Abstract
The presence of multiple variants for many mRNAs is a major contributor to protein diversity. The processing of these variants is tightly controlled in a cell-type specific manner and has a significant impact on gene expression control. Here we investigate the differential translation rates of individual mRNA variants in embryonic stem cells (ESCs) and in ESC derived Neural Precursor Cells (NPCs) using polysome profiling coupled to RNA sequencing. We show that there are a significant number of detectable mRNA variants in ESCs and NPCs and that many of them show variant specific translation rates. This is correlated with differences in the UTRs of the variants with the 5’UTR playing a predominant role. We suggest that mRNA variants that contain alternate UTRs are under different post-transcriptional controls. This is likely due to the presence or absence of miRNA and protein binding sites that regulate translation rate. This highlights the importance of addressing translation rate when using mRNA levels as a read out of protein abundance. Additional analysis shows that many annotated non-coding mRNAs are present on the polysome fractions in ESCs and NPCs. We believe that the use of polysome fractionation coupled to RNA sequencing is a useful method for analysis of the translation state of many different RNAs in the cell.
Introduction
Embryonic stem cells (ESCs) possess the unique ability to self-renew and differentiate into all the cells of the body. Tight regulation of gene expression is essential for cells to maintain their self-renewal state. Upon differentiation, specific cascades of gene expression changes are implemented resulting in conversion of the cells to specific cell types. These gene expression changes are regulated and coordinated on multiple levels including transcription, RNA stability, translational control as well as protein function and degradation. This ensures a tight control over protein expression which is required for self-renewal or differentiation. Changes in transcriptional control are a major driving force for differentiation and much work has focused on mapping the binding sites of transcription factors in ESCs.[1, 2] More recently, post-transcriptional control has been shown to play a major role in ESC function. A number of miRNAs are known to promote ESC self-renewal while others are important for directed differentiation. [3–5]
It is now well established that a single gene can give rise to many different mRNA variants, the majority of which will have different protein products. These variants can arise through a number of mechanisms including alternate transcription start sites (TSS), alternative splicing and alternative poly(A) tail site selection. Alternate TSSs are a major contributor to 5’UTR diversity between transcripts from the same gene. In addition there is a large amount of tissue specific usage of alternative transcription start sites and a number of examples have been shown of TSS selection being developmentally regulated [6, 7]. Alternate TSSs can lead to altered 5’UTR sequences and different start codons resulting in alterations or truncations in the N terminus of the protein. Inappropriate usage of TSSs has been associated with cancer progression highlighting the importance of tight control over TSS usage [8].
Splicing is the most significant contributor to transcript diversity and it is estimated that over 95% of genes produce alternatively spliced mRNAs. Splicing is highly regulated and many tissues show very specific splicing patterns with some genes producing up to 25 different spliced isoforms in different tissues. Advances in sequencing technologies have confirmed the widespread prevalence of alternative splicing in different tissues yet the role of the majority of variants is still unknown [9–11]. Alternative splicing is regulated by specific RNA binding proteins that bind to the pre-RNA to control the function of the splicing machinery [4]. A number of ESC specific splicing events have been characterised demonstrating the importance of splicing control in ESC maintenance [12–16].
Variant mRNA isoforms can arise from utilisation of alternative polyadenylation (APA) sites which affect the length of the 3’end of the RNA. The majority of APA sites are present in the 3’UTR so it affects 3’UTR length. [6, 17] It has been estimated that approximately 70% of genes have APA sites with each gene having an average of two. [6, 18] During development the proximal polyA sites are frequently used causing mRNAs to have shorter UTRs. [19] Highly proliferative cells have also been shown to have shorter 3’UTRs [20]. Longer UTRs have a greater number of cis-acting regulatory sites and so are likely under greater post-transcriptional control resulting in changes in protein level [6, 21, 22].
Alternate TSSs, splicing and alternate polyA site selection can all result in changes in the protein product produced from an mRNA variant. In addition they can affect sequences within the 5’ and 3’UTRs of an mRNA. The UTRs harbour many of the control elements that dictate when, where and at what rate an mRNA is translated. They also regulate the stability of an mRNA. This is mediated by many trans-acting factors in the form of RNA binding proteins and miRNAs that bind to the UTRs of target mRNAs. These post-transcriptional controls can greatly impact on protein levels in a cell. Recently the translation rate of different mRNA isoforms was compared using RNAseq in a human kidney cell line. It was reported that approximately 30% of mRNA isoforms are differentially loaded with ribosomes suggesting they are translated at a different rate. [23] Recent studies in mouse ESCs demonstrated that different splice variants of SERCA2 can be differentially targeted by miRNAs in ESCs and embryoid bodies due to differences in UTR sequences [16]. These and other studies highlight the importance of isoform specific UTR sequences in post-transcriptional control of gene expression [24].
We sought to address the impact of alternate UTRs on translation rate of different variants in mouse embryonic stem cells (ESCs) and ESC derived neural precursor cells (NPCs). We have separated mRNAs based on ribosomal load and assessed the relative translation rate of individual variants using RNA seq. We find that there are a significant proportion of variants that show differential translation rates in ESCs and NPCs. We further show that this altered translation rate correlates with processing events that affect 5’ and 3’ UTR sequences with 5’UTR sequences having the greater role. Our data suggests that analysing the translation state of mRNA variants is essential to get a more accurate read out of the protein levels in a cell.
