Abstract
A bacterial isolate (SCU-B244T) was obtained in China from crickets (Teleogryllus occipitalis) living in cropland deserted for approximately 10 years. The isolated bacteria were Gram-negative, facultatively anaerobic, oxidase-negative rods. A preliminary analysis of the 16S rRNA gene sequence indicated that the strain belongs to either the genus Erwinia or Pantoea. Analysis of multilocus sequence typing based on concatenated partial atpD, gyrB and infB gene sequences and physiological and biochemical characteristics indicated that the strain belonged to the genus Erwinia, as member of a new species as it was distinct from other known Erwinia species. Further analysis of the 16S rRNA gene showed SCU-B244T to have 94.71% identity to the closest species of that genus, Erwinia oleae (DSM 23398T), which is below the threshold of 97% used to discriminate bacterial species. DNA-DNA hybridization results (5.78±2.52%) between SCU-B244T and Erwinia oleae (DSM 23398T) confirmed that SCU-B244T and Erwinia oleae (DSM 23398T) represent different species combined with average nucleotide identity values which range from 72.42% to 74.41. The DNA G+C content of SCU-B244T was 55.32 mol%, which also differs from that of Erwinia oleae (54.7 to 54.9 mol%). The polyphasic taxonomic approach used here confirmed that the strain belongs to the Erwinia group and represents a novel species. The name Erwinia teleogrylli sp. nov. is proposed for this novel taxon, for which the type strain is SCU-B244T (= CGMCC 1.12772T = DSM 28222T = KCTC 42022T).
Introduction
Resistance to pesticides in insects is a serious concern worldwide [1] and often occurs approximately 10 years after the introduction of a new pesticide [2]. The resistance mechanisms have been attributed to evolutionary changes in insect genomes, such as the alteration of drug target sites, up-regulation of degrading enzymes and the enhancement of drug excretion. Kikuchi et al. have shown that infection with an insecticide-degrading bacterial symbiont immediately establishes insecticide resistance in insects [1], indicating bacteria play an important role in pesticide resistance.
In a recent study, we focused on the culturable strains associated with crickets (Teleogryllus occipitalis), a common pest living in a deserted cropland in China. Our aim was to explore the relationship between pesticide resistance and symbionts. Among 274 isolates cultured from Teleogryllus occipitalis, 27 strains of genera Lysinibacillus, Pseudomonas, Sphingobacterium, Exiguobacterium and Staphylococcus could evidently degrade chlorpyrifos, a common insecticide used in this field for many years. One isolate (SCU-B244T) that could degrade chlorpyrifos was cultured on TSA (tryptone soy agar) medium in August 2012 and could not be identified to the species level. A polyphasic taxonomic approach was used to investigate the strain, with the results suggesting that SCU-B244T represents a novel species of the genus Erwinia.
Materials and Methods
Isolation procedures, culture conditions and initial microbiological characterization
Three crickets (collected near 30°33’N, 103°58’E; altitude 495 m) were added to 100 mL sterile 0.85% (w/v) NaCl solution in a 250 mL flask and shaken at 220 rpm for 30 min. The supernatant which contained bacteria was plated onto TSA plates (Tryptone 1.5%, Soy Peptone 0.5%, NaCl 0.5%, Agar 1.5%, w/v, pH 7.2) and subsequently incubated at 37°C for 5 days. Glycerol stock at -80°C was adopted for long term preservation of the isolates. Exponential phase cells cultured in TSB (Tryptone 1.5%, Soy Peptone 0.5%, NaCl 0.5%, pH 7.2) medium with shaking (200 rpm, 16 h) at 37°C were harvested for DNA G+C content and ANI analysis. Phase contrast and transmission electron microscopy were used to examine cellular morphology and motility after growth on TSA medium at 37°C for 24 h. Gram staining was performed as described by Gerhardt et al. [3]. Oxidase and catalase activity were tested according to methods described by Smibert & Krieg [3]. TSB medium with different NaCl concentration and pH were used to test tolerance to NaCl and pH range, and the growth was determined by OD 600. To confirm the results, each experiments mentioned above were performed three times. Erwinia oleae (DSM 23398T) was used as reference strain in this study.
16S rRNA gene sequence analysis
DNA extraction from strain SCU-B244T, PCR amplification, primers used and DNA sequencing conditions of 16S rRNA gene were performed as previously described [4]. The strain was analysed using the EzTaxon server [5] (www.ezbiocloud.net/eztaxon) by comparison with 16S rRNA gene sequence data. A neighbour-joining phylogenetic tree was constructed using the method of Saitou and Nei [6] with MEGA 5.2 software [7]. Similarities were calculated using the Kimura 2-parameter [8, 9] in MEGA 5.2. Maximum-likelihood phylogenetic trees were also constructed using Kimura 2-parameter [8, 9] model and the method of Felsenstein [10] with MEGA 5.2 software. Robustness of the phylogenetic trees was evaluated by using the bootstrap resampling method of Felsenstein [11], with 1000 replicates.
