Abstract
Poultry is known to be a major reservoir of Salmonella. The use of lactic acid bacteria has become one of successful strategies to control Salmonella in poultry. The purpose of this study was to select lactic acid bacteria strains by their in vitro immunomodulatory properties for potential use as probiotics against Salmonella infection in broiler chicks. Among 101 isolated lactic acid bacteria strains, 13 strains effectively survived under acidic (pH 2.5) and bile salt (ranging from 0.1% to 1.0%) conditions, effectively inhibited growth of 6 pathogens, and adhered to Caco-2 cells. However, their in vitro immunomodulatory activities differed significantly. Finally, three strains with higher in vitro immunomodulatory properties (Lactobacillus plantarum PZ01, Lactobacillus salivarius JM32 and Pediococcus acidilactici JH231) and three strains with lower in vitro immunomodulatory activities (Enterococcus faecium JS11, Lactobacillus salivarius JK22 and Lactobacillus salivarius JM2A1) were compared for their inhibitory effects on Salmonella adhesion and invasion to Caco-2 cells in vitro and their antimicrobial effects in vivo. The former three strains inhibited Salmonella adhesion and invasion to Caco-2 cells in vitro, reduced the number of Salmonella in intestinal content, spleen and liver, reduced the levels of lipopolysaccharide-induced TNF-α factor (LITAF), IL-1β, IL-6 and IL-12 in serum and increased the level of IL-10 in serum during a challenge study in vivo more efficiently than the latter three strains. These results suggest that in vitro immunomodulatory activities could be used as additional parameters to select more effective probiotics as feed supplements for poultry.
Introduction
Salmonella is one of the major causes of food-borne illnesses in humans. Poultry is known to be a major reservoir of Salmonella. Vaccination and bio-security have been adopted to control Salmonella in poultry production. In addition, lactic acid bacteria (LAB) based probiotics have been used to control Salmonella in poultry [1]. LAB probiotics mainly consist of Lactobacillus, Pediococcus and Enterococcus. These bacteria can competitively exclude pathogens by adherence to the host intestinal epithelium [2], enhance immune functions and improve the intestinal barrier of hosts [3,4].
Probiotics have usually been selected using conventional selection parameters including tolerance to acids and bile salt, antimicrobial activities, adhesion to epithelial cells and inhibition of pathogen adhesion to epithelial cells [5]. Some probiotics screened by conventional indicators have been shown to be beneficial to poultry health. For example, a single dose of Lactobacillus salivarius reduced the rate of Salmonella infection in 3-day old chicks [6]. Oral administration of 106 or 108 cfu of Lactobacillus-based probiotic culture significantly reduced Salmonella Enteritidis recovered from cecal tonsil of neonatal chicks [7]. However, other probiotics selected by conventional indicators are not effective. For example, Yamawaki et al. [8] showed that L. acidophilus, L. fermentum and L. salivarius did not decrease Salmonella Enteritidis colonization of chick ceca. Also, pre-treatment with Lactobacillus acidophilus, Bifidobacterium bifidum and Streptococcus faecalis did not change IL-6 and IL-10 gene expression in cecal tonsils of chicks [9]. The inconsistent results demonstrate the necessity of additional parameters to screen LAB for probiotics.
In vitro immune-modulating effects of LAB have received increasing attention as potential selection parameters for probiotics recently. The ability of LAB to induce IL-12 production was used to examine the immune-enhancing activity of LAB [10]. In addition, Tsai et al. [2] evaluated the ability of 12 LAB strains to stimulate production of TNF-α by mouse RAW 264.7 macrophage cells before they used LAB in a Salmonella challenge study in mice. Similarly, Chen et al. [4] evaluated production of TNF-α by the same mouse RAW 264.7 macrophage cells for selection of LAB to be used in a Salmonella challenge study in day-old chicks. Chen et al. [3] further included production of IL-12 protein by mouse RAW 264.7 macrophage cells as a parameter to select LAB for a Salmonella challenge study in mice. Gene expression of several cytokines by real-time quantitative PCR was significantly increased when mononuclear cells isolated from spleens of chicks were co-cultured with probiotic LAB for poultry [11]. However, measurement of cytokine production in chicks has been hampered by lack of commercial antibodies. Considering the species difference, we hypothesized that it would be better to use tissues or cells from chickens, instead of mouse RAW 264.7 macrophage cells, to select LAB as probiotics in chicks. TNF-α has not been found in the chicken genome. Instead, lipopolysaccharide-induced TNF-α factor (LITAF) has been cloned (Hong et al., 2006) and is one of the most important monitoring indexes for evaluating inflammatory response. With preparation of specific antibodies against LITAF and IL-12 in chicks in our laboratory, therefore, the objective of this study was to test the hypothesis whether expression levels of LITAF and IL-12 at the protein levels in spleen mononuclear cells in vitro could be used as additional selection parameters to select LAB as candidate probiotics for chickens.
Materials and Methods
Ethics statement
The study was approved by Institutional Animal Care and Use Committee of Northwest A&F University (Permit Number: NWAFAC1019). All chicks were euthanized via cervical dislocation, and all efforts were made to minimize suffering.
Bacterial isolates, culture media, and growth conditions
Lactic acid bacteria were isolated from intestinal contents of healthy free-range laying hens, according to the method described by Brisbin et al. [12]. Briefly, laying hens were purchased from Yangling, Shaanxi, China, and euthanized via cervical dislocation. Two hundred fifty mg of intestinal content from duodenum, jejunum, ileum or cecum were diluted with 10 ml phosphate-buffered saline (PBS, pH 7.4) and plated onto DeMan, Rogosa, and Sharpe (MRS) plates (Becton Dickinson, Mississauga, ON, Canada) for selecting lactic acid bacteria, [13]. The plates were grown at 37°C under anaerobic conditions (85% N2, 10% CO2, and 5% H2) for 48 h, and individual colonies were selected and inoculated into MRS broth, cultured at 37°C under anaerobic conditions for 18 h and subcultured twice. The V3 region of the 16S rRNA gene was amplified by PCR using the specific primers and sequenced as described in a previous study [13] and sequence comparisons were performed using the Basic Local Alignment Search Tool (BLAST) program (http://www.ncbi.nlm.nih.gov/BLAST/).
Pathogenic bacteria used in this study to measure antibacterial activities of probiotics included Staphylococcus aureus ATCC 29213, Escherichia coli K88, 25922 and 1569, Salmonella Enteritidis ATCC 13076 and Salmonella Typhimurium ATCC 14082. To culture these bacterial strains, one colony of each strain was inoculated into 5 ml tryptic soy broth and incubated with shaking at 37°C for 12 h.