Results
Translationally regulated mRNAs on ESC to NPC differentiation
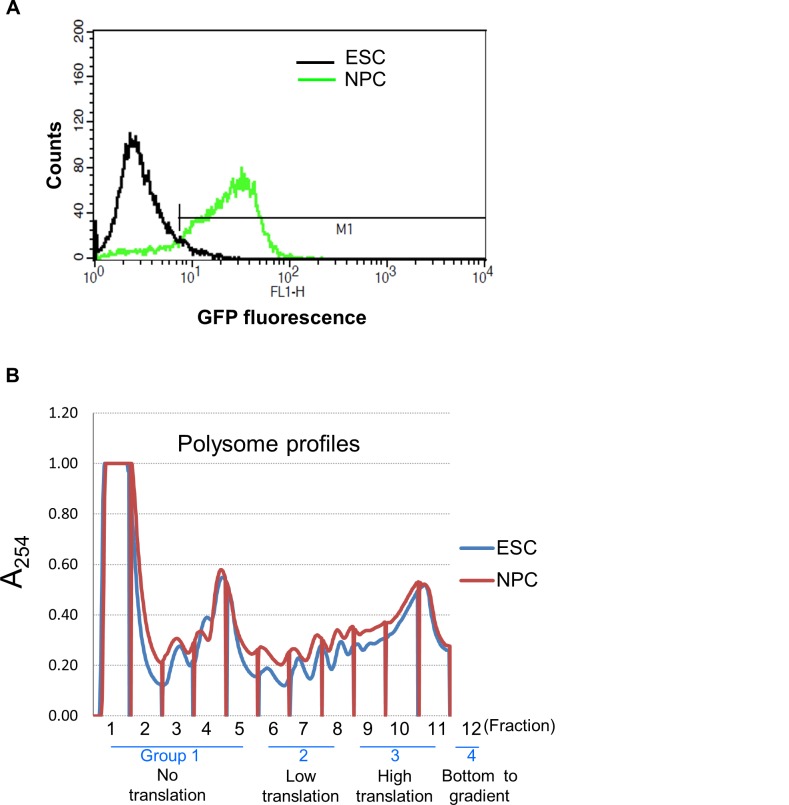
We set out to determine the role of splicing in the regulation of translation rate in ESCs and ESC derived NPCs. ESCs were differentiated into NPCs in a SOX1-GFP reporter line in N2B27 media for six days [25–27]. SOX1 is a marker for NPCs enabling us to confirm efficient differentiation with over 80% of cells expressing SOX1-GFP (Fig 1A). To determine the translation state of RNAs in the two cell types, polysome profiling was performed to separate the differentially translated RNAs across a sucrose gradient. RNAs were pooled together as non-translated, low-translation and high-translation depending on their ribosomal load (Fig 1B). RNAs with a greater ribosomal load sediment in the heavier sucrose fractions and are considered to be more highly translated. RNAs with few or no ribosomes are found in the lighter fractions and are considered to be non- or inefficiently translated. RNAs were also collected from the bottom of the gradient. To determine the translation rate of individual splice variants, we subjected each group of RNAs to RNA sequencing using the ABI SOLiD sequencing platform. Sequences were mapped to the UCSC mm10 reference genome and data was normalised to a pool of four bacterial poly(A) spike-in mRNAs. The location in the polysome profile of a selection of RNAs was validated by qRT-PCR and found to have a good correlation (R2-0.7134), suggesting our RNA sequencing gave an accurate read out of the polysome association of individual mRNAs. (S1 Fig)
Fig 1. Polysome profiling of ESCs and NPCs.
(A) Efficiency of NPC differentiation. Mouse Sox1-GFP ESCs were differentiated to NPCs for 6 days. Flow cytometry of the SOX1-GFP signal showing over 80% SOX1-GFP positive cells after differentiation. (B) ESCs and NPCs were subjected to polysome profiling and 12 fractions were collected into four groups. No translation F1-5), Low translation (F6-8), High translation (F9-11) and the bottom of the gradient (F12).
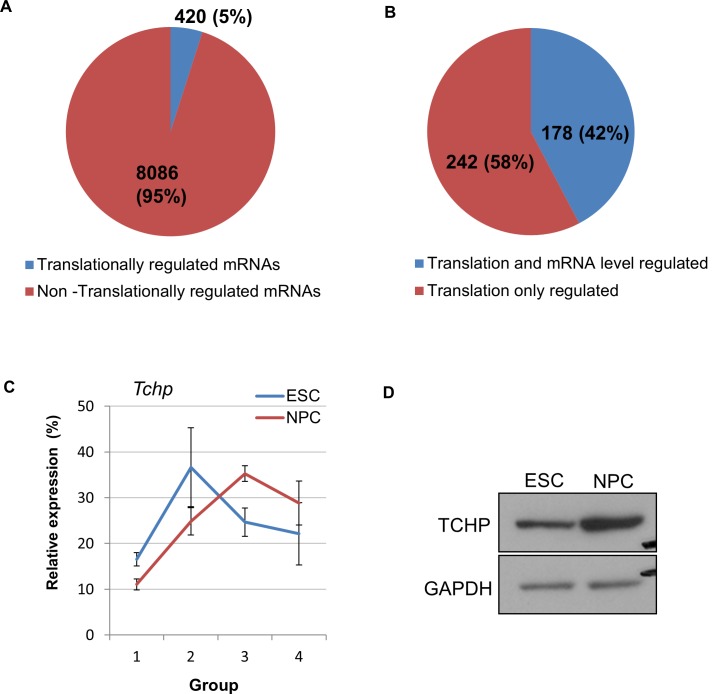
A shift in ribosomal load between ESCs and NPCs represents a likely change in translation rate. We saw that 5% of mRNAs showed differential translation rates between ESCs and NPCs with a translation shift of over 20% (Fig 2A). 26% of mRNAs showed a greater than 2 fold change in total RNA levels confirming earlier observations that the transcriptome is dramatically reorganised on ESC differentiation [28]. Interestingly, the majority of translationally regulated RNAs were not co-regulated at the mRNA level. 58% of translationally regulated mRNAs showed less than 2 fold change in RNA levels suggesting they are predominantly translationally regulated (Fig 2B). We used RT-PCR to confirm the polysome association of one translationally regulated mRNA Tchp. The increased translation was further indicated from the increased protein levels seen by western blot in NPCs compared to ESCs (Fig 2C and 2D).
Fig 2. Transcriptional and translational changes upon differentiation of ESCs to NPCs.
(A) Pie chart showing the degree of translational shift of mRNAs during ESC to NPC differentiation. 5% of mRNAs display a translation shift greater than 20% upon NPC. (B) Pie chart showing transcriptional changes in the sub-fraction of translationally regulated genes. 58% of these genes are purely regulated translationally and show less than 2 fold changes in total mRNA levels. (C) Real-time PCR analysis of Tchp showing increased enrichment in the heavy polysome fractions in NPCs. (D) Western blot of TCHP protein in ESCs and NPCs. GAPDH is shown as a loading control.