The results of 16S rRNA gene sequence alignment on the EzTaxon server revealed that strain SCU-B244T belongs to the family Enterobacteriaceae. Related 16S rRNA gene sequences for initial analysis were taken from the top 66 hits on the EzTaxon server (on May 2014, each representing a different species), with the highest similarity scores being 96.1% to 94.1%. Neighbour-joining and maximum-likelihood phylogenetic trees were constructed using the methods described above.
To refine the taxonomic position of SCU-B244T, other neighbour-joining and maximum-likelihood phylogenetic trees based on 16S rRNA gene sequences were constructed, including the strain SCU-B244T and all culturable type strains of the genera Erwinia (Candidatus Erwinia dacicola) and Pantoea, based on the same methods described above.
Multilocus sequence analysis
Multilocus sequence analysis (MLSA) of concatenated partial atpD, gyrB, infB and rpoB gene sequences enables the differentiation of the phylogenetically related genera Erwinia, Pantoea and Tatumella. A good congruence has previously been observed between DNA-DNA hybridization and MLSA [12–16]. In the present study, to confirm the results observed with 16S rRNA gene sequencing data, three housekeeping genes, atpD, gyrB, and infB, were amplified and sequenced with the primers described by Brady et al. [12]. Neighbour-joining and maximum-likelihood phylogenetic trees based on concatenated partial atpD, gyrB and infB gene sequences were constructed using the same method as described above. Strains from genera Erwinia, Pantoea and Tatumella were used in MLSA analysis, including SCU-B244T. The partial sequence of atpD, gyrB and infB gene of related strains were obtained from GenBank and the accession numbers are indicated on the figures.
DNA-DNA hybridization
DNA-DNA hybridization between SCU-B244T and Erwinia oleae DSM 23398T was conducted as described by De Ley et al. [17] with hybridization temperature set at 67°C. Genomic DNA (OD260/280 = 1.8 to 1.9) was extracted using the Omega Bacterial DNA kit (E.Z.N.A.®)
DNA G+C content
The DNA G+C mol% content of strain SCU-B244T was determined by whole genome sequencing data.
Physiological and biochemical analysis
API 20E, API 50CHE acid production tests and API ZYM enzymatic characteristics test (BioMérieux) were performed according to the manufacturer’s instructions. BIOLOG GN2 carbon-source utilization analysis was conducted according to the instruction manual. Cellular fatty acids analysis was also performed. Biomass for fatty acid analysis was prepared by scraping growth from TSA plates after 24h incubation at 37°C. Lipids were extracted using the method of Folch et al.[18]. Total lipids were converted to fatty acid methyl esters (FAMEs) with 4 mol/L HCl in 55% (V/V) methanol at 80±1°C for 10 mins. FAMEs were analyzed by GC-MS (Trace DSQII, Thermo Fisher), and detailed conditions of the fatty acids analysis by GC-MS have been previously described [19], The compounds were identified using NIST 05 database (NIST Mass Spectral Database, PC-Version 5.0, 2005, National Institute of Standardisation and Technology, Gaithersburg, MD, USA). To confirm the results, each experiment mentioned above was performed three times.
Genome Sequencing and Average Nucleotide Identity
For the genome sequencing of SCU-B244T, Next Generation Sequencing (Illumina Miseq) was conducted at Majorbio Inc. and the data was used to calculate the Average Nucleotide Identity (ANI). DNA extraction method and purity were described above. ANI is a similarity measure between two genome sequences and it correlates well with DNA-DNA hybridization values. [20] A value of 70% DNA-DNA hybridization corresponds to about 95–96% ANI. [21] Genome data were taken from NCBI Genome and EzGenome web site. (www.ezbiocloud.net/ezgenome/browse_db). We used Orthologous Average Nucleotide Identity Tool (OAT software, Lee et al. 2015, Manuscript submitted, www.ezbiocloud.net/sw/oat) to calculate orthologous ANI values. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession LLXO00000000. The version described in this paper is version LLXO01000000.
Results and Discussion
In total 274 isolates belonging to 29 genera were cultured from Teleogryllus occipitalis and a bacterial isolate (SCU-B244T) is herein described. Initial microbiological characterization of the strain revealed that the cells were Gram-negative, oxidase-negative, rod-shaped, catalase-positive and facultatively anaerobic, suggesting that the strain belongs to the family Enterobacteriaceae [22].
Neighbour-joining (S1 Fig) and maximum-likelihood (S2 Fig) phylogenetic trees of the first 66 hit with similarity values ranging from 96.1% to 94.1% revealed that strain SCU-B244T and strains of the genera Erwinia and Pantoea cluster together. The diagram shows the phylogenetic relationship between SCU-B244T and genera Erwinia and Pantoea.
