Abstract
Polymorphisms in the apolipoprotein C-III (APOC3) gene have been reported to be associated with coronary heart disease (CHD), but the data so far have been conflicting. To derive a more precise estimation of these associations, we performed a meta-analysis to investigate the three main polymorphisms (SstI, T-455C, C-482T) of APOC3 in all published studies. Databases including PubMed, Web of Science, Wanfang, SinoMed and CNKI were systematically searched. The association was assessed using odds ratios (ORs) with 95% confidence intervals (CIs). The statistical analysis was performed using Review Manager 5.3.3 and Stata 12.0. A total of 31 studies have been identified. The pooled odds ratio (OR) for the association between the APOC3 gene polymorphisms and CHD and its corresponding 95% confidence interval (95% CI) were evaluated by random or fixed effect models. A statistical association between APOC3 SstI polymorphism and CHD susceptibility was observed under an allelic contrast model (P= 0.003, OR = 1.14, 95% CI = 1.05-1.24), dominant genetic model (P= 0.01, OR = 1.14, 95% CI = 1.03-1.26), and recessive genetic model (P= 0.02, OR = 1.35, 95% CI = 1.06-1.71), respectively. A significant association between the APOC3 T-455C polymorphism and CHD was also detected under an allelic contrast (P < 0.0001, OR = 1.19, 95% CI = 1.10-1.29), dominant genetic model (P= 0.0003, OR = 1.24, 95% CI = 1.11-1.39) and recessive genetic model (P= 0.04, OR = 1.30, 95% CI = 1.01-1.67). No significant association between the APOC3 C-482T polymorphism and CHD was found under an allelic model (P= 0.94, OR = 1.00, 95% CI = 0.93-1.08), dominant genetic model (P= 0.20, OR = 1.07, 95% CI = 0.97-1.18) or recessive genetic model (P= 0.13, OR = 0.90, 95% CI = 0.79-1.03). This meta-analysis revealed that the APOC3 SstI and T-455C polymorphisms significantly increase CHD susceptibility. No significant association was observed between the APOC3 C-482T polymorphism and CHD susceptibility.
Keywords: APOC3, Coronary heart disease, Polymorphism, Meta-analysis
Coronary heart disease (CHD) is one of the leading causes of death globally. Apolipoproteins are lipid-binding proteins involved in lipid transport. Thus, the abnormal structure and synthesis of apolipoproteins affects lipid metabolism and leads to CHD. Apolipoprotein C-III (APOC3) is an 8.8kd glycoprotein comprised of 79 amino acid residues. It is synthesized in the liver and is the major protein constituent of the plasma very low density lipoproteins (VLDLs) and chylomicrons (CMs). Transgenic animal models show that APOC3 directly impacts the level of plasma triglycerides (TGs); if the APOC3 gene is overexpressed, the levels of plasma APOC3 will be elevated, thus inhibiting the clearance of TGs and leading to high triglyceride levels [1]. In vitro studies have shown that APOC3 inhibits the activity of lipoprotein lipase (LPL), delays the clearance of TG-rich lipoproteins and leads to the elevation of plasma TGs [2]. High TG levels are known to increase the risk of atherosclerosis and may also increase the susceptibility to cardiovascular disease [3, 4]. Five polymorphisms have been found in the APOC3 gene promoter. The most extensively studied are the SstI, T-455C and C-482T polymorphisms and a C¬G substitution at nucleotide 3238 (rs5128) in the 3′ untranslated region of SstI, which generates two alleles: S1 and S2. Two promoter polymorphisms, T-455C and C-482T, have also been studied more extensively because they are located on the insulin-responsive element (IRE). Insulin exerts its action by down-regulating APOC3 gene expression transcriptionally. The presence of mutant sequences may reduce the inhibitory effects of the hormone [5]. The relationship between APOC3 gene polymorphisms and genetic susceptibility to CHD has attracted significant clinical and epidemiological research interest in recent years, but the reported results have been inconsistent. We therefore conducted a meta-analysis of the existing published studies on this topic to evaluate the strength of the association between the three main APOC3 polymorphisms (SstI, T-455C, C-482T) and CHD.
MATERIALS AND METHODS
Literature Search
All studies that reported the association between the APOC3 gene polymorphisms and CHD were identified by comprehensive computer-based searches of PubMed (from 1980 to June 2014), the Web of Science, Wanfang, the China Biological Medicine Database (SinoMed) and the China National Knowledge Infrastructure (CNKI). These computer searches were limited to English and Chinese language articles published before June 2014, and did not include reviews and editorials. The following keywords were used for the search: “apolipoprotein C-III” OR “APOC3” AND “polymorphism” OR “mutation” OR “genotype” OR “variant” AND “coronary heart disease” OR “CHD” OR “coronary artery disease” OR “CAD” OR “myocardial Infarction” OR “MI” OR “Acute coronary syndrome” OR “ACS”.