Many splice variants are differentially translated
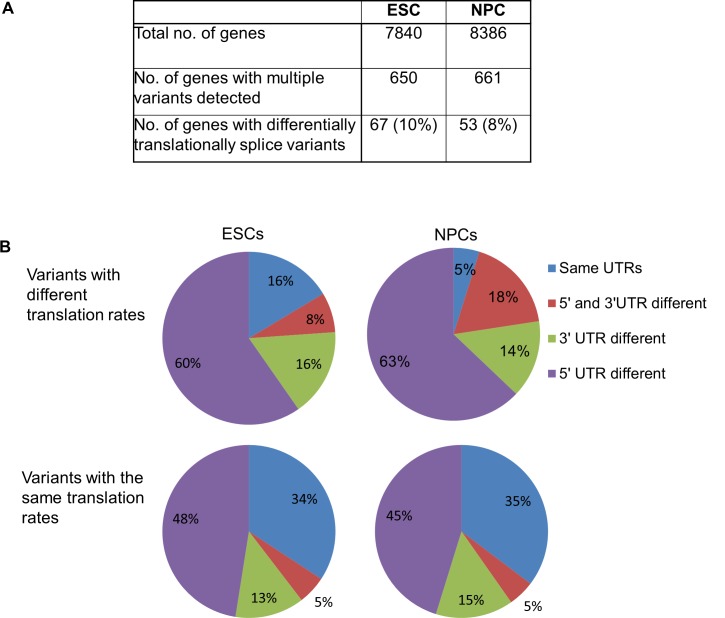
We next sought to determine if splice variants displayed translation rate differences within the same cell type. We identified multiple splice variants for 650 genes in ESCs and 661 genes in NPCs (Fig 3A). Of these, 10% and 8% of splice variants were enriched in different polysome pools in ESCs and NPCs respectively (S1 Table). This suggests the splice variants are being translated at different rates within the same cell type. We next assessed the correlation between altered 5’ or 3’ UTRs and different translation rates between the variants. Interestingly, we saw an enrichment for variants with altered UTRs in the differentially translated group of variants compared to variants that show similar rates of translation (Fig 3B). In ESCs, for variants that showed different translation rates, 84% of variants had different UTRs (95% in NPCs). This is in contrast to variants that do not show changes in translation where only 66% of variants displayed altered UTR sequences (65% in NPCs). The majority of variations in both cases were in the 5’UTR with over 60% (63% in NPCs) of translationally regulated splice variants having different 5’UTR sequences. 16% (14% in NPCs) had different 3’UTR sequences and 8% (18% in NPCs) had different 5’ and 3’ UTRs. We analysed the 5’UTRs of mRNA variants that showed differences in translation rate and altered 5’UTRs. We found no significant difference in the GC content of the UTRs however there was an enrichment for longer UTRs in the more translationally repressed variants (S2 Fig). It is likely that the longer sequences contain regions that inhibit ribosome scanning and thus repress translation. There was also a correlation between translation rate and 5’UTR free energy levels such that variants that were more highly translated had a greater free energy level in their 5’UTRs. A higher free energy generally results in less stable RNA structures that could be more conducive to increased ribosomal scanning and thus increased translation (S2 Fig)
Fig 3. Translational regulation of splice variants in ESCs and NPCs.
(A) Analysis of the translation rate of all mRNAs with more than one variant in ESCs and NPCs. (B) Charts showing the correlation between variants with different translation rates and altered UTRs.
Interestingly 16% (5% in NPCs) of variants that showed different translation rates did not have an altered 5’ or 3’ UTR suggesting the regulatory region for translation rate resides in variant regions within the coding sequence for these mRNAs. An example of this is Ctage5 which has three variants. Variant 1 and 3 have the same UTR sequences but differ only in the inclusion of exons 5 and 6 in variant one. Inclusion of these exons in variant 1 results in a significantly decreased ribosomal load compared with variant 3 suggesting it is being translated at a slower rate. Variant 2 excludes these exons in addition to having a different 3’UTR and its translation is significantly stronger than variant 1 (S3 Fig). Exon 4 and 5 do not have an enrichment of rare codons suggesting the translational repression seen is not due to codon usage. It is likely that exon 5 and 6 contain cis-acting regulatory sequences that might bind to miRNAs or RBPs to repress translation of that variant.
mRNAs with long ORFs may be bound by a higher number of ribosomes which could contribute to the differences in ribosomal load we see between splice variants in one cell type. To see if ORF length was responsible for the altered ribosomal load seen on the different splice variants, we determined the ORF lengths for all variants showing a polysome shift and looked for a correlation with direction of the shift. We saw no strong correlation between longer ORFs and higher ribosomal load (Table 1) (S1 Table). For variants that were bound by more ribosomes less than half had longer open reading frames. This suggests that altered ORF length is not the predominant cause of the altered ribosomal load we see in different splice variants.
Table 1. Relationship between ORF length and translation rate.
| ESC | NPC | |
|---|---|---|
| Genes with multiple variants that show different translation rates correlating with ORF size. | 33 | 20 |
| Genes with multiple variants that show different translation rates not correlating with ORF size. | 34 | 33 |
| TOTAL | 67 | 53 |
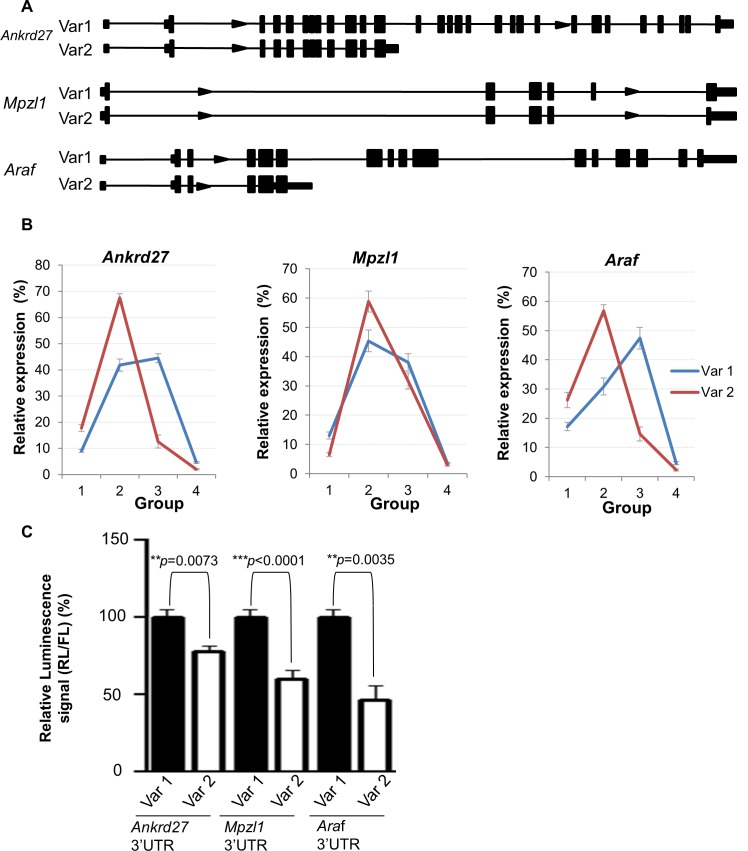
In order to validate the splice variant changes in translation we identified we first performed qRT-PCR with variant specific primers across three biological replicates. We selected three genes that have variant 3’UTRs, Ankrd27, Mpzl1 and Araf (Fig 4A). qRT-PCR on the different polysomal groups confirmed that the variants are differentially loaded with ribosomes (Fig 4B). To determine if this difference in translation is mediated through the 3’UTR we cloned the different 3’UTRs downstream of the luciferase reported gene in the psiCHECK-2 vectors. On transfection into ESCs we determined the luciferase levels of each variant. We saw that in all three cases the variant that displayed the lower ribosomal load had the lower luciferase expression levels (Fig 4C). These data strongly suggest that sequences within the UTRs of these genes are responsible for the different translation rates displayed by the variants.
Fig 4. Translational regulation of splice variants with different 3’UTR in ESC.