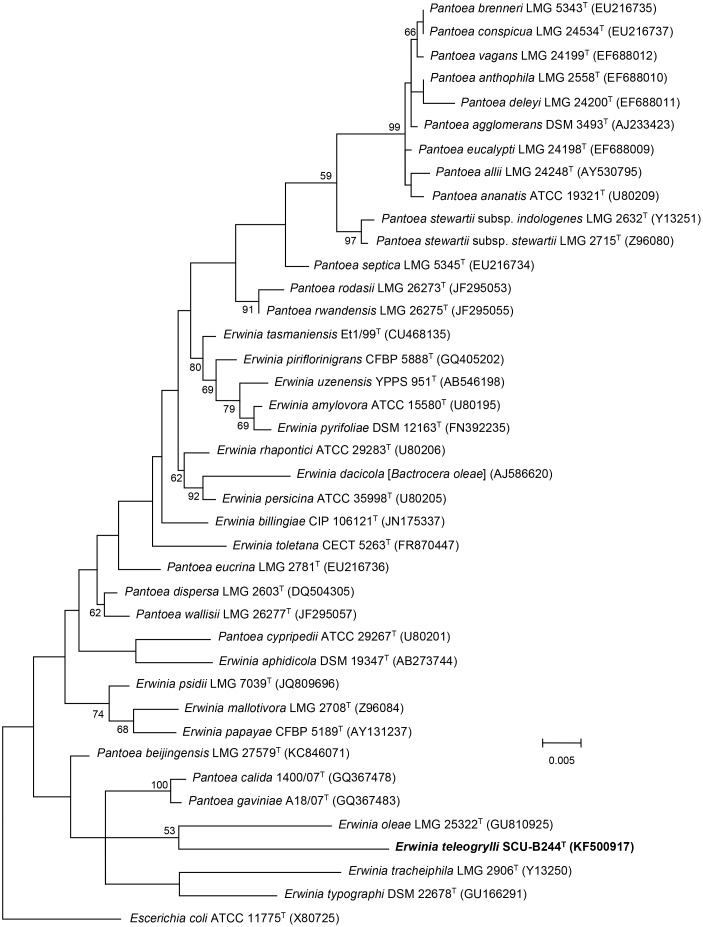
Further neighbour-joining (S3 Fig) and maximum-likelihood (Fig 1) phylogenetic trees, based on 16S rRNA gene sequences from members of the genera Erwinia and Pantoea, indicated that strain SCU-B244T belongs to the genus Erwinia and is most closely related to Erwinia oleae (DSM 23398T), which is consistent with the results of the initial phenotypic analysis.
Fig 1. Maximum-likelihood tree based on partial 16S rRNA gene sequences of the genera Erwinia and Pantoea.
The diagram shows the phylogenetic relationship between Erwinia teleogrylli sp. nov. and related type species of the genera Erwinia and Pantoea except Candidatus Erwinia dacicola. Escherichia coli ATCC 11775T was used as the outgroup. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
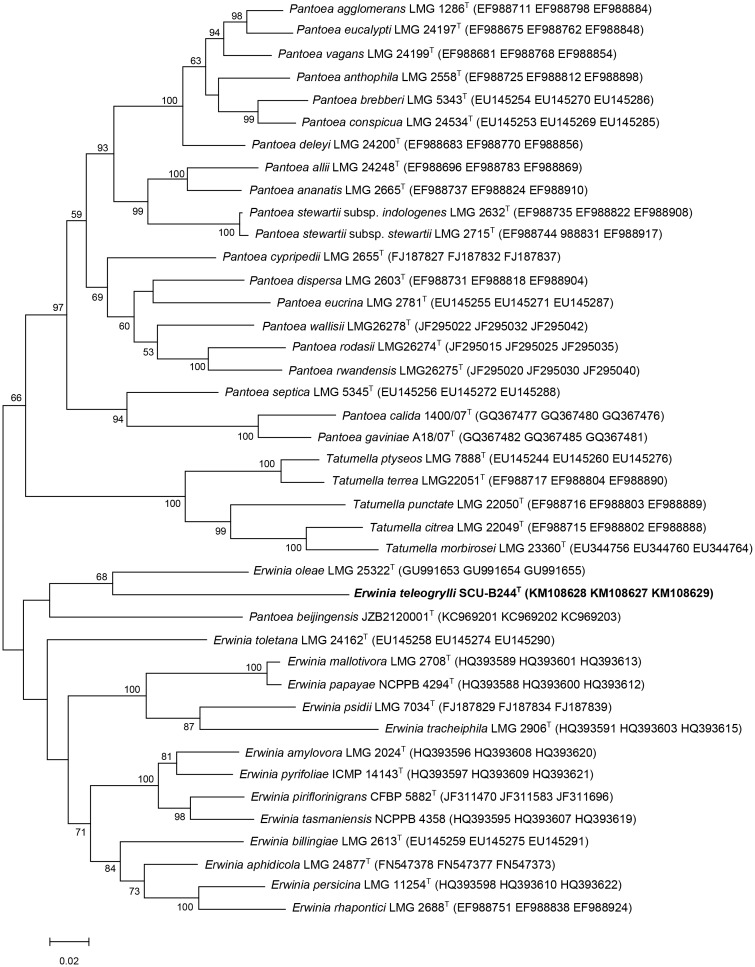
In neighbour-joining (S4 Fig) and maximum-likelihood (Fig 2) phylogenetic trees based on concatenated partial atpD, gyrB and infB gene sequences, strain SCU-B244T and Erwinia oleae (DSM 23398T) cluster together on a single branch, consistent with the previous results, suggesting strain SCU-B244T is most closely related to Erwinia oleae (DSM 23398T).