Culture of cell lines
Caco-2 cells (ATCC HTB-37), a human colon adenocarcinoma cell line, were maintained in Dulbecco’s modified Eagle’s medium (DMEM)/F12 media (Gibco, USA) supplemented with 10% (v/v) inactivated (30 min, 56°C) fetal bovine serum, 20 U/ml penicillin and 100 μg/ml streptomycin. Cells were cultured at 37°C in a 5% CO2/95% air atmosphere using a humidified CO2 incubator. Cells were used at post-confluence after 21 days of culture. For adhesion assays as well as inhibition of intestinal cell adhesion and invasion by pathogens, monolayers of Caco-2 cells were prepared in 24-well tissue culture plates (Costar 3524, Corning Inc. NY, USA). Cells were seeded at a concentration of 5 × 104 cells/ml in the DMEM-F12 medium without penicillin and streptomycin.
Preparation of spleen mononuclear cells
Spleens were harvested sterile from four adult Arbor Acres chickens, which were obtained from Yangling, Shaanxi, China. After euthanizing via cervical dislocation, the spleens were rinsed in 1 × Hanks’ balanced salt solution (HBSS) and then minced with sterile scalpels. The tissue was further disrupted with the flat end of a 10-ml syringe plunger and filtered through a 40-μm nylon cell strainer to obtain a single-cell suspension. The suspension was then overlaid onto a Histopaque-1077 (Sigma, Oakville, ON, Canada) density gradient and centrifuged at 400 × g for 30 min. Mononuclear cells at the interface were collected and washed twice in 1 × HBSS and then suspended in RPMI 1640 (RPMI containing 10% fetal bovine serum, 2% chick serum, 0.146 g L-glutamine, 1.6 mM 2-mercaptoethanol). Cells were counted by the trypan blue dye exclusion assay before being resuspended in RPMI 1640 [12].
Resistance of LAB to acid and bile and antimicrobial activities of LAB
To evaluate acid resistance among the LAB strains, methods described by Tsai et al. [2] were used. Effects of bile salts on the growth of LAB cells were studied by a method modified from that of Osmanagaoglu et al. [14]. Briefly, for determining the acid tolerance, 150 μl of each culture containing about 108−109 cfu/ml of LAB suspension was added to 4.85 ml MRS that had been adjusted to pH 2.5 by 0.1 N HCl. To test the bile salt resistance, the same inoculum was added to normal MRS supplemented with 0.1%, 0.3%, 0.5% or 1.0% bile salts (Sigma, Saint Louis, Missouri, USA). Each mixture was incubated at 37°C for 3 h. After incubation, viable bacterial counts were determined by plating serial dilutions (with PBS, pH 7.4) on the MRS agar under anaerobic conditions at 37°C for 48 h. These assays were performed in triplicate for each of 3 independent experiments.
Antibacterial activities of LAB strains were studied using an agar diffusion test. Strains of LAB were grown overnight (20 h) in MRS broth at 37°C. Pathogenic bacterial strains were grown in the Luria–Bertani (LB) agar. Then, wells were hollowed out of the LB plates using an Oxford cup and 200 μl of the spent culture suspension (LAB-SCS) was added into each well. The culture was incubated at 37°C for 14 h before determination of antimicrobial activities. LAB strains with inhibition zones <11, 11–16, 17–22 and >23 mm were classified as strains of no (-), mild (+), strong (++), or very strong (+++) inhibition, respectively [2].
Adhesion of LAB to intestinal cell lines
The adhesion study of LAB strains was performed by following the procedures described by Bianchi et al. [15] and Tsai et al. [2]. LAB strains were stained with fluorescein isothiocyanate (FITC, Sigma, USA) and kept in darkness at 37°C for 2 h, then washed three times with antibiotic-free PBS solution (pH 7.4) to remove the unlabeled FITC and resuspended with PBS. One hundred μl of each suspension (5×108 cfu/ml) was transferred to the 24-well multidish containing the Caco-2 cells and incubated for 2 h. After incubation, non-adherent bacteria were removed by washing three times with PBS. Two hundred μl of trypsin/EDTA-Na2 were used to digest the cells and adherent bacteria for 5 min before 600 μl of PBS were added into the wells. After mixing, a 200 μl mixture containing cells and bacteria was added to 96 well plates. This fraction contained lysed bacteria attached to Caco-2 cells or within Caco-2 cells and it was reported as the adherent fraction. The fluorescence was read on a Perkin-Elmer LS55 Spectrophotometer (λex = 492 nm; λem = 517 nm). Six independent experiments were performed for each microbial strain on Caco-2 cells. The results of adhesion assay were expressed as the adhesion percentage of adherent bacteria over added bacteria per well.
Production of polyclonal antibodies against LITAF, IL-1β, IL-6, IL-10 and IL-12
Polyclonal antibodies against LITAF, IL-1β, IL-6, IL-10 and IL-12 (antigens) were obtained by immunizing female New Zealand white rabbits (2–2.5 kg body weight) with LITAF, IL-1β, IL-6, IL-10 and IL-12 as previously described elsewhere [16].
In order to prepare antigens for immunization, plasmids pET32a-LITAF, pET32a-IL-1β, pET32a-IL-6, pET32a-IL-10 and pET21a-IL-12 were prepared. Polymerase chain reactions (PCR) were performed on total cDNA of chick using specific primers:
LITAF-F 5’-GGGGTACCATGTCTGCTCCTAGTGGCTTT-3’ sense primer and LITAF-R 5’-CCAAGCTTCTACGCTCCTGACTCATAGCAGAG-3’ antisense primer (GenBank: AY765397); IL-1β-F 5’-GGGGTACCATGGCGTTCGTTCCCG-3’ sense primer and IL-1β-R 5’-CCAAGCTTTCAGCGCCCACTTAGCTT-3’ antisense primer (GenBank: DQ393267); IL-6-F 5’-GGGGTACCGGAGAGGTTGGGCTGGAG-3’ sense primer and IL-6-R 5’-CCAAGCTTTCAGGCACTGAAACTCCTGG-3’ antisense primer (GenBank: HM179640); IL-10-F 5’-GGGGTACCTGCTTGGAGCCCACCT-3’ sense primer and IL-10-R 5’-CCAAGCTTTCACTTCCTCCTCCTCATCA-3’ antisense primer (GenBank: EF554720). IL-12-F 5’-GGGGATCCAAAGAGCCAAGCAAGACG-3’ sense primer and IL-12-R 5’-CCAAGCTTGAAAGTCAAAGGGAAGTAGGA-3’ antisense primer (GenBank: DQ202328). The recombinant plasmids were subsequently introduced into BL21 (DE3) or Transetta (DE3). After induction by IPTG, the target proteins were purified. The rabbits were initially immunized with two intramuscular injections of LITAF, IL-1β, IL-6, IL-10 or IL-12 (300 μg antigens) dissolved in 1 ml ice-cold (4°C) PBS and complete Freund’s adjuvant in a ratio of 1:1. Immunization was repeated three times at 2 week intervals using the same amounts of antigens in the incomplete Freund’s adjuvant.
