Abstract
Butyrate is a nutritional element with strong epigenetic regulatory activity as a histone deacetylase inhibitor. Based on the analysis of differentially expressed genes in the bovine epithelial cells using RNA sequencing technology, a set of unique genes that are activated only after butyrate treatment were revealed. A complementary bioinformatics analysis of the functional category, pathway, and integrated network, using Ingenuity Pathways Analysis, indicated that these genes activated by butyrate treatment are related to major cellular functions, including cell morphological changes, cell cycle arrest, and apoptosis. Our results offered insight into the butyrate-induced transcriptomic changes and will accelerate our discerning of the molecular fundamentals of epigenomic regulation.
Keywords: butyrate, bovine cells, histone modification, gene regulation, transcriptomic
Introduction
Butyrate is a nutritional element produced by bacterial fermentation of dietary fibers in the digestive track of all mammals. Ruminant species metabolize the short-chain fatty acids (acetate, propionate, and butyrate) to fulfill up to 70% of their nutrient energy requirements. It is now well established that microbial fermentation in the gastrointestinal tract contributes to the energy balance of all mammalian species.1,2 This can be of great advantage to animals and human beings, since no digestive enzymes exist for breaking down cellulose or other complex carbohydrates. The large number of bacterial species, the complex nature of their interactions, and the end products of their fermentation processes are all likely to have significance in human and animal health.
Butyrate appears to be involved in metabolism in ways that differ from its role as a nutrient. It has also proven to be a strong regulator of genomic activities. Butyrate participates in metabolism both as a nutrient and as an inhibitor of his-tone deacetylases (HDACs). The major biochemical change that occurs in cells treated with butyrate is the global hyperacetylation of histones. In our previously reported studies, the potential biological roles of butyrate were also investigated, using the established Madin-Darby bovine kidney (MDBK) epithelial cell line. This study focused on determining whether normal bovine cells in a standard cell culture condition were sensitive to the growth inhibitory effects of butyrate. Butyrate not only induced apoptosis but also induced cell cycle arrest at the G1/S boundary and G2/M in MDBK cells. The cell responses were concentration dependent.3
Histone modifications, especially histone acetylation and methylation, play a dominant role in epigenomic regulation. Butyrate is an important player in inducing histone acetylation. For many years, it has been recognized that consumption of dietary fibers has many positive metabolic health effects, such as increasing satiety and lowering blood glucose and cholesterol levels, as well as inducing cell differentiation and apoptosis. These effects may be associated with short-chain fatty acids, particularly propionic and butyric acids. In the past decade, the multiple beneficial effects of butyrate at the intestinal and extraintestinal levels have been demonstrated. Butyrate has different mechanisms of action, and many of these involve epigenetic regulation of gene expression through the inhibition of HDACs. There is a growing interest in butyrate because its impact on epigenetic mechanisms could lead to more specific and efficacious therapeutic strategies for the prevention and treatment of different diseases, ranging from genetic/metabolic conditions to neurological degenerative disorders, as well as cancer.4 Butyrate as a cancer prevention and therapeutic reagent has been extensively studied and reviewed5–8
At physiologic concentrations, butyrate regulates the expression of individual genes by inhibition of histone deacetylation and attendant chromatin remodeling.4,9,10 Utilizing global gene expression profiling, recent studies indicate that butyrate induces significant changes in the expression of genes associated with the regulatory pathways critical to cell growth, immune response, and signal transduction.3,11,12 A majority of these genes is repressed by butyrate and is associated with cell cycle control. Next-generation sequencing (NGS) has provided unique tools for transcriptomic profiling and genome analysis.13–15 As a fundamental step toward an ample understanding of the molecular mechanisms of butyrate-induced acetylation and its biological effects, NGS technology was utilized to give a more complete categorization of the RNA transcripts of MDBK cells.16 In this study, we performed further bioinformatics analysis of the entire transcriptome of cell populations with or without butyrate treatment and identified a unique group of genes that only become activated after butyrate treatment. In order to further understand the process, we analyzed the transcriptome alterations induced by butyrate and discovered a set of genes uniquely expressed only after butyrate treatment. We also conducted a complementary bioinformatics analysis of the functional category, pathway, and integrated network using Ingenuity Pathways Analysis (IPA, http://www.ingenuity. com) of those uniquely expressed genes to identify the possible mechanisms underlying gene expression regulation induced by butyrate.