Fig 2. Maximum-likelihood tree based on concatenated partial atpD, gyrB and infB sequences from the genera Erwinia, Pantoea and Tatumella.
The diagram shows the phylogenetic relationship between Erwinia teleogrylli sp. nov. and related species of the genera Erwinia, Pantoea and Tatumella. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
The highest 16S rRNA gene identity of SCU-B244T with species of genera Erwinia and Pantoea were 95.43% (Erwinia psidii LMG 7039T) and 95.42% (Pantoea calida 1400/07T) respectively, phylogenic closest strain was Erwinia oleae (DSM 23398T) and the identity was 94.71%, which is lower than the 97% threshold that has been established to discriminate species. Strains showing less than 97% 16S rRNA gene identity are unlikely to have more than 60 to 70% DNA—DNA relatedness [23], and this level of rRNA sequence identity strongly suggests that SCU-B244T is a novel species. The DNA—DNA relatedness between SCU-B244T and Erwinia oleae (DSM 23398T) was 5.79 ± 2.52%. The value is mean of six hybridizations ± SD, which is significantly lower than the 70% value considered to be the threshold for the delineation of bacterial species [24]. ANI values between SCU-B244T and related species are listed in Table 1. The values range from 72.42% to 74.41% which are lower than 95–96% ANI.
Table 1. OrthoANI values between SCU-B244T and related species.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9* | 10* | 11* | 12* | 13* | 14* | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | ||||||||||||||||||
| 2 | 73.82 | 100 | |||||||||||||||||
| 3 | 73.62 | 77.41 | 100 | ||||||||||||||||
| 4 | 72.74 | 76.02 | 78.14 | 100 | |||||||||||||||
| 5 | 73.72 | 90.53 | 77.72 | 76.30 | 100 | ||||||||||||||
| 6 | 73.62 | 84.78 | 77.75 | 76.25 | 85.86 | 100 | |||||||||||||
| 7 | 74.03 | 77.14 | 78.05 | 76.23 | 77.45 | 76.86 | 100 | ||||||||||||
| 8 | 72.42 | 75.29 | 76.89 | 79.16 | 75.51 | 75.29 | 75.20 | 100 | |||||||||||
| 9 | 74.07 | 76.72 | 78.62 | 77.06 | 76.98 | 76.91 | 76.94 | 76.09 | 100 | ||||||||||
| 10 | 73.40 | 85.32 | 77.21 | 75.99 | 86.52 | 89.28 | 76.74 | 75.24 | 76.52 | 100 | |||||||||
| 11 | 73.75 | 77.26 | 83.21 | 77.94 | 77.46 | 77.55 | 77.56 | 76.94 | 78.91 | 77.10 | 100 | ||||||||
| 12 | 73.68 | 75.33 | 75.88 | 74.85 | 75.72 | 75.43 | 76.48 | 74.11 | 75.79 | 75.14 | 75.84 | 100 | |||||||
| 13 | 72.68 | 74.69 | 74.98 | 74.09 | 74.99 | 74.66 | 75.88 | 73.54 | 74.62 | 74.46 | 74.75 | 77.58 | 100 | ||||||
| 14 | 73.11 | 75.05 | 75.71 | 74.26 | 75.53 | 74.96 | 76.40 | 73.76 | 75.31 | 74.75 | 75.34 | 78.43 | 85.98 | 100 | |||||
| 15 | 73.23 | 75.20 | 76.05 | 74.62 | 75.28 | 75.17 | 76.25 | 73.92 | 76.14 | 74.83 | 75.37 | 87.99 | 77.63 | 78.29 | 100 | ||||
| 16 | 72.79 | 74.68 | 74.83 | 74.18 | 74.75 | 74.44 | 75.02 | 73.68 | 74.73 | 74.27 | 74.62 | 79.31 | 76.60 | 77.34 | 79.08 | 100 | |||
| 17 | 74.41 | 76.12 | 76.23 | 75.36 | 76.42 | 75.93 | 77.05 | 74.34 | 76.32 | 75.84 | 76.41 | 79.37 | 82.36 | 83.36 | 79.05 | 77.80 | 100 | ||
| 18 | 73.11 | 74.26 | 75.05 | 74.16 | 74.65 | 74.48 | 75.38 | 74.45 | 74.90 | 74.41 | 75.00 | 79.29 | 76.92 | 77.32 | 78.94 | 84.08 | 77.85 | 100 | |
| 19 | 73.43 | 75.34 | 75.74 | 74.75 | 75.72 | 75.50 | 76.44 | 74.08 | 75.69 | 75.02 | 75.65 | 87.77 | 77.76 | 78.61 | 90.67 | 79.14 | 79.23 | 79.31 | 100 |
Taxa: 1, SCU-B244T; 2, Erwinia amylovora NBRC 12687T; 3, Erwinia billingiae Eb661; 4, Erwinia mallotivora BT-MARDI; 5, Erwinia pyrifoliae DSM 12163T; 6, Erwinia tasmaniensis Et1 99T; 7, Erwinia toletana DAPP-PG 735; 8, Erwinia tracheiphila PSU-1; 9, Erwinia oleae DAPP-PG531T; 10, Erwinia piriflorinigrans CFBP 5888T; 11, Erwinia typographi M043b; 12, Pantoea anthophila 11–2; 13, Pantoea rodasii ND03; 14, Pantoea rwandensis ND04; 15, Pantoea agglomerans DAPP-PG734; 16, Pantoea ananatis LMG 2665T; 17, Pantoea dispersa EGD-AAK13; 18, Pantoea stewartii LMG 2715T; 19, Pantoea vagans C9-1
* Data from NCBI Genome, other data except 1 were taken from EzBioCloud
The DNA G+C content of strain SCU-B244T was 55.32 mol% which could be discriminated from Erwinia oleae (DSM 23398T), which has a G+C content of 54.7 to 54.9 mol%.