Assays for LITAF and IL-12 production by spleen mononuclear cells after stimulation with LAB strains
Spleen cells were cultured in triplicate, at a density of 1×106 cells/ml of RPMI-1640 medium without penicillin or streptomycin, in 24-well tissue culture plates. LAB cells were centrifuged at 8,000 g for 5 min and the pellet was resuspended in RPMI-1640 medium containing spleen cells to a final concentration from 108 to 109 cfu/ml. Lipopolysaccharide (10 μg/ml) from E. coli O26:B6 (Sigma, USA) was used as a positive control. After 24 h and 48 h, LITAF and IL-12 produced in these culture supernatants were analyzed.
Cytokines were measured using an enzyme-linked immunosorbent assay (ELISA) method. Ninety-six-well Immuno-Maxisorp plates (Nunc) were coated with polyclonal antibodies for LITAF and IL-12 (1: 1,000) in coating buffer (0.05 M Carbonate Buffer, pH 9.6) overnight at 4°C. Plates were blocked and washed. Culture medium was added to the plates and they were incubated for 2 h at room temperature. Plates were then washed again, and goat-anti-rabbit secondary antibody conjugated horseradish peroxidase (1: 20,000) was added, followed by incubation for 1 h at room temperature and another wash. The chromogenic reactions were developed with the 3, 3’, 5, 5’-tetramethylbenzidine substrate at 37°C for 30 min. The reactions were terminated with 50 μl of 2N H2SO4 and the absorbance at A450 nm was measured. Equivalent levels of LITAF and IL-12 were calculated by comparison with reference curves generated using LITAF and IL-12 standards. The results were expressed as the concentration of the cytokines in the culture medium (ng/ml).
Inhibition of Salmonella adhesion and invasion of Caco-2 cells by LAB strains
Each LAB strain (5×107 cfu per well) was added to Caco-2 cells in a fresh tissue culture medium without Penicillin-Streptomycin and incubated at 37°C for 2 h in a 5% CO2/95% air atmosphere incubator before 2 h of incubation with 100 μl of FITC labeled Salmonella (5×108 cfu/ml). Salmonella were labeled with Fluorescein isothiocyanate (FITC, Sigma, USA) in darkness at 37°C for 2 h. After incubation with FITC labeled Salmonella, non-adherent bacteria were washed away three times with PBS.
To determine the relative adhesion of Salmonella to Caco-2 cells, one 24-well multidish was added with 200 μl trypsin to digest the cells and adherent bacteria for 5 min before adding 600 μl of PBS. After mixing, 200 μl of mixture containing cells and bacteria were added to each well of 96-well plates to measure the strength of fluorescence. To determine the relative invasion of Salmonella to Caco-2 cells, another 24-well multidish was lysed with 200 μl of 1% Triton X-100 for 10 min before trypsin digestion. Six independent experiments were performed for each LAB strain.
The relative adhesion or invasion of S. Enteritidis ATCC13076 to Caco-2 cells was expressed as a percentage using the following formula: relative of adhesion or invasion = 100 x A1/A2, where A1 and A2 were the percentages of adhesion or invasion by S. Enteritidis ATCC13076 in the presence and absence of LAB strains, respectively.
Challenge study
The challenge study was performed by following the procedures described by Chen et al. [4] with few modifications. One hundred ninety-two hatched 1-d-old healthy Arbor Acres male broilers were obtained from a commercial hatchery at Xianyang, Shaanxi, China. Chicks were reared in two layer metal cages, with an average stocking density of 16.7 birds per square meter, and the brooding temperature was 31 to 33°C throughout the experiment. Chicks had free access to water and a commercial starter diet without supplementation of antibiotics. The chicks were randomly divided into 8 groups (6 repeats/group, 4 chicks/repeat): In the negative control group (group 1), the chicks were only given sterile PBS buffer (pH 7.4) (0.2 ml/chick) via the intragastric route once every day throughout 4 experimental days. In the positive control group (group 2), chicks were challenged with Salmonella on d4 (0.2 ml/chick, 108 cfu per 0.2 ml) and were given sterile PBS buffer (pH 7.4) (0.2 ml/chick) during d1-d3. For treatment groups 3–5, 3 LAB strains with higher in vitro immunomodulatory properties (L. plantarum PZ01, L. salivarius JM32 and P. acidilactici JH231) were used, while 3 LAB strains with lower in vitro immunomodulatory properties (E. faecium JS11, L. salivarius JK22 and L. salivarius JM2A1) were used for treatment groups 6–8. For treatment groups 3–8, chicks were gavaged with LAB (0.2 ml/chick, 109 cfu per 0.2 ml) once every day for 3 consecutive days, then challenged with Salmonella Enteritidis ATCC 13076 on d4 (0.2 ml/chick, 108 cfu per 0.2 ml).
Broiler chickens were euthanized via cervical dislocation. Samples for blood, spleens, livers and cecum contents of six randomly selected chicks in each group were taken at 1, 3 and 5 days post Salmonella challenge. Salmonella Enteritidis ATCC 13076 was selected for the challenge study, due to the invasive characteristic previously described by Dawoud et al. [17].
Assessment of LITAF, IL-1β, IL-6, IL-10 and IL-12 in chick serum and enumeration of the Salmonella cells invaded in chick liver and spleen, and colonized in the cecum
For all groups, blood samples were collected from the carotid artery of 6 chicks at 1, 3 and 5 days post Salmonella challenge. Blood serum was obtained after incubation for 1 h at room temperature followed by 2,000 g for 10 min. Sera were stored at -80°C until tested. The cytokines LITAF, IL-1β, IL-6, IL-10 and IL-12 were measured by the ELISA method with preparation of specific antibodies of LITAF, IL-1β, IL-6, IL-10 and IL-12 in our laboratory. Samples for spleens, livers and cecum contents of six randomly selected chicks in each group were taken at 1, 3 and 5 days post Salmonella challenge. Salmonella that had invaded the spleens and livers and colonized in the cecum of chicks were enumerated by cfu method [4]. Spleens and livers were homogenized and without serial dilution, and cecum contents were diluted with PBS (pH 7.4). All the samples were incubated on selected Brilliant Green agar (Difco) containing 50 μg/ml novobiocin (Sigma, USA) for 24 h at 37°C before counting the number of Salmonella.