Methods and Materials
Cell culture and butyrate treatment
Bovine epithelial cells (MDBK epithelial cells, American Type Culture Collection, Catalog No. CCL-22) were used in this study. The culture condition and butyrate treatment were described in our previous report.3 Briefly, at ~50% confluence, the cells were treated with 10 mM sodium butyrate (Calbiochem) for 24 hours. A total of eight samples (four replicate flasks of control cells and four replicates of butyrate-treated [BT] cells) were used for the RNA sequencing technology (RNA-seq) experiments.
RNA extraction and sequencing using RNA-seq
Total RNA was extracted using Trizol (Invitrogen). Any potential DNA contamination was cleaned by DNase digestion and Qiagen RNeasy column (Qiagen), as previously described.11 The RNA integrity was verified using an Agilent Bioanalyzer 2100 (Agilent). An Illumina TruSeq RNA sample preparation kit was used to prepare a high-quality RNA sequencing library (RNA integrity number [RIN] >9.0) by following the manufacturer’s instruction (Illumina). RNA sequencing was performed at 50 bp/sequence using an Illumina HiSeq 2000 sequencer, as described previously.17 Approximately 67.5 million reads per sample (mean ± standard deviation [SD] = 67,527,111 ± 8,330,388.6) were generated.
Bioinformatics and data analysis
Raw sequence reads were first through the quality control pipeline. TopHat18 was used to align trimmed reads to the bovine reference genome (Btau 4.0). Mapped reads were normalized based on the upper-quartile normalization method and Cuffdiff was used to model the variance in fragment counts across replicates using the negative binomial distribution as described previously. The mean number of sequence reads generated per sample was 67,527,111 ± 8,330,388.58 (±SD). A total of eight replicates were used in the two groups, BT and phosphate-buffered saline treated (control [CT]; N = 4 for each group). Raw sequence reads were deposited to the NCBI Sequence read Archive (SRA accession number 051007). Uniquely expressed genes were defined as those RNAs that were only detected in all four replicates of butyrate-treated data sets.
IPA (Ingenuity® Systems, and www.ingenuity.com) was used to further evaluate molecular processes, molecular functions, and genetic networks of differentially expressed genes.19–23 IPA is a web-based software application that allows analysis, integration, and understanding of data from gene expression. Data analysis and search capabilities help in understanding the significance of data, specific targets, or candidate biomarkers in the context of larger biological or chemical systems. The software is supported by the Ingenuity Knowledge Base of highly structured, detail-rich biological, and chemical findings.