API 20E and API 50CHE (BioMérieux) tests were also carried out and the results were compared to related strains (Table 2). The results revealed that SCU-B244T strain can be discriminated from each recognized species of the genus Erwinia by at least three characteristics, and from the phylogenetically most closely related species, Erwinia oleae, by 10 API 50CHE characteristics (Table 3).
Table 2. Comparison of phenotypic characteristics between strain SCU-B244T, Erwinia oleae (DSM 23398T), and other related species.
| Characteristic | 1 | 2 | 3* | 4* | 5* | 6* | 7* | 8* | 9* | 10# | 11* | 12* | 13* | 14* | 15* | 16## |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nitrate reduction | + | + | - | + | + | - | - | + | - | ND | - | + | - | - | - | ND |
| Fermentation of (API 50-CHE): | ||||||||||||||||
| Aesculin | + | + | - | + | + | - | + | + | + | - | - | + | - | + | - | + |
| L-Arabinose | + | + | + | + | + | - | + | + | + | + | + | + | + | + | + | + |
| D-Arabitol | - | + | - | - | + | - | - | - | - | - | - | - | - | + | + | - |
| Potassium Gluconate | + | + | - | + | - | - | + | - | - | + | - | - | - | - | - | + |
| Glycerol | + | - | - | + | + | - | - | + | + | + | + | + | + | + | + | + |
| Inositol | - | - | + | + | + | - | - | + | - | + | + | + | + | + | - | + |
| Potassium 2-Ketogluconate | + | + | - | + | - | - | - | - | - | - | - | - | - | - | - | + |
| D-Mannose | + | + | - | + | + | + | + | + | + | - | - | + | - | + | + | + |
| L-Rhamnose | + | + | - | + | + | - | - | + | + | - | - | + | - | - | - | + |
| D-Sorbitol | + | - | + | - | + | - | - | + | - | - | + | - | - | - | - | + |
| Sucrose | - | - | + | + | - | + | + | + | + | + | + | + | + | - | + | + |
| Xylitol | - | - | - | + | - | - | - | - | - | - | - | + | + | - | - | - |
1, Erwinia teleogrylli sp. nov. SCU-B244T; 2, E. oleae DSM 23398T; 3, E. amylovora LMG 2024T; 4, E. aphidicola LMG 24877T; 5, E. billingiae LMG 2613T; 6, E. mallotivora LMG 2708T; 7, E. papayae CFBP 5189T; 8, E. persicina LMG 11254T; 9, E. psidii LMG 7039T; 10, E. piriflorinigrans CECT 7348T; 11, E. pyrifoliae ICMP 14143T; 12, E. rhapontici LMG 2688T; 13, E. tasmaniensis LMG 25318T; 14, E. toletana CFBP 6631T; 15, E. tracheiphila LMG 2707T; 16, Pantoea gaviniae LMG 26250T.
+, positive; -, negative; ND, not determined.
# Data from López et al. [25].
* Data from Moretti et al. [22].
## Data from Alexandra et al. [26]
Table 3. Differential phenotypic characteristics between SCU-B244T and its closest phylogenetic neighbour, Erwinia oleae DSM 23398T.
| Characteristic | SCU-B244T | Erwinia oleae DSM 23398T |
|---|---|---|
| API 20E: | ||
| β-galactosidase activity | - | + |
| Citrate utilization | + | - |
| API 50CHE: | ||
| Glycerol | + | - |
| D-Xylose | + | - |
| L-Sorbose | + | - |
| D-Sorbitol | + | - |
| Methyl-α-D-glucopyranoside | + | - |
| D-Cellobiose | + | - |
| D-Maltose | + | - |
| D-Melibiose | + | - |
| Gentiobiose | + | - |
| D-Arabitol | - | + |
| Potassium 5-ketogluconate | + | - |
+, Positive; -, negative.
API ZYM enzymatic characteristics and BIOLOG GN2 carbon-source utilization results are presented as supplementary data (S1 and S2 Tables).