Inclusion Criteria
The diagnosis of CHD was determined based on examination results including coronary arteriography, clinical symptoms, echocardiography, the treadmill exercise test, electrocardiogram results, and myocardial perfusion imaging in Emission Computed Tomography. The inclusion criteria were as follows: (1) studies were limited to three main (SstI, T-455C, C-482T) polymorphisms of the APOC3 gene and CHD, (2) all studied were independent case-control studies using either a hospital-based or a population-based design, (3) studies were selected using the literature research methods, (4) the existing literature provided us with a comprehensive statistical index and sufficient data for estimating an odds ratio (OR) with 95% confidence interval (CI). Studies were excluded from analysis when (1) it was not possible to extract data from the published results, (2) the reported appropriate outcomes were excluded, or (3) they contained republished data.
Data Extraction
Two authors (JZ Zhang and CF Dai) independently extracted data from the studies. Disagreement was resolved by consensus. If these two authors could not reach a consensus, the result was reviewed by a third author (X Xie). The extracted data consisted of the following items: the first author’s name, the publication year, the total number of cases and controls, the allele frequency (cases), the methods, the population (ethnicity), the selection criteria, the percentage of the male sex, and the age (in years).
Quality assessment
To determine the methodological quality of each study, we used the Newcastle-Ottawa scale (NOS), which uses a “star” rating system to judge the quality of observational studies [6]. The NOS ranges between zero stars (worst) and nine stars (best). Studies with a score equal to or greater than seven were considered to be of high quality. Two investigators (GT Yin and MM Zhang) independently assessed the quality of the included studies, and the results were reviewed by a third investigator (YT Ma). Disagreement was resolved by discussion.
Statistical analysis
The associations between the SstI, T-455C and C-482T polymorphisms of the APOC3 gene and CHD were compared by using the odds ratio (OR) corresponding to 95% confidence interval (CI) by using Review Manager 5.33 (Cochrane Collaboration, The Nordic Cochrane Centre, Copenhagen). Heterogeneity between the studies was assessed using the I2 statistic; P<0.10 and I2>50% indicated evidence of heterogeneity [7,8]. If heterogeneity existed among the studies, the random effects model was used to estimate the pooled OR (DerSimonian and Laird method) [9]. Otherwise, the fixed effects model was adopted (Mantel-Haenszel method) [10]. For the APOC3 gene polymorphisms, we investigated associations between the genetic variants and CHD risk in allelic contrast, recessive and dominant genetic models. The Z test was used to determine the pooled OR, and significance was set at P < 0.05. The Hardy-Weinberg equilibrium (HWE) for each nucleotide polymorphism was assessed for the controls in each study using X2 test at a significant level of P < 0.05. The potential publication bias was investigated using a funnel plot. Egger’s test (P < 0.05) was also considered to be representative of statistically significant publication bias [11], which was conducted with the Stata 12.0 software.
RESULTS
Study Characteristics
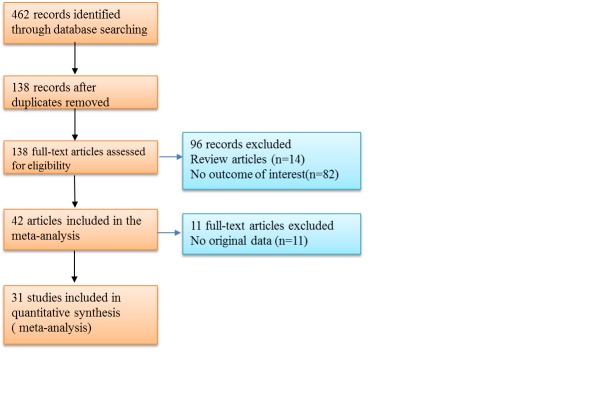
A total of 31 studies [12-43] were included in the final meta-analysis, based on the above inclusion criteria. These studies consisted of 19 studies for SstI, 7 for T-455C and 11 for C-482T, for a total of 8311, 5653 and 6828 samples for the 3 polymorphisms, respectively. All included studies were approved by the various Ethics Committees of their institutions. Figure 1 shows the process of literature retrieval. Table 1 shows the main characteristics of the studies. In all studies, the genotype frequencies in the control population were consistent with the HWE. The NOS results showed that the methodological quality was generally good.
Figure 1.
Flow diagram of the study identification.
Table 1.
