Abstract
Background and Aims
The proportion of serum carnosinase (CN-1) recognized by RYSK173 monoclonal antibody negatively correlates with CN-1 activity. We thus hypothesized that the epitope recognized by RYSK173 is accessible only in a catalytically incompetent conformation of the zinc dependent enzyme and we mapped its position in the CN-1 structure. Since patients with kidney failure are often deficient in zinc and other trace elements we also assessed the RYSK173 CN-1 proportion in serum of these patients and studied the influence of hemodialysis hereon in relation to Zn2+ and Cu2+ concentration during hemodialysis.
Methods and Results
Epitope mapping using myc-tagged CN-1 fragments and overlapping peptides revealed that the RYSK173 epitope directly contributes to the formation of the dinuclear Zn center in the catalytic domain of homodimeric CN-1. Binding of RYSK173 to CN-1 was however not influenced by addition of Zn2+ or Cu2+ to serum. In serum of healthy controls the proportion of CN-1 recognized by RYSK173 was significantly lower compared to end-stage renal disease (ESRD) patients (1.12 ± 0.17 vs. 1.56 ± 0.40% of total CN-1; p<0.001). During hemodialysis the relative proportion of RYSK173 CN-1 decreased in parallel with increased serum Zn2+ and Cu2+ concentrations after dialysis.
Conclusions
Our study clearly indicates that RYSK173 recognizes a sequence within the transition metal binding site of CN-1, thus supporting our hypothesis that metal binding to CN-1 masks the epitope. The CN-1 RYSK173 proportion appears overall increased in ESRD patients, yet it decreases during hemodialysis possibly as a consequence of a relative increase in transition metal bound enzyme.
Introduction
Serum carnosinase (CN-1) (UniProt identifier Q96KN2) is abundantly expressed in the liver from where it is secreted into the circulation [1]. Based on structural similarity, CN-1 has been classified as metallopeptidase belonging to the M20 family of clan MH. CN-1 is composed of two structural domains of which one adopts an α/ß/α sandwich fold that features a dinuclear zinc-binding site [2]. The other, smaller domain is inserted into the middle of the metal-binding domain and, as in most M20 family enzymes, mediates homodimerization of CN-1. Two active sites per dimer are located at the interface between one metal-binding domain and the two associated dimerization domains, respectively. In CN-1 (MEROPS accession number MER015142), H478 and E200 chelate zinc 1, and H132 and D228 chelate zinc 2. D165 acts as a bridging ligand and the catalytic water molecule completes the tetrahedral coordination sphere for both zinc ions. Mutation of H132, D165, or E200 would lead to the loss of CN-1 activity, indicating the importance of metal-binding for enzyme activity [3]. Previously we have demonstrated that serum CN-1 concentration and activity are genetically determined by the (CTG)n polymorphism [4, 5] and by N-glycosylation of CN-1 [6]. In addition CN-1 hydrolytic activity can be modulated by divalent metal ions, such as Cd2+, Co2+, Fe2+, Ni2+ [3], and by competing substrates, such as anserine and homocarnosine [1, 7].
In the past years the CNDP1 gene, encoding CN-1, has attracted much attention as susceptibility locus for diabetic nephropathy (DN) in type 2 diabetic patients [4, 8]. It is believed that genotypes that are associated with low serum CN-1 concentrations may afford protection against DN as a consequence of reduced carnosine degradation. Yet, it should be emphasized that irrespective of the CNDP1 genotype carnosine concentrations are extremely low or undetectable in human serum or plasma. Carnosine can be detected in serum only transiently after oral carnosine supplementation in individuals with low serum CN-1 concentrations [9].
We have developed two ELISA assays for detection of human serum CN-1 [10]. Quantitative assessment of serum CN-1 concentrations using the ATLAS monoclonal antibody based ELISA, reveals a good correlation with CN-1 activity. The other ELISA is based on the so-called RYSK173 monoclonal antibody and only detects a certain proportion of the total serum CN-1 concentration. The RYSK173 proportion can be increased by addition of EDTA or serum denaturation [10]. Hence the RYSK173 based ELISA assesses CN-1 quality rather than quantity. While in the majority of individuals the proportion of total CN-1 that is recognized by RYSK173 is low (0.1 to 2%), we have reported that individuals with a high proportion of this conformation (>15%) have low CN-1 activities [10]. Since metal ions at the active center of CN-1 are contributing to its enzyme activity, the proportion of CN-1 that is recognized by RYSK173 might be partly lacking these ions. Because formal proof for the assumption that RYSK173 distinguishes between apoenzyme and transition metal bound CN-1 is lacking, we sought to probe the position of the RYSK173 epitope in relation to the metal binding site of CN-1.