Fatty acid composition analysis of SCU-B244T and related species of genera Erwinia and Pantoea is shown in Table 4. Strain SCU-B244T contained C16:0 (28.2%), Summed feature 4 (12.4%), C11:0 3-OH (11.6%), Summed feature 7 (10.5%), Summed feature 3 (10.0%), C14:0 (6.4%) as the major fatty acids, while strain DSM 23398T contained C16:0 (38.8%), Summed feature 4 (32.7%), Summed feature 7 (9.1%), C12:0 (7.7%), C16:0 Δ9 cyclo (4.6%) as the major fatty acids (S3 Table). Summed feature 3 (10.0%), C14:0 2-OH (3.7%) and C18:0 (3.7%) were detected from strain SCU-B244T while those were not detected from strain DSM 23398T. Fatty acid composition of SCU-B244T and reference strains data from Erwinia and Pantoea showing that most of the compositions were in the range of Erwinia and Pantoea [27], which indicates that SCU-B244T belongs to group Erwinia and Pantoea.
Table 4. Fatty acid composition of Erwinia teleogrylli SCU-B244T and reference strains from Erwinia and Pantoea.
This table lists fatty acids including all fatty acid detected at a level higher than 0.5%. Summed feature 3 contained one or more of the following fatty acids: iso-C16:1, C14:0 3-OH, C12:0 aldehyde and an unknown fatty acid ECL 10.928. Summed feature 4 contained one or more of the following fatty acids: C16:1 Δ9 cis and iso-C15:0 2-OH. Summed feature 7 contained one or more of the following fatty acids: C18:1Δ11 cis, C18:1Δ9 trans and C18:1 Δ6 trans [27].
| Fatty acid composition (%) | |||
|---|---|---|---|
| Erwinia teleogrylli | Other Erwinia sp.* | Pantoea sp.* | |
| SCU-B244T | Range | Range | |
| C8:0 2-CH2CH3 | 0.6 | ND | ND |
| C11:0 3-OH | 11.7 | ND | ND |
| C12:0 | 4.3 | 3.2–5.9 | 3.3–4.4 |
| C14:0 | 6.4 | TR-5.8 | TR-5.8 |
| C14:0 2-OH | 3.7 | ND | ND |
| C14:1 Δ11 | 1.1 | ND | ND |
| C15:0 | 0.5 | ND-TR | ND-1.6 |
| C16:0 | 28.2 | 26.9–33.4 | 27.3–31.3 |
| C17:0 | ND | ND-1.2 | ND-2.7 |
| C17:0 cyclo | 4.9 | ND-13.6 | ND-13.2 |
| iso-C18:0 | 3.7 | ND | ND |
| C18:0 14-methyl | 1.9 | ND | ND |
| Summed feature 3* | 10.0 | ND-12.2 | 8.5–10.5 |
| Summed feature 4* | 12.4 | 22.7–33.7 | 10.8–24.4 |
| Summed feature 7* | 10.5 | 6.3–16.9 | 12.6–36.9 |
ND: Not Detected, TR: Trace amount (< 1.0%).
*Data from Mergaert et al. [27]
Based on the genotypic data (16S rRNA gene sequence analysis and MLSA), it is clear that SCU-B244T is a member of genus Erwinia. DNA-DNA hybridization results, ANI values and phenotypic data obtained in this study can separate this isolate from related species. In conclusion the bacterial strain isolated from a cricket (genus Teleogryllus) represents a novel species for which the name Erwinia teleogrylli sp. nov. is proposed. Strain SCU-B244T (= CGMCC 1.12772T = DSM 28222T = KCTC 42022T) is the type strain.
Description of Erwinia teleogrylli sp. nov.
Erwinia teleogrylli (teleogryl'li. N.L. gen. n. teleogrylli of Teleogryllus, the insect from which the species was isolated).
The colonies are milky white, circular and convex with entire margins. Cells are short Gram-negative, facultatively anaerobic rods that are oxidase-negative, catalase-positive and able to grow in 6% (w/v) NaCl. Growth pH range is 6 to 9, with optimum growth at 7. Nitrate is reduced to nitrite.
Results obtained with API 20E tests (BioMérieux) gave positive results for acid production from citrate, glucose, mannitol, rhamnose, melibiose, amygdalin and arabinose, and negative results for β-galactosidase, gelatinase, urease, arginine digydrolase, lysine decarboxylase and ornithine decarboxylase activity, H2S and indole production, acid production from sorbitol and sucrose. According to API 50CHE tests (BioMérieux), acid is formed from glycerol, L-arabinose, D-ribose, D-xylose, D-galactose, D-glucose, D-fructose, D-mannose, L-sorbose, L-rhamnose, D-mannitol, D-sorbitol, methyl-αD-glucopyranoside, N-acetylglucosamine, arbutin, esculin ferric citrate, salicin, D-cellobiose, D-maltose, D-melibiose, D-trehalose, gentiobiose, potassium gluconate, potassium 2-ketogluconate, potassium 5-ketogluconate. No acid is produced from erythritol, D-arabinose, L-xylose, D-ardonitol, methyl-β D-xylopyranoside, dulcitol, inositol, methyl-αD-mannopyranoside, amygdalin, D-lactose, D-saccharose, inulin, D-melezitose, D-raffinose, amidon, glycogen, xylitol, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol. Results obtained with API ZYM tests (BioMérieux) gave positive enzymatic activity results for alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase, naphthol-AS-B1-phosphohydrolase, β-galactosidase, β- glucosidase and N-acetyl-β- glucosaminidase, and negative results for lipase (C14), cystine arylamidase, trypsin, α-chymotrypsin, β-glucuronidase, α-glucosidase, α-mannosidase and α-fucosidase.