(A) Diagram showing the gene structure for three genes, Ankrd27, Mpzl1 and Araf, whose variants are differentially loaded with ribosomes in ESCs. (B) qRT-PCR analysis of the polysome distribution of two variants of three genes in ESCs. (C) Luciferase assays showing the different 3’UTRs of Ankrd27, Mpzl1 and Araf variants mediate translational control.
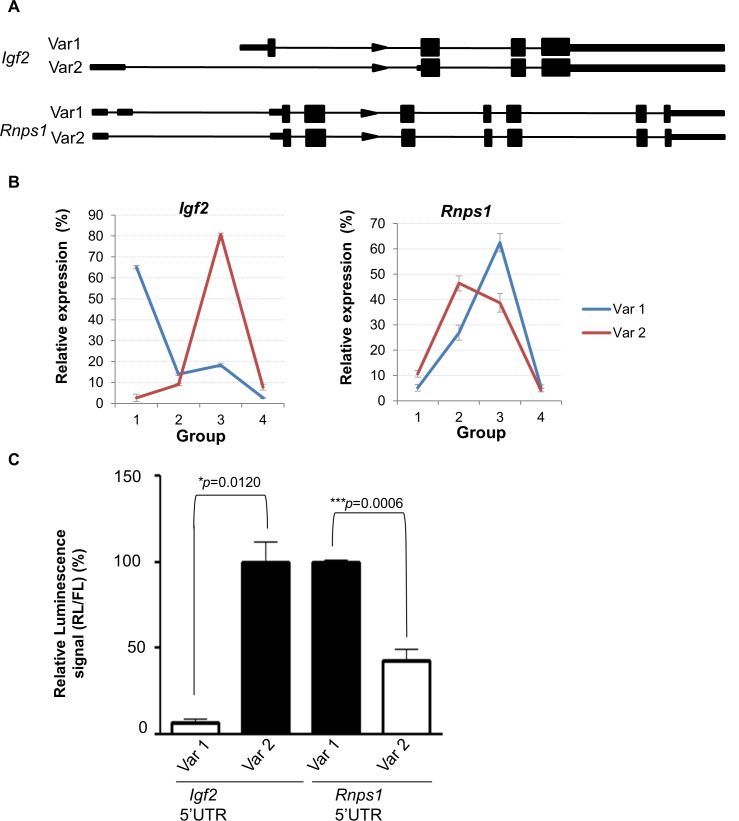
We validated two variants that showed altered 5’UTRs (Fig 5A). qRT-PCR confirmed the differential ribosomal load on polysome fractions (Fig 5B). Igf2 variant 1 (Igf2v1) was enriched in the non-translated fractions while Igf2 variant 2 (Igf2v2) was enriched in the highly translated fractions. For Rnps1, variant 1 (Rnps1v1) was in the heavy polysome fractions while variant 2 (Rnps1v2) was in the low translation fractions. When the 5’UTRs of these mRNAs were cloned upstream of the luciferase reporter gene they caused a decrease in luciferase activity correlating with the changes in ribosomal load seen by PCR (Fig 5C). These data suggest that the Igf2v2 5’UTR promotes translation compared to the Igf2v1 5’UTR, and the Rnps1v2 5’UTR promotes a reduction in translation compared to variant 1. The exclusion of exon 2 in Rnps1v2 results in the creation of an 81 nucleotide long upstream open reading frame (uORF) that may function to repress translation of that variant. Interestingly, the ORF for Rnps1 variants 1 and 2 are identical, so expression of different isoforms in different cell types has the potential to influence protein production through the presence of the uORF. Rnps1 forms part of a pre- and post-splicing RNA bound complex that plays a role in splicing, nuclear-cytoplasmic shuttling and RNA stability [29–31]. These data suggest that the alternative UTR sequences seen in different mRNA isoforms can greatly impact the translation rate of mRNA variants.
Fig 5. Translational regulation of splice variants with different 5’UTR in ESC.
(A) Diagram showing the gene structure for two genes, Igf2 and Rnps1, whose variants are differentially loaded with ribosomes in ESCs. (B) qRT-PCR analysis of the polysomes distribution of two variants of Igf2 and Rnps1 in ESCs. (C) Luciferase assays showing the different 5’UTRs of Igf2 and Rnps1variants mediate translational control.
Variant specific translation likely plays an important role in ESC differentiation. We detect 31 genes that have at least one variant that is selectively translationally regulated on differentiation of ESCs to NPCs (S2 Table). These variants are regulated independently from the other present variants of that gene. One example is Rnps1 (discussed above) where variant1 is highly translated in ESCs and NPCs. Variant 2 is more translationally repressed in ESCs and its polysome association is increased in NPCs indicative of increased translation (S4 Fig).
Non-coding mRNAs are enriched on polysomes
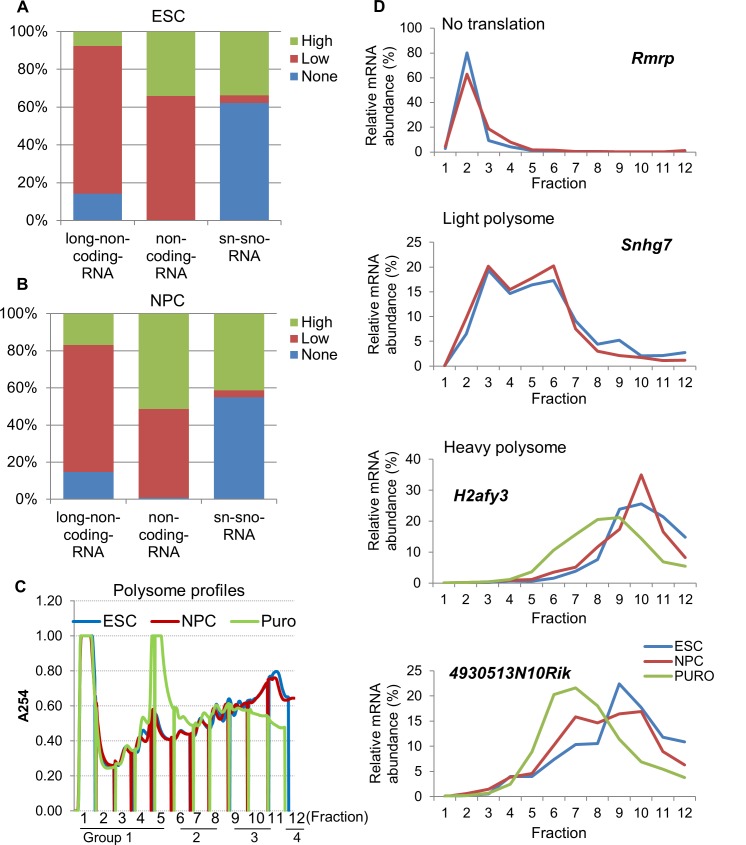
Recently it has been demonstrated that many non-coding mRNAs are present on polysomes. A significant percentage of annotated non-coding mRNAs have ribosomal footprints following ribosomal footprint analysis suggesting they are engaged by the translating ribosome.[32] More recent studies have confirmed that a significant percent of non-coding mRNAs are on the polysomes.[33] Based on this, we determined the polysome association of annotated non-coding mRNAs in our datasets. We identified over 260 GenBank annotated non-coding RNAs in ESCs (280 in NPCs) that include long non-coding RNAs (lnc-RNAs), non-coding RNAs (ncRNAs), small nuclear and small nucleolar RNAs. The lnc-RNAs include predominantly large intergeneic non-coding RNAs (lincRNAs) while the non-coding RNAs include predominantly pseudogenes. In agreement with previous studies we see a large proportion of these non-coding RNAs associated with the light and heavy polysome fractions (Fig 6A and 6B). Over 70% of lnc-RNAs were associated with the low or high translation fractions in both ESCs and NPCs while over 90% of non-coding RNAs were associated with polysomes. We also saw a significant percentage of sn- and sno- RNAs on the polysomes. qRT-PCR validation of a selection of non-coding RNAs showed that Rmrp was enriched in the non-translated fractions. Rmrp is a component of the ubiquitously expressed RNA-processing endoribonuclease [34]. Snhg7, a small nucleolar RNA host gene was enriched in the light polysome fractions and H2afy3, a non-coding pseudogene, and 4930513N10Rik were enriched in the heavy polysome fractions. To confirm that these RNAs were really associated with polysomes and not other heavy RNP particles we treated cells with puromycin to selectively breakdown polysomes (Fig 6C and 6D). H2afy3 and 4930513N10Rik were seen to shift from the heavy fractions to the lighter fractions in the presence of puromycin suggesting they are associated with polysomes. These data therefore confirm previous observations that many non-coding RNAs are enriched on polysomes.
