Abstract
Research in recent years have illuminated data on the mechanisms and targets of phosphonic acid antibiotics and herbicides, including fosfomycin, glyphosate, fosmidomycin and FR900098. Here we review the current state of knowledge of the structural and biochemical characterization of resistance mechanisms against these bioactive natural products. Advances in the understanding of these resistance determinants have spurred knowledge-based campaigns aimed towards the design of derivatives that retain biological activity but are less prone to tolerance.
Introduction
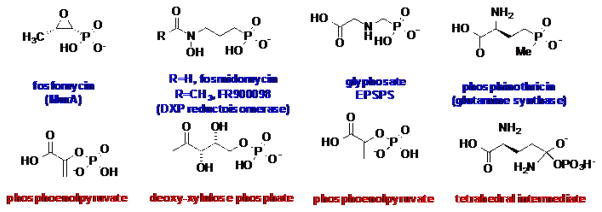
Microbially produced natural products, or secondary metabolites, encompass an astounding array of chemically diverse small molecules with unique, and often medically relevant, properties. Phosphonic and phosphinic acids, bearing an inert C-P linkage, constitute one such group of bioactive small molecules with great pharmaceutical potential. Despite their prevalence throughout numerous phyla, phosphonates and phosphinates are relatively underexploited scaffolds in medicinal chemistry. Among the most commonly known examples are the antibiotic fosfomycin (the only FDA approved drug for treatment of acute cystitis during pregnancy), the potent anti-malarial fosmidomycin, and the widely used herbicides glyphosate and phosphinothricin (Figure 1).
Figure 1.
The top row shows the chemical structures of select phosphonic acid antibiotics and herbicides, along with the names of the enzymes that these compounds target. The bottom row lists the actual substrates that the phosphonates mimic.
Phosphonic acids are structural analogs of labile phosphate-esters and carboxylic acids, but contain a chemically inert C-P bond, in place of the labile O-P or O-C bond found in the latter compounds. Many phosphonates/phosphinates act as mimics of phosphate-esters or carboxylic acids, and compete with their structural analogs for binding to enzymes that utilize the corresponding compounds as substrates (Figure 1). Due to the presence of the inert C-P linkage, phosphonates/phosphinates cannot be further processed and consequently function as potent enzyme inhibitors. Many enzyme targets for phosphonates are metabolically essential and, consequently, enzyme inhibition results in growth inhibition or cell death. As acyl or phosphoryl-transfer chemistry is prevalent throughout metabolism, phosphonates have the potential for targeting a wide-range of processes essential for growth.
The utilization of phosphonates as candidate antibiotics and herbicides has resulted in the unfortunate emergence of multiple mechanisms of resistance against this chemical scaffold. The proliferation of resistance has resulted from either the propagation of determinants from the producing organism or through novel mechanisms, both of which have been disseminated via horizontal gene transfer. Here, we review both the mechanism of action of several of the most widely used phosphonate antibiotics, along with the emerging resistance mechanisms. Further attempts at diversification of these molecules must take into account corresponding resistance mechanisms that may counter the efficacy of these compounds.
Fosfomycin
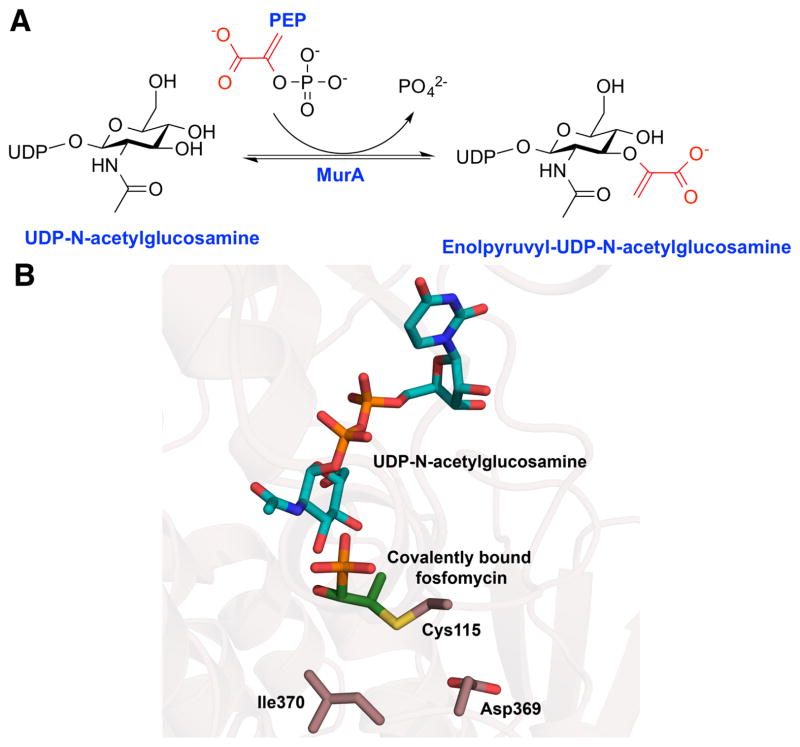
Fosfomycin (phosphomycin) was first isolated in 1969, as a joint effort between Merck and CEPA, from three strains of Streptomyces (Figure 1).1,2 Structural analysis showed fosfomycin to be the sole member of a new class of antibiotic that was effective against both Gram-positive and Gram-negative bacteria.2 Mechanistic studies have shown that fosfomycin functions by targeting cell wall biosynthesis, specifically, through inhibition of the UDP-N-acetylglucosamine enolpyruvyl transferase MurA, the first committed step in peptidoglycan biosynthesis (Figure 2A).3
Figure 2.
(A) The chemical transformation catalyzed by the enoylpyruvyl transferase MurA in the first committed step of peptidoglycan biosynthesis. (B) The phosphonate antibiotic fosfomycin irreversibly modified MurA by forming a covalent linkage with Cys115. The structure of MurA indicates that resistance mutations are located near the Cys115 (PDB: 1UAE).