Characteristics of studies reporting the distribution of three APOC3 polymorphisms (SstI,-455T/C,-482C/T) in CHD cases and controls
| Study [Reference] | Year | Eligible subjects | Allele frequency (case) | Method | Population | Criteria | Gender (male), % | Age (years) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| case | control | S2 | T | C | Case | Control | Case | Control | |||||
| Aalto-Setälä [12] | 1987 | 39 | 61 | 0.115 | - | - | PCR-RFLP | Finnish | CHD | 100 | 50.8 | 38-68 | 18-52 |
| Bai [13] | 1995 | 90 | 68 | 0.328 | - | - | PCR-RFLP | Japanese | CHD | 77.8 | 57.4 | 63±12 | 58±12 |
| Berkinbayev [14]a | 2014 | 161 | 112 | 0.242 | - | - | PCR | Kazakhstan(Kazakh) | MI | 100 | 100 | 46.4±1.9 | 40.7±2.1 |
| Berkinbayev [14]b | 2014 | 80 | 95 | 0.285 | - | - | PCR | Kazakhstan(Uyghur) | CHD | 100 | 100 | 47.3±0.92 | 41.2±1.02 |
| Bi [15] | 2005 | 312 | 317 | - | - | 0.452 | PCR-RFLP | Chinese(Han) | CAD | 67 | 60.3 | 60.2±13.3 | 58.9±14.3 |
| Che [16] | 2012 | 78 | 78 | - | - | 0.647 | PCR-RFLP | Chinese | CAD | 57.7 | 52.6 | 45.3±4.7 | 44.3±7.6 |
| Chen [17] | 2006 | 310 | 499 | - | 0.477 | 0.455 | PCR-RFLP | Chinese(Han) | CHD | 70.6 | 62.7 | 60.56±11.14 | 58.87±9.11 |
| Chhabra [18] | 2004 | 158 | 151 | 0.342 | - | - | PCR | India | CAD | 88 | 92 | 53.25±10.25 | 52.45±10.85 |
| Dallongeville [19] | 2006 | 442 | 475 | - | - | 0.263 | PCR | French | CHD | 100 | 100 | 35-64 | 35-64 |
| Dan [20] | 1995 | 43 | 60 | 0.221 | - | - | PCR-RFLP | Chinese | CHD | N/A | N/A | 57.6-6.9 | 55.4-4.3 |
| Ding [21] | 2012 | 229 | 254 | - | 0.548 | - | PCR | Chinese(Han) | ACS | 55.9 | 53.9 | 59.1±9.4 | 59.3±9.6 |
| Han [22] | 2011 | 275 | 289 | - | - | 0.481 | PCR-RFLP | Chinese(Han) | CHD | 56.4 | 56.4 | 59.2±9.8 | 61.6±10.7 |
| Izar [23] | 2003 | 112 | 112 | 0.136 | - | - | PCR | Brazil | CAD | 58 | 59 | M<45, W<55 | N/A |
| Kee [24] | 1999 | 614 | 761 | 0.093 | - | - | PCR | Europe | MI | 100 | 100 | 25-64 | N/A |
| Li [25] | 2006 | 47 | 104 | 0.274 | - | - | PCR-RFLP | Chinese(Han) | CAD, MI | 72.3 | 60.6 | 62.5±12.1 | 60.0±8.0 |
| Liu [26] | 2004 | 385 | 373 | 0.116 | - | - | PCR-RFLP | Caucasian | MI | 100 | 100 | 60±9 | 59±9 |
| Liu [27] | 2005 | 483 | 502 | 0.301 | 0.534 | 0.449 | PCR-RFLP | Chinese(Han) | CHD | 59 | 63.1 | 54.2±6.3 | 58.2±6.2 |
| Liu [28] | 2005 | 267 | 491 | 0.301 | - | - | PCR-RFLP | Chinese(Han) | CHD | 70.4 | 63.7 | 60.3±8.9 | 58.8±9.2 |
| Martinelli [29] | 2007 | 669 | 244 | - | - | - | PCR | Italy | CAD, MI | 81.3 | 75 | 60.7±9.3 | 58.7±12.7 |
| Muendlein [30] | 2008 | 332 | 225 | - | - | 0.297 | PCR | Caucasian | CAD | 79.5 | 54.7 | 62.5±10.3 | 61.5±10.2 |
| Olivieri [31] | 2002 | 549 | 251 | - | 0.416 | - | PCR | Northern Italy | CAD | 81.8 | 66.9 | 60.4±9.4 | 57.6±12.6 |
| Ordovas [32] | 1991 | 202 | 145 | 0.125 | - | - | PCR-RFLP | Northern Italy | CAD | N/A | N/A | 47.2±8.8 | 49.6±6.9 |
| Paulweber [33] | 1988 | 106 | 118 | 0.104 | - | - | PCR-RFLP | Austrian | CAD, MI | 100 | 100 | 47.3±5.7 | 49.7±5.4 |
| Rigoli [34] | 1995 | 62 | 62 | 0.169 | - | - | PCR-RFLP | Southern Italy | CHD | 69.4 | 67.7 | 58.2±7.1 | 57.6±7.7 |