Results and Discussion
Butyrate-induced differential expression of genes and unique genes detected with deep transcriptomic sequencing
High-throughput RNA sequencing has opened a new horizon for revealing global profiles in gene expression allowing the detection of differential transcription, transcript isoforms, alternative splicing, and genomic structural variation. To decipher the gene regulatory mechanisms induced by butyrate treatment, we used untransformed normal bovine epithelial cells (MDBK) exposed to 10 mM butyrate for 24 hours, which is an established model for butyrate stimulation.3,11,12 Using high-throughput RNA sequencing, we generated 57,303,693 to 78,933,744 sequencing reads per sample (NIH NCBI Sequence Read Archive [SRA] 051007; database accession numbers SRX130570, SRX130571, SRX130572, SRX130573, SRX130576, SRX1305680, SRX130582, and SRX130583). There were 24,526 genes that were detected in at least one sample. There were 16,212 genes that were detected in all the samples and these were accounted for as the core transcriptome of the bovine epithelial cell. In the BT group, 19,477 ± 155 (mean ± SD) genes were identified, while 17,626 ± 124 (mean ± SD) genes were identified in the control group. Figure 1 summarizes the results of RNA sequencing analysis. RNA-seq data from BT cells and control cells allow for the detection of differentially expressed genes of biological significance.16 Figure 1A shows the volcano plot of differential RNA expression between the control group and BT cells. Among those differentially expressed genes, we observed expression changes in over 11,000 genes at a strict false discovery rate of 0.05 (Fig. 1B). The principal component analysis shows the gene expression patterns of control cells and butyrate-treated cells (four replicates each) as two distinctive components (Fig. 1C). To generate a unique set of bovine genes induced by butyrate, cuffcompare was implemented in this study. We detected 588 genes expressed in BT cells of four replicates but not in the normal control cells of the four replicates (Supplementary Table 1). Very interestingly, among these 588 genes, we detected 241 novel genes that have not been annotated. Some of these novel genes are abundantly expressed with butyrate treatment (Supplementary Table 1). The Manhattan plots in Figure 2 show the total number of differentially expressed genes (lower panel) and unique genes in BT cells with genomic coordinates on 29 autosomal chromosomes as well as the X chromosome.
Figure 1.
Summary of RNA sequencing data. (A) Volcano plot of differentially expressed genes between control and butyrate-treated cells (B) Heat map of all differentially expressed genes, legend: −log 2-fold changes. (C) The principal component analysis shows gene expression patterns of control cells and butyrate-treated cells (four replicates each) as two distinctive components. Transcriptomics data were visualized using CLC Genomics Workbench software.
Figure 2.
Manhattan plots of differentially expressed genes chromosomal distribution. Lower panel shows all differentially expressed genes and upper panel shows the genes only expressed with butyrate treatment.
Comprehensive function and pathway analyses of unique genes activated by butyrate treatment
In order to further systematically discover more about the biological function of these unique genes, the function and pathway analysis of uniquely activated genes in cells treated with butyrate were investigated using the IPA. The biological functions that were most significantly enhanced in the data set were identified. When a functional analysis of the genes was performed, genes from the data sets that were associated with biological functions were cataloged, and P values were calculated with Fischer’s exact test. The top molecular and cellular functions, as determined by P-value, are shown in Figure 3. These functions may represent, at least partially, the mechanisms related to the important biological effects of butyrate treatment. These functions, including cell-to-cell signaling, cellular movement, antigen presentation, and nucleic acid metabolism, were directly related to the phenomena induced by butyrate treatment, such as cell cycle arrest, cell morphology changes, and apoptosis.3,12 The perturbation of these functions is likely responsible for the cell cycle arrest at the G1, as a result of butyrate treatment.3
Figure 3.
Global functional analysis. The top molecular and cellular functions altered by butyrate-activated genes were summarized. Data sets were analyzed by the IPA software (Ingenuity® Systems, www.ingenuity.com). The significant value associated with a function in global analysis is a measure for how likely it is that genes from the data set file under investigation participate in that function. The significance is expressed as a P-value, which is calculated using the right-tailed Fisher’s exact test.