The following substrates were utilized according to BIOLOG GN2 carbon-source utilization analysis: dextrin, N-acetyl-D-glucosamine, L-arabinose, D-fructose, D-galactose, α-D-glucose, maltose, D-mannitol, D-mannose, D-melibiose, β-methyl-D-glucoside, D-psicose, L-rhamnose, D-trehalose, turanose, pyruvic acid methyl ester, citric acid, formic acid, D-galactonic acid lactone, D-galacturonic acid, D-gluconic acid, p-hydroxy phenylacetic acid, α-keto glutaric acid, propionic acid, D-saccharic acid, L-alanine, L-alanyl-glycine, L-asparagine, L-aspartic acid, L-glutamic acid, glycyl-L-aspartic acid, glycyl-L-glutamic acid, L-proline, L-serine, inosine, uridine, thymidine, glycerol, D,L-α-glycerol phosphate, glucose-1-phosphate, glucose-6-phosphate.
Major fatty acids are C16:0 (28.2%), Summed feature 4 (12.4%), C11:0 3-OH (11.6%), Summed feature 7 (10.5%) and Summed feature 3 (10.0%), with other minor components, including C14:0 (6.4%), C17:0 Δ9 cyclo (4.9%), C12:0 (4.3%), C14:0 2-OH (3.7%), iso-C18:0 (3.6%), C18:0 14-methyl (1.9%), C14:1 Δ11 (1.1%), C8:0 2-CH2CH3 (0.6%), C15:0 (0.5%). The DNA G+C content is 55.32 mol%.
The type strain, SCU-B244T (= CGMCC 1.12772T = DSM 28222T = KCTC 42022T), was isolated from crickets (genus Teleogryllus) sampled in Chengdu (30°33’ N, 103°58’ E, altitude 495m), Sichuan Province, China.
Supporting Information
(PDF)
(PDF)
The diagram shows the phylogenetic relationship between Erwinia teleogrylli sp. nov. and the first 66 hit strains at the EzTaxon server within the family Enterobacteriaceae. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
(TIF)
The diagram shows the phylogenetic relationship between Erwinia teleogrylli sp. nov. and the first 66 hit strains at the EzTaxon server within the family Enterobacteriaceae. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
(TIF)
The diagram shows the phylogenetic relationship between Erwinia teleogrylli sp. nov. and taxa related type species of the genera Erwinia and Pantoea except Candidatus Erwinia dacicola. Escherichia coli ATCC 11775T was used as the outgroup. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
(TIF)
The diagram shows phylogenetic relationship between Erwinia teleogrylli sp. nov. and taxa related species of genera Erwinia, Pantoea and Tatumella. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are grateful to Shu-Kun Tang for his help with several of the experiments and Prof. Zhao-Bin Song for his help with identification of crickets (Teleogryllus occipitalis). The authors wish to acknowledge Aidan Parte for the help with naming SCU-B244T in Latin.
Data Availability
All sequences files are available from the NCBI nucleotide database with accession numbers for the sequence of 16S rDNA gene, atpD, gyrB and infB: KF500917, KM108628, KM108627 and KM108629, respectively. Other relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1.Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci USA 2012; 109(22): 8618–8622. 10.1073/pnas.1200231109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moretti C, Hosni T, Vandemeulebroecke K, Brady C, De Vos P, Buonaurio R, National Research Council The Future Role of Pesticides in U.S. Agriculture (National Academies Press, Washington, DC: ); 2000. [Google Scholar]
- 3.Gerhardt P, Murray RGE, Costilow RN, Nester EW, Woods WA, Krieg NR, et al. Manual of Methods for General Bacteriology. Washington DC: American Society for Microbiology; 1981. [Google Scholar]
- 4.Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, Xu LH, et al. Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol. 2007; 57(7): 1424–1428. [DOI] [PubMed] [Google Scholar]
- 5.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 2012; 62(Pt 3): 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- 6.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4(4): 406–425. [DOI] [PubMed] [Google Scholar]
- 7.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28(10):2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980; 16(2): 111–120. [DOI] [PubMed] [Google Scholar]
- 9.Kimura M. The neutral theory of molecular evolution. Cambridge University Press; 1984. [Google Scholar]
- 10.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981; 17(6): 368–376. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985; 783–791. [DOI] [PubMed] [Google Scholar]
- 12.Brady C, Cleenwerck I, Venter S, Vancanneyt M, Swings J, Coutinho T. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst Appl Microbiol. 2008; 31(6): 447–460. [DOI] [PubMed] [Google Scholar]