MurA utilizes phosphoenolpyruvate (PEP) as a donor to facilitate enolpyruvyl transfer to the peptidoglycan precursor UDP-N-acetylglucosamine. This addition-elimination reaction involves the unusual cleavage of a C-O bond, rather than the more labile P-O bond, in the substrate PEP (Figure 2A). Within the MurA active site, a nucleophilic Cys115 forms a reversible covalent adduct with the PEP substrate,4 and this residue appears to be critical for product release.5 While the native substrate PEP forms a reversible adduct with the Cys115 of MurA, fosfomycin inhibits the enzyme by forming a chemically inert species. Nucleophilic attack onto the β-carbon of fosfomycin by Cys115 opens the epoxide to form an irreversible active site modification that kills activity.3,6
Initial in vitro work showed that fosfomycin resistance could be readily obtained artificially.7 However, clinically observed resistance has remained both low and constant throughout the 40 years fosfomycin has been used to primarily treat urinary tract infections.8
A 2003 study that examined fosfomycin resistance in over 3,000 clinically isolated E. coli strains from Europe showed only a 1% occurrence of resistance in these isolates.9 Notably, there did not appear to be a significant difference in the emergence of resistance in countries that utilized fosfomycin clinically and those that did not.9 These data seem to indicate that clinical use of fosfomycin did not increase the frequency of resistant isolates of uropathogenic E. coli. Variants that had some level of resistance had to maintain a sufficient growth rate to prevent being cleared from the bladder, and the growth rates of these resistant isolates lagged between 10–15% slower than those for wild type strains.9 Additionally, the resistant strains grew at an even lower rate in the presence of fosfomycin. Modeling of the growth rate in the bladder suggests that the resistant isolates could not establish a stable infection.9 These loss of fitness results help explain the low incidence of fosfomycin resistance observed in the clinic.
The underlying basis for resistance in the 1% of clinical isolated mutants of E. coli that were impervious to the drug was borne out by genome sequencing. In these strains, the majority of mutations were localized to proteins responsible for import of fosfomycin into the cell.9,10 E. coli contains two transport systems that enable fosfomycin to enter the cell, namely GlpT and UhpT.3 The GlpT antiporter is a member of the major facilitator super family (MFS) that is commonly responsible for import of glycerol-3-phosphate (G3P) coupled with inorganic phosphate export. Reconstitution experiments with GlpT confirmed a role in fosfomycin import under clinically relevant concentrations of fosfomycin.9,11 Clinical isolates resistant to fosfomycin contain truncations to the glpT gene, resulting in an inactive GlpT transport system.10,12 Strains with these mutations in glpT are unable to grow under glycerol-3-phosphate as a sole carbon source.10
In addition to explicit mutations to the glpT gene, in vitro selected resistant isolates were also observed to alter regulation of GlpT expression 9,10. Although GlpT is constitutively expressed, its abundance is regulated by cyclic AMP (cAMP) levels in the cell. Cells produce high levels of cAMP under carbon limiting conditions, resulting in increased expression of many import systems, including GlpT. Isolated in vitro mutants were observed to have deletions or insertions that inactivated proteins responsible for cAMP formation, namely adenylate cyclase and phosphoenolpyruvate-protein phosphotransferase.9,13 Such mutations that decrease cAMP production levels result in the decreased expression of GlpT, which, in turn, limits import of fosfomycin into target cells. Although these mutants could be selected for in vitro, they were not observed in the clinical isolates, indicating that mutations that dramatically alter cAMP biosynthesis may compromise infectivity.9
UhpT is a second transporter, normally responsible for the transport of glucose-6-phosphate (G6P), which is linked to fosfomycin import. As with GlpT, mutations to UhpT were seen both in clinical isolates and in in vitro selections.9,10 Observed mutations ranged from point variants to deletions of the entire gene, and a specific Glu350→Gln mutation was observed in four of the sequenced isolates.10 Organisms with this mutation were still able to grow when G6P was used as the exclusive carbon source, but this mutation was always found in combination with GlpT mutations, so its true effect is ambiguous.10 In addition, mutations that alter the expression of UhpT were also observed; deletion of its cognate response regulator prevented expression of UhpT as evidenced by the inability of the variant strain to grow on G6P.9
Another, less common contributor to fosfomycin resistance is due to mutation in the MurA target. Sequence analysis of a resistant MurA ortholog from Mycobacterium tuberculosis reveals an Asp substitution at the Cys115 that is the normally the site of covalent modification by fosfomycin.14 Generation of the Cys115→Asp mutant MurA in sensitive strains of E. coli resulted in a complete loss of fosfomycin sensitivity but the mutation also compromised catalytic efficiency by nearly 1,000-fold.14 Conversely, the reverse (Asp→Cys) mutation in the Mycobacterium tuberculosis MurA resulted in a gain of fosfomycin sensitivity.15 Although mutations at the active site Cys would appear to be a rational route towards fosfomycin resistance, to date no sequenced clinical isolate is shown to bear this mutation, presumably due to the corresponding loss of catalytic efficiency. In contrast, clinical isolates are show to bear mutations at other residues in MurA, in particular Asp369→Asn and Leu370→Ile mutations observed in one isolate.10 In the MurA crystal structure, these two residues are located proximal to Cys115, suggesting that they could sterically occlude fosfomycin binding at the active site (Figure 2B).16 E. coli strains that overexpress this variant MurA demonstrate a lowered fosfomycin sensitivity by nearly 1,000-fold relative to the wild-type.10 Therefore, it appears that these mutations may offer at least some level of fosfomycin protection.
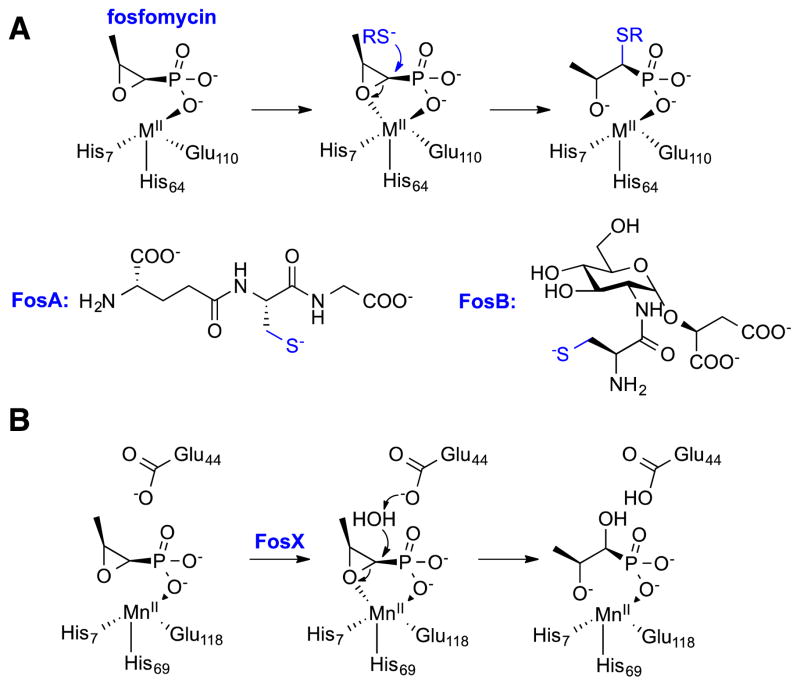
A fosfomycin resistance strategy alternative to modifications of the target or import system involves chemical modification of the drug itself. Bacteria that have evolved mechanisms to inactivate the antibiotic take advantage of the reactivity of fosfomycin, and contain one of three enzymes that can attack the α-carbon to facilitate opening of the epoxide ring (Figure 3).17 These enzymes (FosA, FosB, and FosX) are all members of the Vicinal Oxygen Chelate (VOC) superfamily (Clan CL0104) with each class utilizing a different metal to assist in catalysis.17 FosA is found in Gram-negative bacteria and was initially identified on a resistance plasmid.18 Subsequently, the enzyme was shown to form a glutathione modified fosfomycin product, utilizing Mn2+ as a necessary cofactor.19 FosA activity is greatly improved by monovalent metals such as potassium.20 A second fosfomycin modifying enzyme, FosX, is also found in Gram-negative bacteria. Instead of using glutathione to facilitate ring opening, FosX uses water to hydrolyze the epoxide and does not require any monovalent ions to enhance activity.21 In contrast to both FosA and FosX, FosB is found in Gram-positive bacteria. These bacteria do not generally biosynthesize glutathione, and must import this molecule. As a consequence, FosB has been evolved to utilize the more abundant bacillithiol (BSH) and Mn2+ or L-cysteine and Mg2+ to inactivate fosfomycin 22.