| Sediri [35] | 2011 | 326 | 361 | 0.103 | 0.411 | - | PCR | Tunisia | MI | 100 | 100 | 53.8±8.6 | 51.1±9.5 |
| Tarek [36] | 2013 | 156 | 154 | 0.292 | - | - | PCR-RFLP | Egyptian | CAD | 100 | 100 | 51.5±7.9 | 50.7±9.4 |
| Tarek [37] | 2011 | 200 | 100 | 0.145 | - | - | PCR | Egyptian | MI | 67 | 64 | 50.7±9.5 | 52.9±11.3 |
| Tobin [38] | 2004 | 547 | 505 | - | 0.355 | 0.255 | PCR-RFLP | UK | MI | 68 | 62 | 61.9±9.2 | 58.6±10.7 |
| Weng [39] | 2000 | 50 | 50 | 0.16 | - | - | PCR | Chinese(Han) | CAD, MI | 76 | 74 | 57±6.8 | 55±8.7 |
| Wu [40] | 2000 | 131 | 229 | 0.233 | - | - | PCR | Chinese | CAD | 69.5 | 56.8 | 59.1±10.9 | 51±15.3 |
| Yang [41] | 2008 | 85 | 87 | - | - | 0.412 | PCR-RFLP | Chinese | CHD | 54.1 | 52.9 | 52.79±7.15 | 50.63±7.50 |
| Yi [42] | 2006 | 195 | 181 | - | - | 0.541 | PCR-RFLP | Chinese (Han) | CAD | 56.4 | 56.4 | 69.48±9.21 | 58.63±14.79 |
| Yu [43] | 2011 | 286 | 325 | - | 0.42 | 0.517 | TaqMan | Chinese (Han) | CHD | 74.8 | 52.9 | 56.30±11.57 | 55.79±12.40 |
Main Results, Heterogeneity and Sensitivity Analysis
No significant heterogeneity was observed for the APOC3 SstI polymorphism and its relationship to CHD under the allelic contrast model (I2 = 35%, P= 0.06), dominant genetic model (I2 = 40%, P= 0.04) and the recessive genetic model (I2 = 0%, P= 0.70). Therefore, the Mantel-Haenszel fixed effects model was applied.
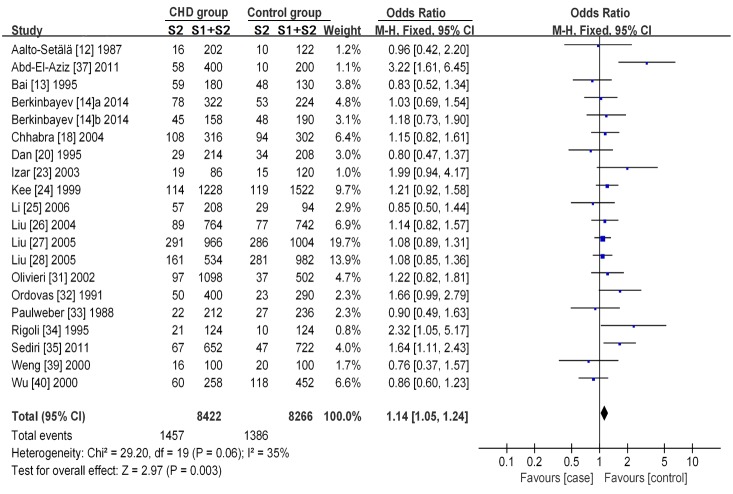
A statistical association between the APOC3 SstI polymorphism and CHD susceptibility was observed under an allelic contrast model (P= 0.003, OR = 1.14, 95% CI = 1.05-1.24), the dominant genetic model (P= 0.01, OR = 1.14, 95% CI = 1.03-1.26), and the recessive genetic model (P= 0.02, OR = 1.35, 95% CI = 1.06-1.71). We used the forest plot of the allelic contrast model as an example (Figure 2), as the dominant and recessive gene models do not fully show the results.
Figure 2.
Forest plot of the association between the APOC3 SstI polymorphism and CHD under the allelic contrast model (S2 vs. S1). The horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of the OR and the 95% CI.
For the APOC T-455C polymorphism and risk of CHD, no significant heterogeneity was found under the allelic contrast model (I2 = 46%, P= 0.09) or dominant genetic model (I2 = 0%, P= 0.53), so we used the Mantel-Haenszel fixed effects model. Heterogeneity was found under the recessive genetic model (I2 = 65%, P= 0.008). The random-effects model (DerSimonian and Laird) was also applied. A significant statistical association was observed between the APOC T-455C polymorphism and CHD under the allelic contrast model (C vs. T, P < 0.0001, OR = 1.19, 95% CI = 1.10-1.29) (Figure 3), the dominant genetic model (CT+CC vs. TT, P= 0.0003, OR = 1.24, 95% CI = 1.11-1.39) and the recessive genetic model (CC vs. CT+TT, P= 0.04, OR = 1.30, 95% CI = 1.01-1.67).
Figure 3.
Forest plot of the association between the APOC3 T-455C polymorphism and CHD under the allelic contrast model (C vs. T). The horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of the OR and the 95% CI.