Methods and Materials
Generation of RYSK173 antibody
Monoclonal antibody RYSK173 was generated as described in [10]. In brief, Balb/c mice were immunized by intraperitoneal injection of recombinant human CN-1. Three days after the last boost splenocytes were collected and fused with SP2/0 myeloma cells. The fused cells were seeded in 96-well plates (1×103 cells per well) and screened for anti-carnosinase antibody in the supernatant by indirect immune fluorescence (IIF) on CN-1 transfected COS7 cells. Positive wells were sub-cloned by limiting dilution. Clone RYSK173 was selected on the basis of a strong staining in IIF.
Cell culture and transfection
COS7 cells (Invitrogen, Karlsruhe, Germany) were cultured in Dulbecco´s modified Eagle´s medium (DMEM/F-12, GlutaMAX(TM), Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin at 37°C and 5% CO2. The cells were transfected with various length of CNDP1 constructs and lipofectamine2000 (Invitrogen, Karlsruhe, Germany) according to the manufacture´s instruction. Five hours after transfection, the medium was replaced by normal DMEM medium. Supernatants and cell lysates were collected after 48 hours. Cells were lysed on ice by lysis buffer with the addition of dithiothreitol (Fluka Chemie GmbH, Buchs, Germany), protease inhibitor (Roche, Mannheim, Germany), and phosphatase inhibitor (Sigma, Steinheim, Germany). Cell lysates were centrifuged for 10 minutes (14000rpm, at 4°C) to remove cell debris.
Human umbilical vein endothelial cells (HUVEC) were isolated from fresh human umbilical cords as previously described [11]. The cells were cultured in endothelial cell growth medium enriched with 2% FCS and 50ng/ml amphotericin B together with 50μg/ml gentamicin at 37°C and 5% CO2. The flasks for HUVECs were coated in 1% gelatin 20 minutes before use. All experiments involving HUVECs were performed at passages 2–6. Umbilical cords were obtained from donors in the obstetric department of Medical Center Mannheim. This was also approved by local ethics committee (Medizinische Ethikkommission II der Medizinischen Fakultät Mannheim) and all donors gave their written informed consent (No. 2015-518N-MA).
Lentivirus production and transduction
Lentiviral transduction was performed to produce HUVECs with stable CN-1 expression. Human CNDP1 cDNA (RZPD, Library 983, entry No.BX094414) was firstly constructed into lentivirus based vector ppM337 which was kindly provided by Prof. P. Maier from Radiology department of Medical Faculty Mannheim of Heidelberg University. In brief, each culture dish was coated with 0.1mg/ml poly-D-lysine (Sigma, Steinheim, Germany) for 5 min and then washed with sterile distilled water. 5×106 HEK293T/17 cells were seeded per dish in DMEM. For each dish, 4.4μg lentiviral plasmid, 3.4μg packaging plasmid pCMV891 and 2.2μg pMD.G were added to cells with 46μl metafectene (Biontex, Munich, Germany). Cell medium was substituted by 14ml DMEM with 10mM Na-butyrate for 8 hours the next day. Lentiviruses containing cell medium was collected and concentrated using Vivaspin20 (Sartorius stedim, Göttingen, Germany) on the third day.
For transduction, HUVECs were incubated in 1:100 diluted virus containing HEK293T/17 supernatant for 48 hours. Cells were lysed either by lysis buffer or by a freeze and thaw cycle in liquid nitrogen. Westernblotting of 20μg total protein from HUVECs supernatant and cell lysates was performed to confirm the transduction.
Construction of CNDP1 variants
CNDP1 cDNA was taken as a template to generate CNDP1 fragments by PCR. PCR amplification was carried out from bp 1 to bp 312, from bp 1 to bp 471 and from bp 313 to bp 471 using the following forward and reverse primers respectively:
1–312: cagcccatcgatatggatcccaaactcaggaga and agctttaaatcgatgatcgggcagctgctgag
1–471: cagcccatcgatatggatcccaaactcaggaga and agctttaaatcgatgtttcccgtctacctccgtcagc
313–471:cagggatccatgggtcagagtcttccaatacctcccg and agctttaaatcgatgtttcccgtctacctccgtcagc.
Amplification products were cloned into the pCSII + mt vector providing a N-terminal 6×myc-tag and subsequently transfected into COS7 cells as previously described.
Synthetic peptides
Three overlapping CN-1 peptides were synthesized (BIOMATIK, Wilmington, USA) that completely covered the coding sequence from bp 313 to bp 471:
313–391: CN1-1 (DGQSLPIPPVILAELGSDPTKGTVCF),
347–440: CN1-2 (LAELGSDPTKGTVCFYGHLDVQPADRGDGWL),
393–471: CN1-3 (HLDVQPADRGDGWLTDPYVLTEVDGK).
Fine epitope mapping was subsequently performed using two peptides that covered the overlapping sequence of CN1-2 and CN1-3, i.e. CN1-4 (HLDVQPAD) and CN1-5 (PADRGDGWL). All peptides were dissolved in sterile distilled water (final concentration: 10mg/ml) and were tested for recognition by RYSK173 by dotblot analysis and/or ELISA.
CN-1 detection
Both the RYSK173 and ATLAS based CN-1 ELISA assays were used for detection of CN-1 in serum samples. CN-1 ELISAs were performed as described [10]. For epitope mapping synthetic peptides were directly coated on the ELISA plates.