Figure 3.
Mechanisms for the enzymatic inactivation of fosfomycin catalyzed by (A) the thiol-S-transferase Mn2+-dependent FosA and the Mg2+-dependent FosB along with the necessary thiol co-substrates (RS−), and (B) the hydrolytic metalloenzyme FosX.
Both FosA and FosB are often contained on plasmids and therefore have the potential to quickly propagate through many bacterial strains. However, the overall prevalence of these genes appears to be fairly low. A 1997 study that examined fosfomycin resistance in Italy, a country that had been using fosfomycin clinically for many years, determined that less than 10% of the resistant isolates contained either of the fosfomycin inactivating fosA or fosB genes.23 Instead, most of the isolates contained mutations to the GlpT transport system.23
Glyphosate
Glyphosate (Figure 1) was identified as a potent herbicide in 1970 by Monsanto24, and initially used to spot treat weeds in non-crop areas. Eventually, its application became broader, and included spot treatment of perennial weeds near cotton and soybean crops. In 1996 Monsanto introduced the Roundup Ready® soybean (Glycine max). This genetically modified plant was able to survive treatment of glyphosate and paved the way for the development of additional glyphosate resistant crops. These plants allowed entire farms to be treated with glyphosate, controlling weed growth while leaving the desired crops unharmed. Today, glyphosate is a widely used herbicide and glyphosate resistant crops make up the majority of soybean (94.4%) and maize (82.2%) planted in the United States.25
The molecular target of glyphosate was not identified until 1980, 10 years after glyphosate’s discovery as an herbicide.26–28 Target determination was aided by the observation that plants treated with glyphosate accumulated the metabolite shikimate.28 Plants and some bacteria utilize a shikimate based pathway to biosynthesize aromatic amino acids, an entirely different pathway than the one found in animals.29 Shikimate is first converted to chorismate using three enzymatic steps, and chorismate is subsequently used as a branch point to produce either Phe and Tyr or Trp. Importantly, plants also use Phe via the phenylpropanoid pathway to produce flavonoids, lignin, coumarins, and tannins.30 Therefore, disrupting the shikimate to chorismate conversion would damage not only the amino acid pool, but also the biosynthesis of important metabolites.30 Ultimately, the second enzyme in the transformation of shikimate to chorismate, 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) was determined to be the target of glyphosate (Figure 4A).26
Figure 4.
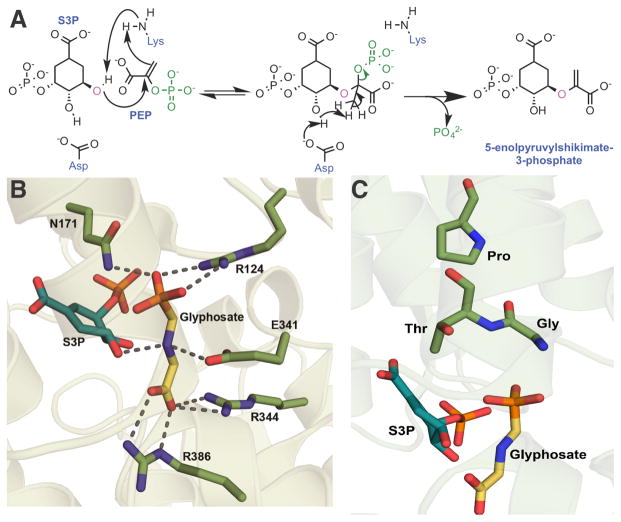
(A) Proposed mechanism of the enolpyruvyl transferase reaction catalyzed by 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), which is the target of glyphosate. MurA is thought utilize the same mechanism, using UDP-N-acetylglucosamine instead of S3P. 32,33,34 (B) Crystal structure of E. coli EPSPS in complex with glyphosate and shikimate-3-phosphate (PDB 1G6S) showing that the positively charged nitrogen of glyphosate in stabilized by Glu341 and the co-substrate S3P. (C) Resistance producing mutations are located near the glyphosate-binding pocket.
EPSPS utilizes phosphoenolpyruvate (PEP) to convert shikmate-3-phosphate (S3P) to 5-enolpyruvylshikimate 3-phosphate (Figure 4A). Upon binding of the two substrates (S3P and PEP), EPSPS undergoes large conformation changes to form a catalytically competent active site.31 Glyphosate functions by mimicking the PEP, and crystal structures of EPSPS with bound S3P and glyphosate suggest that glyphosate acts as a transition state analog (Figure 4B).31 The catalytic mechanism of EPSPS involves the nucleophilic addition of the 5-OH of S3P to the C2 carbon of PEP, followed by the elimination of phosphate. The nitrogen in glyphosate appears to be bound in the active site with a stable positive charge, which may mimic the transient positive charge formed on substrate PEP during the course of the reaction.
