pDMAEMA) is influenced by cell-surface charge and hydrophobicity. J Med Microbiol. 2011;60: 968–976. 10.1099/jmm.0.025619-0 [DOI] [PubMed] [Google Scholar]
- 7.O’Gara JP. ica and beyond: Biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270: 179–188. 10.1111/j.1574-6968.2007.00688.x [DOI] [PubMed] [Google Scholar]
- 8.Cheung GYC, Otto M. Understanding the significance of Staphylococcus epidermidis bacteremia in babies and children. Curr Opin Infect Dis. 2010;23: 208–216. 10.1097/QCO.0b013e328337fecb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzpatrick F, Humphreys H, O’Gara JP. Evidence for icaADBC -Independent Biofilm Development Mechanism in Methicillin-Resistant Staphylococcus aureus Clinical Isolates. J Clin Microbiol. 2005;43: 1973–1976. 10.1128/JCM.43.4.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennig S, Nyunt Wai S, Ziebuhr W. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int J Med Microbiol. 2007;297: 117–122. 10.1016/j.ijmm.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 11.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104: 8113–8118. 10.1073/pnas.0610226104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Büttner H, Mack D, Rohde H. Structural basis of Staphylococcus epidermidis biofilm formation: mechanisms and molecular interactions. Front Cell Infect Microbiol. 2015;5: 1–15. 10.3389/fcimb.2015.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christner M, Franke GC, Schommer NN, Wendt U, Wegert K, Pehle P, et al. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol. 2010;75: 187–207. 10.1111/j.1365-2958.2009.06981.x [DOI] [PubMed] [Google Scholar]
- 14.Houston P, Rowe SE, Pozzi C, Waters EM, O’Gara JP. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79: 1153–1165. 10.1128/IAI.00364-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziebuhr W, Krimmer V, Rachid S, Lössner I, Götz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32: 345–356. 10.1046/j.1365-2958.1999.01353.x [DOI] [PubMed] [Google Scholar]
- 16.Conlon KM, Humphreys H, O’Gara JP. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 2002;184: 4400–4408. 10.1128/JB.184.16.4400-4408.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knobloch JKM, Bartscht K, Sabottke A, Rohde H, Feucht HH, Mack D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: Differential activation mechanisms due to ethanol and salt stress. J Bacteriol. 2001;183: 2624–2633. 10.1128/JB.183.8.2624-2633.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mack D, Davies AP, Harris LG, Rohde H, Horstkotte MA., Knobloch JKM. Microbial interactions in Staphylococcus epidermidis biofilms. Anal Bioanal Chem. 2007;387: 399–408. 10.1007/s00216-006-0745-2 [DOI] [PubMed] [Google Scholar]
- 19.Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012;8 10.1371/journal.ppat.1002626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis SL, Gurusiddappa S, McCrea KW, Perkins S, Höök M. SdrG, a Fibrinogen-binding Bacterial Adhesin of the Microbial Surface Components Recognizing Adhesive Matrix Molecules Subfamily from Staphylococcus epidermidis, Targets the Thrombin Cleavage Site in the Bβ Chain. J Biol Chem. 2001;276: 27799–27805. 10.1074/jbc.M103873200 [DOI] [PubMed] [Google Scholar]
- 21.Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, et al. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151: 1453–1464. 10.1099/mic.0.27534-0 [DOI] [PubMed] [Google Scholar]
- 22.Arrecubieta C, Toba FA, Von Bayern M, Akashi H, Deng MC, Naka Y, et al. SdrF, a Staphylococcus epidermidis surface protein, contributes to the initiation of ventricular assist device driveline-related infections. PLoS Pathog. 2009;5 10.1371/journal.ppat.1000411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahrooei M, Hira V, Stijlemans B, Merckx R, Hermans PWM, Van Eldere J. Inhibition of Staphylococcus epidermidis biofilm formation by rabbit polyclonal antibodies against the SesC protein. Infect Immun. 2009;77: 3670–3678. 10.1128/IAI.01464-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Wijngaerden E, Peetermans WE, Vandersmissen J, Van Lierde S, Bobbaers H, Van Eldere J. Foreign body infection: a new rat model for prophylaxis and treatment. J Antimicrob Chemother. 1999;44: 669–674. [DOI] [PubMed] [Google Scholar]