Gel electrophoresis and western blot were performed using a standard protocol. Dot blot assays were performed by spotting 2 μl of synthetic peptide solution on a nitrocellulose membrane. The membrane was dried and processed similar as has been described for westernblotting [10].
Site mutagenesis
One nucleotide mutation in CN-1 was generated by the QuikChange™XL site-directed mutagenesis kit (Stratagene, Waldbronn, Germany). H132 was exchanged to Q132. The forward and reverse primers were as follows. The sequence of the mutant CNDP1 was confirmed by DNA sequencing.
Forward primer: gcttctacggccagttggacgtgcagc
Reverse primer: gctgcacgtccaactggccgtagaagc
Patients
Thirty one patients on hemodialysis were recruited from our dialysis ward. They all had different causes of end stage renal disease. Renal transplant recipients, patients with urinary tract infection or fever at the time of investigation were excluded. Diabetic patients were also excluded since diabetes per se might potentially influence CN-1 metabolism. Demographic and relevant clinical data of these patients are shown in Table 1. Sera from age and gender matched healthy controls (n = 111) were retrieved from our bio-bank. The study was approved by local ethics committee (Medizinische Ethikkommission II der Medizinischen Fakultät Mannheim) and all patients gave their written informed consent prior to study (No. 0193/2001).
Table 1. Relevant clinical data of ESRD patients.
| ESRD patients (n = 31) | |
|---|---|
| Gender (male) | 20 (65%) |
| Age (year) | 55.0 ± 2.4 |
| Plasma creatinine (mg / dl) | 10.6 ± 0.58 |
| [Zinc] before/after hemodialysis (μmol/L) | 9.5 ± 0.2 / 10.9 ± 0.2 (p < 0.05) |
| [Copper] before/after hemodialysis (μg/L) | 14.1 ± 0.4 / 15.3 ± 0.4 (p < 0.05) |
| Duration of dialysis (months) | 81 ± 82 |
| Causes of disease | |
| Glomerulonephritis | 15 |
| Vascular disease | 4 |
| Pyelonephritis or hydronephrosis | 6 |
| Polycysitic kidney disease | 2 |
| Others (non-diabetes) | 4 |
Trace metal analysis
Copper and zinc in serum were analysed at Laboratory Limbach (Heidelberg, Germany) by colorimetric photometry as described previously [12, 13]. Visibly hemolyzed serum samples were not analysed.
Statistical analysis
Quantitative data are depicted as mean ± SD. Student t test (normal distribution) or Wilcoxon-Mann-Whitney test (non-normal distribution) was used to compare differences between the groups. Correlation between values was evaluated by Pearson correlation coefficients. Significance was defined according to a p-value < 0.05. Statistical analysis was performed with GraphPad Prism 6.0 (GraphPad Software, Inc, La Jolla, California).
Results
Epitope mapping of RYSK173
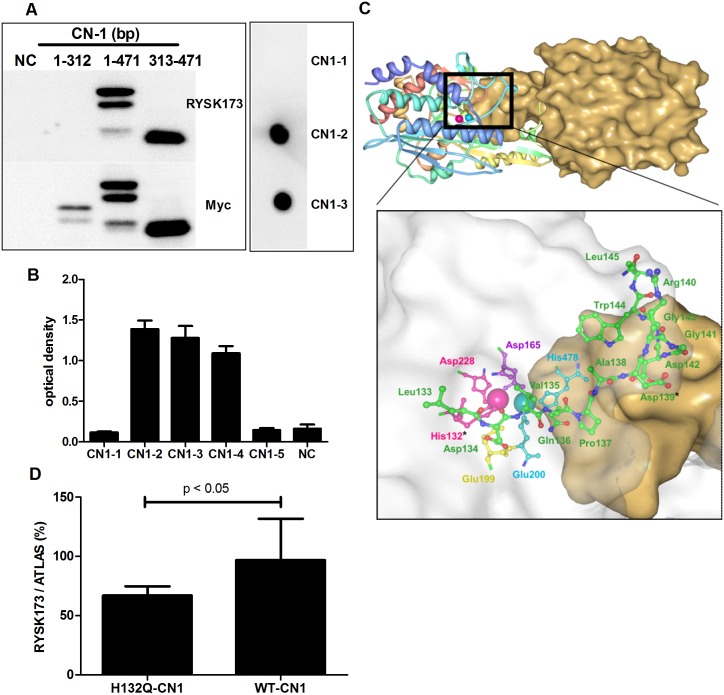
Since we have previously demonstrated that addition of EDTA improved CN-1 detection by RYSK173 [10], we anticipated that the epitope on CN-1 which is recognized by RYSK173 resides within the metal-binding domain of CN-1. Because we also have shown that RYSK173 recognizes a CN-1 fragment when truncated before the first N-glycosylation site [6] we constructed three myc tagged CN-1 fragments that differed in size (Fig 1). While a myc-tagged recombinant fragment of CN-1 spanning bp 1–471 was detected by both RYSK173 and myc antibodies, a shorter fragment (bp 1–312) was only recognized by the myc antibody. Because a recombinant CN-1 fragment corresponding to bp 313–471 was also recognized by both antibodies our data suggest that the RYSK173 epitope resides within this part of the CN-1 protein (Fig 1A, panel on the left). Three overlapping synthetic peptides of approximately 30 amino acids that covered the 313–471 bp fragments were subsequently tested by dot blot analysis for RYSK173 binding. CN1-2 and CN1-3 were clearly recognized whereas CN1-1 was not (Fig 1A, panel on the right). To delineate