As the widespread use of glyphosate as a herbicide would require a resistance determinant in genetically modified plants, initial attempts to engineer resistance focused on using directed evolution and site-directed mutagenesis of EPSPS.35–37 Although these methods were able to identify tolerant EPSPS variants, the mutations reduced activity against PEP too much to be viable. Isolation and characterization of EPSPS orthologs from bacteria naturally tolerant to glyphosate proved to be the most productive source of candidate EPSPSs.38 Ultimately, the EPSPS from Agrobacterium sp. Strain CP4, isolated from the waste feed of a glyphosate product factory, served as the source of the glyphosate tolerant EPSPS used in the Roundup Ready® soybean.38 The crystal structure of the CP4 EPSPS showed the presence of an important active site Ala100, normally a Gly in the plant homologs (Figure 4C). The presence of the β-carbon of Ala100 compresses glyphosate in the active site by 0.6 Å relative to the susceptible plant enzyme.39 This compression displaces the nitrogen of glyphosate, resulting in the loss of a hydrogen bond with an active site Glu. However, this mutation alone does not account for all of the glyphosate tolerance of the Agrobacterium enzyme as the reverted Ala100→Gly variant is over 100 times more resistant to glyphosate than E. coli EPSPS.39 The Ala mutation is also observed in other glyphosate tolerant EPSPS orthologs, such as that from Klebsiella pneumonia.40
Notably, prior to the introduction of transgenic crops, glyphosate was successfully used for 20 years without the appearance of weed resistance. It was even postulated that glyphosate resistance would be unlikely to evolve under normal field conditions.41 Unfortunately, the extensive use of glyphosate over the past two decades has placed a large selective pressure on plants to evolve mechanisms of resistance. So far, three types of resistance have been observed: target mutation, increase of target expression, and modified transport. Modification of the EPSPS target was first observed in 2002 in the prevalent weed goosegrass.42 Sequencing of the resistant weeds showed two point mutations: Pro106→Ser and Pro381→Leu in EPSPS (Figure 4C). Previous mutagenesis studies in petunia EPSPS had identified the Pro106→Ser mutation as one that decreases glyphosate sensitivity but also increased the Km for PEP by nearly 40 fold.36 In the goosegrass EPSPS, the single Pro106→Ser mutation increased the IC50 for glyphosate from 6.3 μM to 38.2 μM, about 5-fold. However, the Km of PEP was not greatly altered, from 3.8 μM to 8.9 μM. The second mutation found in the resistant goosegrass EPSPS, Pro381→Leu, did not produce significant kinetic changes compared to the wild type.42 Based on this data, it appeared that the goosegrass EPSPS was predisposed to a single point mutation, Pro106→Ser, that could allow for glyphosate tolerance, but without a significant loss of activity against PEP. This contrasts with other EPSPS orthologs that compromise the Km of PEP to a much greater degree with the equivalent Pro to Ser mutation.36
Since this initial discovery of the resistant EPSPS from goosegrass, more Pro106 mutants have been isolated from tolerant plants.43–46 These single point mutations conferred protection when provided 2–3 times more glyphosate than the recommended treatment amounts. In 2015, a second mutation of Thr102→Ile was found in conjunction with the Pro106→Ser that allowed even greater resistance (Figure 4C).47 Notably, this Thr102→Ile and Pro106→Ser (TIPS) double mutation had previously been engineered in planta to produce the first commercial glyphosate resistant corn.48 The Thr102→Ile mutation greatly decreases the Vmax of EPSPS and is likely to be toxic to the plant by itself.49 However, this mutation is believed to have evolved in plants that already had the Pro106→Ser containing EPSPS. When these two mutations are combined, the IC50 for glyphosate increased over 2,500-fold compared to the wild type. In contrast, the single Pro106→Ser mutation has only a 4.3-fold increase in IC50. The double mutation comes at a cost though, as the Vmax is about 15-fold lower. In fact, this has a phenotypic effect wherein the plants containing the double mutation have a decreased growth rate 47. Crystallographic studies on the TIPS mutation have showed an alteration of the hydrogen-bonding network that ultimately leads to a narrowing of the active site that would preclude binding of glyphosate.50
In addition to the evolution of resistant EPSPS variants, plants can also utilize overproduction of the target to overcome glyphosate treatment. Amaranthus palmeri plants resistant to glyphosate were isolated from Georgia in 2010 and found to contain a sensitive EPSPS variant. However, closer inspection revealed the resistant plants to have a greatly altered EPSPS expression profile.51 Complementary DNA analysis showed that the resistant plants had between 5 and 160-fold more copies of the EPSPS gene than in plants that are susceptible to glyphosate. Using quantitative PCR, these additional copies in the genome are shown to lead to a higher amount of the corresponding mRNA, and the degree of resistance was directly correlated to the number of additional copies of the gene. For example, plants with only 5 times the number of gene copies accumulated somewhat less shikimate (the EPSPS precursor) than wild type plants when treated with glyphosate. However, plants that contained 65-fold more copies of the EPSPS gene showed almost no accumulation of shikimate, as high level of EPSPS expression may be used as a sponge to soak up glyphosate.51 Hence, target overexpression would allow a minor population of EPSPS molecules to be unperturbed by glyphosate and function normally.
Lastly, alteration of the pathway for efficient glyphosate movement through the plant has been recognized as a method of resistance. Glyphosate is taken up into the plant through the leaves, and this efficient transport is one of the key features that has enabled glyphosate efficacy.52 Therefore, glyphosate’s transport was theorized to be a potential target for generating resistance. Visualization of glyphosate accumulation in different cellular components could be monitored using 31P NMR by exploiting differences in chemical shifts that correlate to the pH changes in different cellular compartments.53–55 Using this technique, a glyphosate resistant horseweed was analyzed for anomalous glyphosate transport, and demonstrated that within 24 hours of treatment, between 65% and 85% of the glyphosate in the cells is moved from the cytoplasm to the vacuoles.55 In sensitive plant cells, no detectable amount of glyphosate could be detected in the vacuole during the same 24 hour period. The molecular basis for this transport is still under investigation, but it has been tentatively linked to increased expression of ATP-binding cassette (ABC) transporters and a tonoplast intrinsic protein (TIP).56–58 It is currently hypothesized that the TIP may be used to pump water into the cell to relieve the glyphosate induced stress while the ABC transporters pump glyphosate into the vacuoles 56.