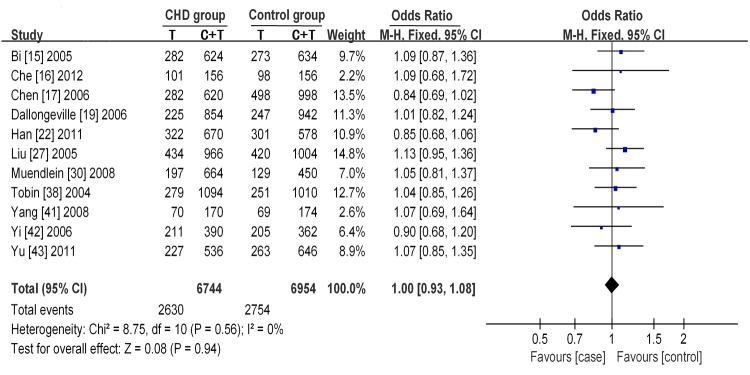
As for the APOC C-482T polymorphism and its relationship to CHD, no significant heterogeneity was found under the allelic contrast model (I2= 0%, P= 0.56), the dominant genetic model (I2 = 0%, P= 0.82) or the recessive genetic model (I2 = 15%, P= 0.30). For this reason, the Mantel-Haenszel fixed effects model was used. No significant statistical association was found under the allelic contrast model (T vs. C, P= 0.94, OR =1.00, 95% CI = 0.93-1.08) (Figure 4), dominant genetic model (TT+TC vs. CC, P= 0.20, OR = 1.07, 95% CI = 0.97-1.18) or recessive genetic model (TT vs. TC+CC, P= 0.13,OR = 0.90, 95% CI = 0.79-1.03).
Figure 4.
Forest plot of the association between the APOC3 C-482T polymorphism and CHD under the allelic contrast model (T vs. C). The horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of the OR and the 95% CI.
Sensitivity analysis
The contribution of each study to the pooled estimate was determined to assess the sensitivity analysis. We excluded individual studies one at a time and recalculated the pooled P or OR estimates for the remaining studies. Ding [21], Liu [27], Martinelli [29] and Olivieri [31] had undue influences on the summary ORs under the recessive genetic model for the T-455C polymorphism. Similarly, Liu [27] had undue influences on the pooled P or OR estimates for the C-482T polymorphism. However, their data did not substantially change the pooled point estimate when converting the random-effects model to the fixed effects model in all studies. Thus, our results are fairly reliable.
Publication Bias
The publication bias of the individual studies was evaluated using a funnel plot and Egger’s test. We also took the figure alleles of the SstI, T-455C and C-482T polymorphisms of APOC3 as representative. No visual publication bias was found in the funnel plot for the SstI (Figure 5A), T-455C (Figure 5B) or C-482T (Figure 5C) gene polymorphisms using the allelic contrast model. We further used Egger's test to assess the whole test publication bias. No statistically significant difference was detected in the Egger’s test by using the allelic contrast model for the SstI (T = -0.56, P = 0.697), T-455C (T = 0.09, P = 0.932) or C-482T (T = -0.29, P = 0.778) polymorphisms in all studies. This indicated that the publication bias was low in the current meta-analysis.
Figure 5.
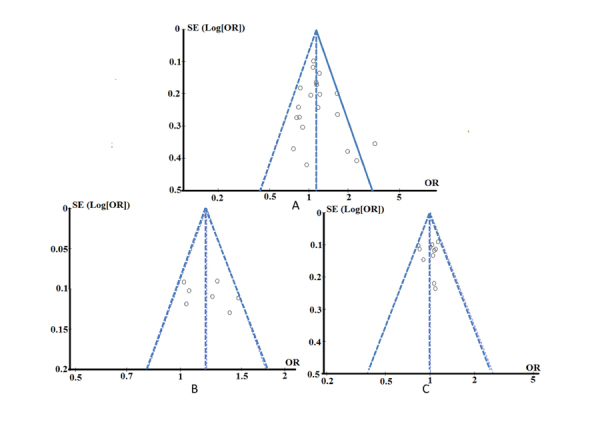
Funnel plot for the publication bias tests. Each point represents a separate study for the indicated association. The horizontal and vertical axis correspond to the OR and confidence limits (OR: odds ratio, SE: standard error). A: SstI polymorphism under the allelic contrast model (S2 vs. S1); B: T-455C polymorphism under the allelic contrast model (C vs. T); C: C-482T polymorphism under the allelic contrast model (T vs. C).
DISCUSSION
CHD is a multi-factorial and polygenic disorder disease, which is thought to result from complex gene-gene and gene-environment interactions [44]. The association between APOC3 gene polymorphisms and the risk of CHD has been intensively studied, but the results remain inconclusive [12-43]. To generate more robust data regarding the APOC3 gene polymorphisms and CHD risk, we conducted a comprehensive genetic meta-analysis on the basis of data from 8311, 5653 and 6828 samples for the SstI, T-455C and C-482T polymorphisms, respectively. The results showed that the APOC3 SstI and T-455C polymorphisms significantly increase CHD susceptibility. No significant association between the APOC3 C-482T polymorphism and an increased risk of CHD was observed.