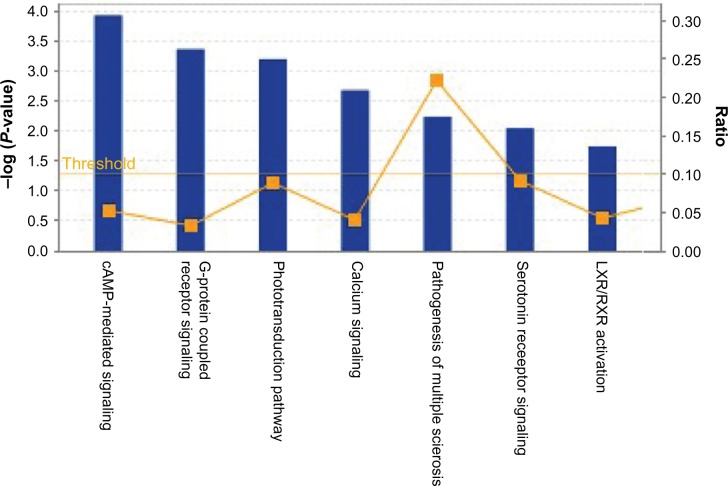
In addition to the functional annotation, IPA analysis also revealed that genes uniquely expressed due to butyrate treatment are involved in some very important canonical pathways (Fig. 4). The canonical pathways include 3′,5′-cyclic adenosine monophosphate (cAMP)-mediated signaling and G protein-coupled receptor (GPCR) signaling. GPCRs are a large family of integral membrane proteins that respond to a variety of extracellular stimuli.24 GPCRs transducer the information provided by these stimuli into intracellular secondary messengers that are interpreted as meaningful signals by the cell. This process involves the coupling of agonist-activated GPCRs to a wide variety of effector systems via their interaction with heterotrimeric guanine nucleotide-binding proteins (G proteins). It is now apparent that GPCR activity and function is regulated by an incredible variety of mechanisms. These mechanisms act at the level of GPCR ligand specificity, G protein activation, and effector regulation. The cAMP-dependent pathway, also known as the adenylyl cyclase pathway, is a GPCR-triggered signaling cascade used in cell communication.25,26 Butyrate treatment induced GPCR and cAMP-dependent pathway activation indicates that these pathways might be involved in the regulation of the biological effects of butyrate. Further investigations are necessary to understand how does butyrate perturb intracellular signal transduction of these signaling pathways.
Figure 4.
Global Canonical Pathway analysis: data sets were analyzed by the IPA software (Ingenuity® Systems, www.ingenuity.com). The significance is expressed as a P-value, which is calculated using the right-tailed Fisher’s exact test.
Upstream regulators and transcription factors
IPA analysis also revealed potential upstream regulators that may be involved in the activation of this group of unique genes. Transcription factors control the transcription of genetic information from DNA to mRNA by binding to specific DNA sequences.27 Transcription factors can function either as activators or as repressors for the expression of specific genes. Table 1 lists the major transcription regulators (factors) and other upstream regulators identified by IPA analysis. Only the upstream regulators with a regulation Z-score, either positive or negative, are listed. A Z-score is a statistical measurement of a score’s relationship to the mean in a set of scores. The positive or negative Z-scores suggest whether the score is above or below the mean and by how many SDs. In addition to these transcription factors, some cell signaling factors, such as cytokines, as well as growth factors, may be involved in the activation of this group of genes. Their regulatory target genes in the data set revealed by IPA analysis are also listed in Table 1. It is worth noting that among all those upstream regulators, LEP, the gene that encodes protein leptin, is itself upregulated. Leptin was initially thought to have a role in energy homeostasis and obesity. More recently, it has also been implicated in the regulation of reproduction, glucose homeostasis, bone formation, wound healing, and the immune system.
Table 1.
Upstream regulators and their targets in the data set.