Fosmidomycin
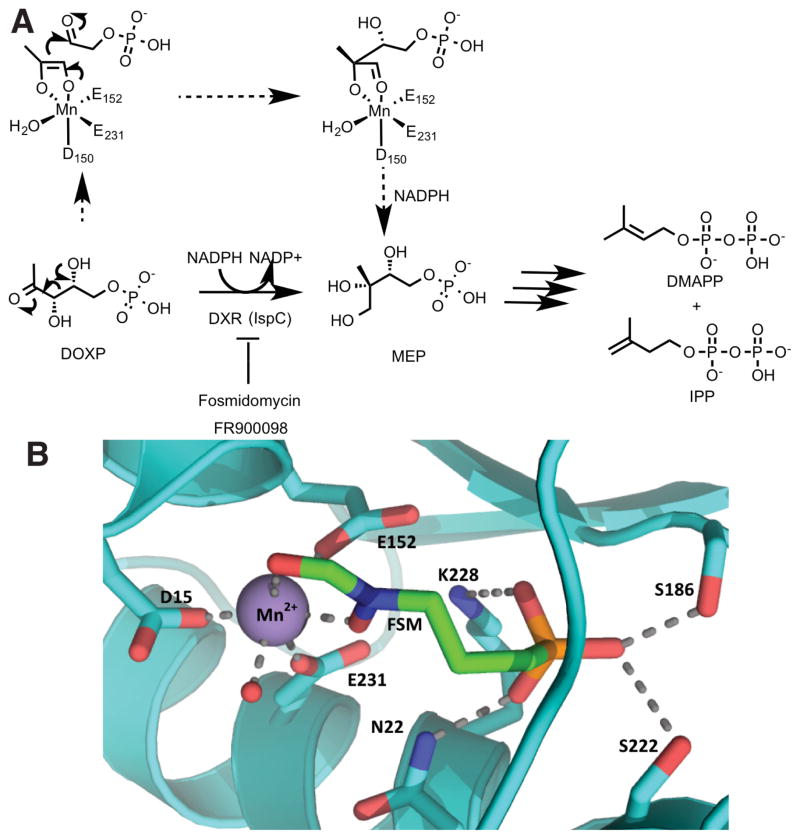
Fosmidomycin (FSM), FR900098, and structurally related analogs represent another class of bioactive phosphonates containing a unique hydroxamic acid moiety (Figure 1).59 These compounds were first discovered in 1980 and initially found to be active against a wide spectrum of Gram-negative bacteria.60,61 The novelty of these compounds has grown as their activities were found to span across many bacteria and extended even to some eukaryotes. This wide-spectrum activity can be attributed to the ubiquitous target of isoprenoid biosynthesis, an essential component across all domains of life. Specifically, FSM and similar compounds have been demonstrated to be inhibitors of D-1-deoxyxylulose-5-phosphate reductoisomerase (DXR), the enzyme responsible for carrying out the first committed step in the mevalonate-independent isoprenoid pathway, or D-methylerythritol-4-phosphate (MEP) pathway (Figure 5A) [also referred to as the D-1-deoxyxylulose-5-phosphate (DOXP) pathway].62,63 As most eukaryotes, including humans, utilize the mevalonate (MVN) dependent pathway for isoprenoid generation, these compounds do not target human enzymes, making them promising antimicrobials for clinical applications. Among the organisms that use the MEP pathway are the clinically relevant pathogens Mycobacterium tuberculosis (the causative agent for tuberculosis) and Plasmodium falciparum (the causative agent for malaria).
Figure 5.
(A) Reaction mechanism of DOXP reductoisomerase (DXR), which is part of the MEP pathway for synthesis of the essential isoprenoids dimethylallyl pyrophosphate (DMAPP) and isopentenyl pyrophosphate (IPP). Fosmidomycin and FR900098 are competitive inhibitors of DXR. (B) Active site of E. coli DXR (PDB 1ONP) bound to fosmidomycin (FSM, in green) and manganese (purple). Metal-coordinated water is represented as a red sphere.
Organisms that utilize the MEP pathway first condense the glycolytic intermediates pyruvate and D-glyceraldehyde-3-phosphate (D-GAP) in a thiamine diphosphate-dependent step to generate DOXP. Here DOXP lies at a metabolic branch point where it can either be directed into biosynthesis of the essential cofactors thiamin diphosphate (ThDP) and pyridoxal phosphate (PLP), or it can serve as a substrate of DXR, the reductoisomerase responsible for the reversible interconversion of DOXP and MEP. Experiments probing the secondary kinetic isotope effect using deuterium labeled DOXP are in support of a retroaldol-aldol type mechanism (Figure 5)64,65. Additionally, Koppisch and co-workers have shown that DXR operates through an ordered mechanism where it must first bind NADPH followed by DOXP, and FSM binds DXR as a slow, tight-binding competitive inhibitor.66 While FSM exhibits promising inhibition of DXR orthologs from a number of organisms in vitro, its mode of action has been overcome in vivo both in the clinic and in the laboratory.
Due to the presence of the negatively charged phosphonate and polar hydroxamic acid, FSM is hydrophilic and must be actively transported across cell walls to reach its target, DXR. Sakamoto et al. have demonstrated, by genetic deletion in E. coli K12, that mutants deficient in the adenylate cyclase gene (cya) are resistant to both FSM and fosfomycin.67 As previously noted, GlpT transporter expression is dependent on elevated levels of cAMP, which is the product of cya. These data suggest that structurally diverse phosphonates might share a similar dependence on GlpT-type proteins for import, and strains that lack these transporters will likely be tolerant. For example, the genome of Mycobacterium tuberculosis (MTB) lacks any polypeptide with primary sequence similarity to the GlpT transporter and it is suggested that FSM and fosfomycin resistance in MTB is due in part to inefficient transport.68 Experimental data show that MTB DXR is both necessary for survival and effectively inhibited by FSM in vitro, suggesting that FSM could be effective against Mycobacteria if properly transported into the cell.68,69 Towards this end, attempts have been made to mask the negatively charged phosphonate to generate phosphonate ester prodrugs that can be hydrolyzed by nonspecific intracellular esterases following active import. For example, the acyloxymethyl phosphonate esters exhibit growth inhibition against Mycobacterium smegmatis in a disc diffusion assay.70 Similarly, a variety of lipophilic ester prodrugs of FR900098 have microbial inhibitory concentrations (MICs) against a panel of Gram-positive bacteria that are greatly improved over those of the free phosphonic acids.71
Given the role of GlpT in fosmidomycin transport, it is not surprising that mutations of the transporter frequently result in tolerant strains. In one such case, resistance towards FSM in the pathogenic Gram-negative bacteria Francisella tularensis implicated the GlpT transporter in its mechanism of resistance when growth of the strain in the presence of FSM or FR900098-soaked discs revealed the presence of breakthrough colonies inside the inhibition zone. Genome sequencing of four selected colonies revealed amino acid deletions, missense mutations, and premature stop codons within the GlpT coding region, all of which are likely to result in translation of a non-functional protein.71 Additionally, transposon insertion mutants at the glpT locus resulted in FSM and FR900098 insensitivity in F. tularensis, consistent with other findings that their GlpT transporter accounts for sensitivity towards phosphonate inhibitors. Moreover, when the resistant strains with a defective GlpT were provided a lipophilic prodrug of FR900098, they regained sensitivity to the DXR inhibitor, supporting the hypothesis that resistance in these strains is solely a result of deficient active transport.72,73