Fig 6. Polysomal association of ncRNA in ESC and NPC.
(A,B) Analysis of the distribution of annotated non-coding RNAs in the different polysome fractions of ESCs and NPCs. (C) Polysome profiles of ESCs, NPCs and puromycin treated ESCs (PURO) to selectively disrupt polysomes. (D) qRT-PCR of representative non-coding RNAs showing their distribution in polysome gradients from ESCs and NPCs with and without the addition of puromycin.
Discussion
We have performed RNA sequencing analysis on mRNA isolated from different fractions of a polysome gradient in order to analyse the translation rate of individual mRNA variants. We find that many mRNAs are translationally regulated during differentiation of ESCs to NPCs. We have validated that a shift in ribosomal load has a consequence for protein production with Tchp. Tchp is translationally upregulated in NPCs resulting in an increase in protein levels. TCHP was originally described as a keratin filament binding protein and plays a role in ciliogenesis and centrosomal function [35, 36]. TCHP has no known role in the nervous system but given its translational upregulation it may be a regulator of early neural differentiation. We identified 31 genes expressing multiple variants that are translationally regulated in a variant specific manner on NPC differentiation. In these cases multiple variants are detected but only one is under translational control on differentiation. In a number of cases there was a switch in the variant that was highly translated so that while the absolute levels of the RNA variants may be the same, their different translation rates will result in the protein products being present at different levels in ESCs and NPCs. Interestingly the role of the majority of these variants is not known so it remains to be determined how this variant specific translation impacts on neural differentiation.
The method of RNAseq analysis of polysomal fractions is different from the alternative approach of ribosomal footprinting which provides information on the mRNA sequence physically bound by ribosomes and is used to infer translation rate [32, 37]. Despite the high resolution of the method the footprint sequences do not provide information on UTRs. In addition, currently the technique is not able to distinguish between different splice variants. Polysome profiling coupled to RNAseq provides information on variant specific ribosomal load and enables the analysis of corresponding UTR sequences. Using polysome profiling coupled with RNAseq, we find that in ESCs ten percent of RNA isoforms display different translation rates. This correlates with differences in the UTR sequences which likely drive the altered translation rate. This suggests that the relative protein levels of the two isoforms will not correlate with their mRNA levels.
Alternate UTRs can arise through a number of different mechanisms including alternate TSS, alternative splicing and alternative polyA site selection. Our datasets were mapped to the RefSeq genome which has annotations for promoter start sites and known alternative splicing events. As such, our studies have focused only on variants arising from alternative transcription start site selection and alternative splicing. Our analysis does not take into account any APA that may be occurring and it is possible that the variants identified in our studies may be further regulated by APA in different cell types. We find that in ESCs 10% of mRNAs with multiple variants display different translation rates for each variant inferred from their ribosomal load. This strongly correlates with the variants having different 5’ and 3’UTR sequences. This represents over 67 genes with different detectable isoforms in ESCs. This demonstrates the importance of analysing the translation rates of RNA variants when studying gene expression patterns. We confirmed the differential ribosomal load of five variants by PCR and performed luciferase assays to confirm that the regulation was mediated through sequences in the UTRs of these mRNAs. We tested two variants that had altered 5’UTRs and three that had altered 3’UTRs. In all cases the UTR sequence was shown to regulate luciferase protein levels in a similar way to that predicted by the polysome profiling data. Where the variant had a decreased ribosomal load following polysome profiling, the UTR promoted a decrease in luciferase activity suggesting a role in translational repression. It is likely that the UTRs that are associated with a decreased ribosomal load have cis-acting sequences that confer translational repression. These could take the form of binding sites for miRNA or RNA binding proteins. Alternate 5’UTRs could also contain uORFs which can function to repress translation [38, 39]. The variants that do not have these sequences are likely not targeted for repression. These different UTR sequences have likely evolved to confer additional levels of regulation on specific RNA variants and have the potential to precisely regulate their translation both spatially and temporally.
Analysis of the ORF size of the translationally regulated variants suggested that for most variants the differences in ribosomal load were not a consequence of altered ORF size. There were a minority of variants that had significantly longer ORFs that correlated with increased ribosomal load and in these cases the ORF length is likely the cause. For variants that did show a correlation between ORF length and ribosomal load the difference in size was not large enough (less than 25% larger) to result in a shift in the polysome fractions in most cases (Table 1). Interestingly we identified a selection of splice variants that had different translation rates but no change in the UTRs. While some of these candidates will have a decreased ribosomal load due to a decreased ORF length it is likely that the remaining transcripts contain cis-acting regulatory sequences within their ORF. miRNAs have been shown to target the ORF of mRNAs [40, 41] so it is possible that the changes in ORF seen in these variants results in altered translational control in addition to altered protein function. Exon 4 and 5 in Ctage5 variant 1 are associated with a decreased ribosomal load compared to variant 3 that skips these exons. While there was no enrichment for rare codons in these exons it is possible that these regions could bind to trans-acting factors or that the sequence itself is inhibiting translation. It will be interesting to determine if these exons can function to regulate translation from within the UTR or if they have to be in the ORF to function.