| UPSTREAM REGULATOR | MOLECULE TYPE | PREDICTED ACTIVATION STATE | ACTIVATION Z-SCORE | P-VALUE OF OVERLAP | TARGET MOLECULES IN DATASET |
|---|---|---|---|---|---|
| IL6 | Cytokine | Activated | 3.202 | 3.05E-03 | CALCA, CCK, CCL24, CCL4, CCR1, CD53, CD79A, CDX2, CHRNB4, EOMES, LEFTY2, LEP, LIN28A, POMC, SCGB2A2, SERPINA1, SP7 |
| CEBPB | Transcription regulator | Activated | 3.044 | 2.24E-03 | ADRB3, AOC1, APLNR, CCL4, CSF3, CYP11A1, FKBP6, LEP, POMC, SEPT3, SERPINA1, TNFAIP6 |
| STAT3 | Transcription regulator | Activated | 2.809 | 2.24E-02 | CCL4, CCR1, CSF3, CXCR2, EOMES, FLRT3, IHH, LEP, NR0B1, POMC, SERPINA1 |
| NFkB (complex) | Complex | Activated | 2.752 | 8.68E-03 | CAMP, CCK, CCL4, CSF3, EOMES, GAD1, IFNA16, MSTN, MT3, SFTPA1, SLPI, TNFAIP6, TNFSF14, TNFSF4 |
| CD44 | Enzyme | Activated | 2.646 | 4.69E-03 | CCL19, CCL4, CD8B, CDX2, CLEC7A, NANOG, SERPINA1 |
| L-triiodothyronine | Chemical-endogenous mammalian | Activated | 2.617 | 7.70E-03 | ALPI, APOA5, CMKLR1, CYP11A1, CYP4A11, DIO3, LEP, MYOD1, NEUROG3, PTGDS |
| IGF1 | Growth factor | Activated | 2.482 | 6.78E-03 | CAMP, CCL4, CYP11A1, INSL3, LEP, MSTN, MYOD1, NEUROG3, PLP1, SLPI, SP7 |
| ERK | Group | Activated | 2.425 | 1.70E-02 | CCK, CCL4, CCR1, DIO3, PIWIL1, SP7, STMN4 |
| LEP | Growth factor | Activated | 2.297 | 4.13E-04 | ADRB3, AQP7, AQP9, CCK, CSF3, CYP11A1, CYP4A11, HCRT, INSL3, LEP, MYOD1, PLP1, POMC, RETN |
| RBPJ | Transcription regulator | Inhibited | −2.236 | 4.29E-03 | CAMP, CSF3, GDF10, GDF5, LEFTY2, MSTN |
| NEUROG3 | Transcription regulator | Activated | 2.236 | 1.50E-03 | CCK, GIP, NEUROD2, NEUROG3, PPY |
| PPIF | Enzyme | Activated | 2.236 | 5.00E-03 | CCL4, CCR1, CMKLR1, CXCR2, GDF5 |
| MEF2C | Transcription regulator | Activated | 2.224 | 5.28E-04 | KCNA5, MYH1, MYL2, MYOD1, SFRP2, SP7 |
| MAP3K8 | Kinase | Activated | 2.213 | 2.78E-02 | ADORA3, CCL4, CCR1, CD70, CSF3 |
| D-glucose | Chemical-endogenous mammalian | Activated | 2.187 | 3.79E-01 | HCRT, LEP, MSTN, PPY, PTGDS, RETN, TRPV6 |
| Butyric acid | Chemical-endogenous mammalian | Activated | 2.055 | 1.17E-01 | ALPI, CAMP, CCK, CCR1, CYP4A11, LEP, MYL2, PTGDS |
| STAT5A | Transcription regulator | Inhibited | −2.000 | 1.78E-02 | CD70, EOMES, MYH1, MYL2, TNFRSF18, TNFSF4 |
| CDKN2A | Transcription regulator | Activated | 2.000 | 1.99E-01 | CCL19, CCL24, CCL4, CXCR2, MYL2 |
| CORT | Other | −1.982 | 4.91E-04 | CCL19, CCL4, CCR1, POMC |
The biologically relevant networks in the uniquely expressed gene data set
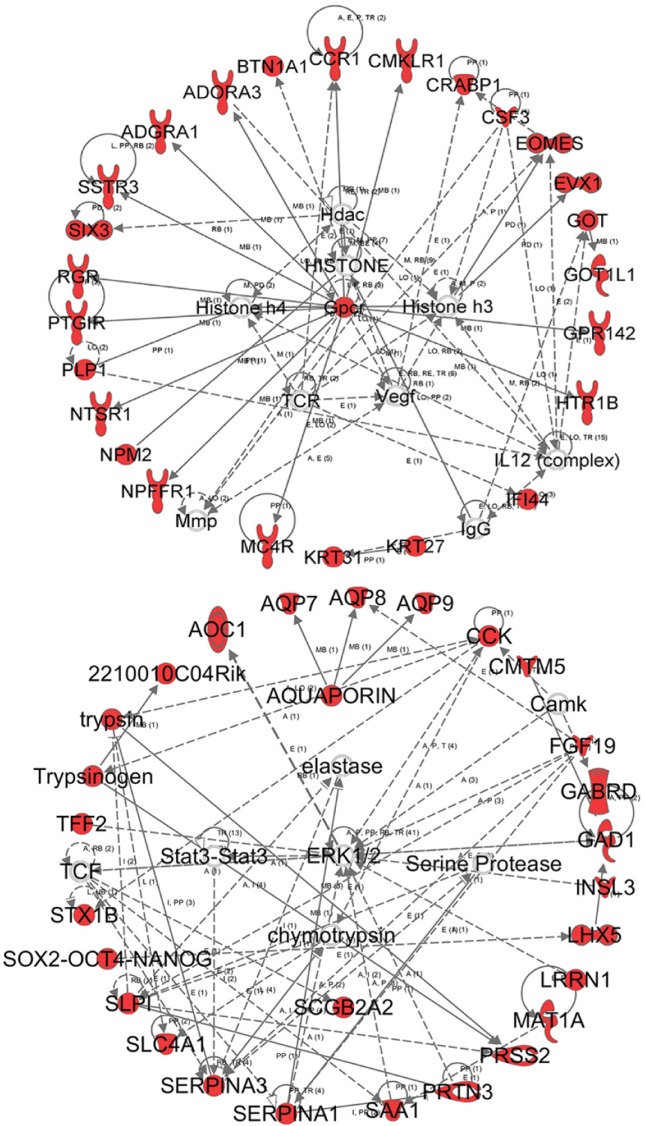