Fosmidomycin tolerance has also been associated with mutations of the target dxr. Recently, error-prone PCR was used to generate expression libraries of dxr mutants in E. coli to identify resistant cells selected on plates containing lethal doses of FSM. Sequencing of the colonies revealed two resistant mutants encoding an identical set of five amino acid changes in dxr. Individual site-directed mutants were generated based on these initial five amino acid changes revealing that a Ser222→Thr substitution was sufficient to incur resistance against FSM.74 In crystal structures of E. coli DXR bound to fosmidomycin and other phosphonates, Ser222 is within hydrogen-bonding distance to the phosphonate moiety of the inhibitors.75,76 An inspection of these structures suggests that a Thr at this position might lead to a sterically induced shift of the side chain hydroxyl away from the phosphonate, thus compromising binding of both inhibitors and the substrate DOXP.. Kinetic analyses of wild type and Ser222→Thr DXR proteins revealed that the mutant has a Km for substrate DOXP that is 7-fold higher than that of the wild-type with no appreciable difference in the Vmax values. Correspondingly, a 30-fold increase in IC50 for FSM was observed in the Ser222→Thr variant. The authors conclude that the benefits incurred by this mutation in lowering the affinity for FSM must outweigh the negative effects of lowering the affinity for its substrate DOXP.74
Mutations in the malaria parasite Plasmodium falciparum that conferred resistance towards FSM were generated by selective growth on media containing FSM. Strains grown under selective pressure achieved resistance through mutations on a member of the haloacid dehalogenase (HAD) superfamily, termed PfHAD1. An InterPro analysis reveals that this protein falls within the Cof-like hydrolase subfamily wherein the majority of characterized proteins are promiscuous phosphatases.77,78,79 PfHAD1 is proposed to be a phosphatase with loose substrate-specificity that is experimentally shown to dephosphorylate intermediates of glycolysis. Consequently, a loss of PfHAD1 activity would lead to an increase in concentration of MEP pathway substrates, and liquid-chromatography mass-spectrometry (LC-MS) analysis detected higher concentrations of MEP pathway intermediates in FSM resistant P. falciparum strains with PfHAD1 mutations.80 Taken together, these findings suggest that PfHAD1 is a HAD member phosphatase that functions as a MEP pathway negative regulator in P. falciparum and inactivation of this protein is sufficient to overcome inhibition of DXR by increasing levels of available substrate to the MEP pathway.
Another example of resistance in P. falciparum was characterized via a high-density tiling microarray approach capable of genome-wide analyses in a single hybridization event. This approach identified copy number variations in P. falciparum genomes and specifically detected an amplification of dxr genes in resistant strains. Quantitative real-time PCR revealed a 3.8-fold increase in transcript level and 2.7-fold increase in copy number of DXR, suggesting that FSM inhibition is overcome by increasing the enzyme concentration in some FSM resistant parasites.81
Conclusions
Research efforts using multi-pronged approaches continue to reveal mechanisms for acquired and spontaneous resistance that has developed against phosphonate antibiotics and herbicides. In some instances, a biochemical understanding for the basis of tolerance has resulted in the development of derivative compounds that can overcome the development of resistance, but other classes of resistance mechanisms are likely to be intractable. As with any class of chemical scaffolds, the best strategy to counter tolerance will be a combination of orthogonal methods, leveraging both the microbiological and biochemical understanding of resistance mechanisms with structure-function attempts focused on improvements in the parent compounds. The benefits of this strategy are borne out by development of derivative herbicides, and as more bioactive phosphonates continue to progress towards clinical trials, similar approaches will likely result in the development of more potent derivatives that are, hopefully, less prone to existing resistance mechanisms.
Acknowledgments
This work has been supported by the National Institutes of Health (PO1GM077596 to S.K.N.). We thank our collaborators Bill Metcalf, Wilfred van der Donk, Huimin Zhao, and their laboratories for their contributions to the work described in this review. We apologize to the numerous investigators in the field of phosphonate enzymology whose work we could not describe here due to space limitations.
Notes and references
- 1.Hendlin D, Stapley EO, Jackson M, Wallick H, Miller AK, Wolf FJ, Miller TW, Chaiet L, Kahan FM, Foltz EL, Woodruff HB, Mata JM, Hernandez S, Mochales S. Science. 1969;166:122–3. doi: 10.1126/science.166.3901.122. [DOI] [PubMed] [Google Scholar]
- 2.Christensen BG, Leanza WJ, Beattie TR, Patchett AA, Arison BH, Ormond RE, Kuehl FA, Albers-Schonberg G, Jardetzky O. Science. 1969;166:123–125. doi: 10.1126/science.166.3901.123. [DOI] [PubMed] [Google Scholar]
- 3.Kahan FM, Kahan JS, Cassidy PJ, Kropp H. Ann N Y Acad Sci. 1974;235:364–86. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown ED, Marquardt JL, Lee JP, Walsh CT, Anderson KS. Biochemistry. 1994;33:10638–10645. doi: 10.1021/bi00201a010. [DOI] [PubMed] [Google Scholar]
- 5.Eschenburg S, Priestman M, Schönbrunn E. J Biol Chem. 2005;280:3757–3763. doi: 10.1074/jbc.M411325200. [DOI] [PubMed] [Google Scholar]