the exact epitope on CN-1 that is recognized by RYSK173, two different peptides corresponding to the overlapping sequence of CN1-2 and CN1-3 (HLDVQPADRGDGWL) were tested by ELISA. The peptides CN1-4 (HLDVQPAD) and CN1-5 (PADRGDGWL) consisted of either the first 8 or last 9 amino acids of the overlapping peptide sequence of CN1-2 and CN1-3 respectively. Synthetic peptides CN1-2, CN1-3 and CN1-4 were all detected in ELISA by RYSK173 whereas CN1-1 and CN1-5 were not (Fig 1B). This demonstrates that the RYSK173 epitope is contained in the sequence spanning the zinc 2-binding residue H132 to D139. Inspection of its spatial position within the metal binding domain of CN-1 reveals its proximity to both metal ions and the homodimer interface (Fig 1C). We also demonstrate that H132 is an integral constituent amino acid of the RYSK173 epitope, since site directed mutation of H132 to Q132 in recombinant CN-1 (rCN-1) expressed in COS7 cells resulted in a significant decrease in detection by RYSK173 (Fig 1D).
Fig 1. Epitope mapping of RYSK173.
A: Myc-tagged recombinant CN-1 fragments were expressed in COS7 cells by transfection. Cells were harvested and the recombinant proteins were detected by Westernblotting using RYSK173 or anti-Myc (Fig on the left). Three overlapping synthetic peptides (CN1-1, CN1-2 and CN1-3) corresponding to bp 313–471 were tested in dotblot analysis (Fig on the right). B: Two additional peptides (CN1-4 and CN1-5) corresponding to the overlapping sequence of CN1-2 and CN1-3 were tested in ELISA for recognition by RYSK173. C: Location of the dinuclear zinc center at the CN-1 homodimer interface. The monomer on the left is depicted as a ribbon drawing and the monomer on the right is rendered as solid surface. Zinc1 and zinc2 in the left monomer are shown as cyan and magenta colored spheres, respectively. The enlarged view details the position of the RYSK173 epitope, which is confined to the amino acid sequence H132 to D139 (both marked with an asterisk). The surface of the left subunit is rendered semi-transparent. The Fig was made with Protein Workshop. D: H132 was exchanged for glutamine (Q) by site directed mutagenesis. The recombinant proteins were detected in transfected cell lysates using the ATLAS and RYSK173 based ELISA. The results are expressed as RYSK173/ATLAS ratio x 100%. NC: negative control, WT: wild type.
Recognition of serum CN-1 and recombinant expressed CN-1 in HUVECs by RYSK173
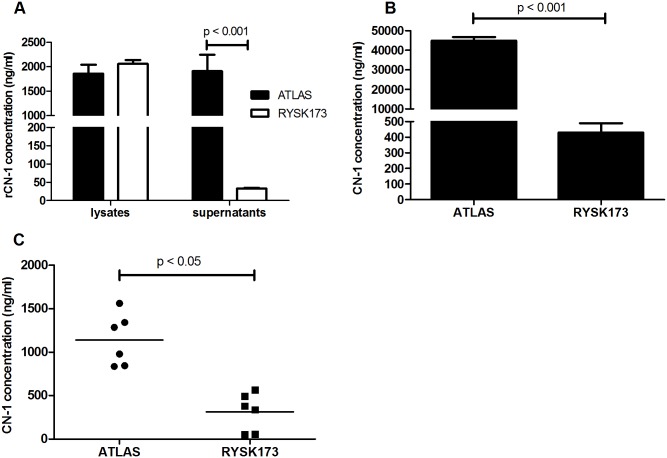
Since our epitope mapping studies have demonstrated that epitope which is recognized by RYSK173 directly contributes to the formation of the Zn dinuclear centres in CN-1, we studied if CN-1 recognition by RYSK173 is changed once CN-1 is secreted. This would indicate that loading of the Zn centres would occur extracellularly. To this end, we first assessed if ectopically expressed rCN-1 in supernatants and cell lysates of CN-1 transduced HUVECs is recognized in the RYSK173- and ATLAS based ELISA, the latter recognizing total CN-1 [10]. While in cell lysates of CNDP1 transfected HUVECs (n = 6) CN-1 was detected to a similar extent by ATLAS and RYSK173, in supernatants only a fraction of total CN-1 was recognized by RYSK173 (Fig 2A). Since the lysis buffer contained both EDTA and DTT, the difference in RYSK173 recognition of CN-1 in cell lysates and supernatants is likely explained by denaturation of the protein [10]. We therefore used freeze thaw cycles as a more gentle way to obtain intra-cellular proteins (n = 6). Although the yield of CN-1 was significantly lower than the previous cell lyses method, similar as observed for serum (Fig 2B), CN-1 concentrations were significantly higher in the ATLAS based as compared to the RYSK173 based CN-1 ELISA (Fig 2C), albeit that the difference was less pronounced as compared to serum.