The IPA analysis was also used to determine the biologically relevant networks among the uniquely expressed genes, as well as the potential association with the uniquely expressed genes induced by butyrate. The top two networks are presented in Figure 5. The first network has associated network functions of cell signaling, nucleic acid metabolism, and small molecular biochemistry. This network also shows the involvement of HDACs and histone 3 and histone 4 in the perturbation of the functional gene network and provides a direct correlation of gene functions and butyrate as an HDAC inhibitor (HDACi). The second network has associated network functions of inflammatory response, cellular movement, and hematological system development and function. The associated network functions underlie the biological effects of the butyrate treatment and shine light on the mechanisms by which histone modification is induced by butyrate treatment.
Figure 5.
The biologically relevant network: the data set of uniquely expressed genes was analyzed by the IPA software (Ingenuity® Systems, www.ingenuity.com). The color indicates the expression level of the genes (red indicating upregulated genes and green indicating downregulated genes). The upper panel shows the first network with 24 focus genes, which are involved in biological functions, such as cell signaling, nucleic acid metabolism, and small molecule biochemistry. The lower panel shows the second network with 24 focus genes, which are involved in biological functions. such as tissue morphology, carbohydrate metabolism, and molecular transport.
Using HDAC inhibitors to study histone acetylation and gene regulation contributed significantly to our knowledge of histone modification and epigenetics regulation. Our previous studies3,11,16 revealed that butyrate participates in metabolism as regulators of histone modification and provides an example of nutrient-induced specific modulation of epigenetic regulation. Development of the NGS technique has presented unique tools for transcriptomic profiling and for genome analysis.15,28 We utilized NGS technology to completely characterize the RNA transcripts of MDBK cells and the effects of butyrate treatment.16 This is a crucial step toward a broad comprehension of the epigenetic system of butyrate-induced hyperacetylation, as well as its biological consequences. The current report is a complementary study to compare the transcriptomes of the normal cells and the cells treated with 10 mM butyrate for 24 hours. Our results uncovered the unique set of genes activated by butyrate treatment. With the technical advantages of NGS deep-sequencing and with an average of >67 million reads per sample, the differential expression of genes induced by butyrate treatment was revealed in much more detail than that of our earlier study that used a microarray. Our previous study, using microarray technique, detected only 450 genes as significantly regulated by butyrate treatment.11 However, using NGS deep sequencing, ~11,408 genes were detected as significantly impacted by butyrate.16 NGS also facilitated the revealing of a unique group of genes activated by butyrate treatment from all differentially expressed genes, as described in this report.