- 6.Marquardt JL, Brown ED, Lane WS, Haley TM, Ichikawa Y, Wong CH, Walsh CT. Biochemistry. 1994;33:10646–10651. doi: 10.1021/bi00201a011. [DOI] [PubMed] [Google Scholar]
- 7.Venkateswaran PS, Wu HC. J Bacteriol. 1972;110:935–44. doi: 10.1128/jb.110.3.935-944.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karageorgopoulos DE, Wang R, Yu XH, Falagas ME. J Antimicrob Chemother. 2012;67:255–68. doi: 10.1093/jac/dkr466. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson aI, Berg OG, Aspevall O, Kahlmeter G, Andersson DI. Antimicrob Agents Chemother. 2003;47:2850–2858. doi: 10.1128/AAC.47.9.2850-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahata S, Ida T, Hiraishi T, Sakakibara S, Maebashi K, Terada S, Muratani T, Matsumoto T, Nakahama C, Tomono K. Int J Antimicrob Agents. 2010;35:333–7. doi: 10.1016/j.ijantimicag.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Santoro A, Cappello AR, Madeo M, Martello E, Iacopetta D, Dolce V. Biochim Biophys Acta - Gen Subj. 2011;1810:1323–1329. doi: 10.1016/j.bbagen.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Lemieux MJ, Song J, Auer M, Wang D. Science. 2003;301:616–20. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 13.Alper MD, Ames BN. J Bacteriol. 1978;133:149–157. doi: 10.1128/jb.133.1.149-157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DH, Lees WJ, Kempsell KE, Lane WS, Duncan K, Walsh CT. Biochemistry. 1996;35:4923–8. doi: 10.1021/bi952937w. [DOI] [PubMed] [Google Scholar]
- 15.De Smet KaL, Kempsell KE, Gallagher A, Duncan K, Young DB. Microbiology. 1999;145:3177–3184. doi: 10.1099/00221287-145-11-3177. [DOI] [PubMed] [Google Scholar]
- 16.Skarzynski T, Mistry A, Wonacott A, Hutchinson SE, Kelly VA, Duncan K. Structure. 1996;4:1465–74. doi: 10.1016/s0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]
- 17.Thompson MK, Keithly ME, Sulikowski Ga, Armstrong RN. Perspect Sci. 2015;4:17–23. [Google Scholar]
- 18.Garcia-Lobo JM, Ortiz JM. J Bacteriol. 1982;151:477–479. doi: 10.1128/jb.151.1.477-479.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arca P, Rico M, Brana AF, Villar CJ, Hardisson C, Suarez JE. Antimicrob Agents Chemother. 1988;32:1552–1556. doi: 10.1128/aac.32.10.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beharry Z, Palzkill T. J Biol Chem. 2005;280:17786–17791. doi: 10.1074/jbc.M501052200. [DOI] [PubMed] [Google Scholar]
- 21.Fillgrove KL, Pakhomova S, Schaab MR, Newcomer ME, Armstrong RN. Biochemistry. 2007;46:8110–8120. doi: 10.1021/bi700625p. [DOI] [PubMed] [Google Scholar]
- 22.Cao M, Bernat Ba, Wang Z, Armstrong RN, Helmann JD. J Bacteriol. 2001;183:2380–3. doi: 10.1128/JB.183.7.2380-2383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arca P, Reguera G, Hardisson C. J Antimicrob Chemother. 1997;40:393–399. doi: 10.1093/jac/40.3.393. [DOI] [PubMed] [Google Scholar]
- 24.3 799 758. US Pat. 1974
- 25.Coupe RH, Capel PD. Pest Manag Sci. 2015 doi: 10.1002/ps.4082. [DOI] [PubMed] [Google Scholar]
- 26.Steinrücken HC, Amrhein N. Biochem Biophys Res Commun. 1980;94:1207–12. doi: 10.1016/0006-291x(80)90547-1. [DOI] [PubMed] [Google Scholar]
- 27.Holländer H, Amrhein N. Plant Physiol. 1980;66:823–829. doi: 10.1104/pp.66.5.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amrhein N, Deus B, Gehrke P, Steinrücken HC. Plant Physiol. 1980;66:830–834. doi: 10.1104/pp.66.5.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda H, Dudareva N. Annu Rev Plant Biol. 2012;63:73–105. doi: 10.1146/annurev-arplant-042811-105439. [DOI] [PubMed] [Google Scholar]
- 30.Vogt T. Mol Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 31.Schönbrunn E, Eschenburg S, Shuttleworth Wa, Schloss JV, Amrhein N, Evans JN, Kabsch W. Proc Natl Acad Sci U S A. 2001;98:1376–80. doi: 10.1073/pnas.98.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berti PJ, Chindemi P. Biochemistry. 2009;48:3699–3707. doi: 10.1021/bi802251s. [DOI] [PubMed] [Google Scholar]
- 33.Lou M, Gilpin ME, Burger SK, Malik AM, Gawuga V, Popović V, Capretta A, Berti PJ. J Am Chem Soc. 2012;134:12947–12957. doi: 10.1021/ja3043382. [DOI] [PubMed] [Google Scholar]
- 34.Eschenburg S, Kabsch W, Healy ML, Schonbrunn E. J Biol Chem. 2003;278:49215–49222. doi: 10.1074/jbc.M309741200. [DOI] [PubMed] [Google Scholar]
- 35.Comai L, Sen LC, Stalker DM. Science. 1983;221:370–371. doi: 10.1126/science.221.4608.370. [DOI] [PubMed] [Google Scholar]
- 36.Padgette SR, Re DB, Gasser CS, Eichholtz Da, Frazier RB, Hironaka CM, Levine EB, Shah DM, Fraley RT, Kishore GM. J Biol Chem. 1991;266:22364–22369. [PubMed] [Google Scholar]
- 37.Eschenburg S, Healy ML, Priestman Ma, Lushington GH, Schönbrunn E. Planta. 2002;216:129–35. doi: 10.1007/s00425-002-0908-0. [DOI] [PubMed] [Google Scholar]
- 38.WO 92/04449. Int Pat. 1992
- 39.Funke T, Han H, Healy-Fried ML, Fischer M, Schönbrunn E. Proc Natl Acad Sci U S A. 2006;103:13010–5. doi: 10.1073/pnas.0603638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sost D, Amrhein N. Arch Biochem Biophys. 1990;282:433–436. doi: 10.1016/0003-9861(90)90140-t. [DOI] [PubMed] [Google Scholar]
- 41.Bradshaw LD, Padgette SR, Kimball SL, Wells BH. Weed Sci Soc Am. 1997;11:189–198. [Google Scholar]
- 42.Baerson SR, Rodriguez DJ, Tran M, Feng Y, Biest NA, Dill GM. Plant Physiol. 2002;129:1265–75. doi: 10.1104/pp.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Q, Cairns A, Powles S. Planta. 2007;225:499–513. doi: 10.1007/s00425-006-0364-3. [DOI] [PubMed] [Google Scholar]
- 44.Wakelin AM, Preston C. Weed Res. 2006;46:432–440. [Google Scholar]
- 45.Ng CH, Wickneswari R, Salmijah S, Teng YT, Ismail BS. Weed Res. 2003;43:108–115. [Google Scholar]
- 46.Sammons RD, Gaines Ta. Pest Manag Sci. 2014;70:1367–1377. doi: 10.1002/ps.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Q, Jalaludin A, Han H, Chen M, Sammons RD, Powles SB. Plant Physiol. 2015;167:1440–7. doi: 10.1104/pp.15.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.6 040 497. US Pat. 2000