Fig 2. Recognition of recombinant and serum CN-1 by RYSK173.
A: recombinant CN-1 was expressed in HUVECs by lentiviral transduction. The recombinant proteins were detected in cell lysates and supernatants using the ATLAS (filled bars) and RYSK173 (open bars) based ELISA. The results of 6 transduction experiments are depicted and expressed as mean CN-1 concentration (ng/ml) ± SD. B: Serum samples of 111 healthy individuals were tested in the ATLAS and RYSK173 based ELISA. C: Cell disruption was performed by repeated freeze thawing. Similar as in serum RYSK173 detected significantly lower amounts of CN-1.
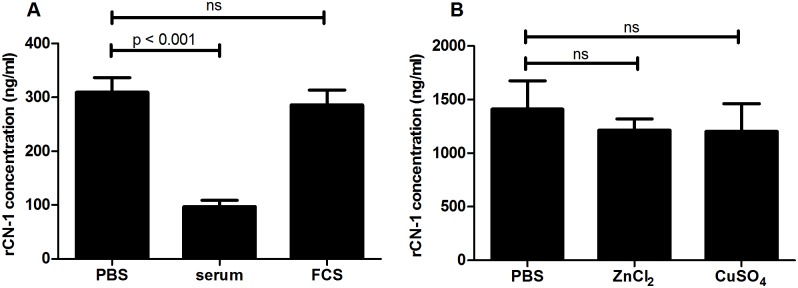
We next tested if recovery of rCN-1 was impaired when spiked in human serum or FCS. While the recovery of rCN-1 significantly decreased in human serum, detection of rCN-1 was not significantly influenced in FCS (Fig 3A). Because the concentrations of the trace metal ions Zn2+ and Cu2+ were different between the two types of serum (Zn2+ concentration: 11.7±0.1 vs. 18.7±0.1 μmol/L and Cu2+ concentration: 822±11.3 vs. 89.5±0.7 μg/L; human serum vs. FCS), we tested if addition of ZnCl2 or CuSO4 influences the recovery of rCN-1 in the RYSK173 based ELISA. Neither addition of 100μM ZnCl2 nor CuSO4 were able to reduce the recovery of rCN-1 in PBS (Fig 3B) or in FCS (data not shown). Additionally, no significant differences were found even if higher concentrations of metal ions were used (data not shown).
Fig 3. Recovery of recombinant CN-1 in serum.
A: Equal amounts of recombinant CN-1 were spiked in PBS, human serum or FCS. While in human serum detection of CN-1 by RYSK173 was strongly diminished this was not observed in FCS. B: The influence of ZnCl2 (100μM) and CuSO4 (100μM) on detection of CN-1 by RYSK173 was tested. Ns: not significant.
The relative proportion of CN-1 recognized by RYSK173 is increased in ESRD patients
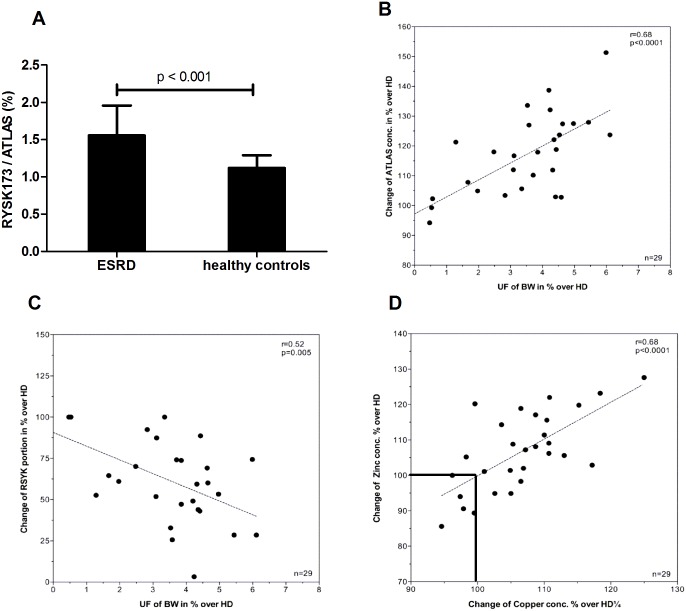
Recent evidence indicates that patients with kidney failure are often deficient in zinc and other trace elements [14]. We therefore assessed to what extent end-stage renal disease (ESRD) patients differ in the proportion of CN-1 that is recognized by RYSK173. As depicted in Fig 4A, the relative proportion of CN-1 that was recognized by RYSK173 was significantly higher in ESRD patients as compared to healthy controls. The serum Zn2+ concentrations in ESRD patients ranged from (6.8 to 12.1 μmol/L) while that of healthy controls were all in the normal range (9.2 to 18.4 μmol/L). Since all ESRD patients were on hemodialysis, we also assessed the influence of hemodialysis on serum CN-1 concentrations in parallel to changes in serum Zn2+ and Cu2+ concentrations. To this end, serum CN-1, measured by both RYSK173 and ATLAS based ELISA, Zn2+ and Cu2+ concentrations were assessed directly before and after one hemodialysis session. While total serum CN-1 concentrations were significantly increased in the post-dialysis samples and correlated to the amount of ultrafiltration (Fig 4B), the proportion of CN-1 recognized by RYSK173 was decreased in these samples (Fig 4C). Zinc and copper concentrations were expressed as percentage (%) change of Zn2+ and Cu2+ concentrations in the post dialysis sample relative to the serum sample obtained directly before hemodialysis. In approximately 5 out of 31 samples the Zn2+ and Cu2+ ion concentration did not change or decreased over hemodialysis (Fig 4D, lower left quadrant), while in the majority of samples either Zn2+, Cu2+ or both were increased after dialysis (Fig 4D).