We assessed the ribosomal load of annotated non-coding RNAs and found that a significant number of long non-coding RNAs (lncRNAs) and non-coding RNAs were associated with the ribosomes. The lnc-RNAs include predominantly long intergenic non-coding RNAs (lincRNAs) while the non-coding RNAs include predominantly pseudogenes. The presence of the pseudogenes on the polysomes is in agreement with recent reports that analysed ribosomal footprinting data from a number of different species. [42] Recent studies have demonstrated the presence of many non-coding RNAs on polysomes. Our studies confirm that many non-coding RNAs are in the polysome fraction but we cannot distinguish between RNAs that are actively being translated by the ribosome and those that may be binding the ribosome or other mRNA molecules that are being translated. It is possible that many non-coding RNAs could be associating with the ribosome in a regulatory capacity and so while associated with the ribosome would show no ribosomal footprint. It has been suggested that non-coding RNAs could be being translated to give short peptides but additional studies are needed to elaborate on this idea [32, 42, 43]. Our data confirms the presence of non-coding RNAs on the ribosome and further investigation is needed to determine why they are there.
Taken together our data illustrates the importance of addressing the translation rate of individual mRNA variants. We demonstrate that different mRNA variants can have very different translation rates. This confirms and expands on previous reports from human cells and demonstrates variant specific translation rates in ESCs and NPCs. Additional work is needed to determine the mechanisms by which these mRNAs are regulated and their significance for ESC self-renewal and pluripotency.
Experimental Procedures
Cell Culture
Sox1-GFP ESCs were a gift from Dr Austin Smith [26, 27]. ESCs were cultured on 0.1% gelatin in DMEM (GIBCO) supplemented with 15% fetal bovine serum (FBS) (GIBCO), 0.2 mM β-mercaptoethanol, 2 mM L-glutamine (GIBCO), 13 MEM nonessential amino acids (GIBCO), and LIF. For NPC differentiation Sox1-GFP ESCs were plated at low density and grown in N2B27 media for 6 days NPC as described [26]. The efficiency of NPC differentiation (>80%) compared to ESC was confirmed by detection of GFP fluorescence signal using flow cytometry.
Polysome Fractionation and RNA extraction
Polysomes were isolated as described previously [5] and equal OD units were loaded onto 10%–50% linear sucrose gradients and centrifuged at 36,000 rpm for 1 hour and 45 min at 8°C in a SW41 rotor (Beckman Coulter). Twelve fractions were collected from the top of the gradient using a piston gradient fractionator (BioComp Instruments). The absorbance at 254 nm was measured with a UV-M II monitor (Bio-Rad).
Following fractionation, 110 μL of 10% SDS and 12 μL of proteinase K (20 mg/mL; Invitrogen) were added to each fraction and incubated with shaking at 1000 rpm for 30 min at 42°C to digest protein debris and to inactivate nucleases. Fractions of 1–5, 6–8, 9–11, 12 were pooled to give groups 1–4, which we classify as no translation, low translation, high translation and mRNAs present at the bottom of sucrose gradient respectively. Spike-in RNAs from the GeneChip Eurkaryotic Poly-A RNA control kit (Affymetrix) were added to each group of fractions (20 μl, 12 μl, 12 μl and 4 μl of 200-fold diluted spike-in RNA into group 1–4 respectively). Polysomal RNA was extracted by phenol chloroform isoamyl alcohol (PCI) and then chloroform isoamyl, further followed by subsequent purifications using sodium chloride (NaCl), lithium chloride (LiCl) and sodium acetate (NaOAC). RNA quality was assessed on the Agilent Bioanalyzer using RNA6000 Nano kit.
Library preparation and sequencing
The twelve polysomal fractions from ESCs and NPCs were pooled and placed into groups that were indicative of their translation rates. Fractions 1–5 were pooled as non-translated mRNAs (non). Fractions 6–8 were pooled into the “low” group containing mRNAs translated at a low rate. Fractions 9–11 were pooled into the “high” group and contained the highly-translated mRNAs. Fraction 12 was collected as the “unknown” group consisting of the cell debris and very large RNPs. Total unfractionated RNA was kept as a control. Each of the pooled groups were subjected to Library preparation and SOLIDTM sequencing. The reads obtained were single-end reads and were 75bp in length.
Alignment, normalization and scaling of the reads
The flowchart of the data analysis is given in S5 Fig. Briefly, the reads for each of the four groups and the total unfractionated RNA for both the ESC and NPC were aligned to the Mus musculus genome (mm10) using LifeScopeTM. The bam files comprising the alignments were then imported into Partek Genomics Suite. The allocation of reads was done through Partek using the “Expectation Maximisation” algorithm. This was followed by RPKM normalisation. The normalized reads were scaled using scaling factors calculated from the polyA spike-in RNAs. The relative proportion of the spike-in RNAs in each polysome group was determined and used to calculate the scaling factor taking into consideration the original sample volume (S6 Fig). The scaled normalised reads were converted into percentage of reads in each pool where the sum of the pools was 100%. Transcripts with an unscaled RPKM value > = 0.5 in each of the four groups and an RPKM value of > = 5 in at least one of the four groups were analysed. The data discussed in this publication has been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and is accessible through GEO Series accession number GSE73467
Analysis of translation rates
To identify mRNAs translationally regulated between ESCs and NPCs transcripts were analysed if they had an unscaled RPKM value > = 0.5 in all polysome fractions in ESCs and NPCs and an RPKM value of > = 5 in any one of the eight groups. mRNAs showing a shift in percentage in any fraction of > = 20 between ESCs and NPCs were considered to be translationally regulated. Total mRNA level was determined by taking the sum total of scaled data in the four groups. A fold change cut-off of 2 fold was applied to obtain transcripts that had different mRNA levels.
To analyse translation rates of splice variants in ESCs or NPCs all genes with multiple detected variants were analysed. The scaled percentages of the variants were compared within the same cell type. Variants predicted to be non-coding were removed to ensure only translational control events were identified. Genes with variants showing a percentage change of > = 20 in any group were classified as having “Translationally Regulated Splice Variants”. Genes with variants not showing a percentage shift > = 20 in any of the groups were classified as genes having “Non Translationally Regulated Splice Variants”
Correlation of ORF and UTR length with translation rate
The ORFs and 5' and 3' UTR sequences were retrieved from the UCSC table browser (Mus musculus genome mm10). The ORF lengths of the translationally regulated splice variants were compared. ORF lengths within 10nt of each other were considered to be the same. These were correlated with the translation rates of the variants. The lengths of the UTRs of different variants were calculated and compared to the translation rate of each variant.