Statistical analysis
All results were expressed as mean ± SD from at least three independent experiments. Statistical analysis was performed using the SPSS for Windows version 17.0 (Chicago, IL, USA). Data were subjected to one-way ANOVA and, where appropriate, the Scheffe test was used for comparison of means. Differences were considered to be statistically significant when the P value was <0.05.
Results
Resistance of LAB to acid and bile, antimicrobial activity and in vitro adhesion assay to Caco-2 cells
Among the 101 isolated LAB strains, 13 strains were better than the remaining 88 LAB strains to survive following exposure to acid (pH 2.5) or bile salts for 3 h (Table 1). These 13 bacteria had a survival rate of over 77% after 3 h at pH 2.5. With the presence of 1.0% of bile salts, the survival rates were more than 60.0% for these 13 strains. Bile salts at the 0.1% level had no significant effect on bacterial survival. As shown in Table 2, the above-mentioned 13 LAB strains were able to inhibit the growth of the pathogenic bacteria. All 13 LAB strains had strong inhibition against Escherichia coli except JM31, JK22, JK231 and JS11 which exhibited mild inhibition against ATCC K88. Similarly, all 13 LAB strains had strong inhibition against Salmonella and Staphylococcus aureus except JM 241 and JH231 which exhibited mild inhibition against Staphylococcus aureus and Salmonella Typhimurium.
Table 1. Resistance to acid and bile of the 13 LAB strains.
| Strainsa | Resistance to acid (% growth) | Resistance to bile (% growth) | |||
|---|---|---|---|---|---|
| pH 2.5 | 0.1% | 0.3% | 0.5% | 1.0% | |
| P. pentosaceus JS233 | 78.9 ± 4.5 * | 98.5 ± 3.5 | 79.2 ± 3.7 * | 77.6 ± 4.6 * | 63.0 ± 5.8 * |
| L. salivarius JM41 | 80.4 ± 5.0 * | 100 ± 3.5 | 82.1 ± 3.8 * | 78.4 ± 5.1 * | 64.7 ± 5.2 * |
| L. plantarum PZ01 | 79.6 ± 1.9 * | 97.0 ± 3.3 | 80.5 ± 3.2 * | 77.5 ± 4.3 * | 64.0 ± 4.3 * |
| L. salivarius JK21V | 79.1 ± 4.8 * | 100 ± 3.2 | 80.2 ± 3.2 * | 78.2 ± 4.1 * | 62.9 ± 3.9 * |
| P. acidilactici JM241 | 78.6 ± 1.6 * | 94.2 ± 3.7 | 80.8 ± 3.7 * | 74.9 ± 4.9 * | 62.7 ± 5.3 * |
| L. salivarius JM31 | 78.9 ± 3.2 * | 93.1 ± 3.4 | 80.4 ± 3.5 * | 73.5 ± 7.2 * | 64.3 ± 7.7 * |
| L. salivarius JS2A | 81.7 ± 5.2 * | 99.4 ± 3.2 | 81.8 ± 3.8 * | 72.4 ± 6.5 * | 60.0 ± 6.8 * |
| L. salivarius JM14 | 77.9 ± 1.4 * | 97.9 ± 3.5 | 81.3 ± 3.5 * | 70.3 ± 3.3 * | 62.7 ± 4.9 * |
| L. salivarius JK22 | 81.4 ± 1.8 * | 98.8 ± 3.2 | 80.7 ± 3.1 * | 70.0 ± 4.2 * | 61.4 ± 5.6 * |
| L. salivarius JM2A1 | 77.7 ± 2.8 * | 98.2 ± 2.6 | 80.5 ± 3.2 * | 78.7 ± 3.5 * | 62.2 ± 3.1 * |
| L. salivarius JM32 | 80.2 ± 1.6 * | 99.2 ± 4.1 | 78.8 ± 4.1 * | 77.9 ± 5.1 * | 64.0 ± 4.8 * |
| P. acidilactici JH231 | 81.2 ± 1.8 * | 93.7 ± 2.8 | 80.0 ± 3.1 * | 73.7 ± 6.2 * | 63.4 ± 5.2 * |
| E. faecium JS11 | 82.3 ± 2.8 * | 93.4 ± 3.5 | 81.4 ± 4.5 * | 74.8 ± 3.0 * | 65.3 ± 6.5 * |
a Values are means ± standard deviation from three experiments. The control was 100%.
* Considered significantly different from the control (P<0.05).
Table 2. Antimicrobial activities of lactic acid bacteria against the growth of pathogenic bacteria in vitro.
| LAB strains | Escherichia coli (EC) | S. aureus ATCC 29213 | S. Enteritidis ATCC 13076 | S. Typhimurium ATCC 14082 | ||
|---|---|---|---|---|---|---|
| ATCC K88 | ATCC 1569 | ATCC 25922 | ||||
| P. pentosaceus JS233 | ++* | ++ | ++ | ++ | ++ | ++ |
| L. salivarius JM41 | ++ | ++ | ++ | ++ | ++ | ++ |
| L. plantarum PZ01 | ++ | ++ | ++ | ++ | ++ | ++ |
| L. salivarius JK21V | ++ | ++ | ++ | ++ | ++ | ++ |
| P. acidilactici JM241 | ++ | ++ | ++ | + | ++ | + |
| L. salivarius JM31 | + | ++ | ++ | ++ | ++ | ++ |
| L. salivarius JS2A | ++ | ++ | ++ | ++ | ++ | ++ |
| L. salivarius JM14 | ++ | ++ | ++ | ++ | ++ | ++ |
| L. salivarius JK22 | + | ++ | ++ | ++ | ++ | ++ |
| L. salivarius JM2A1 | ++ | ++ | ++ | ++ | ++ | ++ |
| L. salivarius JM32 | ++ | ++ | ++ | ++ | ++ | ++ |
| P. acidilactici JH231 | + | ++ | ++ | + | ++ | + |
| E. faecium JS11 | + | ++ | ++ | ++ | ++ | ++ |
* LAB strains with inhibition zones 11–16 mm and 17–22 mm were classified as strains of mild + and strong ++ inhibition, respectively.