Fig 4. Serum CN-1 concentrations in ESRD patients.
A: Sera of 31 ESRD patients were collected and tested in the ATLAS based and RYSK173 based ELISA. Sera of 111 healthy individuals served as control. The results are expressed as RYSK173/ATLAS ratio x 100%. B: Serum was collected directly before or after hemodialysis. The influence of hemodialysis on total CN-1 expression (B), the RYSK173 proportion (C) and changes in Zn2+ and Cu2+ (D) concentrations were assessed. UF: ultrafiltration, BW: body weight, HD: hemodialysis, conc: concentration.
Discussion
In the present study we sought to map the epitope that our anti-CN-1 monoclonal antibody RYSK173 recognizes in CN-1. Our previous finding that chelators of transition metal ions were able to unmask the epitope had led us to expect its location in the vicinity of the metal binding site of CN-1. Our results unambiguously demonstrate that the epitope recognized by RYSK173 indeed is in intimate contact with the dinuclear zinc site in CN-1 and even includes metal ligand H132. Metalloproteases are ubiquitous enzymes able to degrade an array of protein substrates. Mostly these enzymes utilize conserved amino acid residues to generate a scaffold capable of binding one or two metal ions [15]. Their functionality depends on subtle interactions between the electronic properties of the metal ion, dictated by its coordination chemistry, and the stability of protein conformations.
Previous mutational and structural analyses of CN-1 homologs from the M20 family have suggested that these enzymes require both metal sites in their dinuclear zinc centers for optimal protein stability but that zinc 1 is often bound with low affinity [16]. It may thus be easily removed by EDTA, a treatment which we have formerly shown to partially improve CN-1 recognition by RYSK173 [10]. Immediate proximity of the epitope to both zinc sites in CN-1 (Fig 1) agrees with the notion that removal of zinc 1 leads to its exposure due to local protein unfolding. The dinuclear zinc center in CN-1 is formed by H478 and E200 which coordinate with zinc 1 and H132 and D228 coordinating with zinc 2. Site directed mutagenesis of H132 to glutamine significantly impaired binding of RYSK173 indicating that this residue is part of the RYSK173 epitope. It should be emphasized that binding of RYSK173 in the presence of EDTA is still significantly lower compared to binding of the polyclonal antibody ATLAS. Only after protein denaturation binding of RYSK173 to CN-1 equals that of ATLAS. This is in line with our experimental data that no difference in CN-1 binding was observed between RYSK173 and ATLAS in lysates of CNDP1 transduced HUVECs when a DTT and EDTA containing lysis buffer was used.
In line with our previously published data [10], a large difference in CN-1 concentrations are detected in human serum when using the ATLAS or RYSK173 based ELISA (Fig 2B) respectively. This might be explained by the fact that in a fraction of CN-1 the Zn centers are not completely occupied. To elucidate if Zn binding already occurs inside the cells shortly before CN-1 is secreted, we performed ELISA on supernatants and cell lysates using the RYSK173 antibody. Indeed we found a significant lower amount of CN-1 in the RYSK173- as compared to the ATLAS based ELISA in cell lysates (p < 0.05, Fig 2C) that were obtained without the use of denaturation or EDTA. This might indicate that zinc binding to CN-1 already occurs intracellularly and still proceeds extracellularly as the difference between the RYSK173 and ATLAS based ELISA was much more pronounced in supernatants (p < 0.001, Fig 2A) as compared to cell lysates. The ATLAS and RYSK173 based ELISA detected comparable CN-1 concentrations in cell lysates when the lysis buffer contained DTT and EDTA.