Butyrate exerts a very broad range of effects on many biological pathways via its inhibitory ability on HDAC. The genes that were identified (Supplementary Table 1), which fall within a broad range of functional categories, appeared to provide the molecular basis for its biological effects. The pathway analysis on the butyrate-activated genes, using the Ingenuity Pathway Analysis tool, successfully identified remarkable changes in genetic networks, transcription regulators, and functions related to various cell functional activities. These uniquely expressed genes, induced by butyrate treatment, revealed a basis for understanding the molecular mechanisms of the biological effects of butyrate provided by global gene expression profiling using deep RNA sequencing. It is also an expedient step toward understanding the histone modification as an important epigenomic regulatory mechanism.
Supplementary Materials
Supplementary Table 1. Uniquely expressed genes activated by butyrate.
Footnotes
ACADEMIC EDITOR: James Willey, Editor in Chief
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1493 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
CJL, RWL, WL and SW analyzed the data. RLB and LAB contributed to the construction of the manuscript. CJL wrote the manuscript and made critical revisions. All authors reviewed and approved of the final manuscript.
Disclaimer
Mention of a product, reagent, or source does not constitute an endorsement by the USDA to the exclusion of other products or services that perform a comparable function. The US Department of Agriculture is an equal opportunity provider and employer.
REFERENCES
- 1.Andoh A, Tsujikawa T, Fujiyama Y. Role of dietary fiber and short-chain fatty acids in the colon. Curr Pharm Des. 2003;9(4):347–58. doi: 10.2174/1381612033391973. [DOI] [PubMed] [Google Scholar]
- 2.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70(2):567–90. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 3.Li CJ, Elsasser TH. Butyrate-induced apoptosis and cell cycle arrest in bovine kidney epithelial cells: involvement of caspase and proteasome pathways. J Anim Sci. 2005;83(1):89–97. doi: 10.2527/2005.83189x. [DOI] [PubMed] [Google Scholar]
- 4.Myzak MC, Dashwood RH. Histone deacetylases as targets for dietary cancer preventive agents: lessons learned with butyrate, diallyl disulfide, and sulforaphane. Curr Drug Targets. 2006;7(4):443–52. doi: 10.2174/138945006776359467. [DOI] [PubMed] [Google Scholar]
- 5.Khan S, Jena G. The role of butyrate, a histone deacetylase inhibitor in diabetes mellitus: experimental evidence for therapeutic intervention. Epigenomics. 2015;7(4):669–80. doi: 10.2217/epi.15.20. [DOI] [PubMed] [Google Scholar]
- 6.Ong TP, Moreno FS, Ross SA. Targeting the epigenome with bioactive food components for cancer prevention. J Nutrigenet Nutrigenomics. 2011;4(5):275–92. doi: 10.1159/000334585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paparo L, di Costanzo M, di Scala C, et al. The influence of early life nutrition on epigenetic regulatory mechanisms of the immune system. Nutrients. 2014;6(11):4706–19. doi: 10.3390/nu6114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berni Canani R, Di Costanzo M, Leone L. The epigenetic effects of butyrate: potential therapeutic implications for clinical practice. Clin Epigenetics. 2012;4(1):4. doi: 10.1186/1868-7083-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parab S, Nankova BB, La Gamma EF. Differential regulation of the tyrosine hydroxylase and enkephalin neuropeptide transmitter genes in rat PC12 cells by short chain fatty acids: concentration-dependent effects on transcription and RNA stability. Brain Res. 2007;1132(1):42–50. doi: 10.1016/j.brainres.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Della Ragione F, Criniti V, Della Pietra V, et al. Genes modulated by histone acetylation as new effectors of butyrate activity. FEBS Lett. 2001;499(3):199–204. doi: 10.1016/s0014-5793(01)02539-x. [DOI] [PubMed] [Google Scholar]