All 13 tested strains were able to adhere to Caco-2 cells with different adhesion activities (Table 3). JS233, JM41, PZ01 and JK21V showed the strongest adherence to Caco-2 cells (from 19.67 to 10.94%), while JS11 was the least effective one to adhere to Caco-2 cells (2.24 ± 0.24%).
Table 3. Adhesion of lactic acid bacteria to Caco-2 cells.
| Strains | Adhesion (%)* (mean ± S.D) |
|---|---|
| P. pentosaceus JS233 | 19.67 ± 0.48a |
| L. salivarius JM41 | 15.47 ± 0.22b |
| L. plantarum PZ01 | 11.23 ± 0.70c |
| L. salivarius JK21V | 10.94 ± 0.79c |
| P. acidilactici JM241 | 6.49 ± 0.58d |
| L. salivarius JM31 | 6.35 ± 0.24d |
| L. salivarius JS2A | 5.37 ± 0.37e |
| L. salivarius JM14 | 5.30 ± 0.63e |
| L. salivarius JK22 | 5.26 ± 0.64e |
| L. salivarius JM2A1 | 5.24 ± 0.46e |
| L. salivarius JM32 | 4.82 ± 0.19e |
| P. acidilactici JH231 | 4.45 ± 0.27e |
| E. faecium JS11 | 2.24 ± 0.24f |
* Fluorescence values were the ratio between adherent bacteria and added bacteria.
a-f Different superscripts indicate significant differences among different strains (P<0.05).
LITAF and IL-12 production by spleen mononuclear cells in response to recall antigen stimulation
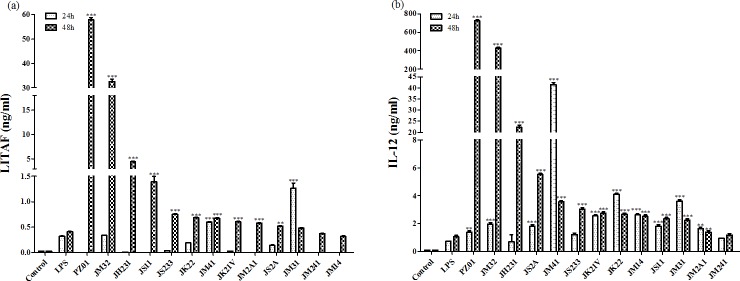
Except JM31, JM241 and JM14, the remaining strains enhanced LITAF production more than the positive control at 48 h post-treatment (P<0.05), especially PZ01 (57.81 ng/ml), JM32 (32.64 ng/ml) and JH231 (4.44 ng/ml). Only JM31 and JM41 produced more LITAF than the positive control group after co-incubation with spleen mononuclear cells for 24 h (P<0.05) (Fig 1A). At 24 h post-treatment, Pediococcus spp. (JH231, JS233 and JM241) did not increase IL-12 production, while the other LAB strains significantly induced IL-12 production in comparison with the LPS positive control group (P<0.05). All 13 strains except JM241 enhanced IL-12 production more than LPS after incubation for 48 h (P<0.05). The highest producers of IL-12 were PZ01 (724.85 ng/ml), JM32 (427.63 ng/ml) and JH231 (22.25 ng/ml) (Fig 1B). These same 3 strains were also the highest producers of LITAF. Thus, they were retained for further assays. Three other strains (JS11, JK22 and JM2A1) were randomly selected to represent lower producers of LITAF and IL-12 for subsequent assays. The remaining 7 LAB strains were not studied further.
Fig 1. (A) LITAF, (B) IL-12 production by spleen mononuclear cells after incubation with different LAB strains.
Spleen cells (1×106 cells/ml) were cultured in a RPMI-1640 medium without penicillin or streptomycin. The RPMI-1640 medium alone was used as a negative control, while LPS (lipopolysaccharide, 10 μg/ml) was used as a positive control. LAB (108 to 109 cfu/ml) were cultured with spleen cells. After 24 h and 48 h of culture, LITAF and IL-12 produced in the culture supernatants were analyzed. Each value represents the mean value ± SD from three independent experiments. * indicates significant differences in comparison with the positive control (*: P<0.05).
Inhibition of pathogen adhesion and invasion to Caco-2 cells by LAB
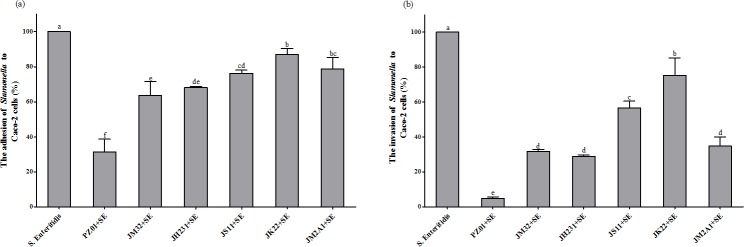
The competitive inhibition of adhesion and invasion of Salmonella Enteritidis to Caco-2 cells by 6 probiotic strains was shown in Fig 2A and 2B, respectively. Strain PZ01 displayed the strongest inhibition of Salmonella Enteritidis adhesion and invasion to Caco-2 cells (Fig 2). On average, 3 strains with higher in vitro immunomodulatory properties reduced adhesion by 45.6% and invasion by 78.3%, while 3 strains with lower in vitro immunomodulatory properties reduced adhesion only by 19.4% and invasion only by 44.4%.
Fig 2. Effects of LAB culture on the adhesion and invasion of Caco-2 cells by Salmonella Enteritidis.
The relative adhesion or invasion of S. Enteritidis ATCC13076 to Caco-2 cells was expressed as a percentage using the following formula: relative adhesion or invasion = 100 x A1/A2, where A1 and A2 were the percentages of adhesion or invasion by S. Enteritidis ATCC13076 in the presence and absence of LAB strains, respectively. Each value represents the mean value ± SD from six trials. Different letters above bars indicate significant differences among treatments within each sampling day (P<0.05).
Inhibitory effects of selected LAB strains on the invasion and colonization of Salmonella in vivo
In order to determine whether LAB strains with higher in vitro immunomodulatory properties had better inhibitory effects against Salmonella in vivo, a Salmonella challenge study was performed. As shown in Table 4, all 6 strains significantly decreased Salmonella Enteritidis in cecum content on sampling time points, compared with the positive control (P<0.05). On average, 3 strains with higher in vitro immunomodulatory properties reduced the bacterial counts by 88.37%, 94.93% and 96.63%, while 3 strains with lower in vitro immunomodulatory properties reduced the bacterial counts only by 61.52%, 73.12% and 76.24% on d1, d3 and d5 post-infection, respectively.