The finding that recovery of rCN-1 by the RYSK173 based ELISA was impaired in human but not in fetal calf serum remains unexplained in our study. While Zn2+ concentrations were slightly different in these two types of serum, the Cu2+ concentration in FCS was only 10% of that found in human serum. Although it has been suggested for bovine lens leucyl aminopeptidases (blLAP) [17] and the aminoacylhistidine dipeptidase (PepD) [18, 19], that the zinc centers might be exchanged by other divalent cations with different exchange kinetics, to our knowledge this has not been reported for CN-1. Addition of CuSO4 did not impair detection of rCN-1 in the RYSK173 based ELISA, suggesting that binding of Cu2+ to CN-1 in human serum is unlikely a cause for the poor recovery by RYSK173.
Although our study does not provide direct evidence that divalent metal ion binding to CN-1 masks the RYSK173 epitope, there seems to be a relation with trace metal ion concentrations in serum. Firstly, we found a significant difference in the proportion of RYSK173 in serum of patients with ESRD as compared to healthy controls. This was paralleled by a lower serum Zn2+ concentration in ESRD patients. Secondly, the proportion of RYSK173 CN-1 was decreased in the post-dialysis samples while in most patients the serum Zn2+ concentration was increased directly after dialysis. Although the Cu2+ concentration increased in parallel in the post-dialysis sample, direct binding of Cu2+ to the metal centres of CN-1 is highly unlikely.
A number of studies have suggested that CN-1 activity, either in serum or cerebrospinal fluid, might be important in the pathology of chronic kidney disease [20], diabetic complications [4, 8, 21], Alzheimer’s disease [22, 23] or dementia [24]. CN-1 activity is not only depending on CN-1 concentrations but also on the presence of competing substrates [1] and the relative proportion of CN-1 that is recognized by RYSK173 [10]. Although the RYSK173 is not intended for implementation in routine diagnostics, it might help understanding why in some patients CN-1 activity is extremely low despite the fact that CN-1 concentrations are normal.
In conclusion our study clearly indicates that RYSK173 recognizes a sequence within the transition metal binding site of CN-1, thus supporting our hypothesis that metal binding to CN-1 masks the epitope. The CN-1 RYSK173 proportion appears overall increased in ESRD patients which might be explained by a relative Zn deficiency. It remains to be assessed if Zn deficiency in general leads to an increase in the CN-1 RYSK173 proportion.
Acknowledgments
We would like to acknowledge Ms. Annette Breedijk (Vth Department of Medicine, University Medical Center Mannheim, University of Heidelberg) for her assistance in performing cell culture. We also extremely appreciate the help from Dr. Frederick Pfister (Nephropathology Department, Universtiy of Erlangen) for his consultation on clinic aspects.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was supported by Deutsche Forschungsgemeinschaft (DFG, URL: http://www.dfg.de/) SA 2143/1-1 to S.H. This author read and revised this manuscript. It was also supported by Graduiertenkolleg (GRK, URL: http://www.umm.uni-heidelberg.de/ag/grk1874/) 1874/1 to B.Y. This author designed, prepared, and decided to publish this manuscript.
References
- 1.Peters V, Jansen EE, Jakobs C, Riedl E, Janssen B, Yard BA, et al. Anserine inhibits carnosine degradation but in human serum carnosinase (CN1) is not correlated with histidine dipeptide concentration. Clin Chim Acta. 2011;412:263–7. 10.1016/j.cca.2010.10.016 [DOI] [PubMed] [Google Scholar]
- 2.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014:D503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teufel M, Saudek V, Ledig JP, Bernhardt A, Boularand S, Carreau A, et al. Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J Biol Chem. 2003;278:6521–31. [DOI] [PubMed] [Google Scholar]
- 4.Janssen B, Hohenadel D, Brinkkoetter P, Peters V, Rind N, Fischer C, et al. Carnosine as a protective factor in diabetic nephropathy: association with a leucine repeat of the carnosinase gene CNDP1. Diabetes. 2005;54:2320–7. [DOI] [PubMed] [Google Scholar]
- 5.Riedl E, Koeppel H, Brinkkoetter P, Sternik P, Steinbeisser H, Sauerhoefer S, et al. A CTG polymorphism in the CNDP1 gene determines the secretion of serum carnosinase in Cos-7 transfected cells. Diabetes. 2007;56:2410–3. [DOI] [PubMed] [Google Scholar]