- 11.Li RW, Li C. Butyrate induces profound changes in gene expression related to multiple signal pathways in bovine kidney epithelial cells. BMC Genomics. 2006;7:234. doi: 10.1186/1471-2164-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li CJ, Li RW, Wang YH, et al. Pathway analysis identifies perturbation of genetic networks induced by butyrate in a bovine kidney epithelial cell line. Funct Integr Genomics. 2007;7(3):193–205. doi: 10.1007/s10142-006-0043-2. [DOI] [PubMed] [Google Scholar]
- 13.Ozsolak F, Milos PM. Transcriptome profiling using single-molecule direct RNA sequencing. Methods Mol Biol. 2011;733:51–61. doi: 10.1007/978-1-61779-089-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat Biotechnol. 2008;26(10):1125–33. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, Li RW, Li W, et al. Transcriptome characterization by RNA-seq unravels the mechanisms of butyrate-induced epigenomic regulation in bovine cells. PLoS One. 2012;7(5):e36940. doi: 10.1371/journal.pone.0036940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li RW, Schroeder SG. Cytoskeleton remodeling and alterations in smooth muscle contractility in the bovine jejunum during nematode infection. Funct Integr Genomics. 2012;12(1):35–44. doi: 10.1007/s10142-011-0259-7. [DOI] [PubMed] [Google Scholar]
- 18.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su YQ, Sugiura K, Woo Y, et al. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302(1):104–17. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdel-Aziz HO, Takasaki I, Tabuchi Y, et al. High-density oligonucleotide microarrays and functional network analysis reveal extended lung carcinogenesis pathway maps and multiple interacting genes in NNK [4-(methylnitrosamino)-1-(3-pyridyle)-1-butanone] induced CD1 mouse lung tumor. J Cancer Res Clin Oncol. 2007;133(2):107–15. doi: 10.1007/s00432-006-0149-x. [DOI] [PubMed] [Google Scholar]
- 21.Pospisil P, Iyer LK, Adelstein SJ, et al. A combined approach to data mining of textual and structured data to identify cancer-related targets. BMC Bioinformatics. 2006;7:354. doi: 10.1186/1471-2105-7-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayburd AL, Martlínez A, Sackett D, et al. Ingenuity network-assisted transcription profiling: identification of a new pharmacologic mechanism for MK886. Clin Cancer Res. 2006;12(6):1820–7. doi: 10.1158/1078-0432.CCR-05-2149. [DOI] [PubMed] [Google Scholar]
- 23.Calvano SE, Xiao W, Richards DR, et al. Inflamm and Host Response to Injury Large Scale Collab. Res. Program. A network-based analysis of systemic inflammation in humans. Nature. 2005;437(7061):1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson SS. Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol Rev. 2001;53(1):1–24. [PubMed] [Google Scholar]
- 25.Hanoune J, Defer N. Regulation and role of adenylyl cyclase isoforms. Annu Rev Pharmacol Toxicol. 2001;41:145–74. doi: 10.1146/annurev.pharmtox.41.1.145. [DOI] [PubMed] [Google Scholar]
- 26.Bruce Alberts AJ, Julian L, Martin R, et al. Essential Cell Biology. New York, NY: Garland Publisher; 2004. [Google Scholar]
- 27.Latchman DS. Transcription factors: an overview. Int J Biochem Cell Biol. 1997;29(12):1305–12. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- 28.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Uniquely expressed genes activated by butyrate.