Table 4. Effects of oral administration of 6 different lactic acid bacteria on reduction of Salmonella cells recovered from the livers, spleens, and cecal content of chicks.
| Item | Spleen | Liver | Cecum content |
|---|---|---|---|
| cfu /organ | cfu /0.3 g | cfu (×104)/0.25 g | |
| 1 d Post-infection | |||
| Salmonella only | 100.67 ± 5.72a | 91.67 ± 8.16 | 237.28 ± 14.07a |
| PZ01 + Salmonella | ND | ND | 26.00 ± 4.75f |
| JM32 + Salmonella | ND | ND | 19.69 ± 1.53f |
| JH231 + Salmonella | ND | ND | 37.13 ± 3.89e |
| JS11 + Salmonella | ND | ND | 132.41 ± 10.82b |
| JK22 + Salmonella | 58.33 ± 4.08b | ND | 59.42 ± 7.17d |
| JM2A1 + Salmonella | ND | ND | 82.03 ± 9.21c |
| 3 d Post-infection | |||
| Salmonella only | 148.33 ± 17.80a | 207.67 ± 9.63a | 245.04 ± 13.49a |
| PZ01 + Salmonella | ND | ND | 12.63 ± 1.70cd |
| JM32 + Salmonella | ND | ND | 6.52 ± 1.16d |
| JH231 + Salmonella | ND | ND | 18.13 ± 3.34c |
| JS11 + Salmonella | 66.33 ± 8.16b | 58.67 ± 8.98c | 67.50 ± 5.00b |
| JK22 + Salmonella | 126.67 ± 10.80a | 158.33 ± 14.72b | 58.98 ± 8.49b |
| JM2A1 + Salmonella | 131.37 ± 16.33a | 171.67 ± 8.16b | 71.13 ± 10.96b |
| 5 d Post-infection | |||
| Salmonella only | 168.33 ± 22.73a | 198.33 ±10.80a | 311.84 ± 15.89a |
| PZ01 + Salmonella | ND | ND | 10.13 ± 5.63ef |
| JM32 + Salmonella | ND | ND | 3.87 ± 0.57f |
| JH231 + Salmonella | ND | ND | 17.50 ± 5.00e |
| JS11 + Salmonella | 144.67 ± 7.12a | 66.67 ± 4.08c | 98.31 ± 10.18b |
| JK22 + Salmonella | 153.33 ± 10.80a | 141.67 ± 10.80b | 52.76 ± 5.23d |
| JM2A1 + Salmonella | 146.67 ± 11.43a | 180.00 ± 14.14a | 71.25 ± 7.50c |
a-f Different superscripts indicate significant differences among different treatments in the same type of sample on each sampling day (n = 6) (P<0.05).
ND: not detectable.
Salmonella was detected in livers and spleens from d1 to d5 post-infection for the positive control group. Three strains with higher in vitro immunomodulatory properties prevented invasion of Salmonella into livers and spleens, with no viable Salmonella detected on d1, d3 and d5. Three strains with lower in vitro immunomodulatory properties reduced invasion of Salmonella in livers and spleens, especially for d1 with no viable Salmonella detected except for strain JK22 in the spleen. On d3, JS11 significantly reduced invasion of Salmonella in livers and spleens, compared to the positive control. Other 2 strains also significantly reduced invasion of Salmonella in livers but they had no effects in spleens. For d5, only JS11 and JM2A1 significantly reduced invasion of Salmonella in livers (P<0.05), while all three strains with lower in vitro immunomodulatory properties had not significantly reduced invasion of Salmonella in spleens (P>0.05).
Assay of the cytokines LITAF, IL-1β, IL-6, IL-10 and IL-12 in chick serum
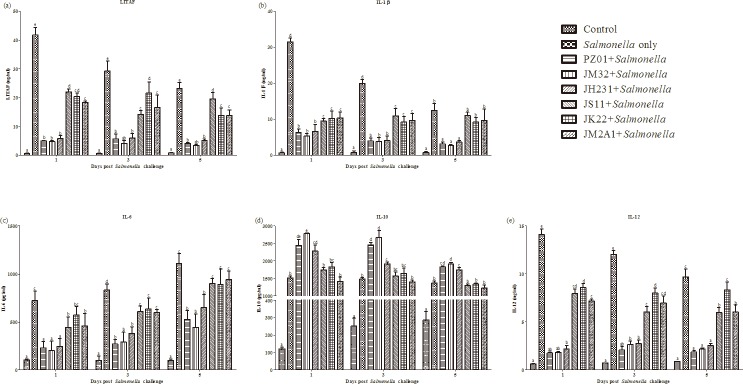
Blood samples were assayed for pro-inflammatory cytokines LITAF, IL-1β, IL-6, IL-12 and anti-inflammatory cytokine IL-10 (Fig 3). For the positive control group, the highest expression levels of LITAF, IL-1β and IL-12 were observed at 1 day post-infection and gradually decreased afterwards. The opposite was true for IL-6 with increasing concentrations after the infection. However, there was no change of IL-10 after infection.
Fig 3. Cytokine levels in sera of chicks gavaged with LAB strains and followed by Salmonella challenge.
The cytokines (A) LITAF, (B) IL-1β, (C) IL-6, (D) IL-10 and (E) IL-12 were measured by the ELISA methods with specific antibodies of LITAF, IL-1β, IL-6, IL-10 and IL-12 in chicks prepared in our laboratory. Each vertical bar represents the mean ± SD (n = 6). Different letters above bars indicate significant differences among treatments within each sampling day (P<0.05).
In comparison with the positive control, three strains with higher in vitro immunomodulatory properties (PZ01, JM32 and JH231) significantly reduced LITAF, IL-1β, IL-6 and IL-12 and increased IL-10 levels at all 3 time points. Three strains with lower in vitro immunomodulatory activities (JS11, JK22 and JM2A1) also significantly reduced levels of LITAF, IL-1β, IL-6 and IL-12 mainly on d1 and d3 but did not affect IL-10 levels in comparison with the positive control. The abilities of PZ01, JM32 and JH231 to reduce LITAF, IL-1β, IL-6 and IL-12 and to increase IL-10 were significantly greater than those JS11, JK22 and JM2A1.