- 6.Riedl E, Koeppel H, Pfister F, Peters V, Sauerhoefer S, Sternik P, et al. N-glycosylation of carnosinase influences protein secretion and enzyme activity: implications for hyperglycemia. Diabetes. 2010;59:1984–90. 10.2337/db09-0868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters V, Kebbewar M, Jansen EW, Jakobs C, Riedl E, Koeppel H, et al. Relevance of allosteric conformations and homocarnosine concentration on carnosinase activity. Amino Acids. 2010;38:1607–15. 10.1007/s00726-009-0367-z [DOI] [PubMed] [Google Scholar]
- 8.Freedman BI, Hicks PJ, Sale MM, Pierson ED, Langefeld CD, Rich SS, et al. A leucine repeat in the carnosinase gene CNDP1 is associated with diabetic end-stage renal disease in European Americans. Nephrol Dial Transplant. 2007;22:1131–5. [DOI] [PubMed] [Google Scholar]
- 9.Everaert I, Taes Y, de Heer E, Baelde HJ, Zutinic A, Yard B, et al. Low plasma carnosinase activity promotes carnosinemia following carnosine ingestion in humans. Am J Physiol Renal Physiol. 2012;302:F1537–44. 10.1152/ajprenal.00084.2012 [DOI] [PubMed] [Google Scholar]
- 10.Adelmann K, Frey D, Riedl E, Koeppel H, Pfister F, Peters V, et al. Different conformational forms of serum carnosinase detected by a newly developed sandwich ELISA for the measurements of carnosinase concentrations. Amino Acids. 2012;43:143–51. 10.1007/s00726-012-1244-8 [DOI] [PubMed] [Google Scholar]
- 11.Yard B, Beck G, Schnuelle P, Braun C, Schaub M, Bechtler M, et al. Prevention of cold-preservation injury of cultured endothelial cells by catecholamines and related compounds. Am J Transplant. 2004;4:22–30. [DOI] [PubMed] [Google Scholar]
- 12.Johnsen O, Eliasson R. Evaluation of a commercially available kit for the colorimetric determination of zinc in human seminal plasma. Int J Androl. 1987;10:435–40. [DOI] [PubMed] [Google Scholar]
- 13.Homsher R, Zak B. Spectrophotometric investigation of sensitive complexing agents for the determination of zinc in serum. Clin Chem. 1985;31:1310–3. [PubMed] [Google Scholar]
- 14.Tonelli M, Wiebe N, Hemmelgarn B, Klarenbach S, Field C, Manns B, et al. Trace elements in hemodialysis patients: a systematic review and meta-analysis. BMC Med. 2009;7:25 10.1186/1741-7015-7-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David A. ''Catalytic mechanisms for metallopeptidases'' in Handbook of Proteolytic Enzyme. 2 ed Barrett AJ, Rawlings ND, Woessner JF, editors. San Diego, Calif, USA: Academic Press; 1998. [Google Scholar]
- 16.Lindner HA, Lunin VV, Alary A, Hecker R, Cygler M, Ménard R. Essential roles of zinc ligation and enzyme dimerization for catalysis in the aminoacylase-1/M20 family. J Biol Chem. 2003;278:44496–504. [DOI] [PubMed] [Google Scholar]
- 17.Cappiello M, Alterio V, Amodeo P, Del Corso A, Scaloni A, Pedone C, et al. Metal ion substitution in the catalytic site greatly affects the binding of sulfhydryl-containing compounds to leucyl aminopeptidase. Biochemistry 2006;45:3226–34. [DOI] [PubMed] [Google Scholar]
- 18.Aoki A, Shibata Y, Okano S, Maruyama F, Amano A, Nakagawa I, et al. Transition metal ions induce carnosinase activity in PepD-homologous protein from Porphyromonas gingivalis. Microb Pathog. 2012;52:17–24. 10.1016/j.micpath.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 19.Wang TY, Chen YC, Kao LW, Chang CY, Wang YK, Liu YH, et al. Expression and characterization of the biofilm-related and carnosine-hydrolyzing aminoacylhistidine dipeptidase from Vibrio alginolyticus. FEBS J. 2008;275:5007–20. 10.1111/j.1742-4658.2008.06635.x [DOI] [PubMed] [Google Scholar]
- 20.Kiliś-Pstrusińska K, Zwolińska D, Grzeszczak W, Group. S. Is carnosinase 1 gene (CNDP1) polymorphism associated with chronic kidney disease progression in children and young adults? results of a family-based study. Arch Med Res. 2010;41:356–62. 10.1016/j.arcmed.2010.07.006 [DOI] [PubMed] [Google Scholar]
- 21.Zhu JM, Wang B, Li J, Chen GM, Fan YG, Feng CC, et al. D18S880 microsatellite polymorphism of carnosinase gene and diabetic nephropathy: a meta-analysis. Genet Test Mol Biomarkers. 2013;17:289–94. 10.1089/gtmb.2012.0341 [DOI] [PubMed] [Google Scholar]
- 22.Perrin RJ, Craig-Schapiro R, Malone JP, Shah AR, Gilmore P, Davis AE, et al. Identification and validation of novel cerebrospinal fluid biomarkers for staging early Alzheimer's disease. PLoS One. 2011;6:e16032 10.1371/journal.pone.0016032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu WW, Chen Z. Role of histamine and its receptors in cerebral ischemia. ACS Chem Neurosci 2012;3:238–47. 10.1021/cn200126p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balion CM, Benson C, Raina PS, Papaioannou A, Patterson C, Ismaila AS. Brain type carnosinase in dementia: a pilot study. BMC Neurol 2007;7:38 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.