Abd El-Aziz [36] and Abd El-Aziz [37] had an undue influence on the summary ORs under the dominant genetic model in the SstI polymorphism; there was no observed effect on the pooled P or OR in the subgroup.
Heterogeneity is a potential problem that may affect the interpretation of the results. Heterogeneity may be attribute to potential confounding resulting from diversity in sample-sizes, design differences, variations in the ethnicities and regions under study, CHD severity, subject gender, methods of genotyping, and/or the interaction with other risk factors. When the study of Abd El-Aziz [37] was included in the Forest plot of the association between APOC3 SstI polymorphism and CHD, significant heterogeneity was found under the allelic contrast and dominant genetic models. Initially, we included two studies conducted by Abd El-Aziz investigating the Severity of Coronary Artery diseases [36] and acute myocardial infarction in diabetic patients [37] in patients with an APOC3 SstI polymorphism. According to the Newcastle-Ottawa scale, these two studies ranked as having six stars, indicating them as possessing medium quality. We found that the publication bias of these two articles was huge, and the heterogeneity analysis found that heterogeneity is large in the study of Apolipoprotein C3 Genes with the Severity of Coronary Artery diseases [36]. Because the two articles were published at a similar time and both investigated the association between Apolipoprotein C3 genes and CHD, we are unsure if they made use of repeated samples. We contacted the corresponding authors by email but did not receive any reply. To increase the credibility of the meta-analysis, we therefore eliminated this article, as it had an excessive impact on the overall publication bias.
To better interpret the results, other limitations of this meta-analysis should also be acknowledged. For one thing, some inevitable publication bias may exist in the results. Only full text articles published in English and Chinese were included in this meta-analysis. Thus, some eligible studies that were unpublished or reported in other languages were likely missed. Cultural background factors can also affect the decision to publish, making researchers more or less likely to report or edit negative results in some areas of research. Furthermore, CHD is a complex disease involving potential interactions among gene-gene and gene-environment factors. However, many eligible studies included in this meta-analysis failed to consider environmental factors, which could also influence the study results.
Despite these limitations or disadvantages, our meta-analysis did have some advantages. First, a systematic review of the association of the APOC3 gene (SstI, T-455C and C-482T) polymorphisms with CHD risk is able to overcome the limitation of the small sample sizes of the study populations by increasing the sample size, thus generating more robust data. Second, the quality of the case-control studies included in our meta-analysis was satisfactory and met our inclusion criteria.
Conclusion
In conclusion, this meta-analysis reveals that the APOC3 SstI and T-455C polymorphisms significantly increased CHD susceptibility. No significant association between APOC3 C-482T polymorphism and the susceptibility of CHD was observed. However, the results should be interpreted with caution because of the discussed study limitations. Further studies with larger sample sizes that consider gene-gene and gene-environment interactions are thus needed to confirm our findings.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81470014) and the Xinjiang Science and Technology Projects (201491181).
References
- [1].Ito Y,Azrolan N,O'Connell A,Walsh A,Breslow JL (1990). Hypertriglyceridemia as a result of human apo CIII gene expression in transgenic mice. Science, 249:790-793. [DOI] [PubMed] [Google Scholar]
- [2].Bi N,Yan SK,Li et al. (2005). Polymorphisms in the apolipoprotein A5 gene and apolipoprotein C3 gene in patients with coronary artery disease. Zhonghua Xin Xue Guan Bing Za Zhi, 33:116-121. [PubMed] [Google Scholar]
- [3].Isaacs A,Willems SM,Bos D, et al. (2013). Risk scores of common genetic variants for lipid levels influence atherosclerosis and incidentcoronary heart disease. Arterioscler Thromb Vasc Biol, 33:2233-2239. [DOI] [PubMed] [Google Scholar]
- [4].Martinelli N,Trabetti E,Bassi A, et al. (2007). The -1131 T>C and S19W APOA5 gene polymorphisms are associated with high levels of triglycerides and apolipoprotein C-III, but not with coronary artery disease: an angiographic study. Atherosclerosis, 191:409-417. [DOI] [PubMed] [Google Scholar]