5’UTR correlation studies
For each gene in the TR sets, the translation efficiency of each variant was correlated with the GC content, Free Energy and length of the 5’UTR. Genes were determined to have a direct correlation when increased translation correlated with increased GC content, free energy or length of the 5’UTRs. Genes were classified as having a direct correlation, opposite correlation or no correlation. For genes where no clear conclusion could be drawn due to the presence of equal numbers of variants showing different trends, they were labelled as undecided.
Non-coding RNA polysomal enrichment analysis
The non-coding RNA transcripts annotated as “NR” were classified according to the pool in which they were most represented. All analysed non-coding RNAs were enriched in one of the NONE, LOW or HIGH translation pools.
Semi-quantitative Real-Time Polymerase Chain Reaction
cDNA was synthesized using a reverse transcription kit (Super-Script III, Invitrogen) according to the manufacturer’s instruction. For fractionated RNA, the same volume of RNA was used, and for total RNA analysis, the same quantity of RNA was used. SYBR Green was used with transcript-specific primers for qRT–PCR on an ABI PRISM 7900 sequence detection system. For polysome fractions, CT values were normalized to spike-in control RNAs dap and thr. Primers were either purchased from Integrated DNA Technologies (IDT) or designed. Expression levels detected by qPCR for each group are presented as percentage of RNA in each fraction relative to the total values of 4 groups.
Luciferase reporter assay
The UTR of candidate mRNAs were cloned into the psiCHECK-2 vector. ESCs were transfected using FuGENE HD (Promega) according to manufacturer’s instructions. Thirty hours after transfection, cells were lysed and Renilla (RL) and firefly (FL) luciferase activities were determined using the Dual-Luciferase Reporter Assay system (Promega). For data analyses, all RL signals were normalized by the non-targeted control FL readings.
Western blot analysis
20ug of protein extract was separated on a NuPAGE 4–12% Bis-Tris Gel and transferred to PVDF membrane. TCHP (ARP60515_P050, Aviva Systems Biology), and GAPDH (Abcam Cat No.ab-9484) antibodies were used at 1:1000 dilution.
Statistical Analysis
Luciferase activities in ES cells transfected with different psiCHECK-2 plasmids were compared using the paired Student’s t-test. For qRT-PCR and luciferase reporter assays, the standard error of the mean is shown. * P<0.05
Supporting Information
The sequencing data shows a high level of correlation with qRT-PCR.
(TIF)
Analysis of the 5’UTRs of variants that showed different translation rates and different 5’UTRs. 5’UTRs were assessed for their level of GC content, free energy and length and compared between the different variants.
(TIF)
(A) schematic showing the different variants of Ctage5. (B) Enrichment of the three Ctage5 splice variants in different polysome fractions.
(TIF)
(A) schematic showing the different variants of Rnsp1. (B) Real-time PCR showing the enrichment of the two Rnsp1 splice variants in different polysome fractions. Variant 2 is translationally more repressed in ESCs but is translationally activated in NPCs. Variant 1 is highly translated in both ESCs and NPCs.
(TIF)
(TIF)
(TIF)
Table showing the different ribosomal loads of variants within ESCs and NPCs. The data is presented as percentage of transcript present in each group. ORF lengths are also shown.
(XLSX)
Table showing the expression levels of different mRNA variants in different polysome fractions on differentiation of ESCs to NPCs. The scaled data is shown along with the percentage of transcript present in each group. All expressed variants are shown with at least one variant showing a change in translation rate in a variant specific manner.
(XLSX)
Acknowledgments
We thank the Agency for Science, Technology and Research for funding and Dr Austin Smith for the Sox1-GFP ESC cell line. We are grateful to the ACRF Biomolecular Resource Facility at Australian National University for the RNASeq library preparation and sequencing. TP acknowledges funding from the Australian Research Council (grant DP1300101928).
Abbreviations
- ESC
Embryonic Stem Cell
- NPC
Neural Precursor Cell
Data Availability
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE73467.
Funding Statement
QWLW, CV, QYL, TYZ, VT and LAV were funded by the Agency for Science, Technology and Research, Singapore. Raymond Luo, a co-author in this study, is employed by Life Technologies, 10 Biopolis Road, Singapore 138670. The funder provided support in the form of salaries for authors [RL], but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. RL was involved in data analysis. The specific roles of the author is articulated in the ‘author contributions’ section. TP acknowledges funding from the Australian Research Council (grant DP1300101928).
References
- 1.Theunissen TW, Jaenisch R. Molecular control of induced pluripotency. Cell Stem Cell. 2014;14(6):720–34. Epub 2014/06/07. 10.1016/j.stem.2014.05.002 S1934-5909(14)00189-1 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeo JC, Ng HH. The transcriptional regulation of pluripotency. Cell Res. 2013;23(1):20–32. Epub 2012/12/12. 10.1038/cr.2012.172 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li MA, He L. microRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. Bioessays. 2012;34(8):670–80. Epub 2012/06/08. 10.1002/bies.201200019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye J, Blelloch R. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell. 2014;15(3):271–80. Epub 2014/09/06. 10.1016/j.stem.2014.08.010 S1934-5909(14)00348-8 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D, Zhao T, Ang HS, Chong P, Saiki R, Igarashi K, et al. AMD1 is essential for ESC self-renewal and is translationally down-regulated on differentiation to neural precursor cells. Genes Dev. 2012;26(5):461–73. Epub 2012/03/07. 10.1101/gad.182998.111 26/5/461 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Klerk E, t Hoen PA. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 2015;31(3):128–39. Epub 2015/02/05. S0168-9525(15)00002-5 [pii] 10.1016/j.tig.2015.01.001 . [DOI] [PubMed] [Google Scholar]
- 7.Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, et al. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–70. Epub 2014/03/29. 10.1038/nature13182 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwal VR, Bulun SE, Leitch M, Rohrich R, Simpson ER. Use of alternative promoters to express the aromatase cytochrome P450 (CYP19) gene in breast adipose tissues of cancer-free and breast cancer patients. J Clin Endocrinol Metab. 1996;81(11):3843–9. Epub 1996/11/01. 10.1210/jcem.81.11.8923826 . [DOI] [PubMed] [Google Scholar]
- 9.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11(1):75–87. Epub 2009/12/19. 10.1038/nrg2673 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–5. Epub 2008/11/04. 10.1038/ng.259 [pii]. . [DOI] [PubMed] [Google Scholar]
- 11.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. Epub 2012/09/08. 10.1038/nature11233 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O'Hanlon D, Sung HK, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147(1):132–46. Epub 2011/09/20. 10.1016/j.cell.2011.08.023 S0092-8674(11)00949-4 [pii]. . [DOI] [PubMed] [Google Scholar]