- 25.Kreiswirth BN, Löfdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS NR. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. [Internet]. Nature. 1983; 709–12. doi:20–26;305(5936):709–12. [DOI] [PubMed] [Google Scholar]
- 26.O’Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, et al. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190: 3835–3850. 10.1128/JB.00167-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. Novel Cassette-Based Shuttle Vector System for Gram-Positive Bacteria Novel Cassette-Based Shuttle Vector System for Gram-Positive Bacteria. Society. 2004;70: 6076–6085. 10.1128/AEM.70.10.6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shahrooei M, Hira V, Khodaparast L, Khodaparast L, Stijlemans B, Kucharíková S, et al. Vaccination with SesC Decreases Staphylococcus epidermidis biofilm formation. Infect Immun. 2012;80: 3660–3668. 10.1128/IAI.00104-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55: 1883–1895. 10.1111/j.1365-2958.2005.04515.x [DOI] [PubMed] [Google Scholar]
- 30.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JK, et al. Genes Involved in the Synthesis and Degradation of Matrix Polysaccharide in. Society. 2004;186: 8213–8220. 10.1128/JB.186.24.8213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh R, Ray P, Das A, Sharma M. Enhanced production of exopolysaccharide matrix and biofilm by a menadione-auxotrophic Staphylococcus aureus small-colony variant. J Med Microbiol. 2010;59: 521–527. 10.1099/jmm.0.017046-0 [DOI] [PubMed] [Google Scholar]
- 32.Hu J, Xu T, Zhu T, Lou Q, Wang X, Wu Y, et al. Monoclonal antibodies against accumulation-associated protein affect EPS biosynthesis and enhance bacterial accumulation of Staphylococcus epidermidis. PLoS One. 2011;6 10.1371/journal.pone.0020918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pintens V, Massonet C, Merckx R, Vandecasteele S, Peetermans WE, Knobloch JKM, et al. The role of σB in persistence of Staphylococcus epidermidis foreign body infection. Microbiology. 2008;154: 2827–2836. 10.1099/mic.0.2007/015768-0 [DOI] [PubMed] [Google Scholar]
- 34.Lazzell AL, Chaturvedi AK, Pierce CG, Prasad D, Uppuluri P, Lopez-Ribot JL. Treatment and prevention of Candida albicans biofilms with caspofungin in a novel central venous catheter murine model of candidiasis. J Antimicrob Chemother. 2009;64: 567–570. 10.1093/jac/dkp242 [DOI] [PubMed] [Google Scholar]
- 35.Herijgers P, Oosterlinck W, Vanderper A, Flameng W. Glucose tolerance and left ventricular pressure-volume relationships in frequently used mouse strains. J Biomed Biotechnol. 2011;7 10.1155/2011/281312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marraffini L a, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70: 192–221. 10.1128/MMBR.70.1.192-221.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: A nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156: 289–297. 10.1016/j.resmic.2004.10.011 [DOI] [PubMed] [Google Scholar]
- 38.Dramsi S, Magnet S, Davison S, Arthur M. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol Rev. 2008;32: 307–320. 10.1111/j.1574-6976.2008.00102.x [DOI] [PubMed] [Google Scholar]
- 39.Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153: 2435–2446. 10.1099/mic.0.2007/006676-0 [DOI] [PubMed] [Google Scholar]
- 40.Otto M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: Staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. BioEssays. 2013;35: 4–11. 10.1002/bies.201200112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta—Biomembr. 2004;1666: 105–117. 10.1016/j.bbamem.2004.04.011 [DOI] [PubMed] [Google Scholar]
- 42.Vergara-Irigaray M, Valle J, Merino N, Latasa C, García B, De Los Mozos IR, et al. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect Immun. 2009;77: 3978–3991. 10.1128/IAI.00616-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarthy H, Rudkin JK, Black NS, Gallagher L, O’Neill E, O’Gara JP. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front Cell Infect Microbiol. 2015;5: 1–9. 10.3389/fcimb.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereira M, Rybarczyk BJ, Odrljin TM, Hocking DC, Sottile J, Simpson-Haidaris PJ. The incorporation of fibrinogen into extracellular matrix is dependent on active assembly of a fibronectin matrix. J Cell Sci. 2002;115: 609–617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.