- [5].Li WW,Dammerman MM,Smith JD,Metzger S,Breslow JL,Leff T (1995). Common genetic variation in the promoter of the human apo CIII gene abolishes regulation by insulin and may contribute to hypertriglyceridemia. J Clin Invest, 96:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wells GA,Shea B,O’Connell D,Peterson J,Welch V (2011). The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses. Ottawa Health Research Institute; Available: www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 2011 October 20. [Google Scholar]
- [7].Ioannidis JP,Patsopoulos NA,Evangelou E (2007). Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One, 2:e841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Berkey CS,Hoaglin DC,Mosteller F,Colditz GA (1995). A random-effects regression model for meta-analysis. Stat Med, 14:395-411. [DOI] [PubMed] [Google Scholar]
- [9].DerSimonian R,Kacker R (2007). Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials, 28:105-114. [DOI] [PubMed] [Google Scholar]
- [10].Mantel N,Haenszel W (1959). Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst, 22:719-748. [PubMed] [Google Scholar]
- [11].Egger M,Davey Smith G,Schneider M,Minder C (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ, 315:629-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aalto-Setälä K,Kontula K,Sane T,Nieminen M,Nikkilä E (1987). DNA polymorphisms of apolipoprotein A-I/C-III and insulin genes in familial hypertriglyceridemia and coronary heart disease. Atherosclerosis, 66:145-52. [DOI] [PubMed] [Google Scholar]
- [13].Bai H,Saku K,Liu R,Imamura M,Arakawa K (1995). Association between coronary heart disease and the apolipoprotein A-I/C-III/A-IV complex in a Japanese population. Hum Genet, 95:102-104. [DOI] [PubMed] [Google Scholar]
- [14].Berkinbayev S,Rysuly M,Mussayev A,Blum K,Baitasova N,Mussagaliyeva A, et al. (2014). Apolipoprotein Gene Polymorphisms (APOB, APOC111, APOE) in the Development of Coronary Heart Disease in Ethnic Groups of Kazakhstan. J Genet Syndr Gene Ther, 5:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bi N,Yan SK,Li GP, et al. (2005). Polymorphisms in the apolipoprotein A5 gene and apolipoprotein C3 gene in patients with coronary artery disease. Zhonghua Xin Xue Guan Bing Za Zhi, 33:116-121. [PubMed] [Google Scholar]
- [16].Che YL,Chou GC (2012). Polymorphsims in the apolipoprotein A5 gene and apolipoprotein C3 gene in patients with coronary artery disease. Labeled Immunoassays and Clinical Medicine, 19:371-372. [Google Scholar]
- [17].Chen SY (2006). Genetic analysis of haplotypes in the apo1ipoprotein A1-C3 gene region: effects on coronary heart disease and type 2 diabetes mellitus. Fujian Medical University, 1-47 [Google Scholar]
- [18].Chhabra S,Narang R,Lakshmy R, et al. (2004). Apolipoprotein C3 SstI polymorphism in the risk assessment of CAD. Mol Cell Biochem, 259:59-66. [DOI] [PubMed] [Google Scholar]
- [19].Dallongeville J,Cottel D,Montaye M,Codron V,Amouyel P,Helbecque N (2006). Impact of APOA5/A4/C3 genetic polymorphisms on lipid variables and cardiovascular disease risk in French men. Int J Cardiol, 106:152-156. [DOI] [PubMed] [Google Scholar]
- [20].Dan QH,Huang YW,Peng ZP (1995). Polymorphsims in the APO A-I/C-III/A-IV gene and apolipoprotein C3 gene in patients with coronary artery disease. National Medical Journal of China, 75:584-586.8697070 [Google Scholar]
- [21].Ding Y,Zhu MA,Wang ZX,Zhu J,Feng JB,Li DS (2012). Associations of polymorphisms in the apolipoprotein APOA1-C3-A5 gene cluster with acute coronary syndrome. J Biomed Biotechnol, 2012:509420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Han TL (2011). A5/C3 Gene cluser and the correlation to the patients with coronary artery disease in ShanDong coastal areas. Shandong University, 1-50 [Google Scholar]
- [23].Izar MC,Fonseca FA,Ihara SS, et al. (2003). Risk Factors, biochemical markers, and genetic polymorphisms in early coronary artery disease. Arq Bras Cardiol, 80:379-395. [DOI] [PubMed] [Google Scholar]
- [24].Kee F,Amouyel P,Fumeron F, et al. (1999). Lack of association between genetic variations of apo A-I-C-III-A-IV gene cluster and myocardial infarction in a sample of European male: ECTIM study. Atherosclerosis, 145:187-195. [DOI] [PubMed] [Google Scholar]
- [25].Li XD,Sun ZH,Yang ZY, et al. (2006). APOAl/C3 gene cluster polymorphisms and risk of coronary artery disease. Chinese Journal of Gerontology, 26:745-747. [Google Scholar]