- 13.Lu X, Goke J, Sachs F, Jacques PE, Liang H, Feng B, et al. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat Cell Biol. 2013;15(10):1141–52. Epub 2013/09/10. 10.1038/ncb2839 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, et al. Characterization of the human ESC transcriptome by hybrid sequencing. Proc Natl Acad Sci U S A. 2013;110(50):E4821–30. Epub 2013/11/28. 10.1073/pnas.1320101110 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K, Dai X, Wu J. Alternative splicing: An important mechanism in stem cell biology. World J Stem Cells. 2015;7(1):1–10. Epub 2015/01/27. 10.4252/wjsc.v7.i1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci U S A. 2010;107(23):10514–9. Epub 2010/05/26. 10.1073/pnas.0912260107 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danckwardt S, Hentze MW, Kulozik AE. 3' end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27(3):482–98. Epub 2008/02/08. 10.1038/sj.emboj.7601932 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. Epub 2010/06/11. 10.1146/annurev-biochem-060308-103103 . [DOI] [PubMed] [Google Scholar]
- 19.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106(17):7028–33. Epub 2009/04/18. 10.1073/pnas.0900028106 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–7. Epub 2008/06/21. 10.1126/science.1155390 320/5883/1643 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Klerk E, Venema A, Anvar SY, Goeman JJ, Hu O, Trollet C, et al. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 2012;40(18):9089–101. Epub 2012/07/10. 10.1093/nar/gks655 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hynes CJ, Clancy JL, Preiss T. miRNAs in cardiac disease: sitting duck or moving target? IUBMB Life. 2012;64(11):872–8. Epub 2012/09/27. 10.1002/iub.1082 . [DOI] [PubMed] [Google Scholar]
- 23.Sterne-Weiler T, Martinez-Nunez RT, Howard JM, Cvitovik I, Katzman S, Tariq MA, et al. Frac-seq reveals isoform-specific recruitment to polyribosomes. Genome Res. 2013;23(10):1615–23. Epub 2013/06/21. 10.1101/gr.148585.112 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clancy JL, Wei GH, Echner N, Humphreys DT, Beilharz TH, Preiss T. mRNA isoform diversity can obscure detection of miRNA-mediated control of translation. RNA. 2011;17(6):1025–31. Epub 2011/04/07. 10.1261/rna.2567611 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollard SM, Benchoua A, Lowell S. Neural stem cells, neurons, and glia. Methods Enzymol. 2006;418:151–69. Epub 2006/12/05. S0076-6879(06)18010-6 [pii] 10.1016/S0076-6879(06)18010-6 . [DOI] [PubMed] [Google Scholar]
- 26.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21(2):183–6. Epub 2003/01/14. 10.1038/nbt780 [pii]. . [DOI] [PubMed] [Google Scholar]
- 27.Aubert J, Stavridis MP, Tweedie S, O'Reilly M, Vierlinger K, Li M, et al. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc Natl Acad Sci U S A. 2003;100 Suppl 1:11836–41. Epub 2003/08/19. 10.1073/pnas.1734197100 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136(14):2311–22. Epub 2009/06/23. 136/14/2311 [pii] 10.1242/dev.024398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res. 2007;35(13):4542–51. Epub 2007/06/26. gkm461 [pii] 10.1093/nar/gkm461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malone CD, Mestdagh C, Akhtar J, Kreim N, Deinhard P, Sachidanandam R, et al. The exon junction complex controls transposable element activity by ensuring faithful splicing of the piwi transcript. Genes Dev. 2014;28(16):1786–99. Epub 2014/08/12. 10.1101/gad.245829.114 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi R, Handler D, Ish-Horowicz D, Brennecke J. The exon junction complex is required for definition and excision of neighboring introns in Drosophila. Genes Dev. 2014;28(16):1772–85. Epub 2014/08/02. 10.1101/gad.245738.114 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. Epub 2011/11/08. 10.1016/j.cell.2011.10.002 S0092-8674(11)01192-5 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, et al. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15(1):R6 Epub 2014/01/08. 10.1186/gb-2014-15-1-r6 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattijssen S, Welting TJ, Pruijn GJ. RNase MRP and disease. Wiley Interdiscip Rev RNA. 2010;1(1):102–16. Epub 2010/07/01. 10.1002/wrna.9 . [DOI] [PubMed] [Google Scholar]
- 35.Ibi M, Zou P, Inoko A, Shiromizu T, Matsuyama M, Hayashi Y, et al. Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. J Cell Sci. 2011;124(Pt 6):857–64. Epub 2011/02/18. 10.1242/jcs.075705 [pii]. . [DOI] [PubMed] [Google Scholar]
- 36.Inoko A, Matsuyama M, Goto H, Ohmuro-Matsuyama Y, Hayashi Y, Enomoto M, et al. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J Cell Biol. 2012;197(3):391–405. Epub 2012/04/25. 10.1083/jcb.201106101 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–23. Epub 2009/02/14. 10.1126/science.1168978 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbosa C, Peixeiro I, Romao L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9(8):e1003529 Epub 2013/08/21. 10.1371/journal.pgen.1003529 PGENETICS-D-13-00138 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106(18):7507–12. Epub 2009/04/18. 10.1073/pnas.0810916106 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–8. Epub 2008/09/23. 10.1038/nature07299 [pii]. . [DOI] [PubMed] [Google Scholar]
- 41.Hausser J, Syed AP, Bilen B, Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23(4):604–15. Epub 2013/01/22. 10.1101/gr.139758.112 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. Long non-coding RNAs as a source of new peptides. Elife. 2014;3:e03523 Epub 2014/09/19. 10.7554/eLife.03523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154(1):240–51. Epub 2013/07/03. 10.1016/j.cell.2013.06.009 S0092-8674(13)00711-3 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequencing data shows a high level of correlation with qRT-PCR.
(TIF)
Analysis of the 5’UTRs of variants that showed different translation rates and different 5’UTRs. 5’UTRs were assessed for their level of GC content, free energy and length and compared between the different variants.
(TIF)
(A) schematic showing the different variants of Ctage5. (B) Enrichment of the three Ctage5 splice variants in different polysome fractions.
(TIF)
(A) schematic showing the different variants of Rnsp1. (B) Real-time PCR showing the enrichment of the two Rnsp1 splice variants in different polysome fractions. Variant 2 is translationally more repressed in ESCs but is translationally activated in NPCs. Variant 1 is highly translated in both ESCs and NPCs.
(TIF)
(TIF)
(TIF)
Table showing the different ribosomal loads of variants within ESCs and NPCs. The data is presented as percentage of transcript present in each group. ORF lengths are also shown.
(XLSX)
Table showing the expression levels of different mRNA variants in different polysome fractions on differentiation of ESCs to NPCs. The scaled data is shown along with the percentage of transcript present in each group. All expressed variants are shown with at least one variant showing a change in translation rate in a variant specific manner.
(XLSX)
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE73467.