- 49.7 723 575 B2. US Pat. 2010
- 50.Funke T, Yang Y, Han H, Healy-Fried M, Olesen S, Becker A, Schönbrunn E. J Biol Chem. 2009;284:9854–9860. doi: 10.1074/jbc.M809771200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaines Ta, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, Nissen SJ, Patzoldt WL, Tranel PJ, Culpepper aS, Grey TL, Webster TM, Vencill WK, Sammons RD, Jiang J, Preston C, Leach JE, Westra P. Proc Natl Acad Sci U S A. 2010;107:1029–34. doi: 10.1073/pnas.0906649107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirkwood RC, Hetherington R, Reynolds TL, Marshall G. Pest Manag Sci. 2000;56:359–367. [Google Scholar]
- 53.Roberts JKM, Ray PM, Wade-Jardetzky N, Jardetzky O. Nature. 1980;283:870–872. [Google Scholar]
- 54.Castellino S, Leo GC, Sammons RD, Sikorski Ja. Biochemistry. 1989;28:3856–3868. [Google Scholar]
- 55.Ge X, d’Avignon DA, Ackerman JJH, Sammons RD. Pest Manag Sci. 2010;66:345–8. doi: 10.1002/ps.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nol N, Tsikou D, Eid M, Livieratos IC, Giannopolitis CN. Weed Res. 2012;52:233–241. [Google Scholar]
- 57.Peng Y, Abercrombie LLG, Yuan JS, Riggins CW, Sammons RD, Tranel PJ, Stewart CN. Pest Manag Sci. 2010;66:1053–1062. doi: 10.1002/ps.2004. [DOI] [PubMed] [Google Scholar]
- 58.Yuan JS, Abercrombie LLG, Cao Y, Halfhill MD, Peng Y, Hu J, Rao MR, Heck GR, Larosa TJ, Douglas R, Wang X, Ranjan P, Johnson DH, Wadl Pa, Scheffler BE, Rinehart Ta, Trigiano RN, Stewart CN, Zhou X, Peng Y, Hu J, Yuan JS, Abercrombie LLG, Cao Y, Halfhill MD, Rao R, Heck R, Larosa TJ, Sammons RD, Wang X, Ranjan P, Johnson DH, Trigiano RN, Stewart CN, Rinehart Ta, Wadl Pa, Scheffler BE. Weed Sci. 2010;58:109–117. [Google Scholar]
- 59.Zinglé C, Kuntz L, Tritsch D, Grosdemange-Billiard C, Rohmer M. Bioorganic Med Chem Lett. 2012;22:6563–6567. doi: 10.1016/j.bmcl.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 60.Mine Y, Kamimura T, Nonoyama S, Nishida M, Goto S, Kuwahara S. J Antibiot (Tokyo) 1980;33:36–43. doi: 10.7164/antibiotics.33.36. [DOI] [PubMed] [Google Scholar]
- 61.Okuhara M, Kuroda Y, Goto T, Okamoto M, Terano H, Kohsaka M, Aoki H, Imanaka H. J Antibiot (Tokyo) 1980;33:13–17. doi: 10.7164/antibiotics.33.13. [DOI] [PubMed] [Google Scholar]
- 62.Zeidler J, Schwender J, Muller C, Wiesner J, Weidemeyer C, Beck E, Jomaa H, Lichtenthaler HK. Zeitschrift fur Naturforsch - Sect C J Biosci. 1998;53:980–986. [Google Scholar]
- 63.Kuzuyama T, Shimizu T, Takahashi S, Seto H. Tetrahedron Lett. 1998;39:7913–7916. [Google Scholar]
- 64.Wong U, Cox RJ. Angew Chemie. 2007;119:5014–5017. [Google Scholar]
- 65.Munos JW, Pu X, Mansoorabadi SO, Kim HJ, Liu HW. J Am Chem Soc. 2009;131:2048–2049. doi: 10.1021/ja807987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koppisch aT, Fox DT, Blagg BSJ, Poulter CD. Biochemistry. 2002;41:236–243. doi: 10.1021/bi0118207. [DOI] [PubMed] [Google Scholar]
- 67.Sakamoto Y, Furukawa S, Ogihara H, Yamasaki M. Biosci Biotechnol Biochem. 2003;67:2030–2033. doi: 10.1271/bbb.67.2030. [DOI] [PubMed] [Google Scholar]
- 68.Brown AC, Parish T. BMC Microbiol. 2008;8:78. doi: 10.1186/1471-2180-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dhiman RK, Schaeffer ML, Bailey AM, Testa Ca, Scherman H, Crick DC. 2005;187:8395–8402. doi: 10.1128/JB.187.24.8395-8402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ponaire S, Zinglé C, Tritsch D, Grosdemange-Billiard C, Rohmer M. Eur J Med Chem. 2012;51:277–285. doi: 10.1016/j.ejmech.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 71.Uh E, Jackson ER, San Jose G, Maddox M, Lee RE, Lee RE, Boshoff HI, Dowd CS. Bioorganic Med Chem Lett. 2011;21:6973–6976. doi: 10.1016/j.bmcl.2011.09.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKenney ES, Sargent M, Khan H, Uh E, Jackson ER, Jose GS, Couch RD, Dowd CS, van Hoek ML. PLoS One. 2012:7. doi: 10.1371/journal.pone.0038167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mackie RS, McKenney ES, van Hoek ML. Front Microbiol. 2012;3:1–12. doi: 10.3389/fmicb.2012.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Armstrong CM, Meyers DJ, Imlay LS, Meyers CF, Odom AR. Antimicrob Agents Chemother. 2015:AAC.00602–15. doi: 10.1128/AAC.00602-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yajima S, Hara K, Iino D, Sasaki Y, Kuzuyama T, Ohsawa K, Seto H. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:466–470. doi: 10.1107/S1744309107024475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mac Sweeney A, Lange R, Fernandes RPM, Schulz H, Dale GE, Douangamath A, Proteau PJ, Oefner C. J Mol Biol. 2005;345:115–127. doi: 10.1016/j.jmb.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 77.Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, Yakunin AF. J Biol Chem. 2006;281:36149–36161. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- 78.Roberts A, Lee SY, McCullagh E, Silversmith RE, Wemmer DE. Proteins Struct Funct Genet. 2005;58:790–801. doi: 10.1002/prot.20267. [DOI] [PubMed] [Google Scholar]
- 79.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, De Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJa, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. Nucleic Acids Res. 2012;40:306–312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guggisberg AM, Park J, Edwards RL, Kelly ML, Hodge DM, Tolia NH, Odom AR. Nat Commun. 2014;5:4467. doi: 10.1038/ncomms5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dharia NV, Sidhu ABS, Cassera MB, Westenberger SJ, Bopp SE, Eastman RT, Plouffe D, Batalov S, Park DJ, Volkman SK, Wirth DF, Zhou Y, Fidock Da, Winzeler Ea. Genome Biol. 2009;10:R21. doi: 10.1186/gb-2009-10-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]