- [26].Liu S,Song Y,Hu FB, et al. (2004). A prospective study of the APOA1 XmnI and APOC3 SstI polymorphisms in the APOA1/C3/A4 gene cluster and risk of incident myocardial infarction in men. Atherosclerosis, 177:119-126. [DOI] [PubMed] [Google Scholar]
- [27].Liu HK (2005). Study on the association of single nucleotide polymorphisms in apolipoprotein APOAl/C3/A4/A5 gene cluster with coronary heart disease and type II diabetes. Sichuan University, 1-116 [Google Scholar]
- [28].Liu HK,Li FX,Zhang SZ, et al. (2005). Association of Sst I Polymorphism in Apolipoprotein C3 Gene with Hypertriglyceridaemia in Coronary Atherosclerotic Heart Disease and Type II Diabetes Mellitus in Chinese Population. ACTA GENETICA SINICA, 32:11-18. [PubMed] [Google Scholar]
- [29].Martinelli N,Trabetti E,Bassi A, et al. (2007). The -1131 T>C and S19W APOA5 gene polymorphisms are associated with high levels of triglycerides and apolipoprotein C-III, but not with coronary artery disease: an angiographic study. Atherosclerosis, 191:409-417. [DOI] [PubMed] [Google Scholar]
- [30].Muendlein A,Saely CH,Marte T, et al. (2008). Synergistic effects of the apolipoprotein E epsilon3/epsilon2/epsilon4, the cholesteryl ester transfer protein TaqIB, and the apolipoprotein C3 -482 C>T polymorphisms on their association with coronary artery disease. Atherosclerosis, 199:179-186. [DOI] [PubMed] [Google Scholar]
- [31].Olivieri O,Stranieri C,Bassi A, et al. (2002). ApoC-III gene polymorphisms and risk of coronary artery disease. J Lipid Res, 43:1450-1457. [DOI] [PubMed] [Google Scholar]
- [32].Ordovas JM,Civeira F,Genest J Jr, et al. (1991). Restriction fragment length polymorphisms of the apolipoprotein A-I, C-III, A-IV gene locus. Relationships with lipids, apolipoproteins, and premature coronary artery disease. Atherosclerosis, 87:75-86. [DOI] [PubMed] [Google Scholar]
- [33].Paulweber B,Friedl W,Krempler F,Humphries SE,Sandhofer F (1988). Genetic variation in the apolipoprotein AI-CIII-AIV gene cluster and coronary heart disease. Atherosclerosis, 73:125-133. [DOI] [PubMed] [Google Scholar]
- [34].Rigoli L,Raimondo G,Di Benedetto A, et al. (1995). Apolipoprotein AI-CIII-AIV genetic polymorphisms and coronary heart disease in type 2 diabetes mellitus. Acta Diabetol, 32:251-256. [DOI] [PubMed] [Google Scholar]
- [35].Sediri Y,Kallel A,Feki M, et al. (2011). Association of a DNA polymorphism of the apolipoprotein AI-CIII-AIV gene cluster with myocardial infarction in a Tunisian population. Eur J Intern Med, 22:407-411. [DOI] [PubMed] [Google Scholar]
- [36].Abd-El-Aziz TA,Mohamed RH,El-Shal AS (2013). Synergistic effect between lipoprotein lipase and apolipoprotein C3 genes in determining the severity of coronary artery disease. J Cardiovasc Transl Res, 6:430-435. [DOI] [PubMed] [Google Scholar]
- [37].Abd El-Aziz TA,Mohamed RH,Hashem RM (2011). Association of lipoprotein lipase and apolipoprotein C-III genes polymorphism with acute myocardial infarction in diabetic patients. Mol Cell Biochem, 354:141-150. [DOI] [PubMed] [Google Scholar]
- [38].Tobin MD,Braund PS,Burton PR,Thompson JR,Steeds R,Channer K,Cheng S,Lindpaintner K,Samani NJ (2004). Genotypes and haplotypes predisposing to myocardial infarction: a multilocus case-control study. Eur Heart J, 25:459-467. [DOI] [PubMed] [Google Scholar]
- [39].Weng GL,Han QQ,Chen HZ, et al. (2000). Relationship of Apolipoprotein AI and CⅢ Loci Gene Polymorphism and Coronary Artery Disease. Journal of Fujian Medical University, 34:96-99. [Google Scholar]
- [40].Wu JH,Kao JT,Wen MS,Lo SK (2000). DNA polymorphisms at the apolipoprotein A1-CIII loci in Taiwanese: correlation of plasma APOCIII with triglyceride level and body mass index. J Formos Med Assoc, 99:367-374. [PubMed] [Google Scholar]
- [41].Yang QR,Ma GS,Chen Z, et al. (2008). The relationship between polymorphism of Apolipoprotein C-III gene and premature coronary heart disease. Practical Journal of Cardic Cerebral Pneumal and Vascular Disease, 16:1-4. [Google Scholar]
- [42].Yi SX (2006). A study on the genes single nucleotide polymorphism of apolipoprotein A5 and C3 in patients with coronary artery disease. Qingdao University, 1-52 [Google Scholar]
- [43].Yu J,Huang J,Liang Y, et al. (2011). Lack of association between apolipoprotein C3 gene polymorphisms and risk of coronary heart disease in a Han population in East China. Lipids Health Dis, 10:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Song C,Chang Z,Magnusson PK,Ingelsson E,Pedersen NL (2014). Genetic factors may play a prominent role in the development of coronary heart disease dependent on important environmental factors. J Intern Med, 275(6):631-639. [DOI] [PMC free article] [PubMed] [Google Scholar]