Discussion
This study was the first to evaluate effects of LAB on LITAF and IL-12 expression by the spleen mononuclear cells of chickens at the protein level. The reason we chose spleen mononuclear cells was because lactic acid bacteria induced significantly more cytokines in spleen cells than in cecal tonsil cells of chickens [11]. LITAF and IL-12 levels detected in our study were generally higher than those from previous studies using mouse macrophages RAW 264.7 cells [2,3]. TNF-α production was significantly increased by lactic acid bacteria (LAB) strains after their co-culturing with RAW 264.7 cells [2]. A further study from the same research group indicated that viable and heat-killed LAB strains, either individually or in mixture, were able to induce the release of TNF-α and IL-12 from RAW 264.7 cells [3]. Considering the species specialization and higher sensitivities of spleen mononuclear cells from chickens than RAW 264.7 cells, it seems reasonable to use spleen mononuclear cells from chickens to evaluate in vitro immunoregulatory activities of lactic acid bacteria for poultry production.
In this study, we demonstrated that cytokines, LITAF, IL-1β, IL-6, IL-10 and IL-12, were involved in the immunity of Salmonella-infected chicks. Three strains with higher in vitro immunomodulatory properties (PZ01, JM32 and JH231) reduced LITAF, IL-1β, IL-6 and IL-12 and increased IL-10 more efficiently than three other strains with lower in vitro immunomodulatory activities (JS11, JK22 and JM2A1). TNF-α is a member of a group of cytokines that stimulate the acute phase reaction in mammal. Although TNF-α has not been found nor described in the chicken genome, LITAF, which is the regulator for TNF-α expression in mammal [18], has been shown to play an important role in the intestinal inflammatory response in chicken [19]. IL-12 is produced by inflammatory myeloid cells and influences the development of TH1 cell responses [20]. Similarly, IL-1β is also a major mediator of inflammation and is produced by monocytes, tissue macrophages, enterocytes and other cells [21]. These three cytokines (LITAF, IL-1β and IL-12) indicate an early inflammatory response [22]. The concentrations of LITAF, IL-1β and IL-12 at the protein level were the highest at d1 and gradually decreased afterwards in the positive control group. Similar changes have been reported at the mRNA level by Chen et al. [4]. IL-6 is a multifunctional cytokine. IL-6 gradually increased after the Salmonella challenge, similar to the previous findings at the mRNA level for chickens [4,23] and at the protein level for mice [3]. In comparison with the positive control, three strains with higher in vitro immunomodulatory activities (PZ01, JM32 and JH231) significantly increased IL-10 levels, while three strains with lower in vitro immunomodulatory activities (JS11, JK22 and JM2A1) did not affect IL-10 levels. In addition, this study showed that three strains with higher in vitro immunomodulatory properties (PZ01, JM32 and JH231) reduced the levels of Salmonella Enteritidis recovered from chick livers, spleens and cecal contents more efficiently than three strains with lower in vitro immunomodulatory activities (JS11, JK22 and JM2A1).
The selected six LAB strains for the in vivo study showed consistent tolerance to acid and bile salts in vitro, suggesting that these six LAB strains could survive the gastrointestinal tract and function effectively [24,25]. Moreover, these six LAB strains were able to inhibit the growth of the pathogenic bacteria. Antimicrobial activities of all six strains might be associated with acidic metabolites such as acetic acid, lactic acid [26] and organic acid [27], or bacteriocins [28,29] and proteinaceous substances [30]. Finally, these six LAB strains were biologically safe due to negative haemolytic activities (data not shown).
In conclusion, compared with strains E. faecium JS11, L. salivarius JK22 and L. salivarius JM2A1 with lower in vitro immunomodulatory properties, strains L. plantarum PZ01, L. salivarius JM32 and P. acidilactici JH231 with higher in vitro immunomodulatory activities were more effective to reduce Salmonella counts in cecal content and decease invasion of Salmonella into livers and spleens. These results suggest that in vitro immunomodulatory activities could be used as additional parameters to select more effective probiotics for poultry.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by funding from an innovation project of science and technology plan project of Shaanxi Province, China (2014KTCL02-21); the Ministry of Agriculture (No. 2013-S16); the thousand talent program to X.Z.; and Natural Science Foundation of China (No. 31402095) and Northwest A&F University PhD Research Start-up funds (No. 2012BSJJ094) to X.Y.
References
- 1.Tellez G, Pixley C, Wolfenden RE, Layton SL, Hargis BM (2012) Probiotics/direct fed microbials for Salmonella control in poultry. Food Res Int 45: 628–633. [Google Scholar]
- 2.Tsai CC, Liang HW, Yu B, Hsieh CC, Hwang CF, Chen MH, et al. (2011) The relative efficacy of different strain combinations of lactic acid bacteria in the reduction of populations of Salmonella enterica Typhimurium in the livers and spleens of mice. FEMS Immunol Med Microbiol 63: 44–53. 10.1111/j.1574-695X.2011.00826.x [DOI] [PubMed] [Google Scholar]
- 3.Chen CY, Tsen HY, Lin CL, Lin CK, Chuang LT, Chen CS, et al. (2013) Enhancement of the immune response against Salmonella infection of mice by heat-killed multispecies combinations of lactic acid bacteria. J Med Microbiol 62: 1657–1664. 10.1099/jmm.0.061010-0 [DOI] [PubMed] [Google Scholar]
- 4.Chen CY, Tsen HY, Lin CL, Yu B, Chen CS (2012) Oral administration of a combination of select lactic acid bacteria strains to reduce the Salmonella invasion and inflammation of broiler chicks. Poult Sci 91: 2139–2147. 10.3382/ps.2012-02237 [DOI] [PubMed] [Google Scholar]
- 5.FAO/WHO (2011) ICMR-DBT guidelines for evaluation of probiotics in food. Indian J Med Res 134: 22–25. [PMC free article] [PubMed] [Google Scholar]