- 13.Brady CL, Venter SN, Cleenwerck, Engelbeen K, Vancanneyt M, Swings J, et al. Pantoea vagans sp. nov., Pantoea eucalypti sp. nov., Pantoea deleyi sp. nov. and Pantoea anthophila sp. nov. Int J Syst Evol Microbiol. 2009; 59(9): 2339–2345. [DOI] [PubMed] [Google Scholar]
- 14.Brady CL, Cleenwerck I, Venter SN, Engelbeen K, De Vos P, Coutinho TA. Emended description of the genus Pantoea, description of four species from human clinical samples, Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov. and Pantoea conspicua sp. nov., and transfer of Pectobacterium cypripedii (Hori 1911) Brenner et al. 1973 emend. Hauben et al. 1998 to the genus as Pantoea cypripedii comb. nov. Int J Syst Evol Microbiol. 2010; 60(10):2430–2440. [DOI] [PubMed] [Google Scholar]
- 15.Brady CL, Venter SN, Cleenwerck I, Vandemeulebroecke K, De Vos P, Coutinho TA. Transfer of Pantoea citrea, Pantoea punctata and Pantoea terrea to the genus Tatumella emend. as Tatumella citrea comb. nov., Tatumella punctata comb. nov. and Tatumella terrea comb. nov. and description of Tatumella morbirosei sp. nov. Int J Syst Evol Microbiol. 2010; 60(3): 484–494. [DOI] [PubMed] [Google Scholar]
- 16.Brady CL, Goszczynska T, Venter SN, Cleenwerck I, De Vos P, Gitaitis RD, et al. Pantoea allii sp. nov., isolated from onion plants and seed. Int J Syst Evol Microbiol. 2011; 61(Pt 4):932–937. 10.1099/ijs.0.022921-0 [DOI] [PubMed] [Google Scholar]
- 17.De Ley J, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970; 12:133–142. [DOI] [PubMed] [Google Scholar]
- 18.Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J biol Chem 1957; 226(1):497–509. [PubMed] [Google Scholar]
- 19.Xiang W, Liu C, Wang X, Du J, Xi L, Huang Y. Actinoalloteichus nanshanensis sp. nov., isolated from the rhizosphere of a fig tree (Ficus religiosa). Int J Syst Evol Microbiol. 2011; 61(5):1165–1169. [DOI] [PubMed] [Google Scholar]
- 20.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007; 57(1): 81–91. [DOI] [PubMed] [Google Scholar]
- 21.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci. 2009; 106(45): 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moretti C, Hosni T, Vandemeulebroecke K, Brady C, De Vos P, Buonaurio R, et al. Erwinia oleae sp. nov., isolated from olive knots caused by Pseudomonas savastanoi pv. Savastanoi. Int J Syst Evol Microbiol. 2011; 61:2745–2752. 10.1099/ijs.0.026336-0 [DOI] [PubMed] [Google Scholar]
- 23.Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994; 44(4): 846–849. [Google Scholar]
- 24.Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987; 37: 463–464. [Google Scholar]
- 25.López MM, Roselló M, Llop P, Ferrer S, Christen R, Gardan L. Erwinia piriflorinigrans sp. nov., a novel pathogen that causes necrosis of pear blossoms. Int J Syst Evol Microbiol. 2011; 61(3): 561–567. [DOI] [PubMed] [Google Scholar]
- 26.Popp A, Cleenwerck I, Iversen C, De Vos P, Stephan R. Pantoea gaviniae sp. nov. and Pantoea calida sp. nov., isolated from infant formula and an infant formula production environment. Int J Syst Evol Microbiol, 2010; 60: 2786–2792. 10.1099/ijs.0.019430-0 [DOI] [PubMed] [Google Scholar]
- 27.Mergaert J, Hauben L, Cnockaert MC, Swings J. Reclassification of non-pigmented Erwinia herbicola strains from trees as Erwinia billingiae sp. nov. Int J Syst Bacteriol. 1999; 49: 377–383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
The diagram shows the phylogenetic relationship between Erwinia teleogrylli sp. nov. and the first 66 hit strains at the EzTaxon server within the family Enterobacteriaceae. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
(TIF)
The diagram shows the phylogenetic relationship between Erwinia teleogrylli sp. nov. and the first 66 hit strains at the EzTaxon server within the family Enterobacteriaceae. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
(TIF)
The diagram shows the phylogenetic relationship between Erwinia teleogrylli sp. nov. and taxa related type species of the genera Erwinia and Pantoea except Candidatus Erwinia dacicola. Escherichia coli ATCC 11775T was used as the outgroup. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
(TIF)
The diagram shows phylogenetic relationship between Erwinia teleogrylli sp. nov. and taxa related species of genera Erwinia, Pantoea and Tatumella. Bar, 0.5% nucleotide substitutions. Numbers at branching points are bootstrap percentage values based on 1000 replications. Only values >50% are shown.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All sequences files are available from the NCBI nucleotide database with accession numbers for the sequence of 16S rDNA gene, atpD, gyrB and infB: KF500917, KM108628, KM108627 and KM108629, respectively. Other relevant data are within the paper and its Supporting Information files.