- 6.Waewdee P, Sukon P, Chaveerach P, Surachon P, Soikum C (2012) Effect of a single dose of Lactobacillus salivarius on prevention of Salmonella enteritidis infection in young broilers. J Anim Vet Adv 11: 955–961. [Google Scholar]
- 7.Higgins SE, Higgins JP, Wolfenden AD, Henderson SN, Torres-Rodriguez A, Tellez G, et al. (2008) Evaluation of a Lactobacillus-based probiotic culture for the reduction of Salmonella enteritidis in neonatal broiler chicks. Poult Sci 87: 27–31. [DOI] [PubMed] [Google Scholar]
- 8.Yamawaki RA, Milbradt EL, Coppola MP, Rodrigues JC, Andreatti Filho RL, Padovani CR, et al. (2013) Effect of immersion and inoculation in ovo of Lactobacillus spp. in embryonated chicken eggs in the prevention of Salmonella Enteritidis after hatch. Poult Sci 92: 1560–1563. 10.3382/ps.2012-02936 [DOI] [PubMed] [Google Scholar]
- 9.Haghighi HR, Abdul-Careem MF, Dara RA, Chambers JR, Sharif S (2008) Cytokine gene expression in chicken cecal tonsils following treatment with probiotics and Salmonella infection. Vet Microbiol 126: 225–233. [DOI] [PubMed] [Google Scholar]
- 10.Hirose Y, Murosaki S, Fujiki T, Yamamoto Y, Yoshikai Y, Yamashita M (2010) Lipoteichoic acids on Lactobacillus plantarum cell surfaces correlate with induction of interleukin-12p40 production. Microbiol Immunol 54: 143–151. 10.1111/j.1348-0421.2009.00189.x [DOI] [PubMed] [Google Scholar]
- 11.Brisbin JT, Gong J, Parvizi P, Sharif S (2010) Effects of lactobacilli on cytokine expression by chicken spleen and cecal tonsil cells. Clin Vaccine Immunol 17: 1337–1343. 10.1128/CVI.00143-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brisbin JT, Gong J, Orouji S, Esufali J, Mallick AI, Parvizi P, et al. (2011) Oral treatment of chickens with Lactobacilli influences elicitation of immune responses. Clin Vaccine Immunol 18: 1447–1455. 10.1128/CVI.05100-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Madboly LA, Abdullah AK (2015) Potent antagonistic activity of Egyptian Lactobacillus plantarum against multiresistant and virulent food-associated pathogens. Front Microbiol 6: 347 10.3389/fmicb.2015.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osmanagaoglu O, Kiran F, Ataoglu H (2010) Evaluation of in vitro probiotic potential of Pediococcus pentosaceus OZF isolated from human breast milk. Probiotics & Antimicro Prot 2: 162–174. [DOI] [PubMed] [Google Scholar]
- 15.Bianchi MA, Del Rio D, Pellegrini N, Sansebastiano G, Neviani E, Brighenti F (2004) A fluorescence-based method for the detection of adhesive properties of lactic acid bacteria to Caco-2 cells. Lett Appl Microbiol 39: 301–305. [DOI] [PubMed] [Google Scholar]
- 16.Kang C, Han DY, Park KI, Pyo MJ, Heo Y, Lee H, et al. (2014) Characterization and neutralization of Nemopilema nomurai (Scyphozoa: Rhizostomeae) jellyfish venom using polyclonal antibody. Toxicon, 86: 116–125. 10.1016/j.toxicon.2014.04.005 [DOI] [PubMed] [Google Scholar]
- 17.Dawoud TM, Hererra P, Hanning I, Kwon YM, Ricke SC (2011) In vitro invasion of laying hen ovarian follicles by Salmonella Enteritidis strains. Poult Sci 90: 1134–1137. 10.3382/ps.2010-01182 [DOI] [PubMed] [Google Scholar]
- 18.Bolcato-Bellemin AL, Mattei MG, Fenton M, Amar S (2004) Molecular cloning and characterization of mouse LITAF cDNA: role in the regulation of tumor necrosis factor-alpha (TNF-alpha) gene expression. J Endotoxin Res 10: 15–23. [DOI] [PubMed] [Google Scholar]
- 19.Husáková E, Spišáková V, Herich R, Kolesárová M, Stašová D, Levkutová M, et al. (2015) Expression of cytokines in chicken peripheral mononuclear blood cells (PMBCs) exposed to probiotic strains and Salmonella Enteritidis. Acta Vet Brno 84: 29–35. [Google Scholar]
- 20.Teng MW, Bowman EP, McElwee JJ, Smyth MJ, Casanova JL, Cooper A, et al. (2015) IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med 21: 719–729. 10.1038/nm.3895 [DOI] [PubMed] [Google Scholar]
- 21.Bar-Shira E, Friedman A (2006) Development and adaptations of innate immunity in the gastrointestinal tract of the newly hatched chick. Dev Comp Immunol 30: 930–941. [DOI] [PubMed] [Google Scholar]
- 22.Withanage GSK, Kaiser P, Wigley P, Powers C, Mastroeni P, Brooks H, et al. (2004) Rapid expression of chemokines and proinflammatory cytokines in newly hatched chickens infected with Salmonella enterica serovar Typhimurium. Infect Immun 72: 2152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Withanage GSK, Wigley P, Kaiser P, Mastroeni P, Brooks H, Powers C, et al. (2005) Cytokine and chemokine responses associated with clearance of a primary Salmonella enterica serovar Typhimurium infection in the chicken and in protective immunity to rechallenge. Infect Immun 73: 5173–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin WH, Yu B, Lin CK, Hwang WZ, Tsen HY (2007) Immune effect of heat-killed multistrain of Lactobacillus acidophilus against Salmonella typhimurium invasion to mice. J Appl Microbiol 102: 22–31. [DOI] [PubMed] [Google Scholar]
- 25.Botta C, Langerholc T, Cencic A, Cocolin L (2014) In vitro selection and characterization of new probiotic candidates from table olive microbiota. Plos One 9: e94457 10.1371/journal.pone.0094457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Servin AL (2004) Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. Fems Microbiol Rev 28: 405–440. [DOI] [PubMed] [Google Scholar]
- 27.Neal-McKinney JM, Lu X, Duong T, Larson CL, Call DR, Shah DH, et al. (2012) Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. Plos One 7: e43928 10.1371/journal.pone.0043928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cotter PD, Hill C, Ross RP (2005) Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3: 777–788. [DOI] [PubMed] [Google Scholar]
- 29.Messaoudi S, Manai M, Kergourlay G, Prévost H, Connil N, Chobert JM, et al. (2013) Lactobacillus salivarius: Bacteriocin and probiotic activity. Food Microbiol 36: 296–304. 10.1016/j.fm.2013.05.010 [DOI] [PubMed] [Google Scholar]
- 30.Liu XM, Liu WY, Zhang QX, Tian FW, Wang G, Zhang H, et al. (2013) Screening of lactobacilli with antagonistic activity against enteroinvasive Escherichia coli. Food Control 30: 563–568. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.